Abstract
The Escherichia coli AlkB protein was recently found to repair cytotoxic DNA lesions 1-methyladenine and 3-methylcytosine by using a novel iron-catalyzed oxidative demethylation mechanism. Three human homologs, ABH1, ABH2 and ABH3, have been identified, and two of them, ABH2 and ABH3, were shown to have similar repair activities to E.coli AlkB. However, ABH1 did not show any repair activity. It was suggested that ABH3 prefers single-stranded DNA and RNA substrates, whereas AlkB and ABH2 can repair damage in both single- and double-stranded DNA. We employed a chemical cross-linking approach to probe the structure and substrate preferences of AlkB and its three human homologs. The putative active site iron ligands in these proteins were mutated to cysteine residues. These mutant proteins were used to cross-link to different DNA probes bearing thiol-tethered bases. Disulfide-linked protein–DNA complexes can be trapped and analyzed by SDS–PAGE. Our results show that ABH2 and ABH3 have structural and functional similarities to E.coli AlkB. ABH3 shows preference for the single-stranded DNA probe. ABH1 failed to cross-link to the probes tested. This protein, unlike other AlkB proteins, does not seem to interact with DNA in its E.coli expressed form.
INTRODUCTION
Cellular DNA is constantly subjected to modifications by intracellular and extracellular chemicals that can result in covalent changes. The widespread alkylating agents are one group of DNA modifiers that introduce damage primarily to the heterocyclic bases of DNA. Much of the alkylated damage, if not repaired, has cytotoxic or mutagenic consequences. Cells have evolved dedicated systems to repair alkylation DNA damage (1,2). Among these repair pathways, direct dealklyation repair represents the simplest and perhaps the most efficient way to remove lesions. The N-terminal domain of Escherichia coli Ada exhibits an interesting function of direct removal of a methyl group on a methyl phosphotriester backbone lesion (3,4). Two other known examples are O6-alkylguanine-DNA alkyltransferases and AlkB proteins that perform direct dealkylation of alkylated base damage (3,5–15).
The involvement of E.coli AlkB in DNA repair was proposed nearly two decades ago. It is one of four genes that are activated in the so-called adaptive response pathway when E.coli is challenged with high levels of alkylating agents (3). Earlier work from several groups, especially Lindahl and Sedgwick, demonstrated that this protein probably repairs single-stranded DNA damage which tends to be induced by SN2-type methylating agents (16–19). The exact function of the protein was unclear at the time. A major clue to its function came from a computational protein-fold analysis of AlkB. This putative DNA repair protein was shown to have a sequence homologous to the 2-ketoglutarate-Fe2+-dependent dioxygenase superfamily (20). Subsequently, two groups discovered the function of AlkB through biochemical studies. It performs an unprecedented oxidative dealkylation of N1-methyladenine and N3-methylcytosine lesions in DNA (7,8). This protein can repair base lesions on single-stranded (ss) and double-stranded (ds) DNA, and even RNA (9). The protein appears to be widely conserved. Three homologs have been found in humans (Fig. 1A) (9,10,18). Among them, the functions of two have been found to be similar to that of E.coli AlkB (9,10).
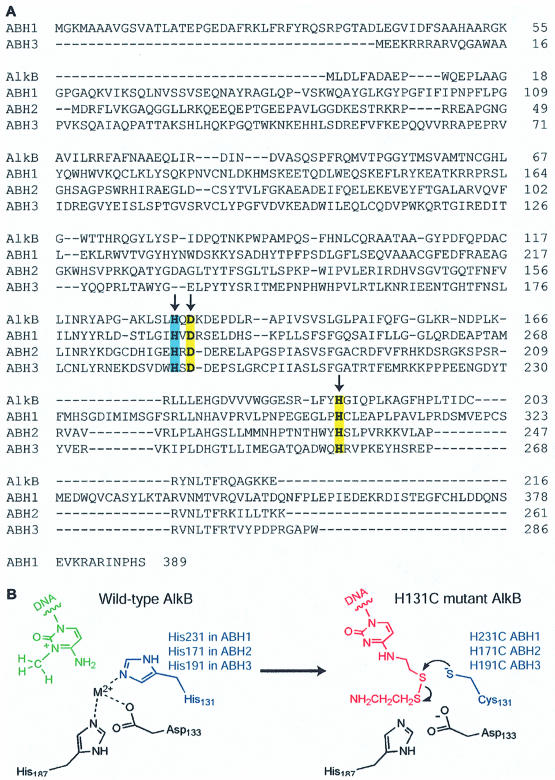
Figure 1.
