SUMMARY
The DNA damage response (DDR) is activated by oncogenic stress, but the mechanisms by which this occurs, and the particular DDR functions that constitute barriers to tumorigenesis, remain unclear. We established a mouse model of sporadic onco-gene-driven breast tumorigenesis in a series of mutant mouse strains with specific DDR deficiencies to reveal a role for the Mre11 complex in the response to oncogene activation. We demonstrate that an Mre11-mediated DDR restrains mammary hyperplasia by effecting an oncogene-induced G2 arrest. Impairment of Mre11 complex functions promotes the progression of mammary hyperplasias into invasive and metastatic breast cancers, which are often associated with secondary inactivation of the Ink4a-Arf (CDKN2a) locus. These findings provide insight into the mechanism of DDR engagement by activated oncogenes and highlight genetic interactions between the DDR and Ink4a-Arf pathways in suppression of oncogene-driven tumorigenesis and metastasis.
INTRODUCTION
The DNA damage response (DDR) network comprises DNA repair, DNA damage signaling, apoptosis, and cell-cycle checkpoint functions (Ciccia and Elledge, 2010). Two lines of evidence support the view that the DDR is a barrier to tumorigenesis. Mutations affecting components of the DDR are frequently associated with predisposition to cancer (Ciccia and Elledge, 2010). Also, indices of DDR activation are evident in preneoplastic lesions or in cultured cells harboring activated oncogenes (Bart-kova et al., 2005; Gorgoulis et al., 2005). Despite supportive genetic data from in vitro and tumor inoculation studies (Bartkova et al., 2006; Di Micco et al., 2006), causal demonstration that the oncogene-induced DDR suppresses tumorigenesis within a tissue context remains limited (Gorrini et al., 2007; Squatrito et al., 2010; Takacova et al., 2012). In certain contexts, the role for ataxia telangiectasia mutated (ATM) in suppressing onco-gene-driven tumorigenesis was relatively minor, although these mouse models were limited by the fact that ATM−/− mice are prone to early spontaneous lymphomagenesis (Efeyan et al., 2009).
The mechanism for DDR activation in response to oncogene expression remains incompletely understood, but the prevailing view posits that oncogene activation leads to replication stress in the form of stalled, and subsequently collapsed, DNA replication forks (Halazonetis et al., 2008). Analysis of the ATRSeckel mouse has indicated that ATR may be required for cell viability upon oncogene activation, suggesting that DNA replication stress may indeed underlie these effects of oncogene activation (López-Contreras et al., 2012; Murga et al., 2011; Schoppy et al., 2012). However, since ATR promotes viability, rather than elimination of the oncogene-expressing cells, this outcome is not consistent with a barrier function for that component of the DDR. The purpose of this study was to delineate the particular aspects of the DDR network that constitute barriers to oncogenesis using a mouse model of sporadic, oncogene-driven breast cancer.
The Mre11 complex is a sensor of DNA double-strand breaks (Stracker and Petrini, 2011). Hypomorphic mutations in this complex, modeled in the mouse after alleles inherited in ataxiatelangiectasia-like disorder (A-TLD) and Nijmegen breakage syndrome (NBS), have facilitated the elucidation of the Mre11 complex's role in the ATM-dependent DDR. Here, we utilize these and other mutant mouse strains, individually and in combination, to define the tumor-suppressive functions of the DDR in mammary epithelium.
RESULTS
A Mouse Model of Sporadic, Oncogene-Induced Mammary Neoplasia
Expression of activated NeuT (Bargmann and Weinberg, 1988), the rodent ortholog of the ERBB2/HER2 oncogene, in the mammary epithelium of adult mice via the RCAS/MMTVTVA system (Du et al., 2006) results in early DDR activation, and oligoclonal tumors with an average latency of 5 months (Reddy et al., 2010). To delineate the aspects of the DDR primarily relevant for tumor suppression in the face of oncogene activation, we interbred MMTV-TVA mice with a variety of mutant mouse strains with established DDR deficiencies. Age-matched cohorts of female animals (12–18 weeks old) were injected with either RCAS-HA-NeuT or control virus via mammary intraductal injection. The genotypes analyzed were Mre11ATLD1/ATLD1, Nbs1ΔB/ΔB, Chk2−/−, Nbs1ΔC/ΔC Chk2−/−, p53515C/515C, p53−/−, and 53BP1−/−, each of which exhibits defects in DNA-damage-induced cell-cycle checkpoint activation, apoptosis, and/or DNA repair (Figures S1A and S1B available online; Liu et al., 2004; Shibata et al., 2010; Stracker et al., 2007, 2008; Stracker and Petrini, 2011; Theunissen et al., 2003; Williams et al., 2002). These mouse strains did not exhibit any histopathological deficits in mammary gland development (data not shown), circumventing the potential problem of differences in mammary tissue among the various genetic backgrounds confounding the analyses.
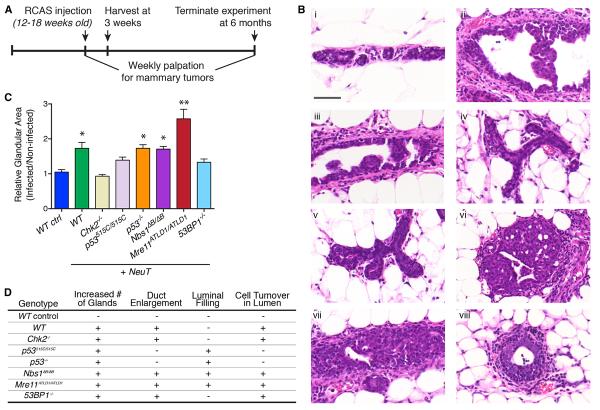
We injected 2 × 106 IU of RCAS virus per mammary gland, which resulted in a low in vivo infection efficiency of 0.04% in MMTV-TVA mammary epithelium, thereby mimicking sporadic oncogene activation within normal tissue (Figure S2). Onco-gene-induced histological changes were evaluated at 3 weeks postinfection, and additional cohorts of mice were monitored for the onset of mammary tumors (Figure 1A). Intraductal injection of RCAS-HA-NeuT into wild-type MMTV-TVA (WT) mice resulted in enlargement of the mammary ducts, with multifocal epithelial hyperplasia at 3 weeks postinjection (Figure 1B). Similar changes were not observed upon injection of a β-actin-expressing virus (Figure 1B). Mammary hyperplasia was correlated with oncogene expression, as the lesions were positive for HA expression (Figure S3). Hyperplasias also expressed estrogen receptor (ER) and were confined within an intact smooth muscle actin (SMA)- and p63-positive myoepithelial layer (Figure S3).
Figure 1. NeuT-Induced Mammary Hyperplasia in DDR Mutant Animals.
(A) The experimental timeline for mammary intraductal RCAS injection and subsequent analyses.
