Abstract
The impact of the Oncotype Dx (ODX) breast cancer assay on adjuvant chemotherapy (ACT) treatment decisions has been evaluated in many previous studies. However, it can be difficult to interpret the collective findings, which were conducted in diverse settings with limited sample sizes. We conducted a systematic review and meta-analysis to synthesize the results and provide insights about ODX utility. Studies, identified from PubMed, Embase, ASCO, and SABCS, were included if patients had ER+, node –, early-stage breast cancer, reported use of ODX to inform actual ACT decisions. Information was summarized and pooled according to: (1) distribution of ODX recurrence scores (RS), (2) impact of ODX on ACT recommendations, (3) impact of ODX on ACT use, and (4) proportion of patients following the treatment suggested by the ODX RS. A total of 23 studies met inclusion criteria. The distribution of RS categories was 48.8 % low, 39.0 % intermediate, and 12.2 % high (21 studies, 4,156 patients). ODX changed the clinical-pathological ACT recommendation in 33.4 % of patients (8 studies, 1,437 patients). In patients receiving ODX, receipt of ACT were: 28.2 % overall, 5.8 % low, 37.4 % intermediate, and 83.4 % high. High RS patients were significantly more likely to follow the treatment suggested by ODX versus low RS patients RR: 1.07 (1.01–1.14). The pooled results are consistent with most individual studies to date. The increased proportion of intermediate scores relative to original estimates may have implications for the clinical utility and cost impacts of testing. In addition, low versus high RS patients were significantly more likely to follow the ODX results, suggesting a tendency toward less aggressive treatment, despite a high ODX RS. Finally, there was a lack of studies on the impact of ODX on ACT use versus standard approaches, suggesting that additional studies are warranted.
Keywords: Gene expression, Breast cancer, Meta-analysis, Adjuvant chemotherapy
Introduction
Historically, the decision to treat estrogen-receptor-positive, lymph node-negative, breast cancer patients with adjuvant chemotherapy (ACT) has been guided by clinical and pathological factors, in conjunction with clinician and patient preferences [1, 2]. In the absence of ACT, ~15 % of patients have a recurrence within 5 years [3]. However, under the traditional clinical-pathological decision-making approach, between 75 and 92 % of women receive ACT [2, 4, 5]. These findings suggest that 60–77 % of patients may be exposed to chemotherapy toxicity and cost with little or no clinical benefit. For this reason, there is a need for better methods to evaluate the recurrence risk and inform ACT treatment decisions. Gene-expression profiling (GEP) is an emerging clinical strategy intended to meet this need by using genomic information to inform risk prediction and treatment selection [6–8].
Several retrospective analyses of clinical trials have shown that GEP testing can provide prognostic and predictive information for patients with estrogen-receptor-positive, lymph node-negative early-stage breast cancer [8–11]. The Oncotype DX™ (ODX) breast cancer assay is the most common GEP in the United States, and was developed by Genomic Health (Redwood City, CA) [12]. The ODX test evaluates the expression levels of 21 genes (16 cancer-related genes and 5 reference genes), which have been shown to be associated with tumor proliferation, invasion, and estrogen signaling [13]. Information about the quantitative levels of gene expression is combined in a mathematical algorithm, along with clinical-pathological data, to provide a recurrence score (RS) that has been demonstrated to be significantly correlated with the likelihood of breast cancer recurrence within 10 years of initial diagnosis [9, 14, 15]. The impact of ODX testing on disease-free and overall survival is currently being evaluated in a prospective clinical trial, but primary results are not expected until 2017 [9, 16, 17]. In the meantime, both American Society of Clinical Oncology (ASCO) and National Comprehensive Cancer Network (NCCN) clinical guidelines recommend the use of ODX in certain situations, and Genomic Health reports the sales of over 200,000 tests since entering the United States market in 2004 [1, 18, 19].
A number of studies have suggested that the ODX test provides better risk discrimination relative to clinical-pathological approaches, and may result in improved clinical outcomes. Further, these improved outcomes may be achieved cost-effectively and may even decrease direct medical costs [9, 15, 20, 21]. There is uncertainty about each of these impacts, and there may be even greater uncertainty when ODX is used in routine clinical practice [7, 22]. For example, if patients and physicians in community settings do not follow the ACT treatment course suggested by the ODX RS, or if the ODX test performs poorly, the health benefits of testing could be attenuated, or there could be clinical harm (relative to standard clinical-pathological decision making strategies). Furthermore, if the ODX test has little to no impact on clinical decision-making in routine clinical practice, the high cost of testing (currently about $4,000) may be incurred without a commensurate health benefit [21].
Many studies have evaluated ODX and its impact on ACT treatment decisions [21, 23–45]. However, it can be difficult for patients, clinicians, and other stakeholders to collectively interpret these findings because the studies were conducted in a variety of different settings, and many include small sample sizes. We conducted a systematic review and meta-analysis to help stakeholders navigate and interpret the ODX literature. Our objective was to provide new insights about the clinical utility of ODX in community practice by summarizing and synthesizing the current ODX evidence base.
Methods
This study was performed in accordance with the PRISMA statement for the conduct of meta-analyses [46]. Below, we address each of the 12 methods-based items specified in the PRISMA group's 2009 statement document [46].
Literature search strategy
We searched MEDLINE and EMBASE electronic databases using the terms: “ODX”, “21-gene profile”, and “Chemotherapy” to identify potential studies for inclusion in the analysis. Additionally, we conducted computerized searches of abstracts presented at the Annual Meetings of the ASCO and San Antonio Breast Cancer Symposium (SABCS) using the same search terms. The references in all reviewed articles were screened to identify additional studies that were not identified by our search strategy. The included MEDLINE, EMBASE, and ASCO studies were published in English between June of 2005 and March of 2012. The included SABCS studies were published between January of 2008 and March of 2012, the full range of electronic abstracts available at the time of this analysis.
