Abstract
CD22 is a transmembrane glycoprotein expressed by mature B cells. It inhibits signal transduction by the B cell receptor and its co-receptor CD19. Recently it was reported that most human lung cancer cells and cell lines express CD22 making it an important new lung cancer therapeutic target (Can Res 72:5556, 2012). The objective of our studies was to independently validate these results with the goal of testing the efficacy of our CD22 immunotoxins on lung cancer cell lines. As determined by qRT-PCR analysis, we found that levels of CD22 mRNA in a panel of human lung cancer cell lines were 200–60,000- fold lower than those observed in the human CD22+ Burkitt’s lymphoma cells, Daudi. Using flow cytometry with a panel of CD22 monoclonal antibodies and Western blot analyses, we could not detect surface or intracellular expression of CD22 protein in a panel of lung cancer cell lines. In addition, the in vitro proliferation of the lung tumor cell lines was not affected by CD22 antibodies or our highly potent anti-CD22 immunotoxin. By contrast, CD22+ Daudi cells expressed high levels of CD22 mRNA and protein and were sensitive to our CD22 immunotoxin. Importantly, primary non-small cell lung cancers from over 250 patient specimens did not express detectable levels of CD22 protein as assessed by immunohistochemistry. We conclude that CD22 is not expressed at measurable levels on the surface of lung cancer cells and that these cells can not be killed by anti-CD22 immunotoxins.
Keywords: lung cancer, CD22 expression
Introduction
Lung cancer is the leading cause of cancer death worldwide (1). There are two major types of lung cancer; non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). Surgical resection remains the main treatment strategy for the former and chemotherapy for the latter (2–4). A major goal in this field is to identify novel targets to treat both types of tumors. (5). Recently, Tuscano et al. (6) reported that CD22, a hallmark marker on B lymphocytes, is expressed on lung cancer cells and might thereby serve as a new target for therapy.
CD22 (Siglec-2) is a 140 kDa B-cell restricted membrane-bound member of the immunoglobulin (Ig) superfamily that binds glycan ligands containing α2,6-linked sialic acid (α2,6Sia) through its two amino-terminal of seven extracellular Ig-like domains (7–9). CD22 inhibits B-cell antigen receptor (BCR) and CD19 signaling, as well as B cell survival, through mechanisms that are both dependent on, and independent of, its ligand-binding activity (10). CD22 has a cytoplasmic domain with six conserved tyrosines, four of which are present within the immunoreceptor tyrosine-based inhibitory motifs. Following cross-linking of BCRs, these tyrosine residues are phosphorylated, allowing the recruitment of Src homology region 2 (SH2) domain-containing protein tyrosine phosphatase-1 (SHP-1) and SH2 domain-containing inositol 5’-phosphatase (SHIP), the subsequent dephosphorylation of BCR-proximal signaling molecules, and the inhibition of BCR signaling (11–13).
Because of its restriction to the B cell lineage and its rapid internalization following antibody-mediated cross-linking, CD22 is an attractive therapeutic target for treating B cell malignancies and autoimmune diseases involving dysregulated B cells (14–22).
If indeed lung cancer cells do express CD22, the agents that target CD22 might be effective in lung cancer patients. Over the last two decades, we and others have tested monoclonal antibodies (MAbs) and immunoconjugates directed against CD22 in mouse xenograft models of human B cell lymphomas and leukemias (14–22). In Phase I and II clinical trials in humans, side effects have been tolerable and efficacy has been encouraging (23–28). Several CD22-based therapies are now moving into advanced clinical trials (23–28). Since the observations of Tuscano and colleagues (6) were encouraging, and since we have developed therapeutics targeting CD22+ cells, we designed studies to determine whether our CD22 immunotoxins (ITs) would kill lung cancer cell lines. Prior to doing so, we repeated the experiments reported by Tuscano et al. (6) to confirm that lung cancer cell lines were indeed CD22+. Using the same methods, cell lines and antibodies, as well as an additional panel of over 30 anti-CD22 MAbs and several cell lines, we failed to confirm that surface CD22 is expressed at measurable levels on lung cancer cell lines and sections of lung tumors, or that CD22 targeted IT kills lung cancer cells including those reported to be CD22+ by Tuscano et al. (6).
Materials and Methods
Cell lines
The human Burkitt’s lymphoma cell line, Daudi (29), the acute T cell leukemia cell line, Jurkat, (30), and the NSCLCs cell lines, Calu-1 (31) and A549 (32), were purchased from the American Type Culture Collection (ATCC, Manassas, VA). Other NSCLC cell lines (H358, H460, HCC827, H1299, H1355, H1650 and H1975) and two SCLC cell lines (H727 and H1184) were obtained from Drs. Minna and Gazdar at UT Southwestern (UTSW) who originally established them (33,34). All cell lines were verified by DNA fingerprinting with the Promega StemElite ID system (Promega, Madison, WI) which consists of 9 short tandem repeat markers, one gender-specific marker (amelogenin) and one mouse-specific locus. These loci collectively provide a genetic profile with a random match probability of 1 in 3 × 109. Fingerprints were compared against the Hamon Center for Therapeutic Oncology Research (HCTOR) at UTSW database of more than 500 reference fingerprints collected from the ATCC and the HCTOR (34). A match was defined as two fingerprints where at least 7 of 10 human markers were identical. This allows for cell culture drift or genomic instability in some cancer cell lines. All cell lines were also confirmed to be free of mycoplasma using the Mycoplasma Tissue Culture-NI kit (Gen-Probe, San Diego, CA). Cell suspensions of Daudi and Jurkat cells, and monolayers of all the adherent lung cancer cell lines were maintained in RPMI 1640 (Sigma, St. Louis, MO) containing 10% heat-inactivated fetal bovine serum (FBS, HyClone, Logan, UT, and Atlanta Biologicals, Lawrenceville, GA), Penicillin (100 U/mL), Streptomycin (100 µg/mL, Gibco, Grand Island, NY, and Mediatech, Herndon, VA) and L-Glutamine (2 mM, Gibco and Mediatech). For passaging, all adherent cells were harvested following treatment with 0.25% trypsin-EDTA solution (Gibco and Sigma) and subcultured at 1:4 to 1:6 split ratios when 80–90% confluence was achieved. Alternatively, adherent cells were harvested using CellStripper® (Mediatech). All cell lines were maintained at 37°C in a humidified chamber of 95% air and 5% CO2. A cryopreserved aliquot of each cell line was thawed every 4 to 5 weeks.
