Abstract
Adoptive T cell therapy has proven effective against melanoma in mice and humans. However, because most responses are incomplete or transient, cures remain rare. To maximize the efficacy of this therapy, it will be essential to gain a better understanding of the processes which result in tumor relapse. We studied these processes using B16ova murine melanoma and adoptive transfer of OT-I T cells. Transfer of T cells as a single therapy provided a significant survival benefit for mice with established subcutaneous tumors. However, tumors which initially regressed often recurred. By analyzing tumors which emerged in the presence of a potent OT-I response, we identified a novel tumor escape mechanism in which tumor cells evaded T cell pressure by undergoing major genomic changes involving loss of the gene encoding the target tumor antigen. Furthermore, we show that these in vivo processes can be recapitulated in vitro using T cell/tumor cell co-cultures. A single round of in vitro co-culture led to significant loss of the ova gene and a tumor cell population with rapidly induced and diverse karyotypic changes. Although these current studies focus on the model OVA antigen, the finding that T cells can directly promote genomic instability has important implications for the development of adoptive T cell therapies.
Introduction
Adoptive T cell transfer is a promising immunotherapy for many types of malignancy, potentially providing patients with a pre-formed, ex vivo optimized anti-tumor immune response which will, in theory, form memory protection against recurrence1–3. Although generating ex vivo cultures of tumor-reactive T cells, and supporting lymphocyte engraftment and function in vivo, have presented clinical challenges4, intensive work in melanoma has resulted in increasing success rates5–8. Despite these encouraging results, the occurrence of incomplete, or transient, responses indicates that current protocols are often unable to eradicate metastases completely and prevent tumor re-growth. To develop therapies which consistently achieve long-term protection, it will, therefore, be necessary to gain a better understanding of the ways in which tumor cells respond to potent T cell therapies and the mechanisms they use to escape them.
Tumors escape from T cell attack in a variety of ways. They can eliminate the T cells9,10, render T cells non-functional11, promote the expansion of suppressive cells12–15, or become resistant to CTL killing16, 17. Loss of target antigens has been repeatedly seen in tumors under T cell attack18,19, associated with loss of RNA expression, which was either irreversible 20 or reversible with drugs or time21,22. Frequently, however, antigen loss is observed at the protein level but the underlying mechanisms remain poorly defined23. In this respect, tumors have been shown to escape immune recognition and clearance by downregulation of MHC expression24–26. Moreover, Garrido and co-workers have described the loss, at the genomic level, of MHC haplotype genes relevant to antigen expression in both pre-clinical and clinical tumor samples25,27.
Here, we show that an adoptively transferred population of OT-I T cells targeting the OVA tumor antigen resulted in tumor escape due to loss of the target antigen gene. Significantly, we show also that OT-I T cell pressure on the target B16ova tumor cells in vitro rapidly promoted the emergence of tumor cells with diverse karyotypes characterized by loss of the gene encoding the target OVA antigen. Together, these data indicate that potent immunotherapies can actively promote tumor evolution. While our system focuses on a surrogate tumor antigen, the number and variety of genomic changes which appeared following OT-I co-culture, as well as the distinct phenotype of escape tumors in vivo, suggests that interaction of tumor cells with anti-tumor T cells may actually help to generate variant tumor cells which are then capable of the evading the initial T cell therapy.
Materials and Methods
Mice, cell lines, antibodies, and reagents
6–8 week old female C57BL/6 mice were purchased from Jackson Laboratories (Bar Harbor, Maine). OT-I mice have been previously described28 and were bred at the Mayo Clinic. The B16ova cell line was derived from a B16.F1 clone transfected with a pcDNA3.1ova plasmid. B16ova cells were grown in DMEM (HyClone, Logan, UT, USA) + 10% FBS (Life Technologies) + 5 mg/mL G418 (Mediatech, Manassas, VA, USA) until challenge. Following in vitro co-cultures or harvest from mice, tumor lines were grown in DMEM + 10% FBS + 1% Pen/Strep (Mediatech). The following antibodies were used for flow cytometry: from BD Biosciences (San Jose, CA, USA) CD45-PerCP (clone 30-F11), H-2Kb-PE (clone AF6-88.5), CD8β.2-PE (clone 53-5.8), Vβ5-PE (clone MR9-4), and Vα2-FITC (clone B20.1), and from eBioscience (San Diego, CA, USA) CD3ε-FITC (clone 145-2C11).
