Abstract
Kohlschütter–Tönz syndrome (KTS) is a rare autosomal recessive disorder characterized by amelogenesis imperfecta, psychomotor delay or regression and seizures starting early in childhood. KTS was established as a distinct clinical entity after the first report by Kohlschütter in 1974, and to date, only a total of 20 pedigrees have been reported. The genetic etiology of KTS remained elusive until recently when mutations in ROGDI were independently identified in three unrelated families and in five likely related Druze families. Herein, we report a clinical and genetic study of 10 KTS families. By using a combination of whole exome sequencing, linkage analysis, and Sanger sequencing, we identify novel homozygous or compound heterozygous ROGDI mutations in five families, all presenting with a typical KTS phenotype. The other families, mostly presenting with additional atypical features, were negative for ROGDI mutations, suggesting genetic heterogeneity of atypical forms of the disease.
Keywords: Kohlschütter–Tönz, ROGDI, amelogenesis imperfecta, epilepsy
Kohlschütter–Tönz syndrome (KTS; MIM #226750), first described in 1974 by Kohlschütter et al. in a Swiss family [Kohlschutter et al., 1974], is a rare autosomal recessive neurodegenerative syndrome characterized by a combination of features that include epilepsy, developmental delay and the most distinctive feature of amelogenesis imperfecta (yellow teeth with abnormal enamel). To date, 15 pedigrees have been reported in the literature, mostly consistent with an autosomal recessive mode of inheritance [Christodoulou et al., 1988; Donnai et al., 2005; Guazzi et al., 1994; Haberlandt et al., 2006; Kohlschutter et al., 1974; Mory et al., 2012; Musumeci et al., 1995; Petermoller et al., 1993; Schossig et al., 2012a; Wygold et al., 1996; Zlotogora et al., 1993]. The major clinical features of KTS include early onset seizures (between birth and 3 years of age) that are usually treatment resistant with various antiepileptic agents, psychomotor delay or regression starting in infancy, spasticity of the upper and lower limbs, and the developmental enamel defect (amelogenesis imperfecta) affecting both primary and secondary dentition. The enamel is thin and rough, and the teeth are yellow and prone to crumble. The teeth abnormalities seen (yellow amelogenesis imperfecta) would seem pathognomonic but similar tooth abnormalities have been reported in association with other complex neurological phenotypes [Linssen, et al., 1994]. Recently, Schossig et al. [2012a] and Mory et al. [2012] identified mutations in ROGDI (MIM #614574), a gene encoding a protein of currently unknown function, in several families with typical features of KTS. We present 10 families (six previously reported) with KTS or a phenotype that was very similar to KTS with the core features of epilepsy, developmental delay, and abnormal teeth affected by amelogenesis imperfecta. A combination of linkage analysis, exome sequencing, and Sanger sequencing identified novel mutations in ROGDI in five of these families.
Written informed consent was obtained from all families before sample collection and institutional ethics approval was also obtained. The family trees are illustrated in Figure 1A. The clinical details from each family are described below.
Family A: This family has been reported in 1988 [Christodoulou et al., 1988]. The parents originate from a small isolated town in Sicily. The affected individuals (six siblings) all had a similar phenotype characterized by delayed neuromotor development, epilepsy (onset between 7 and 22 months), and amelogenesis imperfecta of the hypoplastic rough type.
Family B: This is a new case of KTS originating from Germany. The affected boy was the first child of healthy nonconsanguineous parents; the pedigree is unremarkable. He was born after an uneventful pregnancy and his development was normal during the first half year of his life. At age 7 months, the boy presented with treatment resistant seizures followed by severe delay of motor and cognitive development. The primary dentition revealed yellow teeth with severe enamel defects. At the age of 2 years and 4 months, the boy had small stature (48 cm, −2.3 SD) and microcephaly.
Family C: This family has been reported in 1995 [Musumeci et al., 1995]. Briefly, it consists of two affected siblings (male and female) born from consanguineous parents (first degree cousins) of Sicilian origin. They had their first seizures between the age of 2 and 10 months, developed psychomotor regression starting at 2 years of age, and had yellow teeth with very thin and hypoplastic enamel. Presence of broad thumbs and toes is reported.
