Abstract
Mitochondrial DNA (mtDNA) is organized in discrete protein–DNA complexes, nucleoids, that are usually considered to be mitochondrial-inner-membrane associated. Here we addressed the association of replication factors with nucleoids and show that endogenous mtDNA helicase Twinkle and single-stranded DNA-binding protein, mtSSB, co-localize only with a subset of nucleoids. Using nucleotide analogs to identify replicating mtDNA in situ, the fraction of label-positive nucleoids that is Twinkle/mtSSB positive, is highest with the shortest labeling-pulse. In addition, the recruitment of mtSSB is shown to be Twinkle dependent. These proteins thus transiently associate with mtDNA in an ordered manner to facilitate replication. To understand the nature of mtDNA replication complexes, we examined nucleoid protein membrane association and show that endogenous Twinkle is firmly membrane associated even in the absence of mtDNA, whereas mtSSB and other nucleoid-associated proteins are found in both membrane-bound and soluble fractions. Likewise, a substantial amount of mtDNA is found as soluble or loosely membrane bound. We show that, by manipulation of Twinkle levels, mtDNA membrane association is partially dependent on Twinkle. Our results thus show that Twinkle recruits or is assembled with mtDNA at the inner membrane to form a replication platform and amount to the first clear demonstration that nucleoids are dynamic both in composition and concurrent activity.
INTRODUCTION
Mitochondrial DNA (mtDNA) was first visualized in 1963 (1), and subsequently found associated with the mitochondrial inner membrane (IM) (2). This was confirmed by electron microscopy (3) but to date the nature of mtDNA–membrane association has not been clarified. Microscopic methods have been used to show the organization of mtDNA in distinct structures, nucleoids, within the mitochondrial network (4), but it was not until 2001 when the first specific in situ protein co-localization was shown with the mitochondrial helicase Twinkle and mtDNA (5). The first nucleoid purification method identified two mtDNA binding proteins, mitochondrial single-stranded DNA-binding protein (mtSSB) and transcription factor A (TFAM) (6,7), both shown to co-localize with mtDNA in situ (8–11).
Mammalian mtDNA replication requires the concerted action of several replication factors including the mtDNA polymerase γ (POLG), the mtDNA helicase Twinkle, mtSSB and the transcription and packaging protein TFAM [see e.g. (12) for a review]. A minimal replisome consisting of Twinkle, POLG and mtSSB is capable of synthesizing the equivalent of a full-length mtDNA of 16.5 kb in vitro (13). Although overexpressed Twinkle, as well as endogenous mtSSB and TFAM have been shown in situ to co-localize at least partially with mtDNA, the possible temporal nature of interactions of endogenous mtDNA replication factors has never been demonstrated. Although mtDNA–nucleoids in recent years have been presented as rather static, one might expect many nucleoid-associated proteins such as transcription, replication and repair factors to interact transiently with mtDNA depending on their requirement. This would be reminiscent of many factors that interact with, for example, nuclear DNA in both a spatial and temporal manner. We here set out to ask whether the same applies to mtDNA by examining mtDNA co-localization of two mtDNA replication factors with distinct function, namely Twinkle and mtSSB, and show that their association with mtDNA is indicative of active replication. We previously showed that Twinkle–GFP was present in discrete foci within the mitochondrial network even in the absence of mtDNA in ρ0 cells (5), which we here confirm for endogenous Twinkle. This observation provided us with a handle on the spatial organization of mtDNA replication within the mitochondrial network. We here provide evidence that Twinkle is firmly membrane associated, is one of the proteins of a membrane-associated replication factory and is at least partially involved in mtDNA membrane association.
MATERIALS AND METHODS
Cell culture
Stable cell lines expressing mtDNA maintenance proteins on induction were created as described (14) using the Flp-In™ T-Rex™ 293 host cell line (Invitrogen). The ATAD3-HA expressing cell line was a kind gift of Drs Ian Holt and Hiroshi Sembongi (Cambridge UK). Transgenic cells were grown in Dulbecco's modified Eagle's medium (DMEM; Lonza) supplemented with 10% FCS (PAA laboratories), 2 mM l-glutamine, 1 mM Na pyruvate, 50 µg/ml uridine (Sigma), 100 µg/ml Hygromycin and 15 µg/ml Blasticidin (Invivogen) in a 37°C incubator at 8.5% CO2. Normal HEK293E, U2OS, 143B, 206f and B2ρ° cells were grown under similar conditions but without antibiotics. BJ (ATCC® CRL-2522™) human foreskin derived primary fibroblasts, and other primary human skin fibroblast lines were grown in 4:1 DMEM (Lonza) and M199 (Sigma) containing 15% FCS, 2 mM l-glutamine and 1 mM Na pyruvate. BJ fibroblast lines were used on the basis of availability and because these can be cultured to relatively high passage number without showing senescence, resulting also in no or only a relatively weak autofluorescence at 488 nm excitation. Other fibroblast lines were used on the basis of availability from our diagnostics service and were derived from healthy anonymous donors. These were not used with a passage number higher than 20. All cell lines were frequently checked for mycoplasma infection and found to be negative.
Western blot analysis
Mitochondrial fractions were analyzed by immunoblotting after sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) [(15) & Supplemental Experimental procedures].
Isolation of mitochondria
Cells were collected, resuspended in hypotonic buffer (4 mM Tris–HCl, pH 7.8, 2.5 mM NaCl, 0.5 mM MgCl2 and protease inhibitor complete, Roche Molecular Biochemicals) and subjected to homogenization using a 5-ml chilled Dounce homogeniser until 80% cells were broken. During the testing phase of mitochondrial subfractionations (see below), cells were also disrupted after short cytochalasin treatment (16) and on occasion further purified using sucrose gradient purification as described (15) without noticeable differences in the final results (not shown). With both methods, mitochondria were isolated using differential centrifugation.
Mitochondrial (sub)fractionation
The mitochondrial outer membrane was disrupted by incubation with a digitonin (Sigma Aldrich)/protein ratio ([µg digitonin]/[µg mitochondria]) = 0.2 (unless otherwise indicated) in phosphate buffered saline (PBS) or a buffer containing 225 mM Mannitol, 75 mM sucrose, 10 mM HEPES, pH 7.8, 10 mM EDTA, in either case supplemented with a protease inhibitor. The mitoplasts were obtained by centrifugation at 8000g for 10 min, +4°C. The supernatant was centrifuged at 100 000g for 1 h to obtain intermembrane space supernatant and pellet containing a fraction of outer mitochondrial membrane proteins (see Supplementary Figure S3 and Results). Mitoplasts were suspended in 0.16 mg of Brij58/mg mitoplasts and incubated for 10 min on ice. Membrane (inner + outer) (pellet) and matrix (supernatant) fractions were obtained after centrifugation at 100 000g for 1 h. Proteins from intermembrane space and matrix were precipitated by deoxycholate (DOC)/trichloroacetic acid (TCA) (see below). Equivalent protein concentrations were run on gel for western blot analysis of the various fractions (Supplementary Figure S6).
For digitonin-based fractionation, crude mitochondria from HEK293E or inducible HEK293 Flp-In™ T-Rex™ wt-Twinkle cells were taken up in 1 × PBS (Gibco), the total protein concentration determined with Bradford assays and lysed by addition of digitonin at indicated ratios µg digitonin/µg total mt protein, incubated for 10 min on ice and centrifuged for 5 min at 14 000g and 4°C. Solubilized supernatant fractions were kept separately while pellet fractions were resuspended in volumes equal to the removed soluble fractions. Both supernatant and pellet fractions were brought to a final concentration of 1% SDS.
Crude mitochondria for flotation were further purified over 30% Percoll gradients (30% Percoll, 225 mM sorbitol, 25 mM Tris, pH 7.4, 1 mM EGTA). Purified mitochondria were washed once in 5 volumes of 225 mM sorbitol, 25 mM Tris, pH 7.4, 1 mM EGTA and taken up in 1× PBS. Total mitochondrial protein yield was determined by Bradford assays and the equivalent of 2 mg of total mitochondrial protein was lysed in TN (25 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM DTT, cocktail of protease inhibitors, 10% sucrose) containing either 1% Triton X-100 or digitonin at a ratio of 2.5:1 (w/w) for 30 min on ice. Digitonin-lysed samples were centrifuged for 10 min at 14 000g, the supernatant discarded and the pellet resupended in TN containing 1% Triton X-100. Samples were mixed with cold Optiprep™ to a final concentration of 42.5%, transferred into MLS-55 centrifuge tubes and overlaid with 400 µl of each 40, 37.5, 35, 32.5, 30, 27.5, 25, 20, and 0% Optiprep™ in TN containing 1% Triton X-100. The gradients were centrifuged at 100 000g for 14 h at 4°C. Fractions were collected from top to bottom and aliquots analyzed by western blotting or dot blotting, respectively, as described before.