Cell staining and Flow Cytometry
Tumor cells were analyzed for cell surface CD22 expression by immunofluorescence staining followed by flow cytometric analysis in three different laboratories by five independent investigators. For each staining, samples of 1×106 cells were suspended in 100 µL of stainingbuffer [2% bovine serum albumin (BSA, Sigma) in phosphate buffer saline (PBS)] containing a range of concentrations (0.005 to 5 µg/sample) of primary antibodies (mouse anti-human CD22 MAbs and their corresponding isotype-matched control MAbs for 30 minutes at 4°C in the dark). After washing, a secondary R-phycoerthrin (RPE)-labeled goat anti-mouse IgG (heavy and light chains) antibody adsorbed with human IgG (Southern Biotech Equipment, Birmingham, AL) was added to the cells for 30 minutes at 4°C in the dark. In some experiments a fluorescein isothiocyanate (FITC)-labeled goat anti-mouse IgG was used for primary antibody detection (19). Data were obtained using a FACSCalibur (Beckton Dickinson, San Jose, CA) and analyzed using FloJo 8.5 Software (Tree Star Inc., Stanford, CA). In total, 34 mouse anti-human CD22 MAbs were used that bind different domains of the CD22 molecule, and they originated as follows: HD6, RFB4, UV22-1 and UV22-2 were characterized initially by Shen et al. (17) and Li et al. (35); HB22-2, HB22-5, HB22-7, HB22-12, HB22-13, HB22-15, HB22-17, HB22-18, HB22-19, HBB22-22, HB22-23, HB22-25, HB22-26, HB22-27, HB22-33, and HB22-196 as described previously (36,37); 3G5, 3H4, 4KB128, BC-8, BL-3C4, BU59, EC6 (OKB22A), G28-7, HD39, HD239, HI22, IS7, S-HCL1 (Leu-14) and To15, were obtained from the Fifth International Leukocyte Differentiation Antigen Workshop (Boston, MA) (36,37). Their isotypes and specific CD22 binding domains are summarized in supplementary data, Table 1. The mouse isotype IgG control antibodies included the IgG1s, MOPC-21 (38) and MOPC-31C (39, 40), the IgG2a (G155-178) (BD Biosciences, San Jose, CA, unpublished data) and the IgG2b (MPC-11) (41,42) were purchased from BD Biosciences. All three mouse IgG isotypes were used as negative controls for HB22–33 (IgM) MAb staining.
Western Blotting (WB)
Ten million cells were lysed for 30 minutes on ice in Cell Extraction Buffer (Invitrogen, Camarillo, CA) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF, Sigma) and a protease inhibitor cocktail (Sigma) following the manufacturer’s instructions. The protein concentration of the lysates was determined using a Pierce™ BCA (bicinchoninic acid) Protein Assay Kit (Thermo Scientific, Rockford, IL). Equal amounts of total protein (25 µg) were electrophoresed on 8% sodium dodecyl sulfate polyacrylamide gels (SDS-PAGE) and the proteins were electrotransferred from the gel to an Immun-Blot polyvinylidene difluoride (PDVF) membrane (Bio-Rad Laboratories). After blocking with 5% BSA (Sigma) in Tris Buffered Saline (TBS) with 0.1% Tween-20 (Sigma) for 1 hour at room temperature (RT) with shaking, the membranes were incubated with the specific primary antibodies overnight at 4°C, washed and probed for 1 hour with specific horseradish peroxidase (HRP)-linked goat anti-rabbit Ig (heavy and light chains) antibody (Cell Signaling Technology, Danvers, MA) at a dilution of 1:3000 at RT with shaking. The bands were visualized with Pierce®Enhanced Chemiluminescence (ECL) Western Blotting substrate (Thermo Scientific) and scanned using ScanMaker 4900 (Microtek International Inc., China). As primary antibodies, polyclonal rabbit anti-human CD22 (H-221) (Santa Cruz Biotechnology, Santa Cruz, CA) at a dilution of 1:200 and rabbit anti-human β-actin (Cell Signaling Technology) at a dilution of 1:1000 were used. H-221 was the same anti-CD22 antibody used by Tuscano et al. and purchased from the same company. Each assay was performed by two independent investigators.
Quantification of CD22 transcripts
Two methods were used:
For quantitative real time-polymerase chain reaction (qRT-PCR), mRNA from 1–5 × 106 cells was extracted using QIAcube (Qiagen, Valenica, CA). cDNA was generated from 0.5 µg mRNA using an iScript cDNA synthesis kit (BioRad, Hercules, CA). Gene specific TaqMan probes (Applied Biosystems, Foster City, CA) were utilized for quantitative analyses of mRNA transcript levels (CD22 Hs00233533_m1, GAPDH 02758991_g1). The GAPDH gene was employed as an internal reference to normalize input cDNA. PCR reactions utilized the ABI 7300 Real-time PCR System and Software (Applied Biosystems). The comparative CT method was managed to calculate relative mRNA concentrations.
For PCR mapping of CD22 transcripts, total RNA from 5–10 × 106 cells was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA) following the manufacturer’s directions. Contaminating DNA was removed (Applied Biosystems, Foster City, CA) before performing cDNA synthesis from 1 µg of total RNA (SuperScript III First-Strand Synthesis System; Invitrogen). Primer sequences amplifying three distinct regions of human CD22 were: leader sequence (LS) to the Ig domain I (Ig I): (Forward) 5’ GGGAAGACACGCGGAAACAGG -3’ and (reverse) 5’- GAGGTGCACCGGGTGGATACTC -3’; Ig domain II (Ig II) to Ig domain III (Ig III): (Forward) 5’- GAGGCAGGCTGCTGTCACCTCG -3’ and (Reverse) 5’- CCACGTCATTGGAGACCTGACAGCAGTACTTCC -3’; Ig domain IV (Ig IV) to cytoplasmic domain (Cyto): (Forward) 5’-GGTCAGCCTCCAATGTGACT -3’ and (Reverse) 5’- CTGGCTCTGTGTCCTCTTCC -3’. GAPDH transcripts were amplified as a reference: (Forward) 5’- TCACCAGGGCTGCTTTTA -3’ and (Reverse) 5’- TTCACACCCATGACGAACA -3’. PCR cycle times: 95°C for 30 seconds, 60°C for 45 seconds and 72°C for 45 seconds (30 or 40 cycles).
Cytotoxicity Assays
CD22 MAbs were tested for their ability to kill human lung cancer cell lines vs. the CD22+ Daudi and CD22− Jurkat cell lines. 2 × 104 cells in 100 µL complete RPMI medium were incubated in quadruplicate in 96-well plates at 37°C for 16 hours. Mouse anti-human CD22 MAbs (HB22-7 or RFB4) and chimeric anti-human CD20 MAb, Rituximab™ (Biogen Idec, Inc., San Diego, CA), were added in a final volume of 200 µL at concentrations ranging from 5 to 50 µg/mL (3.33-33.3 × 10−8 M). Two MAbs coupled to deglycosylated ricin A chain (dgA) ITs were used as positive (RFB4-dgA) (16–19) and negative RFT5-dgA (43) controls. Both ITs were added in a final volume of 200 µL at final concentrations ranging from 1 × 10−10 to 1 × 10−13 M. Another nonspecific positive control Triton X-100 (Sigma) at a final concentration of 0.675% in a final volume of 200 µL was used also. Cells were incubated at 37°C for another 72 hours. 20 µL of CellTiter 96 AQueous One Solution (MTS) (Promega) was then added to each well and the plates were incubated for another 3 hours. Absorbance was measured at 492 nm using a Tecan Spectrafluor Plus plate reader (Tecan, Maennedorf, Switzerland) and analyzed with Magellan 2 software (Tecan, Salzburg, Austria). The experiments were repeated at least three times by two independent investigators.
Cytotoxicity was also measured by plating lung cancer cell lines (1 × 104 cells/mL) in triplicate wells of 48-well tissue culture plates in DMEM (Sigma) containing 10% FBS, and allowing them to adhere for 8 hours. Each group of triplicate wells was then treated with 50 µg/mL HB22-7, 50 µg/mL Rituximab™ (negative control), or medium only. After 72 hours, the non-adherent cells were harvested and combined with the remaining adherent cells that were harvested using trypsin-EDTA. The cells were washed in Dulbecco’s PBS (DPBS) (Mediatech), and then analyzed by flow cytometry (FACScan, Becton Dickinson) for the percentage of viable cells using 7-aminoactinomycin D (7-AAD) at a 1:500 dilution for staining (BD Biosciences).