In vivo experiments
All in vivo studies were approved by the Mayo IACUC. Mice were challenged subcutaneously with 5×105 B16ova cells in 100 μL PBS (HyClone). Tumors were measured 3 times per week, and mice were euthanized when tumors reached 10 mm diameter. For adoptive therapy experiments, 1×107 in vitro-activated OT-I T cells were injected intravenously in 100 μL PBS on day 7. In vivo data were analyzed using GraphPad Prism 4 software (GraphPad Software, La Jolla, CA, USA). Tumor volume was calculated as (4π/3)*((larger diameter + smaller diameter)/4)3. Palpable tumors less than 2 mm diameter were recorded as 1 mm diameter.
Flow cytometry
Freshly excised tumors and spleens were dissociated to form a single cell suspension. Red blood cells were lysed with ACK lysis buffer. Total remaining cellular content was stained for flow cytometry. Tumor cell lines were established from some excised tumors. Cells growing in culture were stained for H-2Kb following 48 hour treatment with 500 U/mL murine IFNγ (Peprotech, Rocky Hill, NJ, USA). Data were analyzed using FlowJo software (Tree Star Inc, Ashland, OR, USA).
In vitro T cell activation and co-cultures
Spleens and lymph nodes were harvested from OT-I mice and dissociated to obtain a single cell suspension. Red blood cells were lysed with ACK lysis buffer. Cells were resuspended at 1×106 cells/mL in IMDM (Gibco, Grand Island, NY, USA) + 5% FBS + 1% Pen/Strep + 40 μM 2-ME. Media was supplemented with SIINFEKL peptide at 1 μg/mL and hIL-2 at 50 U/mL. After two days, cells were split into new media supplemented with IL-2. Cells were used for adoptive transfer or in vitro assays following 4 days of activation. Serial co-cultures were performed at 1:1 ratios with 1×106 total cells/mL media. Following overnight co-culture, media and non-adherent cells were removed, and adherent cells were allowed to recover before the co-culture step was repeated. Under these conditions (E:T ratio of 1:1 and 24 hrs of co-culture) typically about 10% of the B16ova tumor cells would survive killing by OT-I T cells, although the number of survivors was variable between experiments. In all cases, additional tumor cell death occurred in the surviving population throughout the 2–3 days following co-culture. For this reason, the surviving population was allowed to recover for 1–3 weeks following co-culture before being subjected to functional, and genomic, analyses, at which point the cells were growing at similar rates to B16ova cultures which had not undergone co-culture.
51Cr cytotoxicity assay
B16ova target cells were treated overnight with 500 U/mL IFNγ. They were loaded with 51Cr for 90 minutes at 37C, washed, and plated at 1×104 cells/well in 96-well v-bottom plates (Nunc, Roskilde, Denmark). OT-I effectors were plated at varying concentrations to obtain a range of E:T ratios. Wells containing targets alone were used to determine spontaneous release. Wells containing targets and 0.1% Triton-X 100 were used to determine maximum release. Plates were incubated at 37C for 4 hours then spun down. 40 μL of supernatant/well was transferred to LumaPlates (PerkinElmer, Shelton, CT, USA) and allowed to dry overnight before being read on a PerkinElmer Topcount machine. Percent specific lysis is defined as (sample cpm-spontaneous cpm)/(maximum cpm-spontaneous cpm) x 100.
ELISA
2×106 total OT-I splenocytes were cultured for 24 hours in 2 mL with 10 μg SIINFEKL peptide. Supernatants were collected and analyzed for IFNγ by ELSIA according to the manufacturer’s instructions (BD OptEIA Mouse IFNγ ELISA Set; BD Biosciences Pharmingen, San Diego, CA, USA).