Treatment of isolated mitochondria with carbonate or KCl
For carbonate extraction, isolated mitochondria were resuspended in a 0.1 M Na2CO3 buffer (pH 11.0) and incubated on ice for 30 min; the pellet was recovered by centrifugation (100 000g, 1 h, 4°C). For salt-wash experiments, mitochondria were diluted 10-fold in buffers consisting of either 30 mM KCl or 500 mM KCl in 30 mM Tris–HCl (pH 7.4) and sonicated at 40% power 3 mm probe 3× 10 s per cycle. The pellet was recovered by centrifugation (100 000g, 1 h, 4°C). Proteins from the resulting supernatants were concentrated by DOC/TCA precipitation: lysates were treated with 0.02% DOC for 30 min on ice before addition of 10% TCA, incubated at +4°C over night and precipitated samples were centrifuged at 15 000g for 15 min at +4°C. Pellet and precipitated supernatant were finally re-solubilized in equal volumes and the same volume loaded on gel for SDS-PAGE and western blot analysis.
Treatment of isolated mitochondria by sonication and nucleases
Mitochondria were resuspended in enzyme-buffer (50 mM Tris–HCl, pH 7.5, 50 mM NaCl, 3 mM CaCl2, 2 mM MgCl2), sonicated on ice at 40% power for three times 20 s before addition of the enzymes as indicated DNase I (Fermentas) 10U, RNAse A (Fermentas) 20 µg, Micrococcal nuclease (Fermentas) 50 U and Benzonase nuclease (Sigma) 50U, and incubated at +37°C for 30 min. Where appropriate, lysates were further subjected to carbonate extraction as described above.
Dot-blot analysis of mtDNA content
For mtDNA analyses, samples of supernatant and pellet lysates [see Mitochondrial (sub)fractionation] were suspended in 2× SSC, boiled for 15 min at 95°C and dot blotted in triplicates onto positively charged nylon membranes. Dot blots were detected using nonradioactively labeled cytb probes using Dig-labeling (Roche). Hybridizations at 48°C and dig-antibody incubations were carried out using Easy-Hyb (Roche) according to the manufacturer’s protocol. ECL detection was performed with CSPD (Roche) and visualized with a ChemiDoc (Biorad). Quantifications of resulting ECL signals were performed with ImageQuant (Ge Healthcare).
Transfections, fluorescence microscopy, ddC treatment, EdU and BrdU labeling
Immunofluorescence (IF) detection of proteins was done as described previously (8) with minor modifications (for detailed procedures, see Supplement). MtDNA depletion in U2OS cells used 100 µM 2′,3′-dideoxycytidine ddC for 48 h. For Twinkle knockdown, cells were transfected in six-well plates (for IF) or 10-cm cell culture dishes (for biochemical fractionation experiments) with a mixture of three Stealth™ siRNA duplex oligonucleotides (C10Orf2 HSS125596, HSS125597, HSS125598, Invitrogen) against Twinkle, at a concentration of 20 pmol each, using Lipofectamine™2000. As controls we used Stealth™ Universal negative controls. Cells were fixed and analyzed 36–72 h after transfection. Transient transfection of a Twinkle–Myc expression construct (5) used TransIT-LT1 (Mirus, Madison, WI) according to the manufacturer’s instruction. Twinkle knockdown followed by biochemical fractionation involved a short exposure to Lipofectamine™2000 (4 h) after which medium was replaced with regular cell culture medium and replaced again 24 h prior to cell isolation.
MtDNA labeling using Click-iT™ EdU (5-ethynyl-2′-deoxyuridine) imaging kits with either AlexaFluor 488 or 568 azide (Invitrogen) was initially done as described by the manufacturer except that we used 50 µM EdU to detect mtDNA label incorporation. Only for the experiment shown in Figure 4C examining EdU, Twinkle, mtSSB co-localization we modified the procedure: the Click-iT™ buffer additive was replaced by 50 mM ascorbic acid and the reaction was done twice for 25 min with a freshly prepared labeling mix. This increased the signal and signal-to-noise ratio (17). Following EdU labeling and detection, we proceeded with IF as above. 5-bromo-2′-deoxyuridine (BrdU) labeling used the BrdU Labeling and Detection Kit I (Roche) using manufacturers protocols except that we used 50 µM BrdU and Alexa 568 anti-mouse for BrdU-antibody detection. Fixation used acid-ethanol resulting in significantly reduced mtSSB antibody staining compared to paraformaldehyde fixation. Both for EdU and BrdU labeling, mtDNA/EdU (or mtDNA/BrdU) and EdU/Twinkle (or BrdU/mtSSB) positive foci were scored manually by first marking all mtDNA/EdU or BrdU foci [using Image Pro Plus 6 ‘create point feature’ (Media Cybernetics) or using the ‘Event Marker’ tool using Axiovision 4.8 software] and overlaying the Twinkle/mtSSB IF, counting all double positives and using both numbers to calculate relative percentages. Twinkle–mtDNA positive foci were similarly scored using Image Pro Plus 6. In all cases, experiments were repeated several times as indicated and in each experiment multiple cells were scored to obtain final numbers. Very rare cytoplasmic EdU or BrdU spots that did not appear to co-localize with mtDNA were not considered. MtSSB/BrdU positive foci were only judged positive with clear position overlap and a distinct focal mtSSB signal on the basis of the strong focal mtSSB presence in a subpopulation of mtDNA foci in paraformaldehyde fixed cells. BrdU or EdU foci in the vicinity of the nucleus could not be assigned positive for mtDNA and/or BrdU/Edu on the basis of the often strong nuclear signal and were therefore not used in the quantification. Intensity line scans were made with the ‘Profile tool’ (Axiovision 4.8 software).
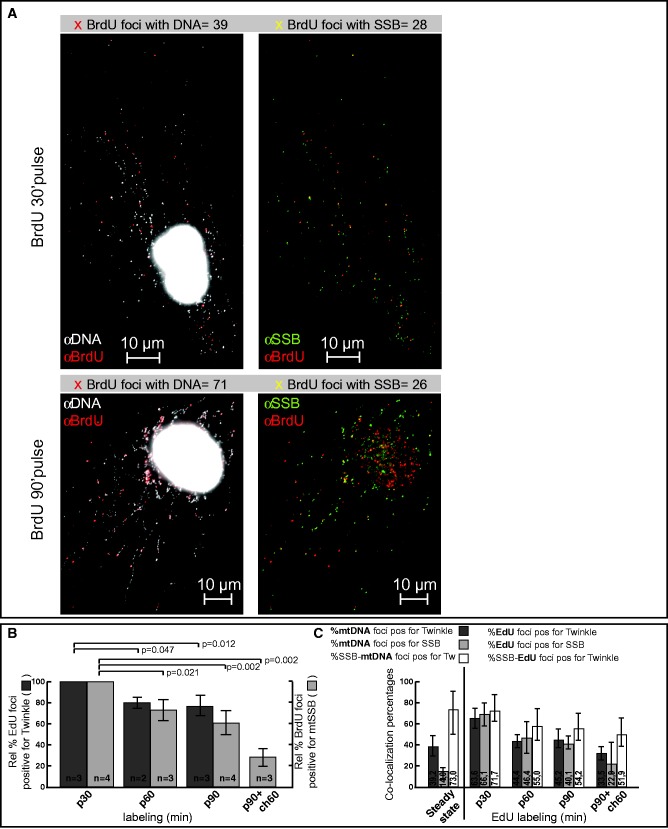
Figure 4.
Twinkle and mtSSB are enriched in mtDNA foci showing de novo mtDNA synthesis. (A) Fibroblasts were labeled for the indicated times with BrdU and processed for BrdU, mtSSB and mtDNA detection. (B) Fibroblasts were labeled for the indicated times with EdU (also see Supplementary Figure S5) or BrdU and processed for EdU/BrdU, Twinkle/mtSSB and mtDNA detection. The graph shows the relative percentage of Edu/Twinkle and BrdU/mtSSB positive foci, with the 30’ time point set to 100%. In reality, this time point showed 73 ± 8% of all EdU foci to be Twinkle positive and 69 ± 8% of all BrdU foci to be mtSSB positive, both significantly higher not only compared with the number of Twinkle positive EdU foci at 60’ and 90’ or mtSSB positive BrdU foci at 90’ or at 90’ pulse (p) + 60’ chase (ch) (paired t-test) but also to the steady-state percentage of Twinkle-positive or mtSSB-positive mtDNA foci (see ‘Results’ section and panel C). Error bars show SD. (C) Fibroblasts were labeled for the indicated times for EdU and slides processed for EdU detection using Alexa Fluor 555, Twinkle detection using Alexa Fluor 488 and mtSSB detection using Alexa Fluor 647 and co-localization determined. At the same time, a parallel slide from the same six-well plate was processed for Twinkle, mtSSB and mtDNA detection to obtain steady-state co-localization of Twinkle and/or mtSSB with mtDNA. For each slide, 10 images were taken and co-localization percentages determined, the bars indicate the upper and lower limits of these percentages, i.e. the range, for each experiment. Numbers in each bar show the average percentage of each experiment.