Immunohistochemistry (IHC)
Formalin-fixed paraffin embedded (FFPE) samples from surgically resected lung cancer specimens were obtained from the Tissue Procurement Resource at UTSW and the Lung Cancer Specialized Program of Research Excellence Tissue Bank at The University of Texas MD Anderson (UTMDA) Cancer Center (Houston, TX). Detailed information regarding the pathologic diagnosis and staging, demographic data (gender and race), smoking history, treatment, overall survival, and time of recurrence for the majority of the clinical cases were obtained and are presented in the supplementary data, Tables 2–4. The tissue-procurement and distribution were approved by the Institutional Review Board of both universities.
Paraffin embedded sections from 10 lung cancer specimens (5 squamous cell carcinomas and 5 adenocarcinomas) (supplementary data, Table 2) were sectioned at 5 µm and affixed to silane-coated microscope slides. Serial sections were de-paraffinized, hydrated, and subjected to antigen unmasking techniques specific to each marker: for CD22, 10 minutes heating at 95°C in 1 mM EDTA at pH 8.0; for thyroid transcription factor-1 (TTF-1), 10 minutes heating at 95°C in 1 × Antigen Retrieval Citrate at pH 6.0 (BioGenex Laboratories, Fremont CA); and for pan cytokeratin, 15 minutes RT incubation with 0.1% Pronase E (Sigma). Following antigen-retrieval, endogenous peroxidase activity was blocked by adding 0.5% H2O2 (Sigma) followed by 3% normal horse serum (Vector, Burlingame, CA). Primary mouse anti-human MAbs against CD22 (FPC1) (dilution 1:30), pan cytokeratin 5/6/18 (LP34) (dilution 1:30), or TTF-1 (SPT24) (dilution 1:50), from Leica Biosystems (Newcastle Upon Tyne, UK), were diluted in PBS applied to sections, and incubated overnight at 4°C. Subsequent biotin/streptavidin horseradish-peroxidase detection of bound primary MAb was conducted according to a previously described immunoperoxidase method using reagents from Vector (44,45) and 3,3'-Diaminobenzidine (DAB) chromogen (Dako, Carpinteria, CA). Sections were counterstained with hematoxylin, dehydrated, cleared and cover slips were affixed with permanent mounting medium. Bright-field photomicrography of resulting stains was performed on a Leica DM2000 compound microscope (Buffalo Grove, IL), equipped with an Optronics Microfire CCD Camera (Goleta, CA). Images were acquired with Pictureframe 2.0 software and saved as TIFF files.
Alternatively, tumor tissues specimens from 217 NSCLCs (supplementary data, Table 3) were used to build a tissue microarray (TMA) using triplicates of 1.5 mm cores from each sample. Additionally, 20 whole section specimen slides (10 squamous cell carcinomas and 10 adenocarcinomas) from NSCLC surgical resections were examined (supplementary data, Table 4). Tonsil tissue, which is rich in B-cell lymphocytes, was used as a positive control for CD22 immunostaining. Moreover, the B cells were present in some of the tumor tissue specimens and were used as an “internal” CD22 positive control. For a negative control, PBS instead of primary antibody was added. For these IHC experiments, heat induced epitope retrieval was performed in a pressure cooker using pH 6.0 Target Retrieval Solution (Dako). The IHC staining was performed in an Autostainer Plus automated immunostaining machine (Dako), using the mouse anti-human CD22 MAb (Leica Biosystems) at a 1:20 dilution as primary antibody, incubated for 60 minutes at RT, and as secondary system for detection a Dako EnVision + Dual Link System-HRP (DAB+) (Dako) was used according to the manufacturer’s recommendations.
Results
Some lung cancer cell lines express very low levels of CD22 transcripts
We first analyzed CD22 mRNA expression in the lung cancer cell lines. Using a CD22 gene-specific TaqMan probe for quantitative analysis by qRT-PCR, we found that the CD22 mRNA levels were 200–60,000- fold lower than those observed in Daudi cells (Table 1). To validate these data, we used PCR primers that amplify three distinct regions of human CD22 transcripts and span introns to avoid DNA amplification. After 30 cycles of amplification, we observed weak corresponding-sized bands only for HCC827, H727, H1650 and H1975 cells relative to Daudi cells. Appropriately-sized bands were readily visualized after 40 cycles of amplification (Figure 1). Furthermore, the PCR amplification product from H727 cell cDNA generated using Ig IV to Cyto primers was sequenced using both forward and reverse primers. The PCR product was a perfect match for the human CD22 cDNA sequence. Thus, H727 cells expressed CD22 transcripts at levels that were ≤ 0.5% of those observed in Daudi cells.
Table 1.
Relative fold expression of CD22 mRNA as compared to GAPDH in the cell lines included in the study, as measured by qRT- PCRa
| Cell line | Mean fold change in gene expression ± SDb |
|---|---|
| Daudi | 1.0084 ± 0.059 |
| Jurkat | 0.000008 ± 0.0000001 |
| Calu-1 | 0.00054 ± 0.00009 |
| H460 | NDc |
| A549 | 0.00014 ± 0.00007 |
| H727 | 0.0027 ± 0.0013 |
| HCC827 | 0.0057 ± 0.0043 |
| H1184 | 0.000016 ± 0.000006 |
| H1299 | 0.00012 ± 0.00001 |
| H1355 | 0.000043 ± 0.00002 |
| H1650 | 0.00049 ± 0.000023 |
| H1975 | 0.0014 ± 0.0008 |
Average of 3–6 experiments performed for each cell line
Data analysis using 2−ΔΔCT algorithm
ND, not detected.
Figure 1. Lung cancer cell lines express a low level of CD22 transcripts.
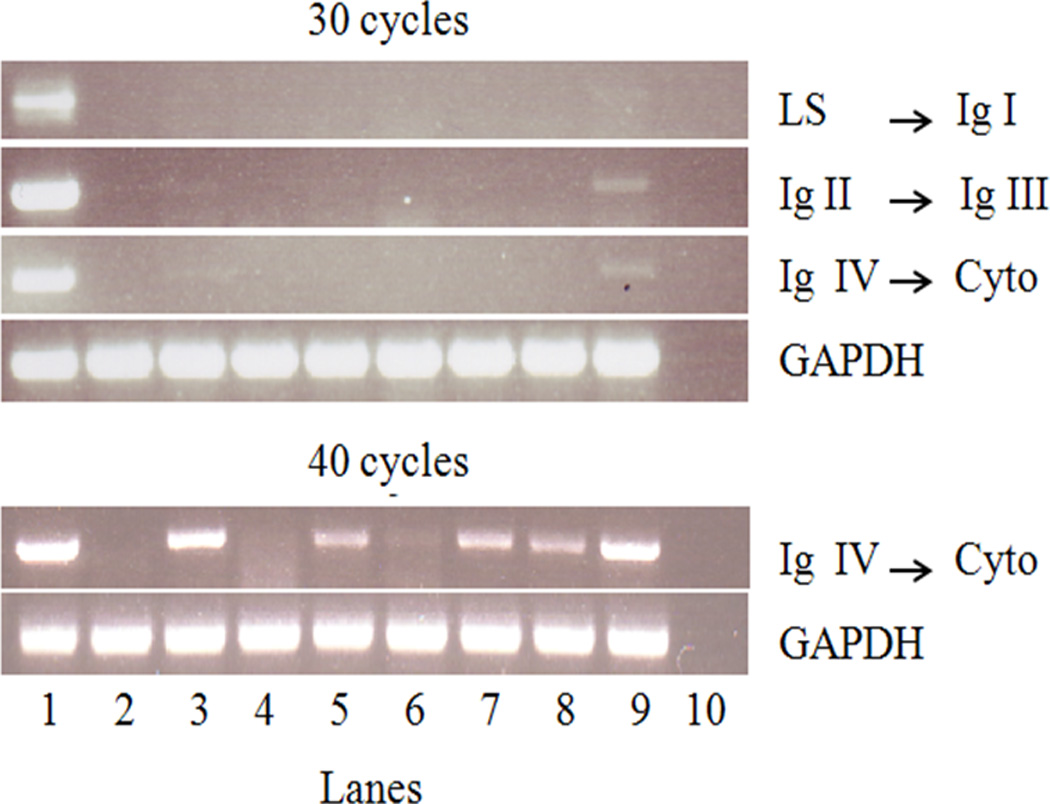
Representative PCR amplification of three distinct CD22 transcript regions using cDNA generated from the designated cell lines (1- Daudi; 2 - Jurkat; 3 - HCC827; 4 - H1355; 5 - H1975; 6 - A549; 7 - Calu-1; 8 - H1650; 9 - H727; 10 - dH2O). Daudi and Jurkat cells served as positive and negative controls, respectively. After 30 cycles of amplification, faint bands corresponding to CD22 transcripts were observed for Calu-1, H727, HCC827, H1650 and H1975 relative to the control (Daudi cells). Similar results were obtained in three or more experiments.