Family D: This family has been reported by Petermoller et al., [1993]. Briefly, the parents in this family are unrelated and of German origin. They had two affected children. Both had their first epileptic seizures at the age of 8 months, yellow teeth with amelogenesis imperfecta, and regression of psychomotor development starting at birth. One of the children died at the age of 21 years.
Family E: This family has been reported in 2012 [Schossig et al., 2012b]. The affected boy was the first child of healthy, unrelated parents of German origin and has two healthy sisters. He was born at term after an uneventful pregnancy and his development was normal until he had his first seizures at the age of 11 months. After that he showed delay of psychomotor development and partial regression of motor skills. His epilepsy was difficult to treat. His teeth showed a brownish discoloration.
Family F: This family from the USA has not been previously reported. The child presented at 8 months of age with seizures and was found to have infantile spasms. MRI of the brain was normal. He went on to have profound developmental handicaps, developed spasticity in all four extremities, and had intractable mixed focal onset and generalized myoclonic/tonic epilepsy. He was noted to have amelogenesis imperfecta as his teeth erupted and they were yellow. Spasticity has been treated with baclofen and he required bilateral femoral osteotomies for hip dislocations and spinal rod placement for scoliosis. At the age of 15, he was inconsistently visually interactive and has no expressive language; he was unable to sit or roll over.
Family G: This family of Scilian origin has been reported in 1994 [Guazzi et al., 1994]. This pedigree was complicated, with four different phenotypes occurring concurrently: amelogenesis imperfecta only, delayed psychomotor development, ataxia, and abnormal EEG, as well as neurological disorder or seizures and amelogenesis imperfecta. This family was atypical for KTS, given the later age at onset and the teeth affected by amelogenesis imperfecta being not truly yellow with missing enamel. Additionally, the inheritance looked dominant in this family but on the contrary, affected individuals from the last generation are born from first-degree cousins parents. It is possible that more than just one disorder is responsible for the complex phenotypes in this pedigree as there were individuals suffering only from amelogenesis imperfecta.
Family H: This English family has been previously reported by Donnai et al. [2005], and comprised of two affected siblings from healthy unrelated parents. Both siblings developed seizures at the age of 5 weeks (male) and 11 weeks (female), delayed psychomotor development starting at the age of 1 year, and yellow teeth with hypoplastic and hypomineralized enamel. They also had feeding problems with multiple food intolerances. The affected female was also clinically and genetically diagnosed with neurofibromatosis type 1.
Family I: This patient from the Netherlands is the second of two children born to healthy unrelated parents. He followed a delayed motor and cognitive development. Both primary and secondary dentition showed absent enamel of several but not all teeth. A brain MRI showed mild atrophy of the cranial part of the vermis and small pons, but was otherwise normal. Although he had periods of unusual quiet behavior and sleepiness thereafter true convulsions were not recognized. EEG studies showed multiple epileptiformic activities, mainly frontal and frontotemporal. Unusual morphological features were deeply set eyes, horizontal eyebrows, short nose, concave nasal ridge, anteverted nares, irregularly placed teeth, many of which without enamel, small ears, and mild hypermobility of the small joints. He had multiple warts.
Family J: This patient originating form the Netherlands showed a mildly delayed global development after normal pregnancy and delivery. Parents are unrelated. The girl had several episodes suggestive of epilepsy from the age of 18 months. A definitive diagnosis of epilepsy was made at the age 6 years. She had a variety of seizures types including atypical absence and myoclonic types. EEG showed continuous spike waves during sleep. Epilepsy was resistant to therapy. Her teeth were first white but discolored some time after eruption. The diagnosis of amelogenesis imperfecta was made by a pediatric dentist. Currently she has mild intellectual disability. Neurological examination shows no abnormalities besides mild clumsiness.
Figure 1.