(B) Representative H&E staining of mammary glands from (Bi and Bii) WT, (Biii). Chk2−/−, (Biv) p53515C/515C, (Bv) p53−/−, (Bvi) Nbs1ΔB/ΔB, (Bvii) Mre11ATLD1/ATLD1, or (Bviii) 53BP1−/− mice, 3 weeks after injection with (Bi) 2 × 3 106 IU RCAS-HA-β-actin or (Bii–Bviii) 2 × 3 106 IU RCAS-HA-NeuT. Scale bar = 50 μm.
(C) Percent glandular area in the injected mammary gland #4 relative to the percent glandular area in the noninjected mammary gland #5, with at least three injected glands per genotype analyzed. Error bars indicate SEM. *p < 0.05 in comparison to WT control, and **p < 0.05 in comparison to WT + NeuT, using an unpaired t test.
(D) Summary of NeuT-induced qualitative changes in mammary gland morphology across the various genotypes.
The Mre11 Complex Restrains the Hyperplastic Response to Oncogene Expression in Mammary Epithelium
To analyze NeuT-induced hyperplasias in the various genotypes, we performed whole-slide digitization of at least three independent mammary glands for each genotype, harvested 3 weeks after mammary intraductal injection with RCAS-HA-NeuT, with corresponding matched control mammary glands derived from the same animals. We were thus able to analyze the effect of oncogene expression on mammary hyperplasia across geno-types. Representative images of hematoxylin and eosin (H&E)-stained tissue sections of control and NeuT-injected mammary glands are shown in Figure 1B. We performed digital quantification of glandular structures relative to total cellular content in the oncogene-expressing mammary glands and normalized this value to the glandular content observed in the matched control mammary glands (Figure 1C). In addition to these quantitative changes, there were also qualitative differences in morphology that distinguished the NeuT-induced hyperplasias across the various genotypes. These variations in mammary ductal enlargement, luminal filling, cellular turnover, and glandular density across the different genotypes are summarized in Figure 1D.
NeuT expression in Chk2−/− and Nbs1ΔC/ΔC Chk2−/− mammary epithelium produced hyperplasias that were only modestly dissimilar from WT (Figures 1B–1D; data not shown), suggesting that apoptosis and the intra-S phase checkpoint—diminished in both mutants (Stracker et al., 2008)—do not mediate the early response to oncogene activation. Consistent with that interpretation, p53515C/515C mutants, in which p53-dependent apoptosis is lost (Liu et al., 2004), also exhibited relatively modest hyper-plasia, although some morphological changes were noted (Figures 1B–1D). In contrast, p53−/− mammary glands resembled p53515C/515C morphologically, but exhibited more extensive NeuT-induced hyperplasia (Figures 1B–1D), consistent with additional deficiencies of the null mutant—including, but not limited to, induction of the G1/S checkpoint and senescence pathways. Deficiency of 53BP1, which is a mediator of the DDR that promotes NHEJ repair (Callen et al., 2013), did not have a major effect on the oncogene-induced hyperplastic response, relative to WT.
In contrast to the aforementioned genotypes, oncogene-induced hyperplasia was markedly distinct in Mre11ATLD1/ATLD1 and Nbs1ΔB/ΔB mammary glands relative to WT mammary glands (Figures 1B–1D). The Mre11 complex mutant genotypes exhibited florid hyperplasia in response to oncogene expression that frequently filled the lumen of the enlarged mammary ducts. Quantification of hyperplasia across the entire mammary gland revealed that Mre11ATLD1/ATLD1 was associated with the most significant degree of oncogene-induced proliferative change (Figure 1C). The Mre11ATLD1 and Nbs1ΔB mutations have previously been associated with defects in intra-S and G2/M checkpoints, reduced DDR signaling, and DSB repair defects (Stracker and Petrini, 2011). While there is significant overlap in their functional deficits, the Mre11ATLD1 and Nbs1ΔB mutants are not equivalent. Significantly, while the entire Mre11 complex is destabilized in Mre11ATLD1 mutant cells (Theunissen et al., 2003), the Nbs1ΔB mutant leaves Mre11 expression unperturbed (J.H.J.P., unpublished data), which may contribute to the less severe effect of Nbs1ΔB on oncogene-driven mammary hyperplasia (Figure 1C).
The Mre11 Complex Is Required for Oncogene-Dependent DDR Activation
We examined oncogene-dependent activation of the DDR in WT and Mre11ATLD1/ATLD1 mammary hyperplasias. Consistent with prior reports (Reddy et al., 2010), we observed the formation of γH2AX foci and accumulation of 53BP1 nuclear staining in WT hyperplasias after the introduction of NeuT (Figures 2A and 2B). However, these changes were nearly undetectable after oncogene expression in Mre11ATLD1/ATLD1 hyperplasias (Figures 2A and 2B). To quantify differences among the genotypes examined, we performed volumetric analysis of integral immunofluorescence staining intensity relative to nuclear volume in at least ten independent confocal imaging series of mammary hyperplasias representing at least three animals for each genotype (>1,000 nuclei per genotype). Although this analysis was unable to exclude admixed stromal cells, we observed a highly significant, >2-fold reduction in both NeuT-induced γH2AX foci formation and 53BP1 accumulation within Mre11ATLD1/ATLD1 lesions relative to WT (p < 0.0001; Figures 2A and 2B). In contrast to the effects of Mre11 complex hypomorphism, oncogene-dependent DDR activation was unperturbed in p53−/− mammary glands (Figure 2A; data not shown). These data demonstrate that the Mre11 complex is required for DDR activation upon NeuT expression.
Figure 2. The Oncogene-Induced DDR Is Mre11 Dependent.
(A) Confocal imaging of γH2AX immunofluorescence (IF) in (Ai) WT control, (Aii) WT + NeuT, (Aiii) Mre11ATLD1/ATLD1 + NeuT, and (Aiv) p53−/− + NeuT mammary glands. Scale bar = 20 μm. (Av) depicts volumetric analysis of γH2AX signal per nuclear volume in the various genotypes, with each data point representing a distinct nucleus. Error bars indicate the interquartile range around the median value. ****p < 0.0001, as per Kolmogorov-Smirnov (K-S) test.
(B) 53BP1 IF in (Bi) WT control, (Bii) WT + NeuT, (Biii) Mre11ATLD1/ATLD1 + NeuT, and (Biv) 53BP1−/− + NeuT mammary glands. Scale bar = 20 μm. The inset in (Bii) shows a higher magnification image of the nuclear 53BP1 nuclear signal. (Bv) depicts volumetric quantification of 53BP1 signal per nuclear volume from at least three glands for each genotype. Error bars represent the interquartile range relative to the median value. ****p < 0.0001, as per K-S test.
(C) Confocal imaging of pKAP1-Ser824 IF in (Ci) WT control, (Cii) WT + NeuT, and (Ciii) WT + 5Gy IR (4 hr). Scale bar = 20 μm.