RESULTS
Endogenous Twinkle and mtSSB at steady state are found in a subset of mtDNA–nucleoids and are enriched in replicating mtDNA foci
The mtDNA helicase Twinkle is a low abundant protein [see Supplement of (14)]. For this reason and the lack of a good antibody, the analysis of the cellular functions of Twinkle so far has used overexpression of Twinkle variants that contain C-terminal epitope tags. These analyses have shown a high degree of in situ co-localization of Twinkle with mtDNA and mtDNA-associated proteins such as TFAM (8). However, overexpression of tagged Twinkle might not accurately copy the properties of the endogenous protein, while at the same time the total mitochondrial pool of the protein is considerably increased. Since Twinkle and, for example, also the mtSSB are mitochondrial proteins considered to be essential for mtDNA replication, but each mtDNA molecule is not continuously replicated, we asked whether either of these proteins dynamically associates with mtDNA. To this end we tested antibodies for Twinkle in immunofluorescence (IF) using immortal cell lines and primary fibroblasts. This analysis identified two monoclonal antibodies both recognizing the C-terminus of Twinkle based on peptide mapping (Anu Suomalainen-Wartiovaara & Milla Lampinen, personal communication) able to detect endogenous (Figure 1 and 2 and Supplementary Figure S1A and C) as well as overexpressed Twinkle (Supplementary Figure S1B), while siRNA mediated depletion of Twinkle showed loss of mitochondrial antibody signal (see below). Overexpression of Myc-tagged Twinkle showed that the antibody recognized all Twinkle–Myc/mtDNA foci as the Twinkle monoclonal and a Myc polyclonal antibody showed perfect overlap of signals (Supplementary Figure S1B).
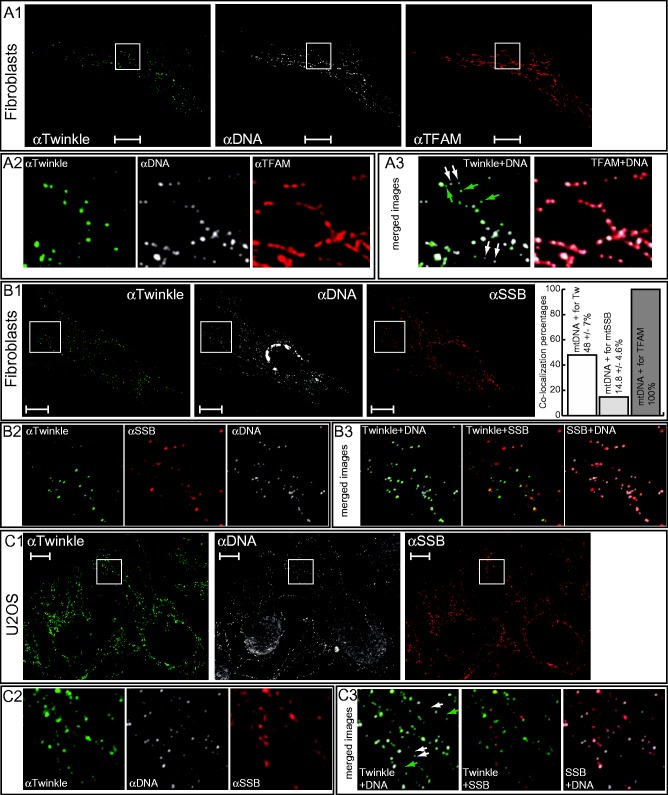
Figure 1.
Endogenous Twinkle is not a constitutive nucleoid protein. (A1) BJ fibroblasts were stained with a mouse IgG monoclonal antibody for Twinkle (green), a mouse IgM monoclonal for mtDNA (white) and a rabbit TFAM antibody (red) and imaged by confocal microscopy. Detailed (A2) and merged (A3) images show that all mtDNA foci were TFAM positive, while ∼50% (see also ‘Results’ section) of mtDNA foci were not positive for Twinkle (some of these foci are indicated with white arrows at the inset). In addition, some Twinkle foci were observed also in the absence of mtDNA (foci indicated with green arrows). (B1) A second primary skin fibroblast line was stained with antibodies for Twinkle (green), mtDNA (white) (same as above) and a rabbit mtSSB polyclonal (red) and imaged in this case using a Zeiss apotome. Detailed (B2) and merged (B3) images show that, similar to BJ fibroblasts, <50% of mtDNA foci were positive for Twinkle, while even fewer mtDNA foci were strongly positive for mtSSB against a weaker more uniform mtSSB staining that nevertheless appears to show some preferential localization with mtDNA. The percentage of mtDNA foci positive for Twinkle or mtSSB was determined in three independent experiments in primary skin fibroblasts, showing only a partial co-localization (see Main text). This is here presented as a small graph in panel B1 (right). The percentage of mtDNA foci positive for TFAM is here set at 100% as we have never seen evidence of any mtDNA foci not also showing a positive TFAM signal. (C1) Similarly, U2OS cells were stained with antibodies (same as above) for Twinkle (green), mtDNA (white) and mtSSB (red) showing multiple mtDNA foci not containing Twinkle and/or high concentrations of mtSSB (detailed in C2 and C3). Scale bars in figures are 10 µm. For additional control experiments see Supplementary Figure S1.
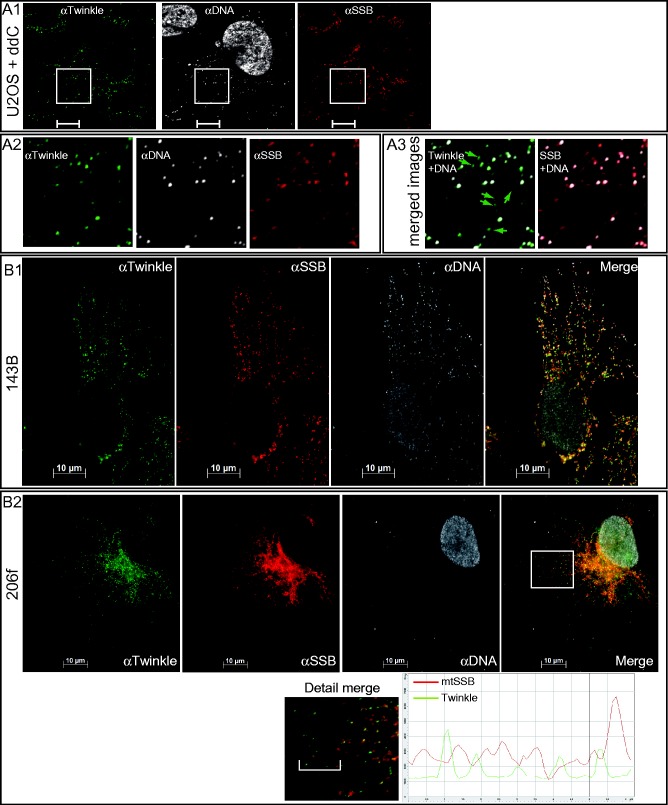
Figure 2.
Twinkle is found in punctate foci within the mitochondrial network in cells depleted of mtDNA and ρ0 cells. (A1) Treatment of U2OS cells with ddC resulted in a severe drop of mtDNA positive foci and revealed multiple Twinkle foci in the absence of mtDNA (detailed in A2 and A3). (B) In mtDNA-less 206f cells, endogenous Twinkle also showed a focal staining, while staining for mtSSB was comparatively uniform, in contrast to these proteins in cells containing mtDNA (8). 206f cells are here also stained for DNA showing a clearly positive nuclear DNA signal but absence of any mtDNA signal confirming the mtDNA-less character of these cells. A detailed section (indicated with a white box) of the merged Twinkle–mtSSB images clearly shows the rather uniform character of the mtSSB staining while Twinkle staining is punctate. This is further illustrated by a profile line scan (region indicated in the ‘Detail merge’ image by a white line) that shows fluctuating mtSSB intensity at levels well above the baseline, whereas Twinkle foci appear as sharper peaks from the baseline.
Analysis of IF images revealed that endogenous Twinkle co-localized only with a subset of mtDNA–nucleoids based on co-staining for mtDNA and/or TFAM (Figure 1A and B shows the results in primary fibroblasts and Figure 1C for osteosarcoma U2OS cells), a result that was also observed using a different fixation and permeabilization method (Supplementary Figure S1A). Similar observations were made in HEK293E cells (Supplementary Figure S1C). An analysis in primary fibroblasts of between 9 and 12 cells in three independent experiments showed the percentage of mtDNA foci that were Twinkle positive to be 48 ± 7% (Figure 1B1). In all, 1.2 ± 0.25 of every 10 Twinkle foci did not appear to co-localize either with mtDNA or TFAM while all mtDNA foci were TFAM positive and vice versa, no TFAM foci were observed that were not also mtDNA positive. These results, apart from showing that less than half of all mtDNA/TFAM foci contained Twinkle, suggested that Twinkle might organize in discrete structures independent of the presence of mtDNA. Earlier we showed that Twinkle–GFP expressed in mtDNA-less (ρ°) cells also shows a punctate mitochondrial fluorescence (5). This was corroborated here for endogenous Twinkle in cells partially depleted of mtDNA using dideoxycytidine (ddC) still showing discrete Twinkle foci both, with and without, mtDNA co-localization (Figure 2A). In 206f- ρ° cells, derived from the 143B osteosarcoma cell line, Twinkle foci are again discrete in an otherwise connected mitochondrial network (Figure 2B1 and B2). In 206f cells (Figure 2B2), endogenous mtSSB was more uniformly distributed over the mitochondrial network as also shown previously (8). This is illustrated by an intensity line profile of a small section of the mitochondrial network showing fluctuating mtSSB intensity at levels well above the baseline, whereas Twinkle foci appear as sharper peaks from the baseline (Figure 2B2). TFAM staining in 206f cells (Supplementary Figure S2A2) showed a much weaker and more uniform signal in comparison to the signal detected in the parental 143B cells (Supplementary Figure S2A1), as previously observed (18).