Lung cancer cell lines do not express measurable levels of cell surface CD22
The indirect cellular staining performed with five CD22 MAbs (HB22-7, HD6, RFB4, UV22-1 and UV22-2) (17,35) vs. their corresponding isotype-matched controls at concentrations ranging from 0.05 to 50 µg/mL revealed no measurable CD22 expression on any of the lung cancer cell lines. As expected, and shown previously (16), Daudi cells expressed high levels of cell surface CD22 (Figure 2A).
Figure 2. Cell surface CD22 can be detected on CD22+ Daudi cells but not on human lung cancer cell lines.
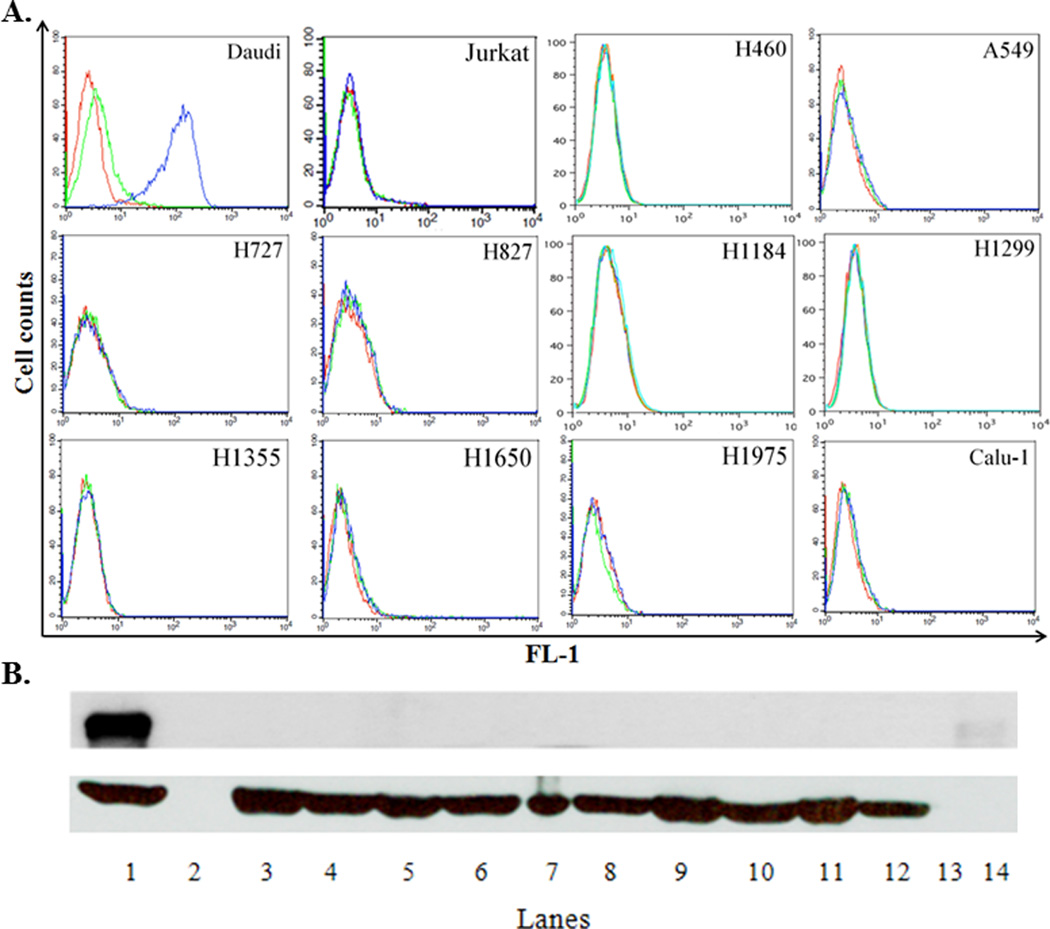
A. Flow cytometric analysis. One million cells from each cell line were incubated with 25 µg/mL of either a mouse isotype control antibody (MPC-11) or mouse antihuman CD22 (clone HB22.7) MAb. After washing out the unbound primary antibody, a FITC-labeled secondary goat anti-human IgG antibody was used. Cells were analyzed in FL-1 using a BD FACSCalibur. Red line – cells without antibodies; green line – cells plus the isotype control antibody and secondary antibody; blue line – cells plus the anti-CD22 antibody and secondary antibody. This is one representative out of 4–6 independent experiments. B. WB analysis. Equal amounts (25 µg) of protein from cell lysates were subjected to 8% SDS-PAGE followed by a WB with a rabbit anti-human CD22 antibody (H-221 clone). Upper panel - CD22 expression; lower panel - β-actin expression (loading control): 1 – Daudi; 2 - no loading; 3 - Calu-1; 4 - H1975; 5 - H1650; 6- H1355; 7 - H1299; 8 - H1184; 9 - HCC827; 10 - H727; 11 - A549; 12 - H460; 13 - no loading; 14 – molecular weight marker for 140 kDa. This is one representative out of 3–5 independent experiments.
Based on the above qRT-PTCR data, where we found that the H727 cells line expressed the highest levels of CD22 transcripts, we examined a comprehensive panel of mouse anti-human CD22 MAbs for binding to the H727 cell line, as well as Daudi (positive control) and Jurkat(negative control) cells. None of the 34 MAbs tested were positive on the H727 cells (supplementary data, Table 1). Because CD22 might be localized in the cytoplasm, we also performed indirect intracellular staining with the HB22-7 MAb on H727 cells. There was no measurable staining relative to Jurkat cells, while Daudi cells were CD22 positive (data not shown).
CD22 protein expression in lung cancer cell lines cannot be detected by WB
To validate the flow cytometry data, we carried out WB for CD22 using cell lysates and a polyclonal antiserum reactive with CD22. As expected, the lysates prepared from Daudi cells showed a very strong band corresponding to 130–140 kDa (the molecular weight of the human CD22 molecule). In contrast, no bands corresponding to the molecular weight of CD22 were observed in the lysates of the lung cancer cells or the negative control, Jurkat cells (Figure 2B). Based on these experiments, we concluded that only Daudi cells express the CD22 protein at detectable levels when 25 µg of cell lysates were examined.