Selection criteria
All studies included in our pooled analyses met the following inclusion criteria: (1) patients had estrogen-receptor-positive, lymph-node-negative, and early-stage breast cancer; (2) reported use of ODX RS to inform clinical ACT decisions; and (3) reported outcomes of interest, including: distribution of ODX RS; impact of ODX on ACT recommendations; impact of ODX on ACT use; and proportion of patients following the treatment suggested by the ODX RS.
Several studies that we identified using the inclusion criteria above were updates of earlier publications, and involved the same patient population. In such cases, we only included the most recent publication results in our analyses.
Data extraction
Data were extracted from each publication by two abstractors (JR, JC) using a data abstraction form developed in Microsoft Excel (Redmond, WA, 2011). For each publication, we recorded: first author's name; publication date; total number of patients in the study sample; proportion with high, intermediate, and low RS; study sponsor; study setting (e.g., academic medical center, community practice, etc.); study country; study design (randomized, prospective observational, and retrospective observational); proportion of clinicians changing ACT recommendations after receiving ODX RS; proportion of patients receiving ACT after receiving ODX RS; and the proportion following the treatment suggested by the ODX RS. In cases where information was not presented, the article's corresponding author was contacted to obtain the information. We classified variables as “not reported” in cases where they were not provided by the publication or author.
Statistical methods
We conducted pooled analyses to evaluate: the proportion ODX classified as high, intermediate, and low-recurrence risk; proportion receiving ACT by ODX RS classification; overall proportion receiving ACT after ODX testing; proportion of clinicians changing ACT recommendation after ODX testing (relative to recommendation based on clinical-pathological factors only); rate ratio (RR) for high versus low, high versus intermediate, and intermediate versus low ODX recurrence risk patients receiving ACT; and RR for patients following ODX test recommendation for ACT in those classified as high versus low recurrence risk. For the latter analyses, the RR was abstracted directly from publications, or calculated using the reported count data. In the analysis of the proportion of patients following the ODX RS treatment “recommendation”, we assumed that the patients with high RS were recommended to receive ACT, and low RS were not recommended to receive ACT.
We calculated the mean proportion and 95 % confidence interval (CI) classified by ODX as high, intermediate, and low by weighting study outcomes by total sample size for the study. Similar approaches were used to calculate the mean proportion receiving ACT and proportion of clinicians changing their ACT recommendation after ODX testing. We calculated the pooled RR and 95 % CI by weighting study outcomes by sample size.
In the analyses of the association between ODX classification and receipt of ACT, a RR of 1 indicates a lack of association between RS classification and receipt of ACT, a RR greater than 1 indicates a greater probability of receiving ACT relative to the referent group, and an RR less than 1 indicates a lower probability of receiving ACT relative to the referent group.
In the analysis of the proportion of patients following the ODX RS treatment “recommendation”, a HR of 1 indicates an equal proportion of patients classified as high and low by ODX following the ACT recommendation, a HR of greater than 1 indicates a greater proportion of patients classified as high by ODX following the ACT recommendation, and a HR less than 1 indicates a greater proportion of patients classified as low by ODX following the ACT recommendation. We did not compare low versus intermediate risk or high versus intermediate risk as the treatment implications of intermediate risk classification are somewhat uncertain, as has been previously discussed [7, 22, 47].
Assessment of heterogeneity
We initially calculated the pooled RR estimates with a fixed effects model. In pooled analyses where the fixed effects p value for the I2 statistic was less than 0.10 (indicating significant heterogeneity across studies), we reported the pooled estimate using a random effects model.
Assessment of publication bias
Each pooled analysis outcome was qualitatively assessed for publication bias using funnel plots. These plots illustrate the relationship between the effect size and standard error in the studies included in the given analysis. We assessed the publication bias by examining whether study outcomes were symmetrically distributed around the pooled effect estimate in the funnel plot. Asymmetric distribution of study outcomes above or below the pooled estimate can indicate that the pooled estimate includes studies with disproportionately positive or negative findings [48].
We did not assess the methodological quality of the included publications, as the quality assessment in meta-analysis is controversial, and results can be misleading [49, 50].
For all tests, a p value lower than 0.05 was considered statistically significant. All statistical tests were two-sided. All statistical analyses were carried out using RevMan 5.0 software (Copenhagen, Denmark, 2008).
Results
Study characteristics
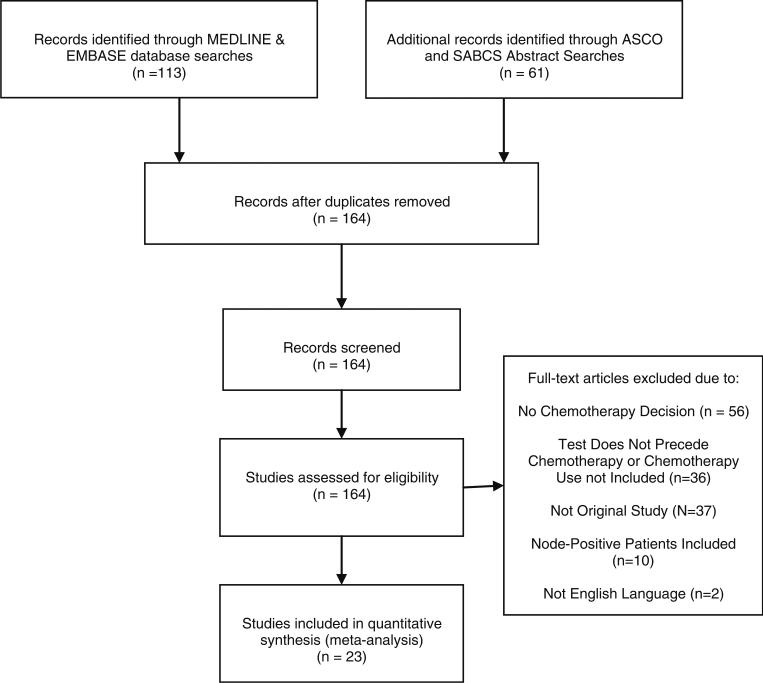
Our MEDLINE electronic database search identified a total of 102 studies, and 14 met inclusion criteria. Our EMBASE identified a total of 11 studies, but only 1 was not already identified in MEDLINE. Additionally, our search of ASCO meeting abstracts (2005–2012) identified 19 abstracts, and 3 met inclusion criteria and were unique studies that were not early reports of those identified through MEDLINE and EMBASE searches. Our SABCS meeting abstract search identified 42 abstracts, and 5 met inclusion criteria and were unique studies that were not early reports of those from our MEDLINE and EMBASE searches, or already captured through our ASCO abstract search. After we applied our inclusion criteria, we were left with 21 studies that were included in our analysis. Additional information about this search strategy is illustrated in flowchart form in Figs. 1 and 2.