(A) Alignment of AlkB, ABH1, ABH2 and ABH3. Arrows indicate the conserved active site ligands to the iron atom. His131 of AlkB, His231 of ABH1, His171 of ABH2 and His191 of ABH3 are highlighted in blue. The other two conserved ligand residues are highlighted in yellow. (B) Disulfide cross-linking strategy. Replacement of one of the ligand residues His131 of AlkB by Cys131 and introduction of a thiol-tethered C* in DNA provides the partners for the formation of a specific disulfide cross-link.
The proposed repair mechanism for AlkB proteins has not yet been subjected to detailed mechanistic and spectroscopic investigations. Escherichia coli AlkB binds both ssDNA and dsDNA very weakly (16). Furthermore, this protein shows only a 2-fold increment of its affinity towards methylated ssDNA compared with unmethylated ssDNA (16). Methods to probe the binding of this protein to different DNA structures are limited due to the labile and non-specific nature of the interaction. Specific protein–DNA complexes of AlkB are difficult to stabilize for structural studies. Recently, we have developed a chemical cross-linking method to probe the structural and functional aspects of E.coli AlkB (11). We mutated the conserved iron ligand residues to cysteine residues and performed cross-linking experiments with both ssDNA and dsDNA probes bearing thiol-tethered cytosine bases (Fig. 1B). We envisaged that AlkB may have a damage-searching mechanism to check bases that resemble the damaged bases in ssDNA or dsDNA in its active site. The modified cytosine (C*) in certain DNA structures may be able to access the substrate-binding pocket if there is enough space to accommodate it, which is possible after the removal of the bound metal ion and co-substrate 2-ketoglutarate. Then, the engineered cysteine residue could attack the disulfide group and form a cross-linked complex. We demonstrated that specific AlkB–DNA complexes can be trapped using this chemical method. Our study confirmed the assignment of the putative metal-binding active site. The results also revealed the potential damage-searching mechanism of E.coli AlkB and support the assigned function of the protein.
Here we report chemical cross-linking studies on the three human homologs of AlkB. Potential DNA substrate preferences of these proteins and E.coli AlkB are investigated by using structurally different thiol-tethered DNA probes. Our results reveal that ABH2 and ABH3 can interact with both ssDNA and dsDNA. ABH1 expressed from E.coli failed to cross-link to our probes. This form of ABH1 may not interact with our DNA probes at all.
MATERIALS AND METHODS
Construction, expression and purification of wild-type and mutant AlkB and ABH3
The cloning, expression and purification of E.coli AlkB and H131C mutant have been described previously (11). The human ABH3 gene was subcloned into HindIII and NcoI sites of a pET28a vector (Novagen) and transformed with E.coli BL21(DE3) onto LB-agar plates containing kanamycin (50 µM). The same procedure to express and purify E.coli AlkB was followed (11). Briefly, overnight pre-cultures were grown aerobically at 37°C and shaken at 190 r.p.m., and they were then used to inoculate 1 l of LB medium and kanamycin (50 µM). The cells were grown until the OD600 was around 0.6. Isopropyl-β-d-thiogalactopyranoside (IPTG; 1 mM) was added and the cells were grown for an additional 3.5–4 h at 30°C. The cells were harvested by centrifugation, frozen in liquid nitrogen and then stored at –80°C. All subsequent steps were performed at 4°C. The cell pellet was resuspended in 30 ml of lysis buffer (100 mM NaCl, 10 mM Tris pH 7.4, 10 mM 2-mercaptoethanol), disintegrated by sonication and centrifuged at 12 000 r.p.m. for 20 min. The supernatant was then centrifuged at 3000 r.p.m. for an extra 20 min with the addition of DEAE-cellulose. The supernatant was later added to four equivalents of buffer A (10 mM Tris–HCl pH 7.4, 5 mM 2-mercaptoethanol), loaded onto an S-Sepharose cation exchange column (Amersham Biosciences) that had been equilibrated with buffer A, and eluted with a linear gradient of NaCl (0.0–1.0 M). The fractions containing the protein were concentrated by ultrafiltration (Centricon YM10 membrane, Amicon) using buffer A to dilute, and purified further with a Mono-S cation exchange column (Amersham Biosciences) using a linear gradient of NaCl (0.0–1.0 M). Point mutation in ABH3 was introduced by megaprimer mutagenesis. The sequences of the wild-type and mutant ABH3 (H191C) were confirmed by sequencing the entire coding sequence. The mutant proteins were eluted from the Mono-S column at the same NaCl concentration as the wild-type protein.