CD22 MAbs and a potent anti-CD22 immunotoxin are not cytotoxic for lung cancer cell lines
Since it had been previously claimed that CD22 MAb alone killed lung cancer cell lines in vitro (6) we repeated the published experiments using a range of concentrations of five anti-CD22 MAbs (HB22-7, HD6, RFB4, UV22-1 and UV22-2) vs. their corresponding isotype MAb controls. We also included our highly potent CD22-specific IT, RFB4-dgA vs. the corresponding isotype-control IT, RFT5 (anti-CD25)-dgA. These treatments had no effect on the viability of the lung tumor cell lines in vitro as measured by the Cell Titer 96® AQueous One Solution assay that measures the functionality of the mitochondrial membrane (a critical parameter of cellular physiology). As expected, only the CD22 IT (but not the isotype-matched IT) was highly effective in killing Daudi cells in vitro (< 10% viability at a molar concentration of 1 × 10−11) (Figure 3). In addition, we also used the chemical 7-AAD, which binds specifically to nuclear DNA following disruption of the cellular membrane, to measure the potential cytotoxic effect of naked CD22 MAb. No differences between the viability of cells treated with HB22-7 vs. untreated cells were observed using 7-AAD staining in any of the cell lines, including lung cancer lines and Daudi B cell lymphoma cells (data not shown).
Figure 3. Proliferation of lung cancer cell lines is not affected by either CD22 MAbs or an anti-CD22 IT.
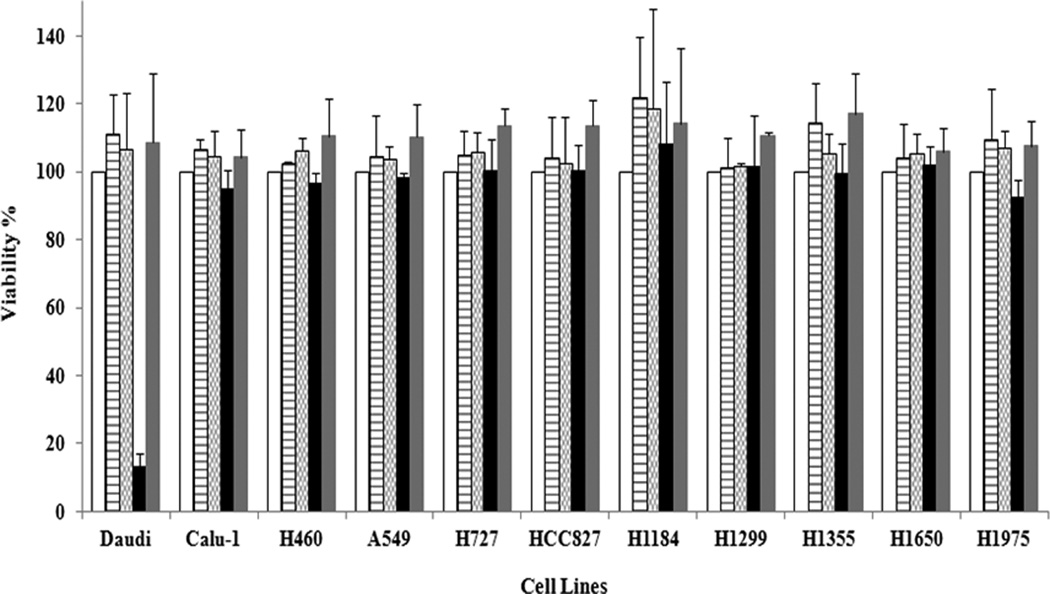
Cell lines were cultured in quadruplicate in 96-well plates at 2 × 105 cells/mL in a volume of 100 µL complete medium. The following day, mouse anti-human CD22 MAbs (HB22.7 or RFB4) were added in a final volume of 200 µL at a final concentration ranging from 5 to 50 µg/mL (3.33 × 10−8 to 3.33 × 10−7 M). Both ITs [RFB4 (anti-CD22)-dgA and RFT5 (anti CD25)-dgA] were added in a final volume of 200 µL at a final molar concentration ranging from 1 × 10−10 to 1 × 10−13 M. Cells were incubated for 72 hours and cell viability was calculated by using CellTiter 96® AQueous One Solution [blank columns - no treatment, media only; horizontally hatched columns - HB22-7 at 3.33 × 10−7 M; checkered columns - RFB4 at 3.33 × 10−7 M; black columns - RFB4 (anti-CD22)-dgA at 1.0 × 10−11 M; light gray columns - RFT5 (anti-CD25)-dgA at 1.0 × 10−11 M]. The figure depicts the average ± SD of cell viability from three independent experiments.
CD22 is not expressed on sections of lung cancer specimens from patients as observed by IHC
Both the primary and secondary antibodies and the detection system were identical to those used by Tuscano et al. We first analyzed 10 FFPE lung cancer patient specimens (5 squamous cell carcinomas and 5 adenocarcinomas) for cytokeratin 5/6/18 and TTF-1, to re-confirm their tissue type. Then, from each FFPE sample, serial sections were investigated for the corresponding cytokeratin 5/6/18 or TTF-1 expression, and for CD22 expression. Lung cancer cells stained positive only for either cytokeratin 5/6/18 (squamous cell carcinoma) or TTF-1 (adenocarcinoma) and the intratumoral B cells stained positive only for CD22 (Figure 4). We also analyzed CD22 expression in a tissue microarray (TMA) from our Lung Cancer SPORE (UTMDA Cancer Center) Pathology Core dataset containing 217 clinically and molecularly annotated NSCLCs (202 evaluable samples, including138 squamous cell carcinomas and 64 adenocarcinomas), as well as 20 whole section specimens (10 squamous cell carcinomas and 10 adenocarcinomas) from NSCLC surgical resections. We observed CD22 expression only on intratumoral B lymphocytes but not on the malignant lung cells (supplementary data, Tables 3 and 4).
Figure 4. Absence of CD22 in lung cancer cells from patients.
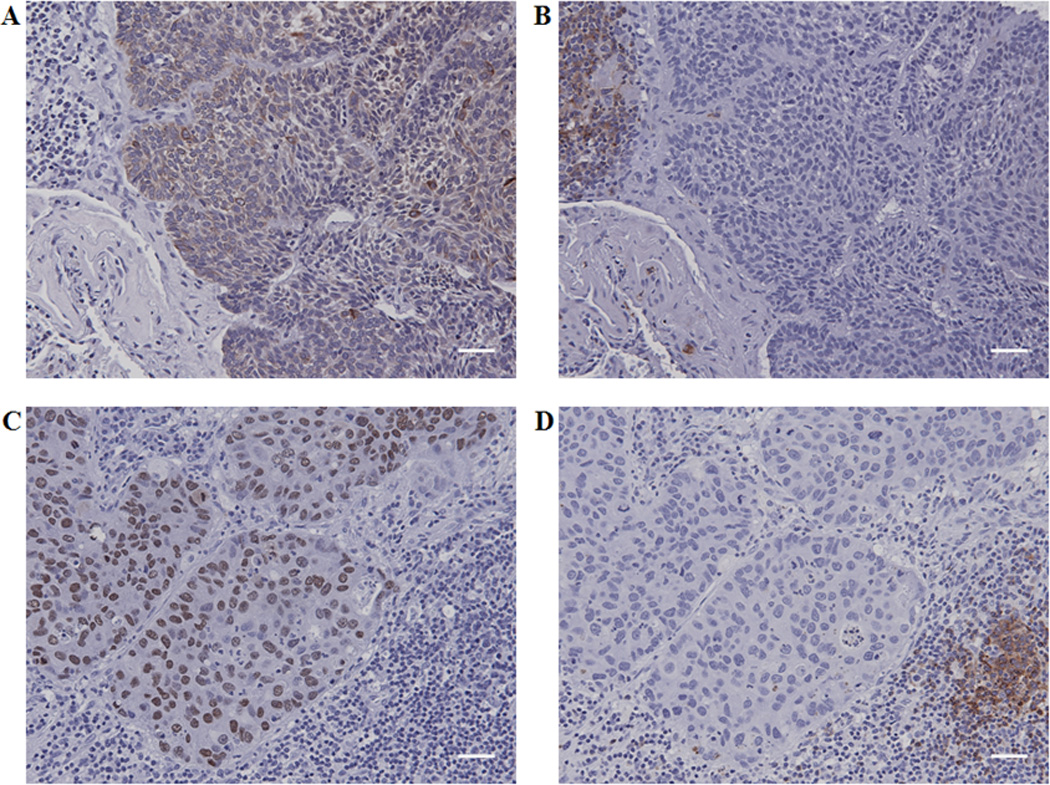
Immunostaining performed on serial sections of lung squamous cell carcinoma shows pan cytokeratin 5/6/18 positive cancer cells (A) and CD22 positive lymphocytes (B). Immunostaining performed on serial sections of lung adenocarcinoma shows thyroid transcription factor-1 positive cancer cells (C) and CD22 positive lymphocytes (D). Scale bars = 40 µm.