The oncogene-driven, Mre11 complex-dependent DDR exhibited dissimilarities from that induced by ionizing radiation (IR). First, oncogene expression in the WT mammary gland resulted in finely punctate 53BP1 staining and did not induce the large foci that develop after irradiation of the mammary gland (Figure S4). In addition, phosphorylation of the ATM target KAP1 at Ser824 was not observed in the oncogene-expressing mammary gland, but was readily detected in IR-treated mammary tissue (Figure 2C). Similarly, we observed significantly less p53 stabilization in mammary epithelial cells after oncogene expression in comparison to irradiated tissue (Figure S4). Hence, the Mre11 complex-mediated response to oncogene activation appears to be qualitatively distinct from the response to clastogen-induced DNA damage.
The Mre11-Dependent DDR Mediates an Oncogene-Induced G2 Arrest
We examined apoptosis and growth arrest—functional outcomes of DDR activation—in hyperplastic lesions. While NeuT expression was associated with increased proliferation and apoptosis rates relative to control mammary glands, we did not observe a statistically significant difference in TUNEL or Ki67 positivity between WT and Mre11ATLD1/ATLD1 oncogene-induced hyperplasias (Figures 3A and 3B). To further characterize the cell-cycle status of oncogene-expressing hyperplasias, we examined expression of phospho-histone H3 serine 10 (pHH3-S10), which is deposited at pericentromeric heterochromatin in late G2 and becomes widely distributed as cells enter mitosis (Hendzel et al., 1997). We observed a 4-fold increase in pHH3-S10 staining in WT versus Mre11ATLD1/ATLD1 hyperplasias (p < 0.001; Figure 3C), which was unexpected given the significantly increased cellularity of Mre11ATLD1/ATLD1 hyperplasias. The pHH3-S10 staining pattern that we observed was punctate, and pHH3-S10-positive nuclei did not exhibit morphological features of mitosis (Figure 3C, inset), suggesting that the pHH3-S10 signal represented pericentromeric staining characteristic of late G2 cells rather than mitotic cells. Consistent with that interpretation, linescan plots of confocal microscopy images revealed colocalization of pHH3-S10 with DAPI-dense chromo-domains, in a manner parallel to that observed for the heterochromatic marker H3 trimethyl-K9 (Figure 4A).
Figure 3. The Mre11 Complex Mediates Oncogene-Induced pHH3-S10 Accumulation.
Immunohistochemistry (IHC) for (A) Ki67, (B) TUNEL reactivity, and (C) pHH3-S10 in (Ci) WT + NeuT and (Cii) Mre11ATLD1/ATLD1 mammary glands 3 weeks after oncogene introduction. Scale bar = 50 μm. Quantification of Ki67 (Aiii) and TUNEL (Biii) IHC signal per mammary glandular area from at least five distinct high-powered fields (HPFs) from three independent experimental animals for WT control, WT + NeuT, and Mre11ATLD1/ATLD1 + NeuT. Error bars indicate SEM, and p values were calculated using a two-tailed t test. The inset in (Ci) depicts a higher-magnification image of the pHH3-S10 nuclear signal observed, which is distinct from a mitotic pattern.
Figure 4. The Mre11 Complex Induces an Oncogene-Driven G2 Arrest.
(A) Representative confocal z slices of DAPI-stained (Ai) pHH3-S10 and (Aii) H3K9me3 IF in WT + NeuT mammary glands. Scale bar = 10 μm. Lower panels depict linescan plots for the indicated lines in (Ai) and (Aii).
(B) Confocal imaging of pHH3-S10 (red), centrin (green), and γ-tubulin (magenta) coIF in a representative pHH3-S10-negative (Bi) or pHH3-S10-positive cell (Bii) in a WT + NeuT hyperplastic mammary gland. Scale bar = 2 μm. (Biii) Quantification of proportion of postduplication (S/G2) and preduplication (G1) centrioles in pHH3-S10-negative (n = 45) or pHH3-S10-positive (n = 32) cells. p value calculated using Fisher's exact test.
(C) Confocal imaging of macroH2A IF in (Ci) WT control, (Cii) WT + NeuT, and (Ciii) Mre11ATLD1/ATLD1 + NeuT mammary glands, 3 weeks after injection. Scale bar = 20 μm. (Civ) Volumetric analysis of macro-H2A signal per nuclear volume. Error bars represent the interquartile range relative to the median value. p values were calculated according to the K-S nonparametric test (****p < 0.0001).
To exclude the possibility that the pHH3-S10-positive cells may have retained the signal even after traversal through mitosis, we performed coimmunofluorescence of pHH3-S10 with centrin and γ-tubulin, which stain the centriole and centrosome, respectively. Centriole duplication was evident in 84% of pHH3-S10-positive cells, compared to only 16% of pHH3-S10-negative cells (p < 0.0001; Figure 4B), indicating a cell-cycle state that is beyond the G1/S transition. These observations collectively suggest that NeuT expression in mammary epithelium activates a Mre11 complex-dependent G2 arrest or accumulation. Notably, this G2 arrest is distinct from the canonical IR-induced G2/M checkpoint, which is also Mre11 dependent (Theunissen et al., 2003). In that context, pHH3-S10 is not induced, suggesting that the heterochromatin-associated accumulation of this marker is oncogene specific.
To determine if the observed NeuT-induced accumulation of pHH3-S10 is associated with additional heterochromatic changes, we performed immunofluorescence for macro-H2a, a variant histone that is enriched in senescence-associated heterochromatin (Kennedy et al., 2010; Kreiling et al., 2011). WT mammary epithelium exhibited a unifocal nuclear staining pattern (Figure 4C) that was consistent with localization of macro-H2a to the inactivated X chromosome in female somatic cells (Nusinow et al., 2007). In contrast, oncogene-expressing WT cells induced macro-H2a at sites of pericentromeric heterochromatin (Figure 4C). Mre11ATLD1/ATLD1 glands, however, did not exhibit any recruitment of macro-H2a to heterochromatin after NeuT expression (Figure 4C). In fact, Barr body localization of macro-H2a was also lost in these tissues, suggesting a possible role for the Mre11 complex in heterochromatin specification during X chromosome inactivation. We again performed volumetric analyses on confocal z stack series (>1,000 nuclei per genotype), which demonstrated over 4-fold reduction in the mean nuclear signal for this marker in Mre11ATLD1/ATLD1 relative to WT mammary epithelium after NeuT expression (Figure 4C, p < 0.0001). Interestingly, senescence-associated β-galactosidase activity was not observed at this time point (data not shown), suggesting that the oncogene-induced Mre11-dependent G2 arrest entails heterochromatin remodeling that may not be equivalent to the stereotypical senescence program.