Fig. 1.
Search strategy flow diagram
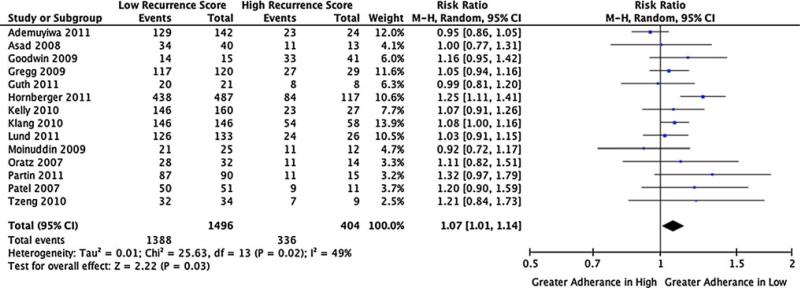
Fig. 2.
Proportion following treatment ODX recommendation, assuming that high RS patients are recommended for adjuvant chemotherapy, and low RS patients are not recommended adjuvant chemotherapy
Table 1 lists the 23 unique studies identified by our search strategy, as well as key study characteristics, and the analyses to which they contributed. The median sample size in the included studies was 135 (IQR: 77–258), 15 (65 %) were published in peer-reviewed journals (vs. conference abstracts), 16 (76 %) were conducted in the United States, and 17 (74 %) were conducted at academic medical centers.
Table 1.
Key characteristics of all studies meeting study inclusion criteria
| Study | Source | Academic medical center | Country | Total sample size |
|---|---|---|---|---|
| Ademuyiwa et al. [23] | Journal | Yes | U.S. | 276 |
| Asad et al. [24] | Journal | Yes | U.S. | 85 |
| de Boer et al. [42] | Conference | Yes | Australia | 101 |
| Geffen et al. [25] | Journal | Yes | Israel | 135 |
| Goodwin et al. [40] | Conference | Yes | U.S. | 105 |
| Gregg et al. [41] | Conference | Mixed | U.S. | 244 |
| Guth et al. [39] | Conference | Mixed | U.S. | 47 |
| Henry et al. [26] | Journal | Yes | U.S. | 29 |
| Holt [43] | Conference | No | U.K. | 142 |
| Hornberger et al. [21] | Journal | Mixed | U.S. | 924 |
| Joh et al. [28] | Journal | Yes | U.S. | 154 |
| Kelly et al. [29] | Journal | Yes | U.S. | 309 |
| Klang et al. [30] | Journal | Yes | Israel | 368 |
| Lo et al. [31] | Journal | Yes | U.S. | 89 |
| Lund et al. [32] | Journal | Yes | U.S. | 272 |
| Moinuddin and Goodman [38] | Conference | No | U.S. | 69 |
| Oratz et al. [33] | Journal | No | U.S. | 68 |
| Partin and Mamounas [34] | Journal | Yes | U.S. | 169 |
| Patel et al. [45] | Conference | Yes | U.S. | 100 |
| Rezai et al. [44] | Conference | Mixed | Germany | 244 |
| Tatarian et al. [35] | Journal | Yes | U.S. | 63 |
| Tzeng et al. [36] | Journal | Yes | U.S. | 68 |
| Wolf et al. [37] | Journal | Mixed | Israel | 300 |
Twenty-one studies and 4,156 patients were included in our pooled analysis of the distribution of ODX RS classifications (Tables 2, 3, 4). Among these studies, the mean proportion classified as low, intermediate, and high risk were 48.8, 39.0, and 12.2 %, respectively.
Table 2.
Pooled mean proportion classified as low, intermediate, and high risk by the ODX assay
| Study | Proportion classified low RS (%) | Low RS study weight | Proportion classified intermediate RS (%) | Intermediate RS study weight | Proportion classified high RS (%) | High RS study weight |
|---|---|---|---|---|---|---|
| Ademuyiwa et al. [23] | 51.4 | 0.07 | 39.9 | 0.07 | 8.7 | 0.05 |
| Asad et al. [24] | 47.0 | 0.02 | 38.0 | 0.02 | 15.0 | 0.03 |
| de Boer et al. [42] | 46.5 | 0.02 | 39.6 | 0.02 | 13.9 | 0.03 |
| Geffen et al. [25] | 50.0 | 0.03 | 38.0 | 0.03 | 12.0 | 0.03 |
| Goodwin et al. [40] | 39.0 | 0.02 | 46.7 | 0.03 | 14.3 | 0.03 |
| Gregg et al. [41] | 49.0 | 0.06 | 39.0 | 0.06 | 12.0 | 0.06 |
| Guth et al. [39] | 44.7 | 0.01 | 38.3 | 0.01 | 17.0 | 0.02 |
| Henry et al. [26] | 59.0 | 0.01 | 34.0 | 0.01 | 7.0 | 0.00 |
| Hornberger et al. [21] | 52.7 | 0.24 | 34.6 | 0.20 | 12.7 | 0.23 |
| Joh et al. [28] | 61.7 | 0.05 | 29.2 | 0.03 | 9.1 | 0.03 |
| Kelly et al. [29] | 52.0 | 0.08 | 39.0 | 0.08 | 9.0 | 0.05 |
| Klang et al. [30] | 39.7 | 0.07 | 44.6 | 0.10 | 15.8 | 0.11 |
| Lo et al. [31] | 42.7 | 0.02 | 47.2 | 0.03 | 10.1 | 0.02 |
| Lund et al. [32] | 48.9 | 0.07 | 41.5 | 0.07 | 9.6 | 0.05 |
| Moinuddin and Goodman [38] | 36.2 | 0.01 | 46.4 | 0.02 | 17.4 | 0.02 |
| Oratz et al. [33] | 47.1 | 0.02 | 32.4 | 0.01 | 20.6 | 0.03 |
| Partin and Mamounas [34] | 53.3 | 0.04 | 37.9 | 0.04 | 8.9 | 0.03 |
| Patel et al. [45] | 51.0 | 0.03 | 38.0 | 0.02 | 11.0 | 0.02 |
| Rezai et al. [44] | 53.7 | 0.06 | 38.9 | 0.06 | 7.4 | 0.04 |
| Tzeng et al. [36] | 50.0 | 0.02 | 36.8 | 0.02 | 13.2 | 0.02 |
| Wolf et al. [37] | 36.3 | 0.05 | 44.7 | 0.08 | 19.0 | 0.11 |
| Pooled Mean | 49.6 | – | 39.4 | – | 13.2 | – |
Table 3.