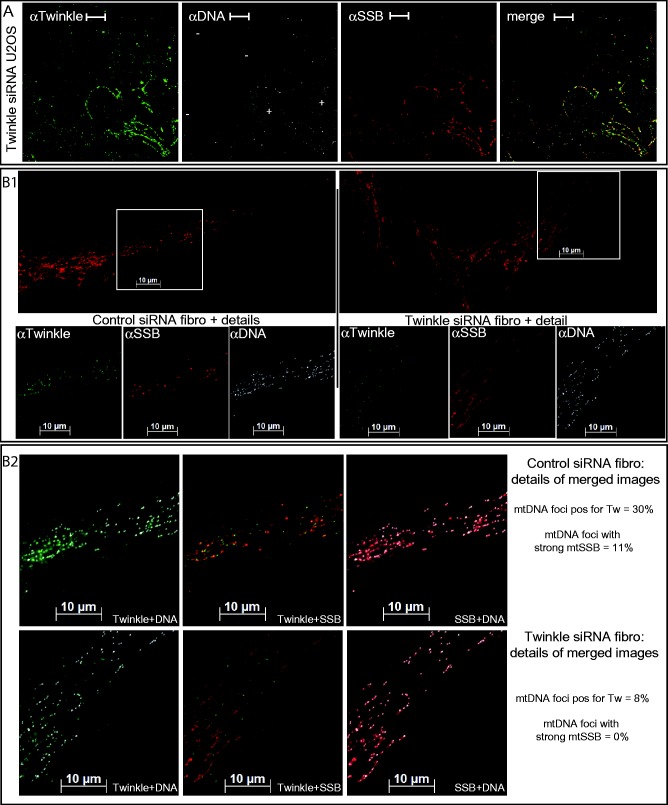
In fibroblasts (Figure 1B1, details in Figure 1B2 and B3) and U2OS cells (Figure 1C1, details in Figure C2 and C3), mtSSB showed both a relatively uniform mitochondrial distribution as well as a focal accumulation of presumably higher concentrations of mtSSB. These intense mtSSB foci co-localized only with a subset of mtDNA foci at a substantially lower percentage (14.8% in fibroblasts) compared with Twinkle. This was observed with two polyclonal mtSSB antibodies that we used in the course of this study. Cells in which Twinkle was depleted using transient (48 h) siRNA transfection only showed a modest decline of detectable mtDNA foci but showed almost complete loss of intense mtSSB foci (Figure 3). This was observed both in U2OS (panels A) and primary skin fibroblasts (panels B1). Detailed images (panels B2) of a small section of control versus knockdown Twinkle fibroblasts show the loss of most mitochondrial Twinkle foci (8% of mtDNA foci are still Twinkle positive compared with 30% in the control cell) while background antibody staining appearing in the cytosol remains similar to that seen in the control. In the whole-cell image (panel B1, upper right), about 10 intense mtSSB foci remain visible with Twinkle knockdown (compared with on average ∼70 in the control), while in the detailed section, 0 remain, compared with 11% in a similar section shown for the control. Similar to U2OS (panels A), various transient knockdown experiments using multiple control fibroblasts lines showed an on-slide correlation between the effectiveness of knockdown in individual cells based on Twinkle IF and the level of loss of intense mtSSB foci (not shown). In further support of these findings, transient expression of strong Twinkle stalling mutants K421A and G575D (14) result in the disappearance of mtSSB foci while these foci remain with expression of wild-type Twinkle or in cells on the same slide in which mutant Twinkle is not expressed (Supplementary Figure S3). In contrast to these findings, a short (48 h) but stronger mtDNA depletion using ddC (Figure 2A and Supplementary Figure S4B) showed a proportion of focal mtSSB co-localizing with mtDNA (Figure 2A3 and Supplementary Figure S4B3) that appeared twice higher than in nontreated U2OS (Supplementary Figure S4A3). These foci were, however, less intense (Supplementary Figure S4C). Taken together, these experiments show that the focal presence of mtSSB at mtDNA foci is dependent on the function of Twinkle, and mtSSB is dynamically recruited to nucleoids during replication (see below and ‘Discussion’ section).
Figure 3.
Twinkle depletion results in a modest decline of mtDNA but a dramatic loss of focal mtSSB. (A) Depletion of Twinkle in U2OS cells using transient (48 h) siRNA transfection. In this image view, several cells with Twinkle knockdown (−) are shown alongside two cells that show unsuccessful knockdown (+) to illustrate the considerable reduction of Twinkle immunofluorescence in knockdown cells and, therefore, showing the specificity of the Twinkle antibody used in this study. Cells with loss of Twinkle immunofluorescence, also show loss of mtSSB at mtDNA foci but only a modest reduction in mtDNA signal. (B) In fibroblasts (fibro), similarly, knockdown of Twinkle after 48 h still showed substantial amounts of mtDNA foci while Twinkle was dramatically reduced compared with a control knockdown experiment using nontargeting siRNA (for a quantification of the indicated section in the periphery of the cell, numbers are indicated on the right in panel (B2) Tw = Twinkle; pos = positive), while only a few intense mtSSB foci remained (10 in the whole cell, none in the detailed section). For clarity we here show detailed sections (regions indicated with a white square in the larger mtSSB image) for the three different antibodies (smaller panels B1) as well as enlarged merged images for both control and Twinkle knockdown (B2). These illustrate the presence of mtDNA foci in Twinkle knockdown cells, the absence of strong mtSSB foci co-localizing with mtDNA and Twinkle, and the loss of mitochondrial Twinkle foci co-localizing with mtDNA, which was quantified as 8% compared with 30% in the section analyzed in the control. The enlarged merged images clearly also illustrate that the knockdown looks deceptive as remaining fluorescence is still clearly visible. Most of this fluorescence, however, is nonmitochondrial background, which is also visible in the control, but as shown here, is not sensitive to Twinkle knockdown. For additional control experiments see Supplementary Figure S3.
The absence of endogenous Twinkle from many mtDNA–nucleoids in primary fibroblasts and the presence of Twinkle foci in the absence of mtDNA suggested that nucleoids might dynamically associate with Twinkle foci (or vice versa) dependent on signals that would indicate the need to replicate mtDNA. Even more dramatically, only a small fraction of mtDNA foci in fibroblasts showed a strong accumulation of mtSSB. To address whether Twinkle and/or mtSSB association with mtDNA showed a positive correlation with ongoing mtDNA replication, we made use of ClickIt–EdU (19) and BrdU labeling to detect de novo mtDNA synthesis. Since mtDNA can incorporate EdU/BrdU at any point during ongoing mtDNA replication, we reasoned that if Twinkle and/or mtSSB temporarily associate with nucleoids to enable replication and would subsequently dissociate or disassemble, short EdU/BrdU pulses would show a relatively larger proportion of labeled nucleoids positive for these proteins, than longer pulses. In initial experiments, we tested Twinkle and mtSSB separately with EdU and BrdU labeling, respectively (Figure 4A and B). The data (Figure 4C and Supplementary Figure S5) indicate that with the shortest possible EdU pulses for unambiguous visualization (30 min), the highest proportion of EdU positive foci were Twinkle positive (73%), while after 60 and 90 min and at steady state (see above and Figure 4C) this percentage was significantly lower, indicating that for EdU incorporation to occur Twinkle needs to be mtDNA associated. Similarly, following BrdU labeling and mtSSB/mtDNA, IF showed 69% strong mtSSB positive BrdU foci after a 30-min pulse that dropped to 41% after 90 min and declined even further to 27% after a 90-min pulse followed by a 60-min chase. These data thus show that core components of the replication machinery and mtDNA dynamically associate with one another to enable replication.
To analyze the dynamics of mtDNA association of both Twinkle and mtSSB in more detail, we examined Twinkle/mtSSB co-localization with EdU-labeled mtDNA. At the same time, parallel slides were processed for mtDNA, Twinkle and mtSSB detection under otherwise identical conditions in order to determine overall levels of mtDNA occupancy by Twinkle and mtSSB. As mtDNA replication occurs throughout the cell cycle, these values represent steady-state co-localization values. Since EdU foci were essentially all mtDNA positive (see Supplementary Figure S5), we chose not to perform EdU labeling with quadruple staining including mtDNA staining because axial chromatic aberration in the ultraviolet range could not be corrected for on the microscope used in these experiments.
The results of the EdU–Twinkle–mtSSB detection (Figure 4C) show the same general kinetics as observed with EdU–Twinkle–mtDNA (Supplementary Figure S5) and BrdU–mtSSB–mtDNA detection as depicted in Figure 4B. In this particular experiment, steady-state Twinkle–mtDNA and mtSSB–mtDNA co-localization were 39 and 14%, respectively (Figure 4C). In contrast, at a short 30-min EdU pulse labeling, Twinkle–EdU and mtSSB–EdU co-localization were 64 and 66%, respectively, and the percentage of EdU–mtSSB foci that were positive for Twinkle at this short pulse was 72%. With longer EdU pulses, co-localization percentages declined to more closely reflect steady-state Twinkle and mtSSB, mtDNA co-localization. At a 90-min pulse plus a 60-min chase, Twinkle–EdU co-localization was somewhat below the steady-state Twinkle–mtDNA co-localization while the mtSSB–EdU co-localization was still somewhat above. The percentage of mtSSB–EdU positive foci that were also Twinkle positive showed slightly different kinetics such that at a 30-min pulse it closely reflected the steady-state level of 73%, but at all later time points, it settled at a relatively stable lower-than-steady-state percentage of 52–55% (see ‘Discussion’ section).