Mre11 Complex Hypomorphism Promotes Oncogene-Driven Mammary Tumorigenesis
The phenotypic severity of Mre11ATLD1/ATLD1 hyperplasias arising 3 weeks after infection presaged an increased risk of progression to malignancy. Whereas all RCAS-NeuT-injected WT mammary glands exhibited early hyperplasia, only 5% of these glands developed mammary tumors in the 6-month follow-up period (Figure 5A). In marked contrast, Mre11ATLD1/ATLD1 mice developed early onset (8–21 weeks postinjection) mammary tumors in 40% of the injected glands (p = 0.009; Figure 5A). Tumor latency in p53515C/515C animals trended slightly earlier than WT but did not achieve statistical significance, whereas Chk2−/− and Nbs1ΔC/ΔC Chk2−/− developed tumors at a rate indistinguishable from WT (Figure 5A; data not shown). p53−/− mice could not be analyzed for mammary tumorigenesis due to early lethality from the development of spontaneous lymphomas. Although hyperplastic lesions in Nbs1ΔB/ΔB were qualitatively similar to Mre11ATLD1/ATLD1 (Figure 1D), progression to tumors in Nbs1ΔB/ΔB was not observed (Figure 5A). While both Mre11ATLD1 and Nbs1ΔB mutants are associated with impaired intra-S and G2/M checkpoint functions, the increased tumorigenesis in Mre11ATLD1/ATLD1 may reflect the more pronounced chromosome fragility and apoptotic defects observed in this genotype (Stracker et al., 2008). Indeed, metaphase analysis of Mre11ATLD1/ATLD1 mammary tumor cells demonstrated a 7-fold increase in the rate of spontaneous chromosomal aberrations relative to cells from a WT mammary tumor (p = 0.0003; Figure S5).
Figure 5. Mre11ATLD1/ATLD1 Animals Are Predisposed to NeuT-Driven Mammary Tumorigenesis.
(A) Kaplan-Meier mammary tumor-free survival curves after introduction of RCAS-HA-NeuT in the various genotypes indicated. p values were calculated using the two-tailed log rank test, relative to the WT genotype.
(B) Mammary tumor growth doubling times for WT* (n = 5) or Mre11ATLD1/ATLD1 (n = 10) mammary tumors. Due to the limited number of tumors in non-Mre11ATLD1/ATLD1 genotypes, they were analyzed collectively under the designation WT*. Error bars indicate SEM, and p value was calculated using an unpaired t test.
(C) Representative H&E-stained sections from Mre11ATLD1/ATLD1 mammary tumors demonstrating aggressive histopathological features. Scale bar = 50 μm.
(D) Representative IHC staining of Mre11ATLD1/ATLD1 mammary tumors for (Di) ER, (Dii) EGFR, and (Diii) cytokeratin 5/6. Scale bars = 50 μm.
NeuT-Induced Mammary Tumors in Mre11ATLD1/ATLD1 Mice Frequently Inactivate the Ink4a-Arf Locus
The variable and prolonged latency of tumor onset in Mre11ATLD1/ATLD1 animals suggests that additional genetic alterations may be required for NeuT-mediated transformation of mammary epithelial cells. We examined p19Arf expression—a well-established oncogene-induced tumor-suppressive pathway (Sherr, 2001)—in the 3-week-old NeuT-expressing mammary hyperplasias from WT and Mre11ATLD1/ATLD1 animals. We observed >10-fold induction of p19Arf after oncogene expression in Mre11ATLD1/ATLD1 relative to control-injected mammary glands (Figure 6A). The extent of p19Arf induction in NeuT-expressing WT mammary glands was <50% of that observed in Mre11ATLD1/ATLD1 (p < 0.007, Figure 6A). Notably, there was no difference in HA-NeuT expression levels between the WT and Mre11ATLD1/ATLD1 mice that could account for the elevated levels of p19Arf (Figure S6A). As expected, p53 levels were modestly elevated in Mre11ATLD1/ATLD1 hyperplasias relative to WT (Figure S6B). This prominent oncogene-dependent activation of p19Arf—further potentiated in the context of Mre11 complex hypomorphism—suggested that the emergent mammary tumors may select for inactivation of this tumor-suppressive pathway.
Figure 6. Oncogene-Associated p19Arf Induction in Mammary Hyperplasias and Tumors.
(A) IHC for p19Arf in (Ai) WT control, (Aii) WT + NeuT, and (Aiii) Mre11ATLD1/ATLD1 + NeuT mammary glands, 3 weeks after intraductal injection. Scale bar = 50 μm. (Aiv) Quantification of p19Arf signal relative to glandular area from at least five HPFs in three independent mammary glands for each genotype.
(B) IHC for p19Arf in NeuT-induced mammary tumors from (Bi) WT*, (Bii) Ink4a-Arf nonmutated Mre11ATLD1/ATLD1, and (Biii) Ink4a-Arf mutated Mre11ATLD1/ATLD1 genotypes. Scale bars = 50 μm. (Biv) Digital quantification of p19Arf IHC relative to total area from at least three independent tumors for each genotype. Error bars indicate SEM, and p values were calculated using an unpaired t test.
To address this possibility, we performed targeted Sanger sequencing of the coding regions for Ink4a-Arf and trp53. Forty percent of the Mre11ATLD1/ATLD1 tumors exhibited alterations in the Ink4a-Arf locus, which were not observed in the WT or p53515C/515C tumors analyzed (Table S1). The observed alterations were frameshift mutations and deletions involving exon 2, predicting disruption of both the INK4a and p19Arf transcripts that are expressed from this locus. As expected, the ensuing mammary tumors that harbored mutations in the Ink4a-Arf locus exhibited loss of p19Arf expression (Figure 6B). Expression of p19Arf was also reduced in the remaining Mre11ATLD1/ATLD1 tumors relative to WT and p53515C/515C tumors, suggesting that alternative modes of p19Arf pathway suppression may be operative in these tumors (Figure 6B). Of note, we did not identify any trp53 mutations in a cohort of ten NeuT-driven tumors arising in Mre11ATLD1/ATLD1 mice, whereas the only WT tumor available for analysis harbored a trp53 mutation (data not shown). These data indicate that in the context of Mre11 complex hypomorphism, there is oncogene-dependent activation of p19Arf that is strongly selected against during mammary tumorigenesis.
NeuT-Driven Mre11ATLD1/ATLD1 Mammary Tumors Are Metastatic to Lung
Mre11ATLD1/ATLD1 mammary tumors exhibited high-grade histo-pathology, frequent mitoses, immune cell infiltration, necrosis, and regions of spindle cell and squamous metaplasia (Figure 5C). Moreover, ER staining was largely absent (Figure 5D), and the basal-like breast cancer markers EGFR and CK 5/6 were present in all Mre11ATLD1/ATLD1 mammary tumors analyzed (Figure 5D). The finding of EGFR overexpression is consistent with prior observations of synergy between Erb-B2/Neu and its heterodimerization partner EGFR in cellular transformation, and may explain why our NeuT-driven model gives rise to mammary tumors that resemble human basal-like breast cancers (Kokai et al., 1989).