Construction, expression and purification of wild-type and mutant ABH1 and ABH2
The human ABH1 gene with an N-terminal His6 tag was expressed and purified as described previously (10). The ABH2 gene and mutants of ABH1 and ABH2 were subcloned into HindIII and NdeI sites of a pET28a vector (Novagen) with an N-terminal His6 tag and transformed with E.coli BL21(DE3) onto LB-agar plates containing kanamycin (50 µM). The same procedure to over-express ABH3 protein was followed. The cell pellet was resuspended in 30 ml of lysis buffer (100 mM NaCl, 50 mM NaH2PO4 pH 6.5, 5% glycerol) with the addition of 5 mM 2-mercaptoethanol, disintegrated by sonication, and centrifuged at 12 000 r.p.m. for 20 min. The supernatant was then centrifuged at 3000 r.p.m. for another 20 min with the addition of DEAE-cellulose. The supernatant was loaded onto a column containing 1 ml of Ni2+-NTA–agarose resin. The resin was washed with 30 ml of wash buffer containing 20 mM imidazole then eluted with 6 ml of elute buffer (100 mM NaCl, 50 mM NaH2PO4 pH 6.5, 400 mM imidazole, 5 mM 2-mercaptoethanol). The protein was then purified further with a Mono-S cation exchange column using a linear gradient of NaCl (0.0–1.0 M). Point mutations in ABH1 and ABH2 were introduced by megaprimer mutagenesis.
Synthetic oligonucleotides
Oligodeoxyribonucleotides were synthesized on an Applied Biosystems 392 DNA synthesizer. Thiol-tethered oligonucleotides were prepared by incorporation of the O4-triazolyl-dU-CE phosphoramidite (Glen Research) at the modified positions during solid-phase synthesis, followed by post-synthetic modification/deprotection by treatment with diamine disulfides (21–23). Modified and unmodified oligonucleotides were purified by denaturing polyacrylamide gel electrophoresis. Concentrations of the oligonucleotides were determined by UV at 260 nm.
Cross-linking reaction and analysis
Purified wild-type and mutant AlkB, ABH1, ABH2 and ABH3 proteins were dialyzed into a buffer containing 10 mM Tris–HCl pH 7.34 and 100 mM NaCl. The same buffer was used for cross-linking reactions. In a typical reaction, all proteins (10 µM) and the thiol-tethered DNA (30 µM for E.coli AlkB and 80 µM for human proteins) were incubated at 4°C for varying periods of time. The reaction was quenched by the addition of 20 mM methyl methanethiolsulfonate, a thiol-capping reagent, for 10 min at 4°C. The final concentration of methyl methanethiolsulfonate was increased to 100 mM in experiments examining disulfide stability using varying amounts of dithiothreitol (DTT), a strong reducing agent. The samples were analyzed by SDS–PAGE under non-reducing conditions. Cross-linking yield was calculated by measuring the intensity of the bands on the gel. Cross-linked protein–ssDNA and protein–dsDNA complexes showed similar mobility on the gel, presumably due to the unwinding of dsDNA under denaturing conditions. We have shown that the complexes for the E.coli AlkB protein do migrate differently on a Mono-Q column (11).
RESULTS AND DISCUSSION
Expression and purification of AlkB proteins
The E.coli AlkB and human ABH1 were expressed and purified as previously published (10,11). The gene encoding ABH2 was subcloned into a pET-28a vector with an N-terminal His6 tag. The ABH3 gene was subcloned into pET-28a vector without any tag. The three proteins were over-expressed in E.coli BL21(DE3) cells. ABH1 and ABH2 were purified by Ni2+-NTA affinity resin followed by a cation exchange Mono-S column. ABH3 was purified twice by cation exchange, as was E.coli AlkB. The ABH1 H231C, ABH2 H171C and ABH3 H191C mutants were prepared by site-specific mutagenesis. The sequences of all protein constructs were confirmed by sequencing the entire coding sequences. All mutant proteins show indistinguishable chromatographic behaviors from those of the wild-type proteins.
Cross-link between E.coli AlkB and DNA
We have shown that by engineering specific cysteine residues to the active site of AlkB, we can set up a specific cross-link between the mutant AlkB proteins and thiol-tethered DNA probes (11). These mutant proteins cross-link with ssDNA-1 and dsDNA probes DNA-3 and DNA-4 in good yields (Fig. 2A). However, they did not cross-link with dsDNA-2, in which C* is stabilized in the duplex structure by hydrogen bonding to the opposite base G. The results support the assigned function of AlkB to repair the N1-methyladenine and N3-methylcytosine lesions since these damaged bases cannot form stable base pairs with the complementary strand in a duplex DNA; AlkB can simply capture the lesioned base that is in an extrahelical conformation. In a ssDNA, the lesioned base is not hidden in a particular structure and may be located by AlkB through a simple scanning process.