Twinkle is firmly membrane associated, enhances mtDNA tethering to the membrane when overexpressed and reduces mtDNA tethering on Twinkle knockdown
Although the first suggestions of mammalian mtDNA membrane association stem from the late 1960s and 1970s (see ‘Introduction’ section), little is known about the nature of this association. The observation that Twinkle forms discrete foci even in the absence of mtDNA raised the possibility that Twinkle is not a matrix-soluble protein, as this would show a uniform staining. In contrast, TFAM and mtSSB lose their localization in discrete foci in ρ° cells and thus require mtDNA for their localization in a discrete complex (see above). We used classical biochemical fractionation to examine the localization and solubility of nucleoid-associated proteins (see legend to Supplementary Figure S6 and ‘Materials and Methods’ section for details). We first used inducible overexpression of tagged proteins, as this required much less material and allowed their detection with tag antibodies. This showed that Twinkle was exclusively present in the mitochondrial membrane fraction as was ATAD3, an IM protein with functions reported in lipid shuttling and mtDNA binding (20,21). In contrast TFAM, mtSSB and POLG1 and 2 were distributed over the membrane and matrix fractions (Supplementary Figure S6). Although Twinkle does not contain any predicted transmembrane helices, these data suggested a strong membrane association. To confirm this, we used two alternative methods to confirm these results for endogenous Twinkle, based either on sodium carbonate fractionation (22) or on 0.5 M KCl extraction (23) (Figure 5A). Both of these methods showed tight association of endogenous Twinkle with mitochondrial membranes. As expected, the same was also observed with overexpressed Twinkle (Figure 5B and C and Supplementary Figure S6). As a control, a combination of TritonX-100 lysis and sodium carbonate extraction showed that the insolubility is not a peculiarity of the Twinkle protein or its overexpression (Figure 5B), as Twinkle is mostly found in the supernatant under these conditions. In contrast to Twinkle, and depending to some extent on the cell line used, substantial proportions of endogenous TFAM and mtSSB could be dissociated using sodium carbonate or 0.5 M KCl extraction. This suggested that the fraction of TFAM or mtSSB that was membrane associated on the basis of the more classical mitochondrial fractionation was mediated mostly by electrostatic interactions with membranes and/or mtDNA. Treatment of isolated mitochondrial membranes with nucleases, following either sonication or carbonate extraction, also released a substantial proportion of TFAM and mtSSB but not Twinkle into the soluble fraction (Figure 5A and C).
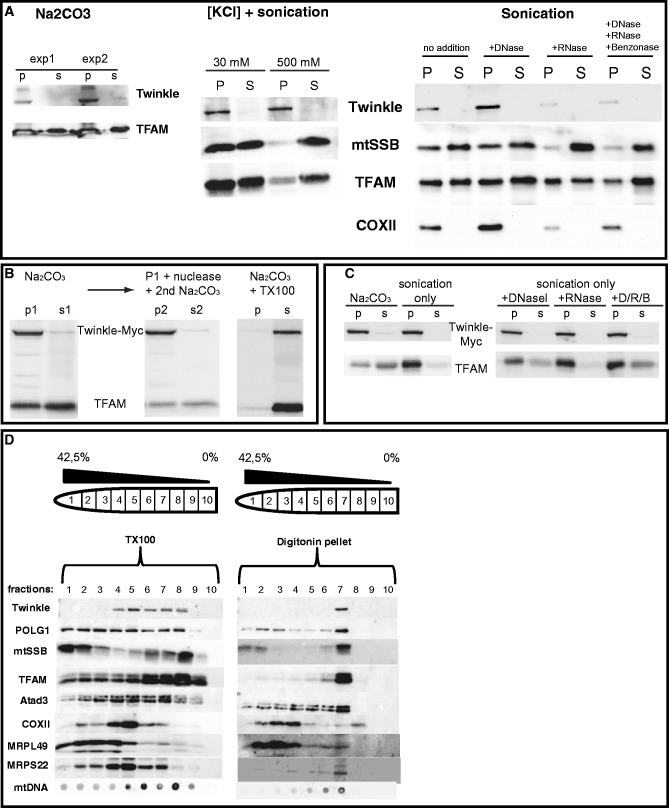
Figure 5.
Twinkle is membrane associated. (A) Isolated mitochondria of HEK293E cells were subjected to either KCl or sodium carbonate extraction (Na2CO3) as described in the main text. Endogenous Twinkle was detected using a monoclonal antibody and blots were re-probed with antibodies for TFAM and mtSSB. Results show that endogenous Twinkle fractionates mostly to the pellet fraction using both methods illustrating its strong membrane association, similar to overexpressed Twinkle (Supplementary Figure S6C), whereas TFAM and mtSSB mostly became soluble, in particular, in combination with 0.5 M KCl and sonication. Please note that to detect Twinkle with confidence, more protein was used for these western blots sometimes resulting in overloading of TFAM. Sonication in combination with DNAseI (D) or DNaseI/RNase A (R, RNase)/Benzonase (B) released more TFAM and mtSSB than sonication alone, showing that a proportion of TFAM and mtSSB can be found in the insoluble fractions of the various experiments by means of their interaction with mtDNA (rightmost blot panel A). (B) Na2CO3 fractionation shows that overexpressed Twinkle–Myc is almost exclusively in the pellet (p) fraction again indicative of tight membrane association, whereas TFAM is partially soluble (s = supernatant) (left panel). Treatment of the Na2CO3 pellet fraction by DNAseI and subsequent re-extraction using Na2CO3 further released a proportion of TFAM (middle panel). Finally, Na2CO3 in combination with Triton-X100 (TX100) treatment solubilized both Twinkle–Myc and endogenous TFAM (right panel). (C) Similar to results shown in panel A, sonication in combination with DNAseI (D) or DNaseI/RNase A (R, RNase)/Benzonase (B) released more TFAM than sonication alone or combined with RNase treatment, showing that a proportion of TFAM can be found in the insoluble fractions of the various experiments by means of its interaction with mtDNA, whereas in this case, overexpressed Twinkle–Myc remains in the pellet fraction with all treatments. (D) Digitonin-based isolation of a mitochondrial membrane fraction again showed the retention of Twinkle and proportions of nucleoid-associated proteins such as TFAM, mtSSB and POLG1 (see main text and Supplementary Figure S6D). This membrane fraction (marked as ‘Digitonin pellet’) was here subjected to flotation by layering a gradient of iodixanol on top of this fraction that was first re-solubilized with 1% TX100 (see ‘Materials and Methods’ for details). The gradient was then subjected to ultracentrifugation. In parallel, a mitochondrial fraction that was directly solubilized by 1% TX100 (marked as ‘TX100’) was subjected to the same procedure. Collected fractions were isolated as indicated and subjected to western blot analysis as well as dot blot analysis to detect mtDNA. The results show that mtDNA and nucleoid-associated proteins in the digitonin-lysis membrane fraction moved up the gradient to a single low-density iodixanol concentration, showing that they likely form a single complex. In contrast, COXII and a marker for the large ribosomal subunit MRPL49 have remained at relatively high-density fractions. In the total mitochondrial TX100 lysate, Twinkle and other nucleoid proteins appear more dispersed as does mtDNA.
On the basis of our digitonin titration experiments (Supplementary Figure S6) it was clear that also the IM could be disrupted by high digitonin concentrations. We reasoned that if many nucleoid-associated proteins are found in both membrane and matrix fractions based on classical mitochondrial subfractionation and only a subset of nucleoids contain Twinkle based on IF, perhaps two pools of mtDNA–protein complexes could also be separated on the basis of their solubility. To test this, we examined the solubility of several mitochondrial marker proteins by titrating the w/w ratio of digitonin/total mt protein, but this time using only one centrifugation step to separate the solubilized components (supernatant) from the digitonin-insoluble (pellet) fraction. This analysis (Supplementary Figure S6D) showed that the IM became somewhat permeable to glutamate dehydrogenase (a matrix localized enzyme) at a 0.5:1 w/w digitonin/protein ratio and was maximally permeable at a 2.5:1 ratio. At this ratio, COX II was still mostly in the pellet fraction, but at a 3.5:1 ratio it also became more soluble. This agrees with supercomplex blue-native PAGE analysis protocols, where a 4:1 ratio is used to solubilize supercomplexes (24).
TFAM and POLG1 behaved similar to glutamate dehydrogenase to the extent that a sizeable proportion became soluble at the lower range of digitonin concentrations. However, a substantial pool was resistant to digitonin solubilization at a 2.5:1 and even a 3.5:1 ratio, as it remained in the pellet. As expected, endogenous Twinkle behaved essentially the same as the transmembrane COX II protein in this assay. Using a 2.5:1 digitonin ratio, we subjected pellet fractions to a combined flotation/fractionation on an iodixanol gradient having re-solubilized the pellet with Triton-X100 and compared this with a total mitochondrial Triton-X100 lysate (Figure 5D). This analysis showed that Twinkle, mtDNA, TFAM, mtSSB and POLG1 all migrated in a single fraction high up the gradient, while, in contrast, for example, COXII or a marker for the large mitoribosomal subunit, MRPL49, migrated only a small distance up the gradient. The small mitoribosomal subunit marker MRPS22 showed the presence of this subunit in the total lysate but it was essentially absent from the digitonin pellet fraction. These results thus showed again that the Twinkle-containing fraction is not an insoluble aggregate of proteins. More importantly, it indicated various proteins that are involved in mtDNA replication and that were insoluble at a 2.5:1 digitonin ratio to co-migrate in a single fraction at low density in the gradient substantiating the idea that they form a single discrete membrane-associated complex (see ‘Discussion’ section).