Tumor growth rates were equivalent in Mre11ATLD1/ATLD1 versus WT mammary tumors (Figure 5B). Nonetheless, 70% (7/10) of the Mre11ATLD1/ATLD1 mammary tumors developed histologically confirmed lung metastases, whereas these were not observed in the other genotypes analyzed (p = 0.026; Figures 7A–7C). Lung metastases have also not been reported in the RCAS-NeuT/MMTV-TVA model as studied in other laboratories (Du et al., 2006; Reddy et al., 2010; Toneff et al., 2010). Of note, Mre11ATLD1/ATLD1 mammary tumors that also had Ink4a-Arf inactivation were uniformly metastatic (4/4, 100%), suggesting a possible role for disruption of this pathway in metastatic progression.
Figure 7. Mre11ATLD1/ATLD1 Mammary Tumors Are Highly Metastatic to Lung.
(A) Gross pathology of lungs from a tumor-bearing Mre11ATLD1/ATLD1 animal, depicting lung metastases.
(B) H&E staining of a representative NeuT-driven Mre11ATLD1/ATLD1 lung metastasis. Scale bar = 100 μm, and the inset shows a higher magnification of metastatic breast cancer cells within the lesion.
(C) Frequency of histologically confirmed lung metastases observed in Mre11ATLD1/ATLD1 and WT* mammary tumors-bearing animals. p value calculated using Fisher's exact test.
(D) Quantification of pulmonary bioluminescence (Di) at different time points in mouse cohorts injected via tail vein inoculation with 5 × 104 WT (n = 5) or Mre11ATLD1/ATLD1 (n = 8) mammary tumor cells. Error bars indicate SEM, and p value was calculated using a Wilcoxon rank sum test. Representative bioluminescence images (Dii) at different time points after cell inoculation, demonstrating lung metastatic outgrowth. Also shown are gross necropsy images of macroscopic lung metastases, corresponding to the associated bioluminescence images.
(E) Oncogene expression induces an Mre11-dependent DDR that constitutes an inducible barrier to tumorigenesis. In Mre11ATLD1/ATLD1 mice, oncogene expression no longer activates the DDR, and there is more extensive oncogene-induced hyperplasia. These hyperplastic lesions are notable for p19Arf upregulation. Upon selection for Ink4a-Arf pathway inactivation, the oncogene-induced lesions progress to genomically unstable breast tumors that metastasize to the lungs.
To clarify the mechanism underlying the observed increase in lung metastases associated with Mre11ATLD1/ATLD1 mammary tumors, we injected luciferase-labeled Mre11ATLD1/ATLD1 and WT primary mammary tumor cells into the tail vein of recipient animals. Serial bioluminescence imaging revealed similar rates of lung colonization for both genotypes of primary mammary tumor cell cultures, which was further confirmed by necropsy and histological assessment (Figure 7D; data not shown). This finding indicates that the augmented lung metastasis potential of Mre11ATLD1/ATLD1 mammary tumor cells is not due to an acquisition of metastatic colonization functions but rather is most likely due to an increased rate of metastatic dissemination from the primary tumor. This finding is in concordance with a previous demonstration that secondary organ colonization is not a rate-limiting step in lung metastasis formation by mouse mammary epithelial cells (Podsypanina et al., 2008).
DISCUSSION
Collectively, the findings presented here indicate that the Mre11 complex constitutes an inducible barrier to oncogene-driven neoplasia. In response to oncogene activation, the Mre11 complex mediates a G2 arrest that appears to be qualitatively distinct from that revealed in previous analyses of Mre11 complex-dependent DDR functions (Figure 7E; Stracker et al., 2004). The arrest is associated with heterochromatin changes, including the appearance of macroH2A and histone H3 (Ser10) phosphorylation. Histone H3 phosphorylation at pericentric heterochromatin begins early in G2 phase and expands as cells enter mitosis (Crosio et al., 2002). That fact, along with the finding that H3 phosphorylation arises in cells that have undergone centriole duplication, indicates that cells in oncogene-expressing hyperplasias accumulate in G2. We cannot exclude the possibility that other NeuT-expressing cells also arrest in G1 without the observed heterochromatic changes. In Mre11ATLD1/ATLD1 mammary epithelium, the NeuT-induced arrest is lost, and macroH2A and histone H3 phosphorylation are not detected in hyperplastic tissue, demonstrating that the G2 accumulation depends on the Mre11 complex.
The Mre11 complex-dependent G2 arrest does not appear permanent, as WT cells are capable at low frequency of progressing to tumors. When the arrest is attenuated, as in Mre11ATLD1/ATLD1, we observe more extensive oncogene-induced mammary hyperplasia, and a significantly greater likelihood of progression to invasive breast cancer. Although previous studies show that the Mre11 complex suppresses genome instability, and thus the risk of spontaneous DNA-damage-associated tumorigenesis (Stracker et al., 2008; Theunissen et al., 2003), this study demonstrates that the Mre11 complex also suppresses oncogene-driven neoplasia and tumorigenesis.
An important question concerns the underlying basis of the response to oncogene activation. Given the importance of the Mre11 complex in sensing DNA double-strand breaks and initiating an ATM-dependent DDR, a parsimonious interpretation is that oncogene activation results in DNA damage. Indeed, there are compelling genetic data supporting the induction of DNA replication stress upon oncogene activation (Bartkova et al., 2006; Campaner and Amati, 2012; Di Micco et al., 2006; Dominguez-Sola et al., 2007; López-Contreras and Fernandez-Capetillo, 2010). DNA replication stress is a common precursor of frank DNA damage when forks collapse (Allen et al., 2011), which would readily account for the induction of DNA damage upon oncogene induction. On the other hand, certain aspects of the data presented deviate from previously defined Mre11 complex responses to clastogen-induced DNA damage. Foremost is the nature of the oncogene-induced Mre11-dependent G2 arrest. As noted, this arrest is qualitatively dissimilar to the Mre11 complex/ATM-dependent G2/M arrest induced by DNA damage. In addition, KAP1 phosphorylation, induced in the mammary epithelium by IR in an ATM-dependent manner, was not observed in hyperplastic lesions. Ex vivo analysis of genomic integrity in oncogene-expressing mammary epithelial cells will be required to illuminate the molecular basis for Mre11 complex activation in this context. Given that individual oncogenes are likely to confer idiosyncratic effects, the significance of specific DDR components may vary according to the oncogene.