PCR
DNA was isolated from freshly excised tumors or cells in culture using the DNeasy kit (Qiagen, Maryland, USA). 0.5 μg of DNA was used in each PCR reaction. RNA was isolated using the RNeasy kit (Qiagen, Maryland, USA). cDNA was made from 1 μg of RNA using the First Strand cDNA Synthesis Kit (Roche, Indianapolis, IN, USA). One tenth of this cDNA was used in each PCR reaction. The following primers were used: ova sense: CACAAGCAATGCCTTTCAGA, ova antisense: TACCACCTCTCTGCCTGCTT, neor sense: TGCTCCTGCCGAGAAAGTAT, neor antisense: AATATCACGGGTAGCCAACG, gapdh sense: TCATGACCACAGTCCATGCC, gapdh antisense: TCAGCTCTGGGATGACCTTG.
Real-time PCR
Applied Biosystems (Carlsbad, CA, USA) Power SYBR green PCR mastermix and standard protocol were used. 10 ng of tumor DNA or 1 ng of cell line DNA was amplified and compared to a standard curve using ova primers sense: AGTGGCATCAATGGCTTCT and antisense: GTTGATTATACTCTCAAGCTGCTCA, gapdh primers sense: GGCAAATTCAACGGCACAGT and antisense: AGAATGGTGATGGGCTTCCC, or apolipoprotein B primers sense: CACGTGGGCTCCAGCATT and antisense: TCACCAGTCATTTCTGCCTTTG. Reactions were run on an ABI 7900 HT Sequence Detection System and analyzed using SDS2.3 software.
Cytogenetic analysis
A biotin-labeled FISH probe of the pcDNAova plasmid was prepared by the cytogenetics shared resource at Mayo Clinic. To assess initial ova content, the pooled B16ova population was probed for pcDNAova using this ova probe and a FITC-avidin secondary probe. For karyotypic analysis, a single-cell-derived population of B16ova was established. Following one round of co-culture with OT-I T cells (10:1 ratio of T cells:tumor cells), the tumor cells were allowed to recover for 23 days, and samples of the input clonal population and the co-cultured population were probed for pcDNAova by FISH as above. The slides were then probed with Mouse SKYPaint (Applied Spectral Imaging, California, USA) to obtain karyotypes of the same metaphase spreads.
Quantification of genomic instability was performed by scoring each karyotype for the number of chromosome pairs which differed from the most common presentation of that pair throughout the 14 karyotypes we obtained. Chromosome losses, gains, or translocations (including loss of the ova signal) were counted as karyotypic changes. A maximum score of 1 per chromosome pair or 20 per cell was possible.
Statistics
Survival curves were analyzed by the Log-Rank test. Differences in karyotypic diversity were quantified by the Mann-Whitney U test. All other data were analyzed by the 2-tailed t-test. Statistical significance was set at p<0.05 for all experiments.
Results
B16ova tumors grow in the presence of a functional anti-ova response
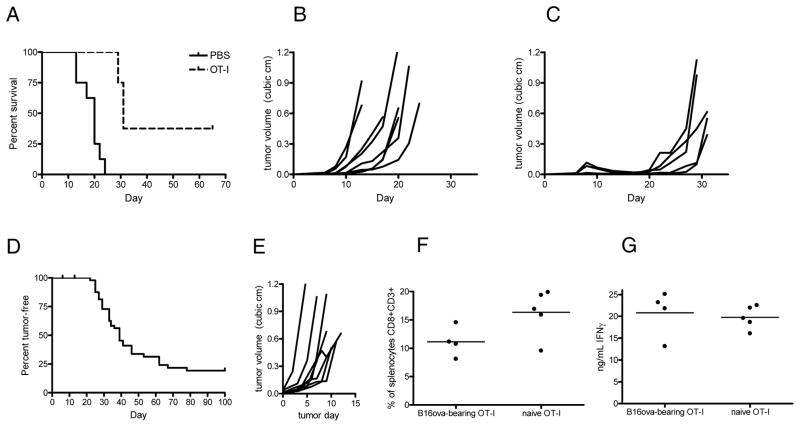
Adoptive transfer of 107 activated OT-I T cells into mice bearing established B16ova tumors resulted in a significant survival benefit over untreated animals (p<0.05) (Fig. 1A–C). Tumors regressed and often became undetectable (Fig. 1C). However, therapy was transient in most mice. Palpable tumors soon re-emerged and grew at least as rapidly as untreated tumors (Fig. 1B and C), which suggested that the recurrent tumors were no longer subject to OT-I pressure.