Having established the relatively simple fractionation procedure based on digitonin lysis, dot-blot analysis was performed for mtDNA and showed that ∼35% of the mtDNA pool was soluble at a 2.5:1 digitonin ratio (see below, Figure 6). This suggested that on mild mitochondrial lysis with digitonin, two pools of mtDNA and associated proteins exist, one that remains in the pellet and contains Twinkle and one that is more soluble and contains little Twinkle.
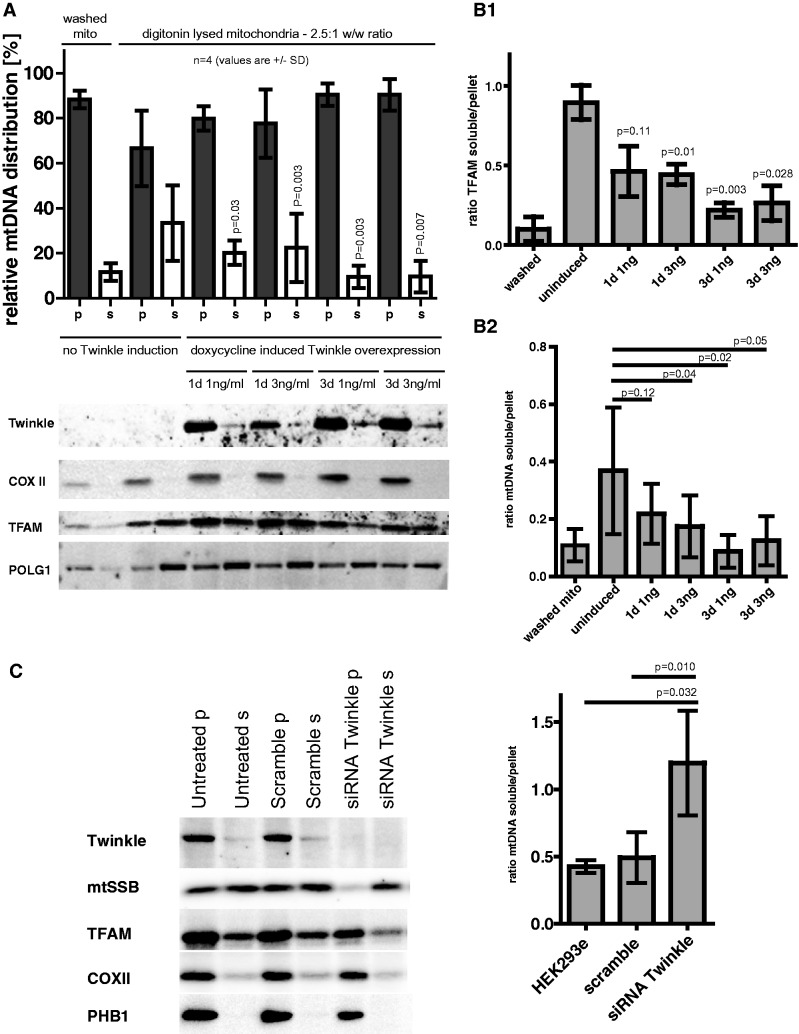
Figure 6.
MtDNA membrane association varies with overexpression or knockdown of Twinkle. (A and B) Mitochondria were isolated from noninduced HEK293 FlpIn™ TRex™, or induced as indicated to express Twinkle. Isolated mitochondria were subsequently lysed under mild conditions (see Supplementary Figure S6 panel D and ‘Results’ section) to release matrix constituents (s) but not membrane components (such as cytochrome c oxidase subunit II—COXII) found in the pellet (p) on centrifugation. Each fraction, including nonlysed mitochondria and their wash solution following centrifugation (also indicated as p and s), were analyzed for mtDNA content using dot-blot analysis (upper graph panel A: combined results of four independent experiments with ratio’s shown in panel B2) and various proteins (Twinkle, COXII, TFAM and POLG1) (lower gel images panel A: one experiment shown). The results show that overexpression of Twinkle results in a redistribution of mtDNA, TFAM (panel B1) and to a lesser extent POLG1 to the pellet fraction. Error bars in the graphs show the standard deviation (not the standard error of the mean), representing inter-experimental scatter. The paired Student t-test was used to account for this scatter showing the results to be highly significant for all but the shortest induction with the lower doxycycline concentration, in which case there was actually less scatter (smaller SD). The presence of some mtDNA, TFAM and POLG1 in the nonsolubilized washed mitochondrial supernatant suggests some mitochondrial damage occurred during the isolation and thus that the amount of soluble mtDNA–nucleoid in the lysed sample is an underestimation. Values in panel B indicate P values of the comparison with uninduced cells. (C) A similar approach as in A and B was used for a 3d Twinkle knockdown experiment in HEK293E cells showing an increase in mtDNA solubility (n = 4) compared with nontreated and nontargeting siRNA. An example western blot on the left shows the efficiency of Twinkle knockdown and, in particular, the redistribution of mtSSB to the soluble fraction. A reduction in the TFAM signal is indicative of the partial mtDNA depletion, which was also evident on the mtDNA dot blots (not shown). PHB1 is prohibitin 1.
Overexpressed Twinkle in cultured cells typically shows a good co-localization with mtDNA [(8) and (Supplementary Figure S1B)]. Given this and the results presented so far we hypothesized that overexpression of Twinkle might increase the fraction of mtDNA in the insoluble pellet fraction. To test this hypothesis, we used inducible expression of wt Twinkle without epitope-tag and tested the distribution of mtDNA and associated proteins using digitonin fractionation. Previously we have shown that the used level of Twinkle induction has little effect on nucleoid structure and has no effect on mtDNA levels, replication, mitochondrial transcript levels or cell growth (25). The results of western- and dot-blot DNA analysis showed that, in contrast to noninduced cells, Twinkle overexpression resulted in almost complete retention of mtDNA and a reproducible redistribution of TFAM to the nonsoluble fraction, in particular, for the 3-day induction period (Figure 6B). The analysis of POLG showed a similar redistribution as TFAM on the blot shown here but was not always as clearly detectable owing to its low abundance, precluding a statistical analysis.
Finally, we performed the same analysis using Twinkle siRNA (Figure 6C). This showed an increase in the amount of soluble mtDNA. Nevertheless, a substantial amount still remained insoluble showing that mtDNA membrane association is not solely dependent on Twinkle (see ‘Discussion’ section). In agreement with the suggested requirement for Twinkle, mtSSB showed a redistribution to the more soluble fraction, corroborating the IF analysis following Twinkle knockdown.
DISCUSSION
In this article we show, using IF and biochemical fractionation, the presence of at least two pools of mtDNA in human mitochondria, one (or more) that is isolated in an insoluble fraction and is likely membrane associated, and one that is more soluble. Similarly, we show that Twinkle behaves as a membrane protein independent of DNA association, whereas mtSSB and TFAM are present with mtDNA and Twinkle in membrane fractions (by biochemical isolation) or with nucleoids (by IF) on the basis of their DNA association and ongoing replication. This is corroborated by IF in cells not containing mtDNA, showing Twinkle in punctate foci, whereas mtSSB and TFAM are more uniformly labeling the mitochondrial network in contrast to their partial punctate co-localization in cells that contain mtDNA. We show, using EdU/BrdU pulse-chase labeling, a clear causative relationship between the presence of Twinkle and mtSSB at mtDNA foci and concurrent mtDNA replication. Finally, using overexpression or knockdown we show a partial dependency of mtDNA membrane association on Twinkle. These findings and the presence in a single fraction of Twinkle, mtDNA and various mtDNA replication factors on flotation of digitonin purified mitochondrial membranes suggest that Twinkle is a core component of a membrane-associated mtDNA replication factory. We thus provide at least a partial explanation for the long-standing observation of an mtDNA membrane connection (see ‘Introduction’ section). Supplementary Figure S7 shows a model that incorporates the major findings of this article.
Twinkle and mtSSB preferentially co-localize with replicating mtDNA; mtSSB is recruited to the replisome in a Twinkle-dependent manner
The steady-state percentages of Twinkle and mtSSB co-localization with mtDNA and the comparison of their co-localization with replicating mtDNA provides insight both into the mechanism of mtDNA replication as well as its dynamics and that of its associated factors. It is clear from the data that Twinkle at steady state co-localizes with a higher number of mtDNA molecules than the number that is being replicated at any given time, as the number of mtDNA foci positive for Twinkle (∼40–50%) is much higher than the number of mtDNA foci that are positive for EdU/BrdU (4–10%) at the shortest labeling time (Supplementary Figure S8). This suggests that mtDNA–Twinkle association in itself is not enough to initiate replication or alternatively that replication following this association is frequently aborted or prematurely terminated. This hypothesis is supported by the observation that at steady state and following a short 30-min EdU pulse, the percentage of mtDNA molecules being positive for mtSSB and also positive for Twinkle was highest (∼70%), whereas at all the other time points of EdU labeling it seemed to have settled at ∼55%. MtDNA copying also involves the frequent synthesis of 7S DNA in the so-called noncoding region. Although the function of this relatively short DNA fragment is currently still unclear, it has a higher synthesis and turnover rate compared with the full-length genome [see e.g. (26) and references herein], which depends on the recently identified mitochondrial genome maintenance exonuclease 1 (MGME1) (27). 7 S DNA synthesis might thus provide one explanation for the observed difference, and it was recently shown that 7 S DNA synthesis is dependent on the Twinkle protein (28). Alternatively, results could indicate, for example, DNA repair processes, with limited EdU incorporation that would fall below the detection limit but require Twinkle/mtSSB association with mtDNA.