Potential crosstalk between the oncogene-induced DDR and the Arf tumor suppressor pathways has recently been described (Evangelou et al., 2013; Monasor et al., 2013; Velimezi et al., 2013). Our data provide direct evidence for a genetic interaction between these pathways during oncogene-driven tumorigenesis. We demonstrate that when Mre11 complex function is impaired, oncogene expression induces Arf expression, and Ink4a-Arf inactivation is commonly observed in the mammary tumors that ensue. The mechanism for how Mre11 hypomorphism promotes oncogene-induced Arf expression remains unclear. One possibility is that genotoxic stress associated with elevated chromosomal instability in Mre11ATLD1/ATLD1 mammary epithelium contributes to Arf induction. However, this explanation appears unlikely, as previous reports have suggested that Arf is not directly responsive to DNA damage (Christophorou et al., 2006). An alternative hypothesis is that Mre11-dependent ATM activation destabilizes Arf, which has been suggested in a recently published report (Velimezi et al., 2013). This would account for elevated Arf levels upon oncogene expression in the context of a hypomorphic Mre11 complex, and would create a strong selective pressure against Arf during oncogene-driven tumorigenesis. Indeed, we observe that 40% of the NeuT-induced mammary tumors that developed in Mre11ATLD1/ATLD1 mice had genetic inactivation of the Ink4a-Arf locus, and the remaining tumors exhibited reduced p19Arf expression, suggesting alternative modes of pathway suppression. These findings provide compelling genetic evidence for the cooperative roles of the Mre11 complex and Ink4a-Arf pathways in the suppression of oncogene-driven tumorigenesis and metastasis.
The breast cancers that develop in the MMTV-TVA/Mre11ATLD1/ATLD1 mouse model exhibit numerous hallmarks of aggressive disease, including high histopathological grade, elevated chromosomal instability, and frequent development of lung metastases. The behavior of the emergent tumors in Mre11ATLD1/ATLD1 mice suggests a link between increased chromosomal instability and an elevated rate of metastatic dissemination from the primary tumor. The observation that all of the Ink4a-Arf mutated mammary tumors were lung metastatic also raises the possibility that Arf loss promotes metastatic progression in the context of Mre11 complex impairment. The mechanisms underlying these associations remain to be explored.
The potential clinical relevance of the Mre11 complex in human breast cancer is highlighted by the observation that a subset of basal-like human breast cancers has markedly reduced expression of Mre11, Rad50, and Nbs1 (Bartkova et al., 2008). Our genetic data suggest that functional hypomorphism of this pathway may be a driver of breast tumorigenesis, genomic instability, and metastasis. Given the profound DDR defects associated with Mre11 complex hypomorphism (Stracker and Petrini, 2011), this subset of human breast cancer may exhibit exquisite DNA damage sensitivities that could be therapeutically exploited to improve clinical outcomes. Future work will explore the translational implications of these findings.
EXPERIMENTAL PROCEDURES
Virus Preparation
RCAS-Y-HA-NeuT, RCAS-HA-β-Actin, and RCAS-GFP were obtained from Y. Li (Baylor College of Medicine, Houston), details of which have been previously described (Du et al., 2006; Reddy et al., 2010). RCAS retroviruses were produced by transfection of viral constructs into DF1 chicken fibroblast cells (ATCC) using Lipofectamine 2000 (Invitrogen). DF1 cells were maintained in DMEM supplemented with 10% FBS, and grown at 39°C in the presence of 5% CO2. Retrovirus from the culture supernatant was concentrated 100-fold by ultracentrifugation at 125,000 × g for 90 min, and stored in aliquots at −80°C for titration and injection. Viral titer was determined by limiting dilution infection of DF-1 cells and assaying for expression of the encoded gene by fluorescence microscopy (GFP), western blot (anti-HA), and/or flow cytometry (anti-HA). The monoclonal rat anti-HA antibody (Roche, catalog number 11867431001) was either detected with an HRP-conjugated secondary antibody or assessed by flow cytometry after treatment with a FITC-conjugated version of the same primary antibody. The same batch of retroviruses was used for all experiments in this study.
Animal Experiments
All animal studies were done in compliance with a protocol approved by the Institutional Animal Care and Use Committee. MMTV-TVA transgenic mice were obtained from the Varmus laboratory. Chk2−/− and p53515C/515C mice were obtained from T. Mak (Ontario Cancer Institute, Ontario) and G. Lozano (The University of Texas MD Anderson Cancer Center, Houston), respectively. Mre11ATLD1/ATLD1, Nbs1ΔB/ΔB, and Nbs1ΔC/ΔC mice have been previously described (Stracker et al., 2007; Theunissen et al., 2003; Williams et al., 2002). Mice were raised in a pathogen-free facility and genotyped by PCR (details available upon request). Mammary intraductal injections were performed essentially as described previously (Behbod et al., 2009), using a 33G bluntend needle on a Hamilton syringe and a dissecting microscope to aid in visualization of the primary mammary duct. Adult virgin female mice, between the ages of 12 and 18 weeks, were injected with 2 × 106 IU of RCAS retrovirus in a 10 μl volume into the fourth mammary ductal tree. The viral suspension was labeled with trace amounts of bromophenol blue to facilitate visualization and to confirm success of the intraductal injection. Survival surgery was conducted under anesthesia with 100 mgkg−1 ketamine/10 mgkg−1 xylazine. Experimental lung metastasis assays were performed by injecting 5 × 104 primary mouse mammary tumor cells into the mouse tail vein in 100 μl of sterile PBS. Primary tumor cells were previously retrovirally transduced with a GFP-luciferase fusion construct, as previously described (Minn et al., 2005). Injected mice were imaged at various time points by introducing 3 mg D-Luciferin (15 mg/mL I PBS) intraperitoneally and bioluminescence detection 10 min later using an IVIS 100 imaging system (Minn et al., 2005).
Primary Cell Culture
Mammary tumors were harvested from tumor-bearing mice, minced with razor blades in sterile PBS, and then incubated for 2 hr at 37°C while shaking in digestion media (DME, 2.5% FBS, 10 mM HEPES, 1 mg/ml collagenase 3 [Worthington], 1 mg/ml hyaluronidase [Worthington]). Dissociated organoids were enriched by pulse centrifugation for 2 s at 600 × g five times. Single-cell suspensions were obtained by trypsinizing (0.5 mg/mL trypsin and 0.2 mg/mL EDTA) for 10 min and then treating with DNase I (0.1 mg/mL [Worthington] in DME) and dispase (1 mg/mL, STEMCELL Technologies) for 5 min. This cellular suspension was filtered through a 40 μm mesh, resuspended in MMEC growth media (DME, 10% FBS, 20 mM HEPES, 10 μg/ml insulin, 1% glutamine, and 1x antibiotic-antimycotic [Gemini]), and coplated with lethally irradiated (40 Gy) LA7 feeder cells (ATCC), as previously described (Ehmann et al., 1984).