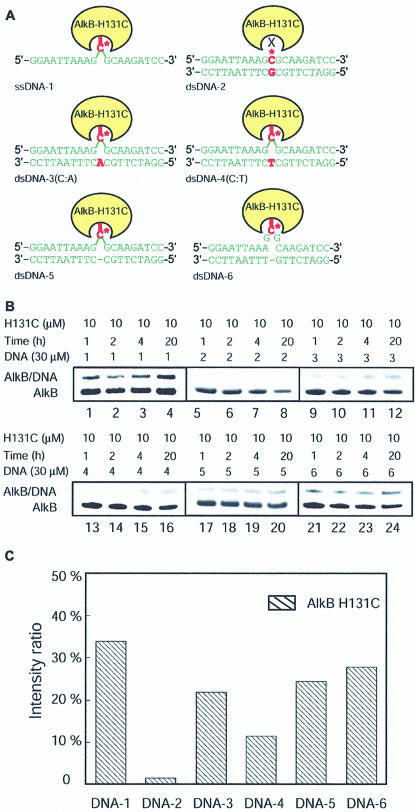
Figure 2.
Cross-linking between AlkB and DNA. (A) Different DNA probes used in the cross-linking study and their formation of a cross-link with AlkB. DNA-1 is a ssDNA probe. DNA-2–4 are dsDNA probes with C* opposite G, A and T, respectively. DNA-5 is a dsDNA probe with a C* bulge. DNA-6 is a dsDNA with C* in a single-stranded loop structure. (B) SDS–PAGE analysis of the time course of the cross-linking reactions between AlkB and DNA probes 1–6. All reactions were performed by incubating 10 µM of AlkB with 30 µM of DNA at 4°C. The reactions were quenched after 1, 2, 4 and 20 h of incubation and analyzed. (C) Comparison of the cross-linked product ratio in the reactions between the AlkB H131 mutant protein and the six different DNA probes after a 20 h reaction.
To compare the preferences of E.coli AlkB for different DNA substrates, two more duplex DNA probes, DNA-5 with a C* bulge and DNA-6 with C* in a loop structure, were prepared. The time course of the cross-linking reactions between the AlkB H131C mutant protein and DNA probes 1–6 was followed under non-reducing conditions at 4°C. The reaction mixtures were analyzed by SDS–PAGE and the results are shown in Figure 2B. ssDNA-1 cross-linked to AlkB H131C efficiently (Fig. 2B, lanes 1–4). The formation of a cross-linked product was observed after a 1 h incubation. After a 20 h incubation, >33% of the cross-linked product was obtained as estimated by integrating the intensities of the bands for the protein and the covalently linked protein–DNA complex (Fig. 2B, lane 4; Fig. 2C, column 1). Consistent with previous observations (11), AlkB H131C forms <2% cross-linked complex with DNA-2 after a 20 h incubation (Fig. 2B, lanes 5–8). AlkB H131C also cross-linked with DNA-3 and DNA-4 in appreciable yields after a 20 h incubation, but these reactions were less efficient compared with the reaction with DNA-1 (Fig. 2B and C). After the first hour of incubation, ∼20% of the AlkB H131C mutant protein cross-linked with DNA-1, but very small amounts of the protein–DNA complexes formed during this period for the reaction between the mutant protein and DNA-3 or DNA-4. AlkB may have an efficient mode to search for C* in ssDNA. In the DNA probes 3 and 4 that adopt a helical duplex structure, AlkB can only capture C* when it samples the extrahelical conformation, which could result in slower cross-link kinetics and a less efficient reaction. The C* in DNA-4 could sample the extrahelical conformation more frequently than that in DNA-3 since the C*·A base pair is less stable than the C*·T base pair (24,25). This explains the observation that AlkB H131C cross-linked with DNA-3 more efficiently than with DNA-4.
The C* in DNA-5 or DNA-6 was designed so that it cannot form any base pair. If AlkB prefers to work on bases that are unpaired and extrahelical in DNA, these probes should cross-link with the AlkB H131C mutant protein. Indeed, this is the case as revealed from the cross-linking studies (Fig. 2B, lanes 17–24). DNA-6, with a loop structure resembling ssDNA, cross-linked to AlkB H131C more efficiently than any other dsDNA probe (Fig. 2B and C). Overall, it seems that AlkB H131C slightly prefers ssDNA substrates over any dsDNA, as it cross-linked to the ssDNA-1 most efficiently and preferred the single-stranded loop in the duplex DNA probes. We may be able to use the finding that the AlkB protein prefers a bulge or a loop structure in dsDNA in order to stabilize a complex of AlkB bound to duplex DNA. Such a complex could be used for structural studies and other characterizations.