The much lower steady-state mtSSB–mtDNA co-localization and the concomitant low percentage of mtSSB that does not co-localize with EdU/BrdU at the shortest labeling pulses shows that mtSSB is a better in situ marker for ongoing full-genome replication than Twinkle. This is supported by siRNA-mediated Twinkle depletion: a 48-h Twinkle knockdown in fibroblasts (as shown in Figure 3) shows a modest reduction in visible mtDNA foci, which agrees with previous siRNA experiments showing a slow decline in mtDNA copy number (29). At the same time, few intense mtSSB positive foci, as seen in untreated cells, remain. Biochemical fractionation shows similarly that mtSSB becomes more soluble on Twinkle siRNA. In contrast, cells that are more strongly depleted of mtDNA following ddC treatment show mtDNA foci containing Twinkle and strongly positive for mtSSB by IF. The use of Twinkle siRNA allows us to conclude that there is a correlation between the level of Twinkle expression and the strong accumulation of mtSSB in distinct foci. As Twinkle is depleted by siRNA treatment, mtDNA replication can no longer initiate and mtSSB will no longer be recruited in large quantities, as single-stranded mtDNA will no longer be generated by the unwinding action of Twinkle. In contrast, in case of ddC mediated depletion, replication can still initiate but will stall or stutter and both Twinkle and mtSSB will be trapped on partially replicated DNA molecules (Supplementary Figure S7). It is noteworthy that it was recently shown in vitro that Twinkle is capable of loading on a circular DNA molecule without the assistance of a helicase loader (30). Combined, the data suggest that also in vivo Twinkle–mtDNA association is an important step in the initiation of mtDNA replication and show that Twinkle mtDNA unwinding is required for the recruitment of larger quantities of mtSSB during ongoing replication.
Does twinkle mark specialized replication factories at the mitochondrial IM?
Since Twinkle does not appear to be a constitutive mtDNA–nucleoid component, nucleoids must dynamically associate with Twinkle at, as we suggest here, specialized foci at the mitochondrial IM. Alternatively, mtDNA might dynamically associate with preexisting platforms at the IM and subsequently recruit Twinkle. The presence of distinct Twinkle foci in rho-zero cells and cells depleted of mtDNA by ddC argues that Twinkle is present at preexisting membrane foci with which mtDNA dynamically interacts. This is substantiated by a considerable decrease of mtDNA membrane association on Twinkle depletion using siRNA. Twinkle–mtDNA membrane association is also substantiated by the observation that GFP-tagged Twinkle was previously observed to show the same velocity and directionality as overall mitochondrial movement (8).
Although siRNA-mediated Twinkle depletion is not 100%, the observation that a considerable fraction of mtDNA remains membrane associated suggest other means by which it associates with the membrane. Other proposed proteins for involvement in mtDNA membrane association include a processed splice variant of OPA1 (31), ATAD3 (32) and prohibitin (33). Although we have not tested all possible candidates in the analysis of mtDNA fractionation, all available data combined suggest that mtDNA membrane association will likely depend on multiple proteins as well as physiological conditions and signals, and is a dynamic process. Current data suggests the existence of a soluble and possibly distinct membrane bound fractions. This explains published nucleoid proteome analyses in which proteins involved in mtDNA replication and repair, transcription, translation and biogenesis have all been identified (33,34). It is also substantiated by high-resolution immunofluorescence that shows partial but noncomplete overlap of signals or closely adjacent signals of mtDNA/TFAM and proteins with functions in protein synthesis, import and biogenesis (35). This has also been observed, for example, for ATAD3 colocalization (20) while both prohibitin and ATAD3 have recently been implicated in mitochondrial protein synthesis and mtDNA membrane association (34). We thus not only support the original suggestions by Iborra and colleagues that nucleoids are found adjacent to sites of mitochondrial biogenesis, but show that distinct and dynamic populations of mtDNA must exist in association with the IM, possibly dedicated to distinct functions such as translation, replication or repair. In contrast to the above, TFAM generally shows a perfect mtDNA co-localization pattern. It has been argued on the basis of super-resolution microscopy and theoretical considerations that perhaps the only permanently mtDNA-associated factor is TFAM (36,37).
Contrary to the current paradigm, we have shown that a substantial nonmembrane-bound or loosely membrane-associated mtDNA–protein fraction also exists in mammalian mitochondria. A second recent super-resolution microscopy study has similarly suggested that not all nucleoids are in direct close contact with the inner mitochondrial membrane (38) and that consequently mtDNA–membrane interactions could be transient in nature. We can agree with these findings using a different, and in this case biochemical, approach, and show that transient interactions include association with Twinkle to facilitate mtDNA replication.
Finally, in yeast, mtDNA replication showed a similar dynamic association of some of its nucleoid proteins including the POLG1 homologue, Mip1p, with mtDNA (39). In the same study, it was demonstrated that some of the core components of the mtDNA maintenance machinery are present as discrete complexes also in the absence of mtDNA. Although yeast does not have a Twinkle homologue (40), it was shown that one of the yeast mtDNA helicases, Pif1p, is membrane associated. Abf2p, the yeast TFAM homologue, could be partly dissociated by nuclease treatment (41), similar to our findings here for Twinkle and TFAM. Our data thus show a strong mechanistic similarity between yeast and mammalian mtDNA organization and replication, and suggest that mtDNA replication factories in close association with the IM exist throughout the Eukaryote lineage.
To conclude, in this article, we present the first clear evidence that human mtDNA replication factors dynamically associate with mtDNA to facilitate replication. Our type of analysis provides a direct handle on the functional characterization of putative mtDNA replication factors as we would predict these factors to co-localize only partially with mtDNA and to be enriched at foci that contain Twinkle and/or mtSSB. In particular, mtSSB seems to provide a good marker for mtDNA replication because it will be positive for fewer foci that are not in the process of replication, and because of its higher abundance, it gives a clearer immunofluorescent read-out when used with high-resolution imaging. On the basis of our results, we can anticipate that similar dynamic protein–mtDNA associations will exist for transcription and mtDNA repair factors. Evidence for the transient interaction of repair proteins includes the organization of base-excision repair proteins in discrete structures distinct from nucleoids (42); the increased co-localization of Cockayne syndrome group B protein with TFAM on menadione treatment (43); and the increased nucleoid co-localization of DNA2 on replication stalling (44). The dynamic nature of protein–mtDNA interactions raises the important question how these interactions are regulated at the molecular level.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Academy of Finland [CoE funding to J.N.S., 110463 and 108380 to P.M., currently Photonics grant 134893]; Tampere University Hospital Medical Research Fund [9J119, 9K126 and 9L097 to J.N.S.]; Netherlands Organization for Scientific Research [NWO: VICI grant 865.10.004]; European Molecular Biology Organization [EMBO LTF co-funded by Marie Curie Actions, EMBO LTF 1066_2011 (including MCA-EMBOCOFUND2010, GA-2010-267146) to J.M.G.]. Funding for open access charge: Netherlands Organization for Scientific Research [NWO].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ian Holt, Howy Jacobs, Anu Suomalainen-Wartiovaara and Brendan Battersby for their interest and discussions.