Discussion
Tuscano et al. have reported that CD22 is expressed on human lung tumors (6). Although CD22 has been previously described to be restricted to B cells (7–9, 10, 13), if indeed it is also expressed on lung cancers, this would be enormously important since it would provide a target for many already-developed CD22-specific targeted therapies (14–22). Thus, the aim of the present study was to further investigate the presence of CD22 on human lung cancer cell lines and lung tumors with the goal of testing our CD22 ITs (16–19) on lung cancer cell lines in vitro. Using the same cell lines, antibodies and assays previously reported by Tuscano et al. (6), we have been unable to reproduce most of their published findings. Puzzled by this, we validated our cell lines and also demonstrated that they did not contain mycoplasma. Alternative methods were also used to identify CD22 transcripts and proteins in and on the cell lines. All the experiments (except WB and IHC) reported here were carried out at least three times by three independent investigators at three universities. In addition, we tested a large panel of more than 30 mouse anti-human CD22 MAbs, 4 additional lung cancer cell lines and over 250 tissue sections from lung tumors and were still unable to identify cell-surface CD22 expression in lung cancer.
In essence, we found that: 1. Some of the lung cancer cell lines expressed low levels of CD22 mRNA transcripts, but these transcripts represented 1% or less of those found in a B lymphoma cell line (Daudi); 2. The lung cancer cell lines did not express CD22 on their surface or internally as determined by flow cytometry or by WB; 3. Using IHC, lung cancer specimens from over 250 patients were found to be CD22 negative; 4. CD22-targeted “naked” MAbs or our anti-CD22 IT (14–18) had no effect on the viability of the lung cancer cell lines or a T cell line but effectively killed CD22+ Daudi cells.
Since we were able to find very low levels of mRNA transcripts in the lung cancer cell lines, we determined whether CD22 expression might be influenced by the promoters of the neighboring genes in lung cancer lines. To this end we investigated genes localized adjacent to the CD22 locus within q13.1 of chromosome 19. The nearest flanking neighbor genes of CD22 are myelin associated glycoprotein (MAG) at 5’ (46) and free fatty acid receptor 3 (FFAR3) at 3’ (47). Furthermore, MAG has been described as a protein with many structural similarities to CD22 (48). However, we were unable to detect either MAG or FFAR3 protein in the lung cancer cell lines by WB (data not shown).
The possibility that truncated CD22 proteins might be expressed on these lung cancer cell lines was tested by using a panel of over 30 mouse anti-human CD22 MAbs which recognize and bind to all 7 different extracellular domain(s) of CD22 molecules (36, 37). None of the MAbs bound to the lung cancer cell lines at measurable levels as observed by flow cytometry analysis. Furthermore, the H727 cell line expressed CD22 transcripts that included both the extracellular plus transmembrane regions of CD22.
IHC on lung cancer samples was performed in three consecutive stages: (a) the validation of the specificity of the primary MAbs vs. control on normal corresponding tissues; (b) the re-confirmation of the initial post-surgical histopathologic diagnosis of lung cancer and B cell lymphoma samples, and (c) evaluation of CD22 expression on lung cancer cell lines. We found that all the human lung cancer specimens from patients were completely CD22 negative. The only CD22 positive signal in these specimens was provided by the infiltrating intratumoral B lymphocytes.
With regard to the report that Tuscano et al. (6) could kill lung cancer cell in vitro with anti-CD22 mAbs, we also investigated the toxicity of the CD22 MAbs and ITs using fluorescent 7-AAD which binds to the intracellular DNA only if the cell membranes are permeable (e.g., damaged) (49). Because some drugs might affect the cell viability without disrupting membrane integrity, we used a second proliferation assay where the read-out was the quantification of formazan produced by the bioreduction of MTS tetrazolium compound in mitochondria (50). Both methods showed that neither CD22 MAb nor its IT had any effect on the viability of the lung cancer cell lines in culture. In contrast, the same CD22 IT effectively killed CD22+ Daudi cells.
In comparing our results to those of Tuscano et al. (6), differences cannot be explained by the use of different antibodies, cell lines or methods. Indeed we extended their studies to a large panel of CD22 MAbs and an IT. We also used many more cell lines and tissue sections, and great care was taken in our studies to avoid problems (including the use of MAb isotype controls, careful WB protein loading, and using mycoplasma free tumor lines that were DNA fingerprinted). We cannot explain the fact that CD22 MAbs in their studies killed cells, although it is possible that their antibodies contained low levels of sodium azide or other toxic chemicals.
While it has been shown that tumor cells can express molecules not found on the corresponding normal tissue, in defining any new or unusual markers on cells, it is essential to carefully control all the experiments. We hope that other laboratories will carry out further studies to confirm our results or those of Tuscano et al. before coming to any final conclusions to use CD22 based reagents as diagnostics or therapeutics for lung cancer.
Supplementary Material
Acknowledgments
We are grateful to Drs. Cheryl Lewis and Kuntal Majmudar from the Tissue Procurement Center at UTSW for providing us with lung cancer specimens. We also want to thank Linda Berry for her assistance in preparing the manuscript.
Grant Support: This work was supported by the SPORE in Lung Cancer (P50CA70907), the Cancer Immunobiology Center, the Horchow Foundation, the Cancer Center Support Grant (5P30CA142543-03), the National Institute of Health grants AI56363 and AI057157, and a grant from The Lymphoma Research Foundation.