It should be noted that non-specific cross-linking between the wild-type AlkB and any of the six DNA probes tested in this study is negligible (<2%). Therefore, all the cross-links reported here between mutant AlkB and the DNA probes are specific for the engineered cysteine residue.
Cross-link between ABH3 and DNA
As a homolog of E.coli AlkB, ABH3 showed similar repair functions to previous reactivity studies. The ligands to the active site iron atom were assigned to be His191, Asp193 and His257 based on the sequence alignment of ABH3 with E.coli AlkB (Fig. 1A). The conserved HXD sequence from His191 to Asp193 is a clear indication that these two residues are involved in binding the active site iron atom. The His191 residue in ABH3 was mutated to a cysteine residue. The ABH3 H191C mutant protein, which is analogous to AlkB H131C, was expressed and purified for the cross-linking studies with DNA probes 1–6.
A large excess of thiol-tethered DNA probes (eight equivalents) was used to cross-link with one equivalent of ABH3 proteins at 4°C. ABH3 H191C cross-linked efficiently with the ssDNA-1. About 40% of the covalently linked product was produced after a 20 h incubation (Fig. 3A, lane 2; Fig. 3D). The mutant protein formed very small amounts of cross-linked product (<2% after 20 h) with both the dsDNA-2 and dsDNA-4 probes (Fig. 3A, lanes 3 and 5; Fig. 3D). Some cross-linked product was formed between the mutant protein and DNA-3 (∼10% after 20 h, Fig. 3A, lane 4; Fig. 3D).
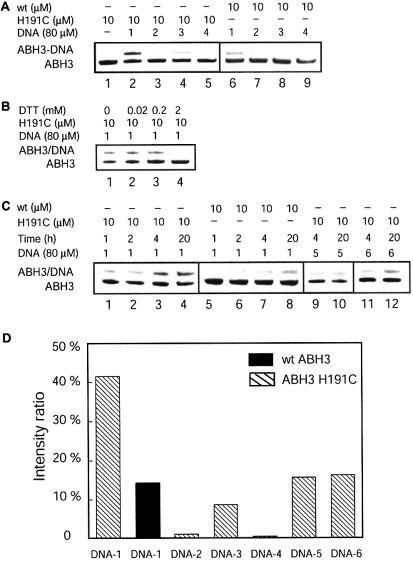
Figure 3.
Cross-linking between ABH3 and DNA. All reactions were performed by incubating 10 µM of ABH3 with 80 µM of DNA at 4°C. (A) SDS–PAGE analysis of the cross-linking reactions between ABH3 and DNA probes 1–4. Lane 1 is a size standard for ABH3. Lanes 2–5: cross-linking results between the ABH3 H191C mutant protein and DNA probes 1–4 after a 20 h reaction. Lanes 6–9: control reactions of wild-type ABH3 with DNA probes 1–4 after a 20 h incubation. (B) Effect of an external thiol (DTT) on the reaction of the ABH3 H191C mutant protein with DNA-1 after a 20 h incubation. (C) Time course of the cross-linking reactions between ABH3 and DNA probes 1, 5 and 6. Lanes 1–4: time course for the reaction between the ABH3 H191C mutant protein and DNA-1. Lanes 5–8: time course for the reaction between wild-type ABH3 and DNA-1. Lanes 9–12: reactions between the ABH3 H191C mutant protein and DNA probes 5 and 6. (D) Comparison of the cross-linked product ratio in the reactions between ABH3 and the six different DNA probes after a 20 h reaction.
Wild-type ABH3 was used to react with the same DNA probes under the same conditions as in the control experiments. Any non-specific cross-link between the wild-type ABH3 and DNA probes could be revealed from these controls. After a 20 h reaction, negligible amounts of products (<2%) were generated between the wild-type ABH3 and the dsDNA probes 2–4. The result indicates that non-specific cross-links between ABH3 H191C mutant and the dsDNA probes are negligible. The cross-links formed between the ABH3 H191C mutant and these probes are mostly specific to the Cys191 residue. However, unlike the wild-type AlkB, ∼15% of the wild-type ABH3 cross-linked with the ssDNA-1. This observation suggests that there are other cysteine residues that can form disulfide bonds to the thiol-tethered ssDNA. The cysteine residues that are responsible for the ‘non-specific’ cross-links may reside close to the DNA-binding surface of the protein. We postulate that ssDNA is less structured; thus, when it binds to ABH3, the flexibility of the DNA structure may allow the thiol tether to react with the cysteine residues close to the DNA-binding surface of the protein. With a dsDNA probe, the thiol-tethered base is much less flexible due to constraints imposed by the helical duplex structure and cannot reach the surface cysteine residues. It is also possible that ABH3 simply prefers to interact with the ssDNA probe.