REFERENCES
- 1.Nass MM, Nass S. Intramitochondrial fibers with DNA characteristics. I. Fixation and electron staining reactions. J. Cell Biol. 1963;19:593–611. doi: 10.1083/jcb.19.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nass MM. Mitochondrial DNA.I. Intramitochondrial distribution and structural relations of single- and double-length circular DNA. J. Mol. Biol. 1969;42:521–528. doi: 10.1016/0022-2836(69)90240-x. [DOI] [PubMed] [Google Scholar]
- 3.Albring M, Griffith J, Attardi G. Association of a protein structure of probable membrane derivation with HeLa cell mitochondrial DNA near its origin of replication. Proc. Natl Acad. Sci. USA. 1977;74:1348–1352. doi: 10.1073/pnas.74.4.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Satoh M, Kuroiwa T. Organization of multiple nucleoids and DNA molecules in mitochondria of a human cell. Exp. Cell Res. 1991;196:137–140. doi: 10.1016/0014-4827(91)90467-9. [DOI] [PubMed] [Google Scholar]
- 5.Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, Tariq M, Wanrooij S, Garrido N, Comi G, Morandi L, et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat. Genet. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 6.Van Tuyle GC, Pavco PA. Characterization of a rat liver mitochondrial DNA-protein complex. Replicative intermediates are protected against branch migrational loss. J. Biol. Chem. 1981;256:12772–12779. [PubMed] [Google Scholar]
- 7.Mignotte B, Barat M. Characterization of a Xenopus laevis mitochondrial protein with a high affinity for supercoiled DNA. Nucleic Acids Res. 1986;14:5969–5980. doi: 10.1093/nar/14.15.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrido N, Griparic L, Jokitalo E, Wartiovaara J, Van Der Bliek AM, Spelbrink JN. Composition and dynamics of human mitochondrial nucleoids. Mol. Biol. Cell. 2003;14:1583–1596. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alam TI, Kanki T, Muta T, Ukaji K, Abe Y, Nakayama H, Takio K, Hamasaki N, Kang D. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 2003;31:1640–1645. doi: 10.1093/nar/gkg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Legros F, Malka F, Frachon P, Lombes A, Rojo M. Organization and dynamics of human mitochondrial DNA. J. Cell Sci. 2004;117:2653–2662. doi: 10.1242/jcs.01134. [DOI] [PubMed] [Google Scholar]
- 11.Kukat C, Wurm CA, Spahr H, Falkenberg M, Larsson NG, Jakobs S. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc. Natl Acad. Sci. USA. 2011;108:13534–13539. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 13.Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004;23:2423–2429. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wanrooij S, Goffart S, Pohjoismaki JL, Yasukawa T, Spelbrink JN. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35:3238–3251. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spelbrink JN, Toivonen JM, Hakkaart GA, Kurkela JM, Cooper HM, Lehtinen SK, Lecrenier N, Back JW, Speijer D, Foury F, et al. In vivo functional analysis of the human mitochondrial DNA polymerase POLG expressed in cultured human cells. J. Biol. Chem. 2000;275:24818–24828. doi: 10.1074/jbc.M000559200. [DOI] [PubMed] [Google Scholar]
- 16.Yasukawa T, Yang MY, Jacobs HT, Holt IJ. A bidirectional origin of replication maps to the major noncoding region of human mitochondrial DNA. Mol. Cell. 2005;18:651–662. doi: 10.1016/j.molcel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl Acad. Sci. USA. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis AF, Ropp PA, Clayton DA, Copeland WC. Mitochondrial DNA polymerase gamma is expressed and translated in the absence of mitochondrial DNA maintenance and replication. Nucleic Acids Res. 1996;24:2753–2759. doi: 10.1093/nar/24.14.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lentz SI, Edwards JL, Backus C, McLean LL, Haines KM, Feldman EL. Mitochondrial DNA (mtDNA) biogenesis: visualization and duel incorporation of BrdU and EdU into newly synthesized mtDNA in vitro. J. Histochem. Cytochem. 2009;58:207–218. doi: 10.1369/jhc.2009.954701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He J, Mao CC, Reyes A, Sembongi H, Di Re M, Granycome C, Clippingdale AB, Fearnley IM, Harbour M, Robinson AJ, et al. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J. Cell Biol. 2007;176:141–146. doi: 10.1083/jcb.200609158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilquin B, Taillebourg E, Cherradi N, Hubstenberger A, Gay O, Merle N, Assard N, Fauvarque MO, Tomohiro S, Kuge O, et al. The AAA+ ATPase ATAD3A controls mitochondrial dynamics at the interface of the inner and outer membranes. Mol. Cell Biol. 2010;30:1984–1996. doi: 10.1128/MCB.00007-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arnoult D, Soares F, Tattoli I, Castanier C, Philpott DJ, Girardin SE. An N-terminal addressing sequence targets NLRX1 to the mitochondrial matrix. J. Cell Sci. 2009;122:3161–3168. doi: 10.1242/jcs.051193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schagger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goffart S, Cooper HM, Tyynismaa H, Wanrooij S, Suomalainen A, Spelbrink JN. Twinkle mutations associated with autosomal dominant progressive external ophthalmoplegia lead to impaired helicase function and in vivo mtDNA replication stalling. Hum. Mol. Genet. 2009;18:328–340. doi: 10.1093/hmg/ddn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antes A, Tappin I, Chung S, Lim R, Lu B, Parrott AM, Hill HZ, Suzuki CK, Lee C-G. Differential regulation of full-length genome and a single-stranded 7S DNA along the cell cycle in human mitochondria. Nucleic Acids Res. 2010;38:6466–6476. doi: 10.1093/nar/gkq493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kornblum C, Nicholls TJ, Haack TB, Scholer S, Peeva V, Danhauser K, Hallmann K, Zsurka G, Rorbach J, Iuso A, et al. Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease. Nat. Genet. 2013;45:214–219. doi: 10.1038/ng.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milenkovic D, Matic S, Kuhl I, Ruzzenente B, Freyer C, Jemt E, Park CB, Falkenberg M, Larsson NG. TWINKLE is an essential mitochondrial helicase required for synthesis of nascent D-loop strands and complete mtDNA replication. Hum. Mol. Genet. 2013;22:1983–1993. doi: 10.1093/hmg/ddt051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tyynismaa H, Sembongi H, Bokori-Brown M, Granycome C, Ashley N, Poulton J, Jalanko A, Spelbrink JN, Holt IJ, Suomalainen A. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum. Mol. Genet. 2004;13:3219–3227. doi: 10.1093/hmg/ddh342. [DOI] [PubMed] [Google Scholar]
- 30.Jemt E, Farge G, Backstrom S, Holmlund T, Gustafsson CM, Falkenberg M. The mitochondrial DNA helicase TWINKLE can assemble on a closed circular template and support initiation of DNA synthesis. Nucleic Acids Res. 2011;39:9238–9249. doi: 10.1093/nar/gkr653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elachouri G, Vidoni S, Zanna C, Pattyn A, Boukhaddaoui H, Gaget K, Yu-Wai-Man P, Gasparre G, Sarzi E, Delettre C, et al. OPA1 links human mitochondrial genome maintenance to mtDNA replication and distribution. Genome Res. 2011;21:12–20. doi: 10.1101/gr.108696.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holt IJ, He J, Mao CC, Boyd-Kirkup JD, Martinsson P, Sembongi H, Reyes A, Spelbrink JN. Mammalian mitochondrial nucleoids: Organizing an independently minded genome. Mitochondrion. 2007;7:311–321. doi: 10.1016/j.mito.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 33.He J, Cooper HM, Reyes A, Di Re M, Sembongi H, Litwin TR, Gao J, Neuman KC, Fearnley IM, Spinazzola A, et al. Mitochondrial nucleoid interacting proteins support mitochondrial protein synthesis. Nucleic Acids Res. 2012;40:6109–6121. doi: 10.1093/nar/gks266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bogenhagen DF, Rousseau D, Burke S. The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 2008;283:3665–3675. doi: 10.1074/jbc.M708444200. [DOI] [PubMed] [Google Scholar]
- 35.Iborra FJ, Kimura H, Cook PR. The functional organization of mitochondrial genomes in human cells. BMC Biol. 2004;2:9. doi: 10.1186/1741-7007-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park CB, Larsson N-G. Mitochondrial DNA mutations in disease and aging. J. Cell Biol. 2011;193:809–818. doi: 10.1083/jcb.201010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kukat C, Wurm CA, Spåhr H, Falkenberg M, Larsson N-G, Jakobs S. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc. Natl Acad. Sci. USA. 2011;108:13534–13539. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown TA, Tkachuk AN, Shtengel G, Kopek BG, Bogenhagen DF, Hess HF, Clayton DA. Superresolution fluorescence imaging of mitochondrial nucleoids reveals their spatial range, limits, and membrane interaction. Mol. Cell. Biol. 2011;31:4994–5010. doi: 10.1128/MCB.05694-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meeusen S, Nunnari J. Evidence for a two membrane-spanning autonomous mitochondrial DNA replisome. J. Cell. Biol. 2003;163:503–510. doi: 10.1083/jcb.200304040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shutt TE, Gray MW. Twinkle, the mitochondrial replicative DNA helicase, is widespread in the eukaryotic radiation and may also be the mitochondrial DNA primase in most eukaryotes. J. Mol. Evol. 2006;62:588–599. doi: 10.1007/s00239-005-0162-8. [DOI] [PubMed] [Google Scholar]
- 41.Cheng X, Ivessa AS. Association of the yeast DNA helicase Pif1p with mitochondrial membranes and mitochondrial DNA. Eur. J. Cell Biol. 2010;89:742–747. doi: 10.1016/j.ejcb.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 42.Stuart JA, Mayard S, Hashiguchi K, Souza-Pinto NC, Bohr VA. Localization of mitochondrial DNA base excision repair to an inner membrane-associated particulate fraction. Nucleic Acids Res. 2005;33:3722–3732. doi: 10.1093/nar/gki683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aamann MD, Sorensen MM, Hvitby C, Berquist BR, Muftuoglu M, Tian J, de Souza-Pinto NC, Scheibye-Knudsen M, Wilson DM, Stevnsner T, et al. Cockayne syndrome group B protein promotes mitochondrial DNA stability by supporting the DNA repair association with the mitochondrial membrane. FASEB J. 2010;24:2334–2346. doi: 10.1096/fj.09-147991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duxin JP, Dao B, Martinsson P, Rajala N, Guittat L, Campbell JL, Spelbrink JN, Stewart SA. Human Dna2 Is a nuclear and mitochondrial DNA maintenance protein. Mol. Cell. Biol. 2009;29:4274–4282. doi: 10.1128/MCB.01834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.