Overall mean proportion receiving adjuvant chemotherapy after ODX testing
| Study | Proportion receiving ACT (%) | Study weight |
|---|---|---|
| Ademuyiwa et al. [23] | 31.9 | 0.08 |
| Asad et al. [24] | 37.6 | 0.03 |
| de Boer et al. [42] | 29.7 | 0.03 |
| Goodwin et al. [40] | 27.6 | 0.03 |
| Gregg et al. [41] | 28.0 | 0.07 |
| Guth et al. [39] | 44.1 | 0.01 |
| Hornberger et al. [21] | 27.6 | 0.28 |
| Kelly et al. [29] | 24.0 | 0.09 |
| Klang et al. [30] | 29.4 | 0.11 |
| Lo et al. [31] | 22.5 | 0.03 |
| Lund et al. [32] | 21.3 | 0.08 |
| Moinuddin and Goodman [38] | 44.9 | 0.02 |
| Oratz et al. [33] | 32.4 | 0.02 |
| Partin and Mamounas [34] | 24.9 | 0.05 |
| Patel et al. [45] | 22.7 | 0.03 |
| Tzeng et al. [36] | 32.3 | 0.02 |
| Pooled mean | 28.0 | – |
Table 4.
Pooled mean proportion of patients receiving adjuvant chemotherapy by ODX risk group
| Study | % High RS receiving ACT | % Intermediate RS receiving ACT | % Low RS receiving ACT | Study weight |
|---|---|---|---|---|
| Ademuyiwa et al. [23] | 96.0 | 47.0 | 9.0 | 0.09 |
| Asad et al. [24] | 84.6 | 46.9 | 15.0 | 0.03 |
| Goodwin et al. [40] | 80.0 | 26.5 | 9.8 | 0.03 |
| Gregg et al. [41] | 93.0 | 40.0 | 2.5 | 0.08 |
| Guth et al. [39] | 100.0 | 65.0 | 5.0 | 0.02 |
| Hornberger et al. [21] | 72.0 | 36.0 | 10.0 | 0.30 |
| Kelly et al. [29] | 85.0 | 39.0 | 9.0 | 0.10 |
| Klang et al. [30] | 93.0 | 33.0 | 0.0 | 0.12 |
| Lund et al. [32] | 92.3 | 23.9 | 5.3 | 0.09 |
| Moinuddin and Goodman [38] | 91.7 | 50.0 | 16.0 | 0.02 |
| Oratz et al. [33] | 78.6 | 31.8 | 12.5 | 0.02 |
| Partin and Mamounas [34] | 73.3 | 43.8 | 3.3 | 0.05 |
| Patel et al. [45] | 80.0 | 34.0 | 2.0 | 0.03 |
| Tzeng et al. [36] | 77.8 | 47.8 | 6.7 | 0.02 |
| Pooled mean | 83.4 | 37.4 | 5.8 | – |
Eight studies and 1,437 patients were included in our pooled analysis of the proportion of patients, where ODX test results changed clinical-pathological ACT physician recommendation (Table 5). Among these studies, ODX changed physician recommendation in 33.4 % of patients.
Table 5.
Pooled mean proportion of physicians changing adjuvant chemotherapy recommendation after ODX testing (vs. clinical-pathological factors only)
| Study | % Changing ACT recommendation after ODX test | Study weight |
|---|---|---|
| Ademuyiwa et al. [23] | 38.0 | 0.19 |
| de Boer et al. [42] | 22.8 | 0.07 |
| Holt [43] | 26.8 | 0.10 |
| Joh et al. [28] | 24.9 | 0.11 |
| Klang et al. [30] | 40.0 | 0.26 |
| Lo et al. [31] | 31.5 | 0.06 |
| Rezai et al. [44] | 30.3 | 0.17 |
| Tatarian et al. [35] | 42.9 | 0.04 |
| Pooled mean | 33.4 | – |
Fourteen studies and 3,104 patients contributed to our pooled analysis of the proportion of low, intermediate, and high-risk patients receiving ACT after receiving their ODX RS classification. In these studies, the mean proportion of low, intermediate, and high-risk patients that received ACT were 5.8, 37.4, and 83.4 %, respectively.
Lastly, 14 studies and 1,900 patients contributed to our analysis of the ratio of patients following the treatment pathway suggested by the low versus high RS classification. In this analysis, we found that low-risk patients were significantly more likely to follow the treatment suggested by ODX (no ACT) versus high-risk patients (receive ACT), RR: 1.07 (1.01–1.14).
Discussion
We performed a systematic review and meta-analysis on the use and impact of the ODX breast cancer assay in early-stage breast cancer. The review focused on the proportion ODX patients classified as high, intermediate, and low-recurrence risk, the overall, and category-specific proportion of patients receiving ACT, the proportion of clinicians changing their ACT recommendation after ODX testing, the comparative use of ACT across categories, and the likelihood that patients received the treatment recommendation suggested by the ODX test. Our review yielded 23 studies (spanning 7 years) for inclusion in our pooled analyses. In addition to increasing the precision in the point estimates for these outcomes, a number of relevant observations warrant further discussion.