The ABH3 H191C mutant protein cross-linked to DNA-1 much more efficiently than the wild-type protein after a 20 h incubation, as indicated in Figure 3D. The time course of these reactions was also followed (Fig. 3C, lanes 1–8). The results show that the specific cross-link is more efficient. Thus, the majority of the cross-linked product formed in the reaction between the ABH3 mutant protein and DNA-1 occurred through the Cys191 residue. The cross-link between the mutant protein and DNA-1 is modestly stable under mild reducing conditions (Fig. 3B). An excess of the strong reducing agent DTT eliminated the cross-link completely, which indicates that the covalent linkage between the protein and DNA-1 was due to disulfide bonds.
ABH3 clearly cross-linked with DNA-1 more efficiently than any of the dsDNA probes 2–4 even if the contribution from the non-specific cross-linking reaction is not considered (Fig. 3D). DNA probes 5 and 6 were tested, and these two probes with unpaired C* cross-linked with ABH3 H191C better than either DNA-3 or DNA-4. The results seem to suggest that ABH3 prefers to interact with ssDNA structures. A similar idea was previously proposed from the activity studies of this protein (9).
Cross-link between ABH2 and DNA
Cross-linking between ABH2 and DNA probes 1–6 was also examined under the same conditions as the cross-linking reactions for ABH3. The ABH2 H171C mutant protein, which is analogous to AlkB H131C and ABH3 H191C, was prepared and purified for the cross-linking assay. The ABH2 H171C mutant protein cross-linked efficiently with the ssDNA-1 (Fig. 4A, lane 2; Fig. 4E). This cross-link is stable under mild reducing conditions and can be cleaved by a treatment with an excess of DTT (Fig. 4B). The ABH2 H171C mutant protein also cross-linked with DNA-3, DNA-4, DNA-5 and DNA-6 in good yields (Fig. 4A, C and E). Introducing a loop or a bulge into DNA probes (DNA-5 and DNA-6) did not seem to significantly alter the cross-linking efficiency.
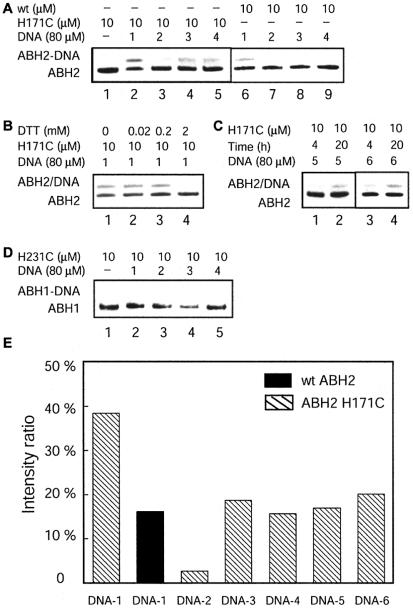
Figure 4.
Cross-linking between ABH2, ABH1 and DNA. All reactions were performed by incubating 10 µM of ABH1 or ABH2 with 80 µM of DNA at 4°C. (A) SDS–PAGE analysis of the cross-linking reactions between ABH2 and DNA probes 1–4. Lane 1 is a size standard for ABH2. Lanes 2–5: cross-linking results between the ABH2 H171C mutant protein and DNA probes 1–4 after a 20 h reaction. Lanes 6–9: control reactions of wild-type ABH2 with DNA probes 1–4 after a 20 h incubation. (B) Effect of external thiol (DTT) on the reaction of the ABH2 H171C mutant protein with DNA-1 after a 20 h incubation. (C) Cross-linking reactions between the ABH2 H171C mutant protein and DNA probes 5 and 6. (D) Cross- linking reactions between the ABH1 H231C mutant protein and DNA probes 1–4. No cross-linked product was observed for these probes. (E) Comparison of the cross-linked product ratio in the reactions between ABH2 and the six different DNA probes after a 20 h reaction.
The wild-type ABH2 did not form appreciable amounts of cross-linked products with all of the dsDNA probes in the control experiments, indicating that the cross-links between the mutant ABH2 and dsDNA probes 3–6 are mostly specific. Like ABH3, the wild-type ABH2 also formed non-specific cross-linked products with DNA-1. This could be due to the flexibility of the ssDNA that may allow the thiol tether on the DNA probe to reach and react with cysteine residues on the surface of ABH2. It is interesting to notice that appreciable amounts of non-specific cross-links only occur between ssDNA-1 and ABH2 or ABH3. Negligible amounts of non-specific cross-links were observed between these proteins and all dsDNA probes. AlkB forms very small amounts of non-specific cross-linked products (<2%) with all DNA probes. The exact reason is not clear to us.