Tissue Harvest and Processing
Mammary glands were surgically resected and either fixed overnight in 4% paraformaldehyde (PFA) and subsequently stored in 70% ethanol, or processed immediately for freezing in O.C.T. solution (Fisher Scientific) on dry ice. PFA-fixed tissues were paraffin embedded, and 5 μm tissue sections were used for immunohistochemistry and/or immunofluorescence (γH2AX). After a 5 min fixation in 4% PFA at room temperature, and 10 min exposure to permeabilization buffer (50 mM NaCl, 200 mM sucrose, 3 mM MgCl2, 10 mM HEPES [pH 7.9], and 0.5% Triton X-100), 53BP1, macro-H2a, pHH3-S10, and H3K9me3 immunofluorescence was performed on 10 μm sections of fresh frozen mammary glands embedded in O.C.T.
Tumor Sequencing
Genomic DNA was purified from liquid N2-flash-frozen mammary tumors using the QIAGEN DNeasy kit. Sanger sequencing was performed using forward and reverse primers providing coverage of the coding exons in the Ink4a-Arf and trp53 genes. Sequence reads were aligned using BLAST to identify regions of deletion or mutation.
Imaging and Quantification
H&E and immunohistochemistry slides were digitally scanned using the Mirax scanner and quantified using standardized color filter settings in the Meta-morph image analysis software. Image analysis represented the complete mammary gland #4 area of at least three mammary glands for each genotype. 53BP1 and macroH2a immunofluorescence was imaged using either 40x or 63x objectives on a SP5 confocal microscope. Z stacks were obtained, and later analyzed volumetrically, using the Imaris data analysis software. In brief, DAPI signal was used to define nuclear volumes, and the sum intensity of secondary antibody fluorescence intensity was normalized to the corresponding DAPI volume. Thousands of nuclei were analyzed from at least three different high-power field z stacks from three or more independent mammary glands for each genotype. Linescan data for pHH3-S10 and H3K9Me3 immunofluorescence were generated using Metamorph analysis of a representative z slice acquired using the 63x objective.
Statistical Analyses
Two-tailed statistical tests were used for all analyses. Survival analyses were performed by the Kaplan-Meier method, and statistical comparisons were made using the log rank test. Pairwise comparisons of nonsurvival data were analyzed using an unpaired t test, Fisher's exact test, Wilcoxon rank sum test, or Komolgorov-Smirnov test. Statistical analyses were performed using the Graphpad Prism software.
Details regarding immunostaining procedures can be found in Supplemental Information.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the Varmus, Y. Li, Pehrson, and M.F. Tsou laboratories for sharing reagents. We would also like to thank the Molecular Cytology Core Facility and M. Akram for providing expert technical assistance. Members of the Petrini laboratory, T. Kelly, and C. Sherr provided helpful comments and suggestions throughout the course of this study. We thank A. Koff, Y. Pylayeva-Gupta, and P. Gupta for critical reading of this manuscript. This work was supported by NIH grant GM59413 (J.H.J.P.) and DOD award W81XWH-10-1-0427 (G.P.G.).
Footnotes
SUPPLEMENTAL INFORMATION Supplemental Information includes six figures, one table, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2013.09.001.
REFERENCES
- Allen C, Ashley AK, Hromas R, Nickoloff JA. More forks on the road to replication stress recovery. J. Mol. Cell Biol. 2011;3:4–12. doi: 10.1093/jmcb/mjq049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI, Weinberg RA. Increased tyrosine kinase activity associated with the protein encoded by the activated neu oncogene. Proc. Natl. Acad. Sci. USA. 1988;85:5394–5398. doi: 10.1073/pnas.85.15.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Horejsí Z, Koed K, Kraämer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Tommiska J, Oplustilova L, Aaltonen K, Tamminen A, Heikkinen T, Mistrik M, Aittomaäki K, Blomqvist C, Heikkilaä P, et al. Aberrations of the MRE11-RAD50-NBS1 DNA damage sensor complex in human breast cancer: MRE11 as a candidate familial cancer-predisposing gene. Mol. Oncol. 2008;2:296–316. doi: 10.1016/j.molonc.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behbod F, Kittrell FS, LaMarca H, Edwards D, Kerbawy S, Heestand JC, Young E, Mukhopadhyay P, Yeh HW, Allred DC, et al. An intraductal human-in-mouse transplantation model mimics the subtypes of ductal carcinoma in situ. Breast Cancer Res. 2009;11:R66. doi: 10.1186/bcr2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E, Di Virgilio M, Kruhlak MJ, Nieto-Soler M, Wong N, Chen HT, Faryabi RB, Polato F, Santos M, Starnes LM, et al. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell. 2013;153:1266–1280. doi: 10.1016/j.cell.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campaner S, Amati B. Two sides of the Myc-induced DNA damage response: from tumor suppression to tumor maintenance. Cell Div. 2012;7:6. doi: 10.1186/1747-1028-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophorou MA, Ringshausen I, Finch AJ, Swigart LB, Evan GI. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature. 2006;443:214–217. doi: 10.1038/nature05077. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosio C, Fimia GM, Loury R, Kimura M, Okano Y, Zhou H, Sen S, Allis CD, Sassone-Corsi P. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 2002;22:874–885. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre' M, Nuciforo PG, Bensimon A, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- Du Z, Podsypanina K, Huang S, McGrath A, Toneff MJ, Bogoslovskaia E, Zhang X, Moraes RC, Fluck M, Allred DC, et al. Introduction of oncogenes into mammary glands in vivo with an avian retroviral vector initiates and promotes carcinogenesis in mouse models. Proc. Natl. Acad. Sci. USA. 2006;103:17396–17401. doi: 10.1073/pnas.0608607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Murga M, Martinez-Pastor B, Ortega-Molina A, Soria R, Collado M, Fernandez-Capetillo O, Serrano M. Limited role of murine ATM in oncogene-induced senescence and p53-dependent tumor suppression. PLoS ONE. 2009;4:e5475. doi: 10.1371/journal.pone.0005475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehmann UK, Peterson WD, Jr., Misfeldt DS. To grow mouse mammary epithelial cells in culture. J. Cell Biol. 1984;98:1026–1032. doi: 10.1083/jcb.98.3.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelou K, Bartkova J, Kotsinas A, Pateras IS, Liontos M, Velimezi G, Kosar M, Liloglou T, Trougakos IP, Dyrskjot L, et al. The DNA damage checkpoint precedes activation of ARF in response to escalating oncogenic stress during tumorigenesis. Cell Death Differ. 2013 doi: 10.1038/cdd.2013.76. Published online July 12, 2013. http://dx.doi.org/10.1038/cdd.2013.76. [DOI] [PMC free article] [PubMed]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr., Kastrinakis NG, Levy B, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, Martinato F, Sardella D, Verrecchia A, Bennett S, et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448:1063–1067. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, Bazett-Jones DP, Allis CD. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromo-some condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- Kennedy AL, McBryan T, Enders GH, Johnson FB, Zhang R, Adams PD. Senescent mouse cells fail to overtly regulate the HIRA histone chaperone and do not form robust Senescence Associated Heterochromatin Foci. Cell Div. 2010;5:16. doi: 10.1186/1747-1028-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokai Y, Myers JN, Wada T, Brown VI, LeVea CM, Davis JG, Dobashi K, Greene MI. Synergistic interaction of p185c-neu and the EGF receptor leads to transformation of rodent fibroblasts. Cell. 1989;58:287–292. doi: 10.1016/0092-8674(89)90843-x. [DOI] [PubMed] [Google Scholar]