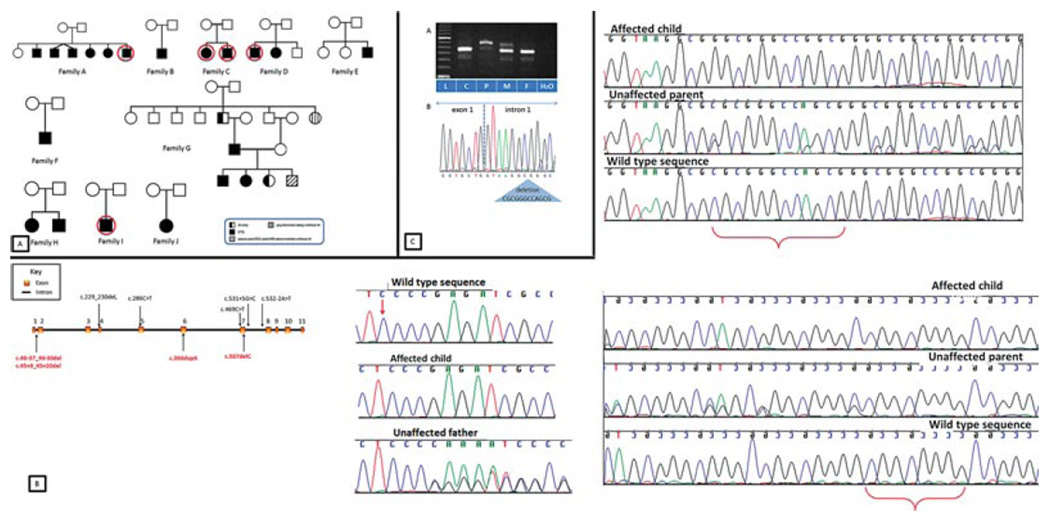
A: Family trees. Circles indicate samples that underwent whole exome sequencing. AI, amelogenesis imperfecta; KTS, Kohlschütter– Tönz syndrome. B: Gene structure with location of mutations and chromatograms showing the mutations reported in this manuscript. Black/above gene: previously reported mutations. Red/below: novel mutations reported in the present manuscript. Curly brackets and red arrow indicate deleted nucleotides in affected members. C: ROGDI transcript analyses in family E. (a) RT-PCR products generated with awild-type specific reversed primer in exon 6. The mother (M) shows a strong 400 bp wild-type amplicon and an aberrant approximately 460 bp band. The wild-type band is also found in the father (F) and the control (C). The affected child (P) shows predominantly the aberrant 460 bp band. All samples have an additional band of approximately 340 bp, which represents a small fraction of transcripts skipping of exon 4 as deduced from sequencing analysis of the RT-PCR products (data not shown). L, size standard. (b) Sequence of the RT-PCR product of the ROGDI transcripts in the affected child. The mutation c.45+9_45+20del-containing intron 1 is retained between the unaltered exons 1 and 2.
Linkage analysis was carried out as part of a previous PhD thesis [Lo, 2009] in families A, C, D, G, and H, and revealed a linkage peak (LOD score = 3.05) at D16S423 (chromosome 16) in parametric linkage analysis for the combined families, considering mode of inheritance as recessive (Supp. Table S1). Further evidence of a disease locus at 16p13.3–13.2 was provided by haplotype analysis of families A, C, and D and also by whole genome SNP genotyping on affected individuals from families A and C, which revealed a shared a region of homozygosity on the telomeric arm of chromosome 16 (data not shown).
Given the large number of genes in the linked region, we performed whole exome sequencing on affected individuals from families A, C, D, and I (Fig. 1; Table 1). All samples were enriched using the Illumina TruSeq exome capture system and sequenced on an Illumina HiSeq 2000 except of the proband from family C who was run using the Nimblegen SeqCap EZ enrichment kit and sequenced on one flowcell on the Illumina Genome Analyzer IIx (Supp. Table S2 for details on whole exome sequencing summary metrics). Using an allele frequency cutoff of 1% (based on both the 1,000 Genomes project and the NHLBI exome variant server), we found a novel homozygous putative pathogenic mutation in the linked region on chromosome 16 within the ROGDI gene in the affected individual from Family A (c.507delC, p.Glu170Argfs*72). The remaining exome sequence data from families C, D, and I did not identify additional rare variants in ROGDI though an in depth analysis showed that some of the ROGDI exons were not well covered (mainly exons 1 and 2). As ROGDI mutations were independently identified in KTS families at that time [Mory et al., 2012; Schossig et al., 2012a], we performed Sanger sequencing of all coding exons, exon–intron boundaries, and the short introns 1, 3, 8, 9, and 10 of ROGDI in all available, unresolved families (primers as published by Schossig et al., 2012). Mutations were named based on the Gen-Bank reference sequences with accession numbers NM_024589.2 and NP_078865.1; nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon (codon 1) in the reference sequence. Novel variants detected were submitted to the Leiden Open Variation database (http://www.lovd.nl/ROGDI).