Figure 1. OT-I T cells incompletely eliminate B16ova tumors.
A, WT C57BL/6 mice (8 mice/group) were challenged subcutaneously with B16ova cells and received PBS or in vitro-activated OT-I T cells intravenously on day 7. OT-I-treated mice survived significantly longer than PBS-treated mice (p<0.05). B and C, growth curves of individual tumors from PBS-treated (B) or OT-I-treated mice (C). D, OT-I mice were challenged subcutaneously with B16ova tumors. The graph indicates days from the time of tumor challenge until tumors became measurable (>2 mm diameter). E, Growth curves of individual tumors in D. Tumor day 0 is the day at which each tumor was first measurable. F and G, spleens were harvested from naive OT-I mice and from OT-I mice with 10 mm tumors. Splenocytes were analyzed by flow cytometry and ELISA. F, average size of the splenic CTL population of naïve and tumor-bearing OT-I mice. G, splenocytes were pulsed with SIINFEKL peptide, and IFNγ production was measured by ELISA of the supernatants. Data are representative of at least three independent experiments.
To focus our study on tumors evading T cell pressure, we seeded B16ova tumors in naive OT-I mice. In these mice, tumors were subjected to an anti-ova T cell response at a much greater level than is achieved through adoptive transfer. OT-I mice survived significantly longer than wild-type mice following challenge (p<0.05), with tumors not appearing for 1–2 months (Fig. 1D). However, once tumors were detectable, they grew rapidly, and most mice had to be euthanized within 2 weeks (Fig. 1E). The delayed emergence of tumors, and their aggressive growth once they became palpable, resembled the pattern seen in mice treated with OT-I adoptive therapy, suggesting that they escaped by the same mechanism. To determine whether tumor growth was due to loss of the OT-I T cells or loss of their function, we analyzed spleens from naive OT-I mice and from OT-I mice with large tumors. Splenic CTL populations were of similar size in naïve and tumor-bearing mice (p=0.0674) (Fig. 1F), and when splenocytes were pulsed with SIINFEKL peptide, there was no difference in IFNγ production (p=0.7088) (Fig. 1G). Thus, escape did not appear to involve systemic suppression of anti-tumor T cells.