The amount of cross-linked product between ABH2 H171C and DNA-1 is similar to that of the mutant protein and DNA probes 3–6 when the non-specific cross-linking is not accounted for. The ABH2 mutant proteins cross-linked well with all dsDNA probes except DNA-2. As discussed earlier, the C* in DNA-2 is stabilized intrahelically and is not expected to cross-link with the ABH2 mutant protein. The fact that ABH2 did not discriminate much between the other four probes seems to suggest that the protein can work on ssDNA and dsDNA equally well.
Cross-link between ABH1 and DNA
The wild-type ABH1 and the ABH1 H231C mutant protein were also expressed from E.coli and purified. The ABH1 H231C mutant protein is analogous to AlkB H131C. Although sufficient amounts of protein were obtained, no cross-link between the ABH1 H231C mutant protein and the single-stranded and dsDNA probes 1–4 was observed after extensive incubation under the same conditions (Fig. 4D). We conclude that this form of ABH1 does not cross-link to our DNA probes. It is interesting that in vitro activity studies also failed to identify the repair function of this protein (10). It is possible that the form of this protein expressed from E.coli lacks certain components or features that allow it to exhibit functions similar to other AlkB proteins. Our result seems to suggest that ABH1 expressed from E.coli interacts with neither ssDNA nor dsDNA.
In summary, we showed that ABH2 and ABH3 have structural and functional features similar to E.coli AlkB. The conserved ligand residues in these proteins are most probably the active site residues. These proteins have a mode where they can search for damaged bases by scanning through ssDNA. Like E.coli AlkB, these proteins can also locate base damage in dsDNA. They cannot flip out bases for damage searching and there is no need for them to do so. The substrates for these proteins do not form stable base pairs in the duplex DNA. The repair proteins simply capture the base lesions that are extrahelical. ABH1 failed to cross-link to our DNA probes and it is likely that this form of protein expressed from E.coli may not interact with DNA at all. The protein may either be missing some components while it was being expressed from bacteria or it may act on substrates that are not nucleic acids.
The substrate preferences of the AlkB proteins are different, as probed from the cross-linking studies. E.coli AlkB has a slight preference for ssDNA. ABH3 seems to prefer interacting with ssDNA. ABH2 works equally well on both ssDNA and dsDNA. The different substrate preferences may reflect the need for these proteins to repair damage occurring on different types of substrates, as also proposed by others (9,10). It is important to point out that we did not compare the results of the cross-linking experiments for these proteins with one another. The active sites of these proteins could be different and the proteins may recognize the thiol-tethered cytosine in a DNA probe differently. However, within each protein, the interaction between the protein and the different DNA probes should not be affected by how the C* is recognized in the active site of the protein. Multiple experiments were performed on each reaction and similar substrate preferences were observed for each protein. That being said, we did notice that the E.coli AlkB mutant protein cross-linked more efficiently than the ABH proteins. Lower amounts of DNA probes were needed for the observation of efficient cross-links for E.coli AlkB.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr T. Lindahl and Dr B. Sedgwick for the generous gifts of the original ABH1, ABH2 and ABH3 genes. We also wish to thank Dr J. Piccirilli for the use of the DNA synthesizer, and Dr N.-S. Li for his assistance in the DNA synthesis. This work is supported by the University of Chicago.