Footnotes
Disclosure of Potential Conflicts of Interest: none
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Bunn PA., Jr Worldwide overview of the current status of lung cancer diagnosis and treatment. Arch Pathol Lab Med. 2012;136:1478–1481. doi: 10.5858/arpa.2012-0295-SA. [DOI] [PubMed] [Google Scholar]
- 3.Cufer T, Ovcaricek T, O'Brien ME. Systemic therapy of advanced non-small cell lung cancer: Major-developments of the last 5-years. Eur J Cancer. 2013;49:1216–1225. doi: 10.1016/j.ejca.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 4.Sgambato A, Casaluce F, Maione P, Rossi A, Sacco PC, et al. Medical treatment of small cell lung cancer: state of the art and new development. Expert Opin Pharmacother. 2013 Aug 1; doi: 10.1517/14656566.2013.823401. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 5.Petrosyan F, Daw H, Haddad A, Spiro T. Targeted therapy for lung cancer. Anticancer Drugs. 2012;23:1016–1021. doi: 10.1097/CAD.0b013e3283585149. [DOI] [PubMed] [Google Scholar]
- 6.Tuscano JM, Kato J, Pearson D, Xiong C, Newell L, Ma Y, et al. CD22 antigen is broadly expressed on lung cancer cells and is a target for antibody-based therapy. Cancer Res. 2012;72:5556–5565. doi: 10.1158/0008-5472.CAN-12-0173. [DOI] [PubMed] [Google Scholar]
- 7.Cyster JG, Goodnow CC. Tuning antigen receptor signaling by CD22: integrating cues from antigens and the microenvironment. Immunity. 1997;6:509–517. doi: 10.1016/s1074-7613(00)80339-8. [DOI] [PubMed] [Google Scholar]
- 8.Tedder TF, Tuscano J, Sato S, Kehrl JH. CD22, a B lymphocyte specific adhesion molecule that regulates antigen receptor signaling. Annu Rev Immunol. 1997;15:481–504. doi: 10.1146/annurev.immunol.15.1.481. [DOI] [PubMed] [Google Scholar]
- 9.Walker JA, Smith KG. CD22: an inhibitory enigma. Immunology. 2008;123:314–325. doi: 10.1111/j.1365-2567.2007.02752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poe JC, Fujimoto Y, Hasegawa M, Haas KM, Miller AS, Sanford IG, et al. CD22 regulates B lymphocyte function in vivo through both ligand-dependent and ligand-independent mechanisms. Nat Immunol. 2004;5:1078–1087. doi: 10.1038/ni1121. [DOI] [PubMed] [Google Scholar]
- 11.Zhu C, Sato M, Yanagisawa T, Fujimoto M, Adachi T, Tsubata T. Novel binding site for Src homology 2-containing protein-tyrosine phosphatase-1 in CD22 activated by B lymphocyte stimulation with antigen. J Biol Chem. 2008;283:1653–1659. doi: 10.1074/jbc.M706584200. [DOI] [PubMed] [Google Scholar]
- 12.Doody GM, Justement LB, Delibrias CC, Matthews RJ, Lin J, Thomas ML, et al. A role in B cell activation for CD22 and the protein tyrosine phosphatase SHP. Science. 1995;269:242–244. doi: 10.1126/science.7618087. [DOI] [PubMed] [Google Scholar]
- 13.Poe JC, Fujimoto M, Jansen PJ, Miller AS, Tedder TF. CD22 forms a quaternary complex with SHIP, Grb2, and Shc. A pathway for regulation of B lymphocyte antigen receptor-induced calcium flux. J Biol Chem. 2000;275:17420–17427. doi: 10.1074/jbc.M001892200. [DOI] [PubMed] [Google Scholar]
- 14.Tuscano JM, O'Donnell RT, Miers LA, Kroger LA, Kukis DL, Lamborn KR, et al. Anti-CD22 ligand-blocking antibody HB22.7 has independent lymphomacidal properties and augments the efficacy of 90Y-DOTA-peptide-Lym-1 in lymphoma xenografts. Blood. 2003;101:3641–3647. doi: 10.1182/blood-2002-08-2629. [DOI] [PubMed] [Google Scholar]
- 15.Leonard JP, Goldenberg DM. Preclinical and clinical evaluation of epratuzumab (anti-CD22 IgG) in B-cell malignancies. Oncogene. 2007;26:3704–3713. doi: 10.1038/sj.onc.1210370. [DOI] [PubMed] [Google Scholar]
- 16.May RD, Vitetta ES, Moldenhauer G, Dörken B. Selective killing of normal and neoplastic human B cells with anti-CD19- and anti-CD22-ricin A chain immunotoxins. Cancer Drug Deliv. 1986;3:261–272. doi: 10.1089/cdd.1986.3.261. [DOI] [PubMed] [Google Scholar]
- 17.Shen GL, Li JL, Ghetie MA, Ghetie V, May RD, Till M, et al. Evaluation of four CD22 antibodies as ricin A chain-containing immunotoxins for the in vivo therapy of human B-cell leukemias and lymphomas. Int J Cancer. 1988;42:792–797. doi: 10.1002/ijc.2910420527. [DOI] [PubMed] [Google Scholar]
- 18.Herrera L, Yarbrough S, Ghetie V, Aquino DB, Vitetta ES. Treatment of SCID/human B cell precursor ALL with anti-CD19 and anti-CD22 immunotoxins. Leukemia. 2003;17:334–338. doi: 10.1038/sj.leu.2402790. [DOI] [PubMed] [Google Scholar]
- 19.Pop LM, Liu X, Ghetie V, Vitetta ES. The generation of immunotoxins using chimeric anti-CD22 antibodies containing mutations which alter their serum half-life. Int Immunopharmacol. 2005;5:1279–1290. doi: 10.1016/j.intimp.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 20.Salvatore G, Beers R, Margulies I, Kreitman RJ, Pastan I. Improved cytotoxic activity toward cell lines and fresh leukemia cells of a mutant anti-CD22 immunotoxin obtained by antibody phage display. Clin Cancer Res. 2002;8:995–1002. [PubMed] [Google Scholar]
- 21.Mansfield E, Amlot P, Pastan I, FitzGerald DJ. Recombinant RFB4 immunotoxins exhibit potent cytotoxic activity for CD22-bearing cells and tumors. Blood. 1997;90:2020–2026. [PubMed] [Google Scholar]
- 22.de Vries JF, Zwaan CM, De Bie M, Voerman JS, den Boer ML, van Dongen JJ, et al. The novel calicheamicin-conjugated CD22 antibody inotuzumab ozogamicin (CMC-544) effectively kills primary pediatric acute lymphoblastic leukemia cells. Leukemia. 2012;26:255–264. doi: 10.1038/leu.2011.206. [DOI] [PubMed] [Google Scholar]
- 23.Leonard JP, Schuster SJ, Emmanouilides C, Couture F, Teoh N, Wegener WA, et al. Durable complete responses from therapy with combined epratuzumab and rituximab: final results from an international multicenter, phase 2 study in recurrent, indolent, non-Hodgkin lymphoma. Cancer. 2008;113:2714–2723. doi: 10.1002/cncr.23890. [DOI] [PubMed] [Google Scholar]
- 24.Amlot PL, Stone MJ, Cunningham D, Fay J, Newman J, Collins R, et al. A phase I study of an anti-CD22-deglycosylated ricin a chain immunotoxin in the treatment of B-cell lymphomas resistant to conventional therapy. Blood. 1993;82:2624–2633. [PubMed] [Google Scholar]