Tumors which grow in OT-I mice lose the ova gene
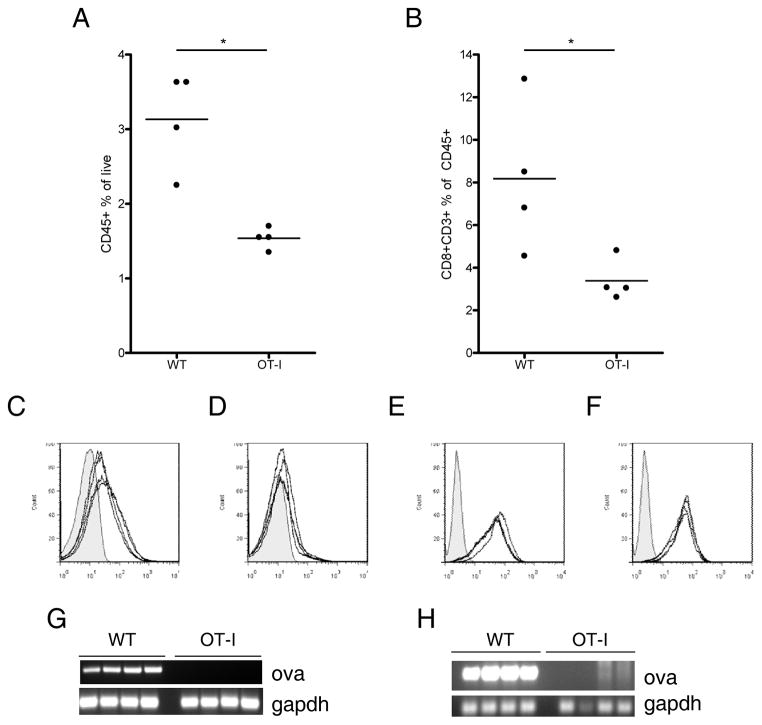
Total CD45+ infiltration within B16ova tumors was significantly lower in OT-I mice than in WT mice (p<0.05) (Fig. 2A). In addition, even within the CD45+ population, the relative number of CTL was significantly reduced in OT-I mice (p<0.05) (Fig. 2B). We also observed significant decreases in expression of both H-2Kb (Fig. 2C and D) and H-2Db (data not shown) (p<0.05) in tumors excised from OT-I mice. However, this decreased expression was not stable, as excised tumors re-established in culture expressed normal levels of MHC upon IFNγ treatment (p=0.4283) (Fig. 2E and F).
Figure 2. B16ova tumors which escape in OT-I mice display several characteristics distinguishing them from tumors in WT mice.
10 mm tumors were harvested from WT or OT-I mice, homogenized, and analyzed by flow cytometry and PCR. A, Tumor homogenates were stained for CD45 to identify infiltrating immune cells. B, CD45+ cells in A were analyzed for expression of CD8 and CD3 to identify tumor-infiltrating CTL. C and D, Expression of H-2Kb on the CD45− cells of tumors freshly excised from WT (C) or OT-I mice (D). Histograms show a shaded isotype control and H-2Kb levels of 4 individual tumors. E and F, tumors were re-established in culture, treated with 500 U/mL IFNγ to induce MHC expression, and stained for H-2Kb. H-2Kb expression on tumor lines which came from WT (E) or OT-I (F) mice. RNA and DNA were isolated from freshly excised tumors and tested for ova and gapdh by RT-PCR (G) or PCR (H). Data are representative of at least three independent experiments.
While all tumors excised from WT mice continued to express OVA mRNA, tumors excised from OT-I mice did not contain detectable OVA message (Fig. 2G). Moreover, when DNA was analyzed, most of the tumors excised from OT-I mice were negative for ova gene sequences (Fig. 2H). To confirm gene loss, we tested for the functional presence of the neor gene which had been transfected into B16 cells on the same plasmid (pcDNA-ova) to generate the B16ova cells. Of tumors recovered from OT-I mice in 10 separate experiments, less than 2% remained resistant to G418. In contrast, all tumors recovered from WT mice grew well in G418.
To determine whether the parental population of B16ova cells was composed of a mixed population of ova-positive and ova-negative cells, we performed FISH for the pcDNAova insertion on B16ova cells in culture. 100 out of 100 cells analyzed contained an ova signal. Additionally, of 20 single cell clones screened by PCR, all were ova positive (data not shown). When OT-I mice were challenged with one of the confirmed ova-positive clones, tumors grew with the same kinetics as previously seen with the pooled B16ova population. Excised tumors were ova-negative by PCR and failed to grow in G418. Furthermore, the same patterns of diminished infiltration and MHC expression were observed whether mice were challenged with the parental B16ova population, or with a single cell clone (data not shown).