REFERENCES
- 1.Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC. [Google Scholar]
- 2.Wood R.D., Mitchell,M., Sgouros,J. and Lindahl,T. (2001) Human DNA repair genes. Science, 291, 1284–1289. [DOI] [PubMed] [Google Scholar]
- 3.Lindahl T., Sedgwick,B., Sekiguchi,M. and Nakabeppu,Y. (1988) Regulation and expression of the adaptive response to alkylating agents. Annu. Rev. Biochem., 57, 133–157. [DOI] [PubMed] [Google Scholar]
- 4.Sedgwick B., Robins,P., Totty,N. and Lindahl,T. (1988) Functional domains and methyl acceptor sites of the Escherichia coli Ada protein. J. Biol. Chem., 263, 4430–4433. [PubMed] [Google Scholar]
- 5.Lindahl T., Demple,B. and Robins,P. (1982) Suicidal inactivation of the E.coli O6-methylguanine-DNA methyltransferase. EMBO J., 1, 1359–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pegg A.E. (2000) Repair of O6-alkylguanine by alkyltransferases. Mutat. Res., 462, 83–100. [DOI] [PubMed] [Google Scholar]
- 7.Trewick S.C., Henshaw,T.F., Hausinger,R.P., Lindahl,T. and Sedgwick,B. (2002) Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature, 419, 174–178. [DOI] [PubMed] [Google Scholar]
- 8.Falnes P.O., Johansen,R.F. and Seeberg,E. (2002) AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature, 419, 178–182. [DOI] [PubMed] [Google Scholar]
- 9.Aas P.A., Otterlei,M., Falnes,P.Ø., Vågbo,C.B., Skorpen,F., Akbari,M., Sundheim,O., Bjørås,M., Slupphaug,G., Seeberg,E. et al. (2003) Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature, 421, 859–863. [DOI] [PubMed] [Google Scholar]
- 10.Duncan T., Trewick,S.C., Koivisto,P., Bates,P.A., Lindahl,T. and Sedgwick,B. (2002) Reversal of DNA alkylation damage by two human dioxygenases. Proc. Natl Acad. Sci. USA, 99, 16660–16665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mishina Y. and He,C. (2003) Probing the structure and function of the Escherichia coli DNA alkylation repair AlkB protein through chemical cross-linking. J. Am. Chem. Soc., 125, 8730–8731. [DOI] [PubMed] [Google Scholar]
- 12.Welford R.W.D., Schlemminger,I., McNeill,L.A., Hewitson,K.S. and Schofield,C.J. (2003) The selectivity and inhibition of AlkB. J. Biol. Chem., 278, 10157–10161. [DOI] [PubMed] [Google Scholar]
- 13.Begley T.J. and Samson,L.D. (2003) AlkB mystery solved: oxidative demethylation of N1-methyladenine and N3-methylcytosine adducts by a direct reversal mechanism. Trends Biochem. Sci., 28, 2–5. [DOI] [PubMed] [Google Scholar]
- 14.Margison G. (2002) A new damage limitation exercise: ironing (Fe(II)) out minor DNA methylation lesions. DNA Repair, 1, 1057–1061. [DOI] [PubMed] [Google Scholar]
- 15.Sedgwick B. and Lindahl,T. (2002) Recent progress on the Ada response for inducible repair of DNA alkylation damage. Oncogene, 21, 8886–8894. [DOI] [PubMed] [Google Scholar]
- 16.Dinglay S., Trewick,S.C., Lindahl,T. and Sedgwick,B. (2000) Defective processing of methylated single-stranded DNA by E.coli alkB mutants. Genes Dev., 14, 2097–2105. [PMC free article] [PubMed] [Google Scholar]
- 17.Kondo H., Nakabeppu,Y., Kataoka,H., Kuhara,S., Kawabata,S. and Sekiguchi,M. (1986) Structure and expression of the alkB gene of Escherichia coli related to the repair of alkylated DNA. J. Biol. Chem., 261, 15772–15777. [PubMed] [Google Scholar]
- 18.Wei Y.-F., Carter,K.C., Wang,R.-P. and Shell,B.K. (1996) Molecular cloning and functional analysis of a human cDNA encoding an Escherichia coli AlkB homolog, a protein involved in DNA alkylation damage repair. Nucleic Acids Res., 24, 931–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samson L., Derfler,B. and Waldstein,E.A. (1986) Suppression of human DNA alkylation-repair defects by Escherichia coli DNA-repair genes. Proc. Natl Acad. Sci. USA, 83, 5607–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aravind L. and Koonin,E.V. (2001) The DNA-repair protein AlkB, EGL-9 and leprecan define new families of 2-oxoglutarate and iron-dependent dioxygenases. Genome Biol., 2, 0007.1–0007.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He C. and Verdine,G.L. (2002) Trapping distinct structural states of a protein–DNA interaction through disulfide crosslinking. Chem. Biol., 9, 1297–1303. [DOI] [PubMed] [Google Scholar]
- 22.Xu Y.Z., Zheng,Q. and Swann,P.F. (1992) Synthesis of DNA containing modified bases by post-synthetic substitution. Synthesis of oligomers containing 4-substituted thymine: O4-alkylthymine, 5-methylcytosine, N4-(dimethylamino)-5-methylcytosine and 4-thiothymine. J. Org. Chem., 57, 3839–3845. [Google Scholar]
- 23.MacMillan A.M. and Verdine,G.L. (1990) Synthesis of functionally tethered oligonucleotides by the convertible nucleoside approach. J. Org. Chem., 55, 5931–5933. [Google Scholar]
- 24.Allawi H.T. and SantaLucia,J.,Jr (1998) Nearest-neighbor thermodynamics of internal A:C mismatches in DNA: sequence dependence and pH effects. Biochemistry, 37, 9435–9444. [DOI] [PubMed] [Google Scholar]
- 25.Allawi H.T. and SantaLucia,J.,Jr (1998) Thermodynamics of internal C:T mismatches in DNA. Nucleic Acids Res., 26, 2694–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]