Our pooled results for the impact of ODX on ACT recommendations are consistent with most individual studies to date. However, the proportion of intermediate RS results is nearly twofold higher than reported in the ODX development studies by Paik et al. [9, 17], which may have implications for the clinical utility and cost impacts of testing. That is, the treatment decision suggested by the ODX test and the clinical and economic impact for ODX is most clear for the high and low-risk score patients. The data and suggested treatment strategy for intermediate patients are less clear and will likely remain so until the TailorRx study reports in 2017 [16]. Therefore, if high and low-risk patients opt against ODX, the impact of testing may be attenuated. This distributional shift may reflect patients that are high and low risk by traditional clinical-pathological approaches opting against ODX testing because they believe it will not change their ACT treatment decision. Another interesting result was that low RS patients were significantly more likely to follow the treatment suggested by ODX relative to high RS patients RR: 1.07 (1.01–1.14). This finding suggests a tendency toward less aggressive treatment, despite a high ODX RS. Finally, there was a lack of studies on the impact of ODX on ACT use versus standard approaches, suggesting that additional studies are warranted.
There have been a number of reviews of GEP outcomes in early-stage breast cancer [8, 51]. However, none to date have included statistical pooling of data from the literature. Marchionni et al. [8, 52] have reviewed the evidence on GEP in early- stage breast cancer, focusing on the ODX, MammaPrint, and Breast Cancer Profiling (BCP or H/I ratio) test. They found that the ODX test is the furthest along the validation pathway, but further evidence is needed to understand the relationship between predicted risk and observed risk (i.e., the impact of the test on real world clinical outcomes—a sentiment we share) [8, 52]. Other reviews have come to similar conclusions regarding the evidence in support of GEP testing and ODX breast cancer testing, specifically that there is good supporting evidence for both accurate risk stratification and the prediction of chemotherapy benefit, but further prospective evidence is desired to fully evaluate the clinical utility [6, 53].
Our systematic review and meta-analysis have several limitations that should be noted. First, we were unable to adjust for potential patient-level confounders (e.g., stage, race/ethnicity, and histology) in our pooled analyses, as our data were at the study level. These covariates likely influence the decisions to undergo testing and follow test recommendations, and should be evaluated in future studies. Another limitation is that our included studies were predominantly conducted in urban academic medical centers in the United States, and therefore may not be representative of practice in other clinical and geographic settings, or generalizable to other countries. Future studies should include patients across a broader array of settings and countries to facilitate the comparison, and provide more readily generalizable results. Our inclusion criteria, which limited our dataset to studies with information on ODX score and chemotherapy use, has the potential to bias the sample used to inform the distribution of ODX scores for patients undergoing testing. However, it is unlikely to contribute substantial bias because the inclusion criteria does not relate to the underlying patient population, but rather the study design, i.e., the data elements collected and the data collection time points. Lastly, many of the studies included in our analysis were abstracts from the ASCO and SABCS annual meetings. Though these are among the most distinguished breast cancer conferences, and submissions are reviewed, it is possible that average study quality is lesser than that of the peer-reviewed literature. However, because the majority of our outcomes utilized simple counts, it is unlikely that our pooled analyses were substantially impacted by any potential abstract issues.
The introduction of ODX testing in clinical practice is a landmark event for genomic medicine, and portends the increasing use of GEP in oncology practice. Recently, additional gene-expression profiles have been developed and marketed for colon, lung, and other cancers [54, 55]. The evidence base for ODX continues to develop primarily through the generation of smaller observational studies. Our review of these studies increases the precision of the relevant estimates of impact and can facilitate informed decision-making. In a broad context, the use of GEP testing holds the promise of improved risk stratification, treatment selection, and the commensurate clinical and economic benefits that follow. However, our enthusiasm should be measured and proceed in step with the generation and evaluation of high-quality supporting evidence, such that our decisions are based not on promise, but on the critical evaluation and rational assessment of the evidence.
Acknowledgements
Carlson JJ is supported by the Agency for Healthcare Research and Quality Mentored Clinical Scientists Comparative Effectiveness Development Program (K12) at University of Washington (HS019482). Roth JA is supported by a National Institute on Aging T32 (AG027677).
Contributor Information
Josh J. Carlson, Pharmaceutical Outcomes Research and Policy Program, University of Washington, 1959 NE Pacific Ave, Box 357630, Seattle, WA 98195-7630, USA Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
Joshua A. Roth, Fred Hutchinson Cancer Research Center, Seattle, WA, USA Group Health Research Institute, Seattle, WA, USA.