- Kreiling JA, Tamamori-Adachi M, Sexton AN, Jeyapalan JC, Munoz-Najar U, Peterson AL, Manivannan J, Rogers ES, Pchelintsev NA, Adams PD, Sedivy JM. Age-associated increase in heterochromatic marks in murine and primate tissues. Aging Cell. 2011;10:292–304. doi: 10.1111/j.1474-9726.2010.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Parant JM, Lang G, Chau P, Chavez-Reyes A, El-Naggar AK, Multani A, Chang S, Lozano G. Chromosome stability, in the absence of apoptosis, is critical for suppression of tumorigenesis in Trp53 mutant mice. Nat. Genet. 2004;36:63–68. doi: 10.1038/ng1282. [DOI] [PubMed] [Google Scholar]
- López-Contreras AJ, Fernandez-Capetillo O. The ATR barrier to replication-born DNA damage. DNA Repair (Amst.) 2010;9:1249–1255. doi: 10.1016/j.dnarep.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Contreras AJ, Gutierrez-Martinez P, Specks J, Rodrigo-Perez S, Fernandez-Capetillo O. An extra allele of Chk1 limits oncogene-induced replicative stress and promotes transformation. J. Exp. Med. 2012;209:455–461. doi: 10.1084/jem.20112147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massagué J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monasor A, Murga M, Lopez-Contreras AJ, Navas C, Gomez G, Pisano DG, Fernandez-Capetillo O. INK4a/ARF limits the expansion of cells suffering from replication stress. Cell Cycle. 2013;12:1948–1954. doi: 10.4161/cc.25017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga M, Campaner S, Lopez-Contreras AJ, Toledo LI, Soria R, Montaña MF, D'Artista L, Schleker T, Guerra C, Garcia E, et al. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat. Struct. Mol. Biol. 2011;18:1331–1335. doi: 10.1038/nsmb.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow DA, Sharp JA, Morris A, Salas S, Plath K, Panning B. The histone domain of macroH2A1 contains several dispersed elements that are each sufficient to direct enrichment on the inactive X chromosome. J. Mol. Biol. 2007;371:11–18. doi: 10.1016/j.jmb.2007.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podsypanina K, Du YC, Jechlinger M, Beverly LJ, Hambardzumyan D, Varmus H. Seeding and propagation of untransformed mouse mammary cells in the lung. Science. 2008;321:1841–1844. doi: 10.1126/science.1161621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy JP, Peddibhotla S, Bu W, Zhao J, Haricharan S, Du YC, Podsypanina K, Rosen JM, Donehower LA, Li Y. Defining the ATM-mediated barrier to tumorigenesis in somatic mammary cells following ErbB2 activation. Proc. Natl. Acad. Sci. USA. 2010;107:3728–3733. doi: 10.1073/pnas.0910665107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoppy DW, Ragland RL, Gilad O, Shastri N, Peters AA, Murga M, Fernandez-Capetillo O, Diehl JA, Brown EJ. Oncogenic stress sensitizes murine cancers to hypomorphic suppression of ATR. J. Clin. Invest. 2012;122:241–252. doi: 10.1172/JCI58928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. The INK4a/ARF network in tumour suppression. Nat. Rev. Mol. Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- Shibata A, Barton O, Noon AT, Dahm K, Deckbar D, Goodarzi AA, Löbrich M, Jeggo PA. Role of ATM and the damage response mediator proteins 53BP1 and MDC1 in the maintenance of G(2)/M checkpoint arrest. Mol. Cell. Biol. 2010;30:3371–3383. doi: 10.1128/MCB.01644-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squatrito M, Brennan CW, Helmy K, Huse JT, Petrini JH, Holland EC. Loss of ATM/Chk2/p53 pathway components accelerates tumor development and contributes to radiation resistance in gliomas. Cancer Cell. 2010;18:619–629. doi: 10.1016/j.ccr.2010.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell Biol. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Theunissen JW, Morales M, Petrini JH. The Mre11 complex and the metabolism of chromosome breaks: the importance of communicating and holding things together. DNA Repair (Amst.) 2004;3:845–854. doi: 10.1016/j.dnarep.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Stracker TH, Morales M, Couto SS, Hussein H, Petrini JH. The carboxy terminus of NBS1 is required for induction of apoptosis by the MRE11 complex. Nature. 2007;447:218–221. doi: 10.1038/nature05740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Couto SS, Cordon-Cardo C, Matos T, Petrini JH. Chk2 suppresses the oncogenic potential of DNA replication-associated DNA damage. Mol. Cell. 2008;31:21–32. doi: 10.1016/j.molcel.2008.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takacova S, Slany R, Bartkova J, Stranecky V, Dolezel P, Luzna P, Bartek J, Divoky V. DNA damage response and inflammatory signaling limit the MLL-ENL-induced leukemogenesis in vivo. Cancer Cell. 2012;21:517–531. doi: 10.1016/j.ccr.2012.01.021. [DOI] [PubMed] [Google Scholar]
- Theunissen JW, Kaplan MI, Hunt PA, Williams BR, Ferguson DO, Alt FW, Petrini JH. Checkpoint failure and chromosomal instability without lymphomagenesis in Mre11(ATLD1/ATLD1) mice. Mol. Cell. 2003;12:1511–1523. doi: 10.1016/s1097-2765(03)00455-6. [DOI] [PubMed] [Google Scholar]
- Toneff MJ, Du Z, Dong J, Huang J, Sinai P, Forman J, Hilsenbeck S, Schiff R, Huang S, Li Y. Somatic expression of PyMT or activated ErbB2 induces estrogen-independent mammary tumorigenesis. Neoplasia. 2010;12:718–726. doi: 10.1593/neo.10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velimezi G, Liontos M, Vougas K, Roumeliotis T, Bartkova J, Sideridou M, Dereli-Oz A, Kocylowski M, Pateras IS, Evangelou K, et al. Functional interplay between the DNA-damage-response kinase ATM and ARF tumour suppressor protein in human cancer. Nat. Cell Biol. 2013;15:967–977. doi: 10.1038/ncb2795. [DOI] [PubMed] [Google Scholar]
- Williams BR, Mirzoeva OK, Morgan WF, Lin J, Dunnick W, Petrini JH. A murine model of Nijmegen breakage syndrome. Curr. Biol. 2002;12:648–653. doi: 10.1016/s0960-9822(02)00763-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.