The ova gene is partially lost following OT-I adoptive therapy
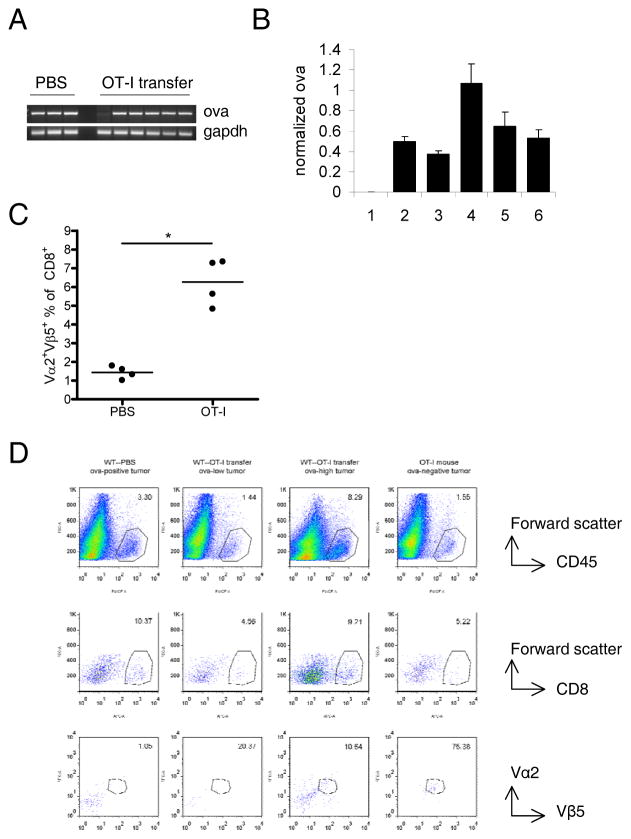
To verify that the escape mechanisms operative in OT-I mice reflected those occurring in mice receiving adoptive therapy, we also studied recurrent tumors from C57BL/6 mice receiving OT-I T cells. Ova loss was incomplete in this setting. By standard PCR, most tumors were still ova positive (Fig. 3A). However, quantitative PCR revealed significantly less ova DNA in these tumors than in tumors from untreated mice (Fig. 3B). Mice which had received OT-I transfer maintained a large splenic population of T cells bearing the Vα2Vβ5 TCR combination utilized by OT-I T cells (Fig. 3C). The size of this population was similar in mice with ova-high and ova-low tumors. There was a wide variation in ova content of escape tumors (for example, tumors 1 & 4 in Fig. 3B). However, similar to tumors from OT-I mice, ova-low tumors in WT mice often had decreased CD45+ and CD8+ infiltration. Similarly, both ova-high and ova-low tumors were enriched with T cells with the OT-I TCR (Fig. 3D).
Figure 3. Tumors which escape OT-I adoptive therapy show similar growth patterns to those seen in OT-I mice.
WT mice were treated as in figure 1. Tumors and spleens were harvested when mice were killed due to tumor burden. A, PCR of DNA from freshly-excised tumors. B, Real-time PCR of the DNA analyzed in A. Ova content of each tumor was normalized to gapdh. Bars represent normalized ova content of tumors from OT-I-treated mice relative to average normalized ova of untreated tumors. C, Percent of splenic CD8+ cells staining positive for the Vα and Vβ chains used by the OT-I TCR. 4 mice per group are shown. D, flow cytometry of representative tumors. Numbers on graphs indicate percent of total cells that are CD45+ (to row), percent of CD45+ cells that are CD8+ (middle row), and percent of CD45+CD8+ cells that are Vα2+Vβ5+ (bottom row).