References
- 1.National Comprehensive Cancer Network NCCN Clinical practice guidelines in oncology: breast cancer V.1. 2012 2012. [Google Scholar]
- 2.Oestreicher N, Ramsey SD, Linden HM, McCune JS, van't Veer LJ, Burke W, Veenstra DL. Gene expression profiling and breast cancer care: what are the potential benefits and policy implications? Genet Med. 2005;7(6):380–389. doi: 10.1097/01.gim.0000170776.31248.75. doi:10.109701.gim. 0000170776.31248.75. [DOI] [PubMed] [Google Scholar]
- 3.Peto R, Davies C, Godwin J, Gray R, Pan HC, Clarke M, Cutter D, Darby S, McGale P, Taylor C, Wang YC, Bergh J, Di Leo A, Albain K, Swain S, Piccart M, Pritchard K. Comparisons between different polychemotherapy regimens for early breast cancer: meta-analyses of long-term outcome among 100,000 women in 123 randomised trials. Lancet. 2012;379(9814):432–444. doi: 10.1016/S0140-6736(11)61625-5. doi:10.1016/s0140-6736(11)61625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365(9472):1687–1717. doi: 10.1016/S0140-6736(05)66544-0. doi:10.1016/s0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 5.Griggs JJ, Hawley ST, Graff JJ, Hamilton AS, Jagsi R, Janz NK, Mujahid MS, Friese CR, Salem B, Abrahamse PH, Katz SJ. Factors associated with receipt of breast cancer adjuvant chemotherapy in a diverse population-based sample. J Clin Oncol. 2012;30(25):3058–3064. doi: 10.1200/JCO.2012.41.9564. doi:10.1200/JCO.2012.41.9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albain KS, Paik S, van't Veer L. Prediction of adjuvant chemotherapy benefit in endocrine responsive, early breast cancer using multigene assays. Breast. 2009;18(Suppl 3):S141–S145. doi: 10.1016/S0960-9776(09)70290-5. doi:10. 1016/S0960-9776(09)70290-5. [DOI] [PubMed] [Google Scholar]
- 7.Arpino G, Generali D, Sapino A, Lucia DM, Frassoldati A, de Laurentis M, Paolo P, Mustacchi G, Cazzaniga M, De Placido S, Conte P, Cappelletti M, Zanoni V, Antonelli A, Martinotti M, Puglisi F, Berruti A, Bottini A, Dogliotti L. Gene expression profiling in breast cancer: a clinical perspective. Breast. 2013 doi: 10.1016/j.breast.2013.01.016. doi:10.1016/j.breast.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 8.Marchionni L, Wilson RF, Wolff AC, Marinopoulos S, Parmigiani G, Bass EB, Goodman SN. Systematic review: gene expression profiling assays in early-stage breast cancer. Ann Intern Med. 2008;148(5):358–369. doi: 10.7326/0003-4819-148-5-200803040-00208. [DOI] [PubMed] [Google Scholar]
- 9.Paik S, Tang G, Shak S, Kim C, Baker J, Kim W, Cronin M, Baehner FL, Watson D, Bryant J, Costantino JP, Geyer CE, Jr, Wickerham DL, Wolmark N. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24(23):3726–3734. doi: 10.1200/JCO.2005.04.7985. doi:10.1200/jco.2005.04.798. [DOI] [PubMed] [Google Scholar]
- 10.Buyse M, Loi S, van't Veer L, Viale G, Delorenzi M, Glas AM, d'Assignies MS, Bergh J, Lidereau R, Ellis P, Harris A, Bogaerts J, Therasse P, Floore A, Amakrane M, Piette F, Rutgers E, Sotiriou C, Cardoso F, Piccart MJ. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006;98(17):1183–1192. doi: 10.1093/jnci/djj329. doi:10.1093/jnci/djj329. [DOI] [PubMed] [Google Scholar]
- 11.Kok M, Linn SC, Van Laar RK, Jansen MP, van den Berg TM, Delahaye LJ, Glas AM, Peterse JL, Hauptmann M, Foekens JA, Klijn JG, Wessels LF, Van't Veer LJ, Berns EM. Comparison of gene expression profiles predicting progression in breast cancer patients treated with tamoxifen. Breast Cancer Res Treat. 2009;113(2):275–283. doi: 10.1007/s10549-008-9939-y. doi:10.1007/s10549-008-9939-y. [DOI] [PubMed] [Google Scholar]
- 12.Genomic Health [6 Jun 2012];Oncotype Dx-Information for patients and care-givers. http://www.oncotypedx.com/en-US/Breast/PatientCaregiver/OncoOverview.aspx.
- 13.Sparano JA, Paik S. Development of the 21-gene assay and its application in clinical practice and clinical trials. J Clin Oncol. 2008;26(5):721–728. doi: 10.1200/JCO.2007.15.1068. doi:10.1200/jco.2007.15.1068. [DOI] [PubMed] [Google Scholar]
- 14.Mamounas EP, Tang G, Fisher B, Paik S, Shak S, Costantino JP, Watson D, Geyer CE, Jr, Wickerham DL, Wolmark N. Association between the 21-gene recurrence score assay and risk of locoregional recurrence in node-negative, estrogen receptor-positive breast cancer: results from NSABP B-14 and NSABP B-20. J Clin Oncol. 2010;28(10):1677–1683. doi: 10.1200/JCO.2009.23.7610. doi:10.1200/jco.2009.23.7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang G, Shak S, Paik S, Anderson SJ, Costantino JP, Geyer CE, Jr, Mamounas EP, Wickerham DL, Wolmark N. Comparison of the prognostic and predictive utilities of the 21-gene recurrence score assay and adjuvant! for women with node-negative, ER-positive breast cancer: results from NSABP B-14 and NSABP B-20. Breast Cancer Res Treat. 2011;127(1):133–142. doi: 10.1007/s10549-010-1331-z. doi:10.1007/s10549-010-1331-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.ClinicalTrials.gov [25 Feb 2011];Hormone therapy with or without combination chemotherapy in treating women who have undergone surgery for node-negative breast cancer (The TAILORx Trial) 2011 http://www.clinicaltrials.gov/ct2/show/NCT00310180?term=oncotype&rank=3.
- 17.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. doi:10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 18.Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, Somerfield MR, Hayes DF, Bast RC., Jr American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25(33):5287–5312. doi: 10.1200/JCO.2007.14.2364. doi:10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 19.Genomic Health [25 Aug 2012];Corporate fact sheet. http://files.shareholder.com/downloads/GHDX/2036901272x0x237157/FFA8A1C1-117B-411E-86A2-FFFDA42F0CFE/FactSheet.pdf.