Table 1.
Summary of Analysis and Results for Each Family Herein Studied
| Family ID | Phenotype | Exome sequencing |
Linkage and homozygosity mapping |
ROGDI Sanger sequencing |
Fragment and cDNA analysis |
ROGDI mutations |
|---|---|---|---|---|---|---|
| Family A | Core features | One affected proband | Linkage | Yes | N/A | Homoz c.507delC |
| Family B | Core features | N/A | N/A | Yes | q RT-PCR | Homoz c.507delC |
| Family C | Core features | Both affected siblings | Linkage and homozygosity mapping | Yes | Fragment analysis | Homoz c.46–37_46–30del |
| Family D | Core features | One affected proband | Linkage | Yes | cDNA and Fragment analysis | Homoz c.45+9_45+20del |
| Family E | Core features | N/A | N/A | Yes | q RT-PCR, cDNA amplification, and sequence analysis | Comp het c.366dupA/c.45+9_45+20del |
| Family F | Core features + spasticity | N/A | N/A | Yes | N/A | None |
| Family G | Atypical KTS | N/A | Linkage | Yes | N/A | None |
| Family H | Core features + feeding problems + neurofibromatosis 1 | N/A | Linkage | Yes | N/A | None |
| Family I | Core features + dysmorphic signs + multiple warts | Single affected member | N/A | Yes | N/A | None |
| Family J | Core features | N/A | N/A | Yes | q RT-PCR, cDNA amplification, and sequence analysis | None |
Core features = epilepsy, developmental delay, and amelogenesis imperfecta.
Numbering of cDNA bases according to the ROGDI reference sequence (GenBank NM_024589.2) with +1 as the A of the ATG translation initiation codon.
Homoz, Homozygous; Comp het, Compound heterozygous.
We identified ROGDI mutations in five out of 10 families analyzed. We confirmed the homozygous 1-bp deletion (c.507delC) in family A and found the same mutation homozygous in the affected child from family B. Both parents were confirmed to be heterozygous. Quantitative RT-PCR in the affected child from family B showed a marked reduction of the amount of ROGDI transcript to about 25% compared with controls (data not shown), indicating nonsense-mediated decay. Affected individuals in families C and D carried two different homozygous deletions in intron 1: c.46−37_46−30del and c.45+9_45+20del, respectively (Fig. 1B). The mutations were found heterozygous in the parents. The presence of these deletions was confirmed through fragment analysis (Supp. Fig. S2). The affected individual from family E was compound heterozygous for the deletion c.45+9_45+20del in intron 1 and a 1-bp duplication c.366dupA predicted to result in a frameshift with a premature stop codon after 19 amino acids, denoted p.Ala123Serfs*19. The father of this patient was heterozygous for c.366dupA and the mother for c.45+9_45+20del. Quantitative RT-PCR with specific primers spanning ROGDI exons 3–5, as previously described [Schossig et al., 2012a], showed a reduction of ROGDI transcripts to about 55% and 60% in both the mother and the affected child, respectively, when compared with controls (data not shown), whereas it did not show any reduction in the father.
To investigate the effect of c.45+9_45+20del on pre-mRNA processing we designed an RT-PCR primer pair that preferentially amplifies transcripts of the maternal allele in the affected child (see Supp. Methods section for details). We obtained an RT-PCR product around 60 bp longer than the wild-type RT-PCR products (Fig. 1C). Sequencing of this longer product showed retention of the mutated 61-bp intron 1 lacking the 12 deleted base pairs (Fig. 1C). This confirms that the deletion c.45+9_45+20del prevents recognition of intron 1 by the splicing machinery and leads to frameshift and creation of a premature stop codon (p.Glu16Valfs*57) in exon 3.