- 25.Schindler J, Gajavelli S, Ravandi F, Shen Y, Parekh S, Braunchweig I, et al. A phase I study of a combination of anti-CD19 and anti-CD22 immunotoxins (Combotox) in adult patients with refractory B-lineage acute lymphoblastic leukaemia. Br J Haematol. 2011;154:471–476. doi: 10.1111/j.1365-2141.2011.08762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wayne AS, Kreitman RJ, Findley HW, Lew G, Delbrook C, Steinberg SM, et al. Anti-CD22 immunotoxin RFB4(dsFv)-PE38 (BL22) for CD22-positive hematologic malignancies of childhood: preclinical studies and phase I clinical trial. Clin Cancer Res. 2010;16:1894–1903. doi: 10.1158/1078-0432.CCR-09-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kreitman RJ, Tallman MS, Robak T, Coutre S, Wilson WH, Stetler-Stevenson M, Fitzgerald DJ, Lechleider R, Pastan I. Phase I trial of anti-CD22 recombinant immunotoxin moxetumomab pasudotox (CAT-8015 or HA22) in patients with hairy cell leukemia. J Clin Oncol. 2012;30:1822–1828. doi: 10.1200/JCO.2011.38.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Advani A, Coiffier B, Czuczman MS, Dreyling M, Foran J, Gine E, et al. Safety, pharmacokinetics, and preliminary clinical activity of inotuzumab ozogamicin, a novel immunoconjugate for the treatment of B-cell non-Hodgkin's lymphoma: results of a phase I study. J Clin Oncol. 2010;28:2085–2093. doi: 10.1200/JCO.2009.25.1900. [DOI] [PubMed] [Google Scholar]
- 29.Klein E, Klein G, Nadkarni JS, Nadkarni JJ, Wiqzell H, Clifford P. Surface IgM-kappa specificity on a Burkitt lymphoma cell in vivo and in derived culture lines. Cancer Res. 1968;28:1300–1310. [PubMed] [Google Scholar]
- 30.Schneider U, Schwenk HU, Bornkamm G. Characterization of EBV-genome negative "null" and "T" cell lines derived from children with acute lymphoblastic leukemia and leukemic transformed non-Hodgkin lymphoma. Int J Cancer. 1977;19:621–626. doi: 10.1002/ijc.2910190505. [DOI] [PubMed] [Google Scholar]
- 31.Fogh J, Wright WC, Loveless JD. Absence of HeLa cell contamination in 169 cell lines derived from human tumors. J Natl Cancer Inst. 1977;58:209–214. doi: 10.1093/jnci/58.2.209. [DOI] [PubMed] [Google Scholar]
- 32.Giard DJ, Aaronson SA, Todaro GJ, Arnstein P, Kersey JH, Dosik H, et al. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J Natl Cancer Inst. 1973;51:1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- 33.Phelps RM, Johnson BE, Ihde DC, Gazdar AF, Carbone DP, McClintock PR, et al. NCI-Navy Medical Oncology Branch cell line data base. J Cell Biochem Suppl. 1996;24:32–91. doi: 10.1002/jcb.240630505. [DOI] [PubMed] [Google Scholar]
- 34.Gazdar AF, Girard L, Lockwood WW, Lam LL, Minna JD. Lung cancer cell lines as tools for biomedical discovery and research. J Natl Cancer Inst. 2010;102:1–12. doi: 10.1093/jnci/djq279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li JL, Shen GL, Ghetie MA, May RD, Till M, Ghetie V, et al. The epitope specificity and tissue reactivity of four murine monoclonal anti-CD22 antibodies. Cell Immunol. 1989;118:85–99. doi: 10.1016/0008-8749(89)90359-6. [DOI] [PubMed] [Google Scholar]
- 36.Engel P, Nojima Y, Rothstein D, Zhou LJ, Wilson GL, Kehrl JH, et al. The same epitope on CD22 of B lymphocytes mediates the adhesion of erythrocytes, T and B lymphocytes, neutrophils, and monocytes. J Immunol. 1993;150:4719–4732. [PubMed] [Google Scholar]
- 37.Engel P, Wagner N, Miller AS, Tedder TF. Identification of the ligand-binding domains of CD22, a member of the immunoglobulin superfamily that uniquely binds a sialic acid-dependent ligand. J Exp Med. 1995;181:1581–1586. doi: 10.1084/jem.181.4.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melchers F. Biosynthesis of the carbohydrate portion of immunoglobulins. Incorporation of radioactive fucose into immunoglobulin G1 synthesized and secreted by mouse plasma-cell tumour MOPC 21. Biochem J. 1971;125:241–247. doi: 10.1042/bj1250241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Percy ME, Baumal R, Dorrington KJ, Percy JR. Covalent assembly of mouse immunoglobulin G subclasses in vitro: application of a theoretical model for interchain disulfide bond formation. Can J Biochem. 1976;54:675–687. doi: 10.1139/o76-098. [DOI] [PubMed] [Google Scholar]
- 40.Potter M, Keff EL. Disorders in the differentiation of protein secretion in neoplastic plasma cells. J Mol Biol. 1964;9:537–544. doi: 10.1016/s0022-2836(64)80225-4. [DOI] [PubMed] [Google Scholar]
- 41.Fahey JL. Physiocochemical characterization of mouse myeloma proteins: demonstration of heterogeneity for each myeloma globulin. J Exp Med. 1961;114:399–413. doi: 10.1084/jem.114.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laskov R, Scharff MD. Synthesis, assembly, and secretion of gamma globulin by mouse myeloma cells. I. Adaptation of the Merwin plasma cell tumor-11 to culture, cloning, and characterization of gamma globulin subunits. J Exp Med. 1970;131:515–541. doi: 10.1084/jem.131.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engert A, Martin G, Amlot P, Wijdenes J, Diehl V, Thorpe P. Immunotoxins constructed with anti-CD25 monoclonal antibodies and deglycosylated ricin A-chain have potent anti-tumour effects against human Hodgkin cells in vitro and solid Hodgkin tumours in mice. Int J Cancer. 1991;49:450–456. doi: 10.1002/ijc.2910490324. [DOI] [PubMed] [Google Scholar]
- 44.Cianga P, Medesan C, Richardson JA, Ghetie V, Ward SE. Identification and function of neonatal Fc receptor in mammary gland of lactating mice. European Journal of Immunology. 1999;29:2515–2523. doi: 10.1002/(SICI)1521-4141(199908)29:08<2515::AID-IMMU2515>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 45.Borvak J, Richardson JA, Medesan C, Antohe F, Radu C, Simonescu, et al. Functional expression of the MHC class I-related receptor, FcRn, in endothelial cells of mice. International Immunology. 1998;10:1289–1298. doi: 10.1093/intimm/10.9.1289. [DOI] [PubMed] [Google Scholar]
- 46.Barton DE, Arquint M, Roder J, Dunn R, Francke U. The myelin-associated glycoprotein gene: mapping to human chromosome 19 and mouse chromosome 7 and expression in quivering mice. Genomics. 1987;1:107–112. doi: 10.1016/0888-7543(87)90002-4. [DOI] [PubMed] [Google Scholar]
- 47.Sawzdargo M, George SR, Nguyen T, Xu S, Kolakowski LF, O'Dowd BF. A cluster of four novel human G protein-coupled receptor genes occurring in close proximity to CD22 gene on chromosome 19q13.1. Biochem Biophys Res Commun. 1997;239:543–547. doi: 10.1006/bbrc.1997.7513. [DOI] [PubMed] [Google Scholar]
- 48.Stamenkovic I, Seed B. The B-cell antigen CD22 mediates monocyte and erythrocyte adhesion. Nature. 1990;345:74–77. doi: 10.1038/345074a0. [DOI] [PubMed] [Google Scholar]
- 49.Liu X, Chen H, Patel D. Solution structure of actinomycin-DNA complexes: drug intercalation at isolated G-C sites. J Biomol NMR. 1991;1:323–347. doi: 10.1007/BF02192858. [DOI] [PubMed] [Google Scholar]
- 50.Berridge MV, Tan AS. Characterization of the cellular reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT): subcellular localization, substrate dependence, and involvement of mitochondrial electron transport in MTT reduction. Arch Biochem Biophys. 1993;303:474–482. doi: 10.1006/abbi.1993.1311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