- 20.Hornberger J, Cosler LE, Lyman GH. Economic analysis of targeting chemotherapy using a 21-gene RT-PCR assay in lymph-node-negative, estrogen-receptor-positive, early-stage breast cancer. Am J Manag Care. 2005;11(5):313–324. [PubMed] [Google Scholar]
- 21.Hornberger J, Chien R, Krebs K, Hochheiser L. US Insurance Program’s experience with a multigene assay for early-stage breast cancer. J Oncol Pract. 2011;7(3 Suppl):e38s–e45s. doi: 10.1200/JOP.2011.000303. doi:10.1200/JOP.2011.000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oakman C, Santarpia L, Di Leo A. Breast cancer assessment tools and optimizing adjuvant therapy. Nat Rev Clin Oncol. 2010;7(12):725–732. doi: 10.1038/nrclinonc.2010.170. doi:10.1038/nrclinonc.2010.170. [DOI] [PubMed] [Google Scholar]
- 23.Ademuyiwa FO, Miller A, O'Connor T, Edge SB, Thorat MA, Sledge GW, Levine E, Badve S. The effects of oncotype DX recurrence scores on chemotherapy utilization in a multi-institutional breast cancer cohort. Breast Cancer Res Treat. 2011;126(3):797–802. doi: 10.1007/s10549-010-1329-6. doi:10.1007/s10549-010-1329-6. [DOI] [PubMed] [Google Scholar]
- 24.Asad J, Jacobson AF, Estabrook A, Smith SR, Boolbol SK, Feldman SM, Osborne MP, Boachie-Adjei K, Twardzik W, Tartter PI. Does oncotype DX recurrence score affect the management of patients with early-stage breast cancer? Am J Surg. 2008;196(4):527–529. doi: 10.1016/j.amjsurg.2008.06.021. doi:10.1016/j.amjsurg.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 25.Geffen DB, Abu-Ghanem S, Sion-Vardy N, Braunstein R, Tokar M, Ariad S, Delgado B, Bayme M, Koretz M. The impact of the 21-gene recurrence score assay on decision making about adjuvant chemotherapy in early-stage estrogen-receptor-positive breast cancer in an oncology practice with a unified treatment policy. Ann Oncol. 2011;22(11):2381–2386. doi: 10.1093/annonc/mdq769. doi:10.1093/annonc/mdq769. [DOI] [PubMed] [Google Scholar]
- 26.Henry LR, Stojadinovic A, Swain SM, Prindiville S, Cordes R, Soballe PW. The influence of a gene expression profile on breast cancer decisions. J Surg Oncol. 2009;99(6):319–323. doi: 10.1002/jso.21244. doi:10.1002/jso.21244. [DOI] [PubMed] [Google Scholar]
- 27.Hornberger J, Chien R, Krebs K, Hochheiser L. US insurance program’s experience with a multigene assay for early-stage breast cancer. Am J Manag Care. 2011;17(5):e194–202. Spec No. [PubMed] [Google Scholar]
- 28.Joh JE, Esposito NN, Kiluk JV, Laronga C, Lee MC, Loftus L, Soliman H, Boughey JC, Reynolds C, Lawton TJ, Acs PI, Gordan L, Acs G. The effect of Oncotype DX recurrence score on treatment recommendations for patients with estrogen receptor-positive early stage breast cancer and correlation with estimation of recurrence risk by breast cancer specialists. Oncologist. 2011;16(11):1520–1526. doi: 10.1634/theoncologist.2011-0045. doi:10.1634/theoncologist.2011-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelly CM, Krishnamurthy S, Bianchini G, Litton JK, Gonzalez-Angulo AM, Hortobagyi GN, Pusztai L. Utility of onco-type DX risk estimates in clinically intermediate risk hormone receptor-positive, HER2-normal, grade II, lymph node-negative breast cancers. Cancer. 2010;116(22):5161–5167. doi: 10.1002/cncr.25269. doi:10.1002/cncr.25269. [DOI] [PubMed] [Google Scholar]
- 30.Klang SH, Hammerman A, Liebermann N, Efrat N, Doberne J, Hornberger J. Economic implications of 21-gene breast cancer risk assay from the perspective of an Israeli-managed health-care organization. Value Health. 2010;13(4):381–387. doi: 10.1111/j.1524-4733.2010.00724.x. doi:10.1111/j.1524-4733.2010.00724.x. [DOI] [PubMed] [Google Scholar]
- 31.Lo SS, Mumby PB, Norton J, Rychlik K, Smerage J, Kash J, Chew HK, Gaynor ER, Hayes DF, Epstein A, Albain KS. Prospective multicenter study of the impact of the 21-gene recurrence score assay on medical oncologist and patient adjuvant breast cancer treatment selection. J Clin Oncol. 2010;28(10):1671–1676. doi: 10.1200/JCO.2008.20.2119. doi:10.1200/jco.2008.20.2119. [DOI] [PubMed] [Google Scholar]
- 32.Lund MJ, Mosunjac M, Davis KM, Gabram-Mendola S, Rizzo M, Bumpers HL, Hearn S, Zelnak A, Styblo T, O'Regan RM. 21-Gene recurrence scores: racial differences in testing, scores, treatment, and outcome. Cancer. 2012;118(3):788–796. doi: 10.1002/cncr.26180. doi:10.1002/cncr.26180. [DOI] [PubMed] [Google Scholar]
- 33.Oratz R, Paul D, Cohn AL, Sedlacek SM. Impact of a commercial reference laboratory test recurrence score on decision making in early-stage breast cancer. J Oncol Pract. 2007;3(4):182–186. doi: 10.1200/JOP.0742001. doi:10.1200/jop.0742001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Partin JF, Mamounas EP. Impact of the 21-gene recurrence score assay compared with standard clinicopathologic guidelines in adjuvant therapy selection for node-negative, estrogen receptor-positive breast cancer. Ann Surg Oncol. 2011;18(12):3399–3406. doi: 10.1245/s10434-011-1698-z. doi:10.1245/s10434-011-1698-z. [DOI] [PubMed] [Google Scholar]
- 35.Tatarian T, Damle S, McSwain AP, Ojong-Ntui M, Tabbara I, Teal CB. Clinical utility and therapeutic implications of oncotype analysis in patients with breast cancer. Ann Surg Oncol. 2011;18:S182. [Google Scholar]
- 36.Tzeng JP, Mayer D, Richman AR, Lipkus I, Han PK, Valle CG, Carey LA, Brewer NT. Women's experiences with genomic testing for breast cancer recurrence risk. Cancer. 2010;116(8):1992–2000. doi: 10.1002/cncr.24990. doi:10.1002/cncr.24990. [DOI] [PubMed] [Google Scholar]