We failed to identify any putative ROGDI mutations in families F, G, H, I, and J. In addition, ROGDI expression levels in the affected from family J and his parents were comparable to those in normal controls and also direct cDNA sequencing did not uncover a ROGDI mutation. Independent genetic analyses in the proband from family I revealed a 2.75Mb de novo duplication on chromosome 7 (hg18 coordinates chr7: 5,100,000–7,855,000) (Supporting Information and Supp. Fig. S2).
In summary, the identification of a novel homozygous frameshift deletion (family A and family B) and the novel intronic short deletions (families C, D, and E) (one of which was confirmed to affect splicing in family E), all segregating with disease, fully supports the fact that bi-allelic loss-of-function variants in the gene ROGDI cause KTS and confirm the results of Schossig et al. [2012a] and Mory et al. [2012]. These families had typical KTS, and three of them had previously been linked to chromosome 16 in the region of the ROGDI locus.
Our results further emphasize the importance of sequencing non-coding regulatory regions, at least to within 20 bp of the exon for the analysis of Mendelian disorders. In particular, the intronic deletion c.45+9_45+20del is interesting as it is not a typical splice site mutation and prediction programs available through ALAMUT did not predict a significant alteration of splicing. The molecular mechanism in which this deletion disrupts the recognition of the already short intron 1 is unclear. Another important lesson from this study is that exome sequencing did not identify these intronic deletions, even though they were 20 bp away from the exon boundaries, perhaps because the region is repetitive and GC rich.
In five KTS families (families F, G, H, I, and J), no mutations were found in ROGDI. Clinically these families had an atypical KTS phenotype as they had additional features, except for the affected of family J who had a rather mild phenotype. Family F had spasticity. Family G presented late, had a likely dominant pattern of inheritance and teeth that were amelogenic but not truly yellow. The affected individuals from family H had all the features of KTS but were unusual in that they also had feeding difficulties, and family I had additional dysmorphic features. These clinical differences reflect that so far ROGDI mutations have been identified only in typical KTS cases. Even though the phenotype is complex, our data indicate that there may be a relatively specific pattern of clinical features associated with mutations in this gene. Although we cannot rule out difficult to detect deep intronic mutations or compound heterozygous exonic copy number variations, this is improbable in families G and Has they did not share the same haplotypes on chromosome 16p13.3–13.2. Additionally, in family J additional RT-PCR sequencing was unremarkable in the affected individual and quantitative RT-PCR showed no reduction of mRNA. Our data indicate that atypical phenotypes of KTS are more likely due to mutations of other genes; indeed, extensive genetic analysis in the proband from family I (whole exome sequencing and CGH array) led to the identification of a de novo 2.75 Mb duplication on chromosome 7, comprising about 60 genes (NCBI mapviewer). It is likely that a dosage effect of one of the genes in this region is causing a KTS-like phenotype, although this was not seen in any of the other families.
To conclude, we have confirmed that a typical KTS phenotype [Schossig et al., 2012b] is generally caused by loss of function mutations in ROGDI and identified intronic deletions in ROGDI that were missed using whole exome sequencing. A number of families with atypical KTS phenotype were negative for ROGDI mutations and failed to show homozygosity in the ROGDI region, suggesting genetic heterogeneity of atypical forms of the disease.
Supplementary Material
Acknowledgments
We would like to thank the patients and families for their essential help with our work. We are grateful to the Medical Research Council (MRC) for their support, the National Organisation for Rare Disorders (NORD), the Dystonia Medical Research Foundation (DMRF), the Brain Research Trust (BRT), and the MSA trust. This work was undertaken at UCLH/UCL, which received a proportion of funding from the Department of Health’s NIHR Biomedical Research Centres funding scheme.
We would like to thank the NHLBI GO Exome Sequencing Project and its ongoing studies which produced and provided exome variant calls for comparison: the Lung GO Sequencing Project (HL-102923), the WHI Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926), and the Heart GO Sequencing Project (HL-103010).
Contract grant sponsor: Intramural Research Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services (Z01 AG 000958–08).
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- Christodoulou J, Hall RK, Menahem S, Hopkins IJ, Rogers JG. A syndrome of epilepsy, dementia, and amelogenesis imperfecta: genetic and clinical features. J Med Genet. 1988;25:827–830. doi: 10.1136/jmg.25.12.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnai D, Tomlin PI, Winter RM. Kohlschutter syndrome in siblings. Clin Dysmorphol. 2005;14:123–126. [PubMed] [Google Scholar]
- Guazzi G, Palmeri S, Malandrini A, Ciacci G, Di Perri R, Mancini G, Messina C, Salvadori C. Ataxia, mental deterioration, epilepsy in a family with dominant enamel hypoplasia: a variant of Kohlschutter-Tonz syndrome? Am J Med Genet. 1994;50:79–83. doi: 10.1002/ajmg.1320500117. [DOI] [PubMed] [Google Scholar]
- Haberlandt E, Svejda C, Felber S, Baumgartner S, Gunther B, Utermann G, Kotzot D. Yellow teeth, seizures, and mental retardation: a less severe case of Kohlschutter-Tonz syndrome. Am J Med Genet A. 2006;140:281–283. doi: 10.1002/ajmg.a.31071. [DOI] [PubMed] [Google Scholar]
- Kohlschutter A, Chappuis D, Meier C, Tonz O, Vassella F, Herschkowitz N. Familial epilepsy and yellow teeth—a disease of the CNS associated with enamel hypoplasia. Helv Paediatr Acta. 1974;29:283–294. [PubMed] [Google Scholar]
- Linssen WH, Van den Bent MJ, Brunner HG, Poels PJ. Deafness, sensory neuropathy, and ovarian dysgenesis: a new syndrome or a broader spectrum of Perrault syndrome? Am J Med Genet. 1994;51:81–82. doi: 10.1002/ajmg.1320510117. [DOI] [PubMed] [Google Scholar]
- Lo C-N. Genetics in epilepsy. PhD thesis. London, UK: University College London; 2009. p. 319. [Google Scholar]
- Mory A, Dagan E, Illi B, Duquesnoy P, Mordechai S, Shahor I, Romani S, Hawash-Moustafa N, Mandel H, Valente EM, Amselem S, Gershoni-Baruch R. A nonsense mutation in the human homolog of Drosophila rogdi causes Kohlschutter-Tonz syndrome. Am J Hum Genet. 2012;90:708–714. doi: 10.1016/j.ajhg.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musumeci SA, Elia M, Ferri R, Romano C, Scuderi C, Del Gracco S. A further family with epilepsy, dementia and yellow teeth: the Kohlschutter syndrome. Brain Dev. 1995;17:133–138. doi: 10.1016/0387-7604(95)00013-2. discussion 142–133. [DOI] [PubMed] [Google Scholar]
- Petermoller M, Kunze J, Gross-Selbeck G. Kohlschutter syndrome: syndrome of epilepsy–dementia–amelogenesis imperfecta. Neuropediatrics. 1993;24:337–338. doi: 10.1055/s-2008-1071567. [DOI] [PubMed] [Google Scholar]
- Schossig A, Wolf NI, Fischer C, Fischer M, Stocker G, Pabinger S, Dander A, Steiner B, Tonz O, Kotzot D, Haberlandt E, Amberger A, et al. Mutations in ROGDI cause Kohlschutter-Tonz syndrome. Am J Hum Genet. 2012a;90:701–707. doi: 10.1016/j.ajhg.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schossig A, Wolf NI, Kapferer I, Kohlschutter A, Zschocke J. Epileptic encephalopathy and amelogenesis imperfecta: Kohlschutter-Tonz syndrome. Eur J Med Genet. 2012b;55:319–322. doi: 10.1016/j.ejmg.2012.02.008. [DOI] [PubMed] [Google Scholar]
- Wygold T, Kurlemann G, Schuierer G. Kohlschutter syndrome-an example of a rare progressive neuroectodermal disease. Case report and review of the literature. Klin Padiatr. 1996;208:271–275. doi: 10.1055/s-2008-1046481. [DOI] [PubMed] [Google Scholar]
- Zlotogora J, Fuks A, Borochowitz Z, Tal Y. Kohlschutter-Tonz syndrome: epilepsy, dementia, and amelogenesis imperfecta. Am J Med Genet. 1993;46:453–454. doi: 10.1002/ajmg.1320460422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.