- 37.Wolf I, Ben-Baruch N, Shapira-Frommer R, Rizel S, Goldberg H, Yaal-Hahoshen N, Klein B, Geffen DB, Kaufman B. Association between standard clinical and pathologic characteristics and the 21-gene recurrence score in breast cancer patients: a population-based study. Cancer. 2008;112(4):731–736. doi: 10.1002/cncr.23225. doi:10.1002/cncr.23225. [DOI] [PubMed] [Google Scholar]
- 38.Moinuddin IYS, Goodman J. Effect of menopausal status and oncotype recurrence score on treatment choice for early-stage receptor-positive breast cancer.. Paper presented at the American Society of Clinical Oncology Breast Cancer Symposium.2009. [Google Scholar]
- 39.Guth AAFS, Fei K, Franco R, Bickell N. Utilization of Oncotype DX to predict chemotherapy use in an innercity population.. Paper presented at the American Society of Clinical Oncology Annual Meeting.2011. [Google Scholar]
- 40.Goodwin MCHJ, Diego M, Frazier TG. Impact of Oncotype DXTM recurrence score and tumor size on making chemotherapy decisions in breast cancer patients.. Paper presented at the Sand Antonio Breast Cancer Symposium.2009. [Google Scholar]
- 41.Gregg XBT, Rowley B, Rees W. Experience with use of the Oncotype DX gene assay test in a multicenter community-based healthcare system.. Paper presented at the San Antonio Breast Cancer Symposium.2009. [Google Scholar]
- 42.de Boer RHBC, Speakman D, Mann B. Australian decision impact study: the impact of Oncotype DX recurrence score (RS) on adjuvant treatment decisions in hormone receptor positive (HR+), node negative (N0) and node positive (N+) early stage breast cancer (ESBC) in the multidisciplinary clinic (MDC).. Paper presented at the San Antonio Breast Cancer Symposium.2011. [Google Scholar]
- 43.Holt SBG, Brinkworth E, Durrani S, Jones S, Khawaja S, Laggner U, Moe M, Pudney D, Pitcher S, Rolles M, Sharaiha Y, Whelan S. Results from a prospective clinical study on the impact of Oncotype DX on adjuvant treatment decision making in a cohort of 142 UK patients.. Paper presented at the San Antonio Breast Cancer Symposium.2011. [Google Scholar]
- 44.Rezai MEW, Kummel S, Kuhn T, Warm M, Friedrichs K, Schneeweiss A, Markmann S, Eggemann H, Hilfrich J, Jackisch C, Witzel I, Eidtmann H, Kaufmann M, Blohmer JU. Impact of the recurrence score on adjuvant decision-making in ER-positive early breast cancer—results of a large prospective multicentre decision impact study in node negative and node positive disease.. Paper presented at the San Antonio Breast Cancer Symposium.2011. [Google Scholar]
- 45.Patel HHK, Kaplan C, Davidson R, DeMichele A, Fox KR, Domcheck SM. Utilization of Oncotype DX in node-negative, ER-positive breast cancer patients.. Paper presented at the American Society of Clinical Oncology Annual Meeting.2007. [Google Scholar]
- 46.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264–269. W264. doi: 10.7326/0003-4819-151-4-200908180-00135. [DOI] [PubMed] [Google Scholar]
- 47.Turaga K, Acs G, Laronga C. Gene expression profiling in breast cancer. Cancer Control. 2010;17(3):177–182. doi: 10.1177/107327481001700306. [DOI] [PubMed] [Google Scholar]
- 48.Duval S, Tweedie R. Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56(2):455–463. doi: 10.1111/j.0006-341x.2000.00455.x. [DOI] [PubMed] [Google Scholar]
- 49.Greenland S. Invited commentary: a critical look at some popular meta-analytic methods. Am J Epidemiol. 1994;140(3):290–296. doi: 10.1093/oxfordjournals.aje.a117248. [DOI] [PubMed] [Google Scholar]
- 50.Juni P, Witschi A, Bloch R, Egger M. The hazards of scoring the quality of clinical trials for meta-analysis. JAMA. 1999;282(11):1054–1060. doi: 10.1001/jama.282.11.1054. [DOI] [PubMed] [Google Scholar]
- 51.Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group Recommendations from the EGAPP Working Group: can tumor gene expression profiling improve outcomes in patients with breast cancer? Genet Med. 2009;11(1):66–73. doi: 10.1097/GIM.0b013e3181928f56. doi:10.1097/GIM.0b013e3181928f56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marchionni L, Wilson RF, Marinopoulos SS, Wolff AC, Parmigiani G, Bass EB, Goodman SN. Impact of gene expression profiling tests on breast cancer outcomes. Evid Rep Technol Assess (Full Rep) 2007;160:1–105. [PMC free article] [PubMed] [Google Scholar]
- 53.Dunn L, Demichele A. Genomic predictors of outcome and treatment response in breast cancer. Mol Diagn Ther. 2009;13(2):73–90. doi: 10.1007/BF03256317. doi:10.2165/01250444-200913020-00002. [DOI] [PubMed] [Google Scholar]
- 54.Kratz JR, He J, Van Den Eeden SK, Zhu Z-H, Gao W, Pham PT, Mulvihill MS, Ziaei F, Zhang H, Su B, Zhi X, Quesenberry CP, Habel LA, Deng Q, Wang Z, Zhou J, Li H, Huang M-C, Yeh C–C, Segal MR, Ray MR, Jones KD, Raz DJ, Xu Z, Jahan TM, Berryman D, He B, Mann MJ, Jablons DM. A practical molecular assay to predict survival in resected non-squamous, non-small-cell lung cancer: development and international validation studies. Lancet. 2012;379(9818):823–832. doi: 10.1016/S0140-6736(11)61941-7. doi:10.1016/s0140-6736(11)61941-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gray RG, Quirke P, Handley K, Lopatin M, Magill L, Baehner FL, Beaumont C, Clark-Langone KM, Yoshizawa CN, Lee M, Watson D, Shak S, Kerr DJ. Validation study of a quantitative multigene reverse transcriptase-polymerase chain reaction assay for assessment of recurrence risk in patients with stage II colon cancer. J Clin Oncol. 2011;29(35):4611–4619. doi: 10.1200/JCO.2010.32.8732. doi:10.1200/jco.2010.32.8732. [DOI] [PubMed] [Google Scholar]