T cells mediate ova gene loss and genomic instability in vitro
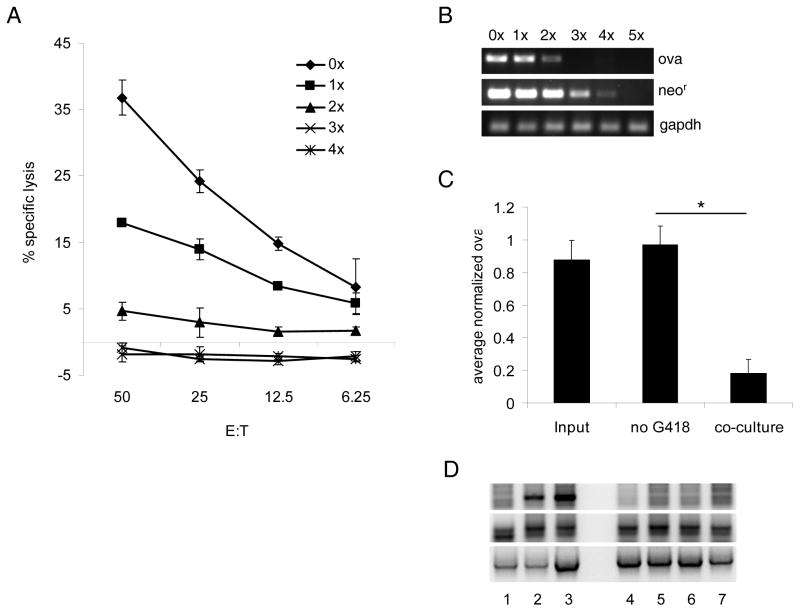
To determine whether T cell attack was sufficient to induce the emergence of ova-loss cells, we performed co-cultures of B16ova and OT-I T cells. After one round of co-culture, tumor cells were significantly less susceptible to OT-I T cell killing, and after 3 rounds there was no detectable killing (Fig. 4A). At the population level, no MHC down-regulation occurred following in vitro co-culture. Instead, H-2Kb surface levels on B16ova cells were increased in the days following co-culture. Within a few weeks following co-culture, MHC expression returned to normal levels (data not shown). DNA extracted from each population showed a progressive loss of ova and neor (Fig. 4B). The ability of OT-I to promote ova gene loss was dependent upon T cell-tumor cell contact and was not replicated in cultures separated by transwells. B16ova populations which had undergone co-culture typically contained between 20% and 80% of the ova levels present in the input population. In contrast, no ova gene loss was detected by quantitative PCR in B16ova cells grown for 4 weeks without G418, excluding the possibility that our results were due to spontaneous (non-T cell mediated) ova loss (Fig. 4C).
Figure 4. T cells directly promote the emergence of ova-loss cells in vitro.
Activated OT-I T cells were co-cultured overnight with B16ova derived from a single-cell clone. Once the B16ova population recovered, the co-culture step was repeated. Tumor cell lines which had undergone zero to five rounds of co-culture were maintained (named 0x–5x). A, Cells from each population were used as targets for OT-I T cells in a cytotoxicity assay. B, DNA was isolated from each population and tested for ova and neor gene content by PCR. C, B16ova cells were grown in vitro without G418. Some cultures were subjected to a single round of co-culture on the first day following removal from G418 as in A and B. Other cultures were left untreated. After 29 days, DNA was isolated from these cells and from untreated cells growing in G418 (named input). Ova content was quantified by real-time PCR. Ova levels in each sample were normalized to apolipoprotein B. Bars represent the average normalized ova content of 3–4 samples receiving each treatment. Data are representative of at least two independent experiments. D. Activated OT-I (lanes 1&4) or Pmel(lanes 2,5&6) T cells, or control C57BL/6 splenocytes (lanes 3&7), were co-cultured with B16ova (lanes 1–3) or B16 (lanes 4–7) tumor cells as described in A. above. To increase T cell activation, target B16 tumor cells were also co-cultured with Pmel T cells in the presence of the KVPRNQDWL (hgp10025–33) peptide (lane 6). DNA isolated from tumor cells which had undergone five rounds of co-culture was tested for the ova, gp100 and GAPDH genes by PCR.
We also investigated the effects of co-culture of B16ova cells with a different transgenic T cell population. For this, we used Pmel T cells, which express a transgenic T cell receptor specific for the KVPRNQDWL peptide from the human, melanocyte-specific gp100 antigen (hgp10025–33) in the context of the murine H-2Db MHC Class I molecule expressed by C57BL/6 mice29. Both B16ova, and B16, tumor cells express the murine homologue of gp100 which is recognized, but at lower efficiency than the human epitope, by the Pmel T cells29.
As before, co-culture of OT-I T cells with B16ova cells induced loss of the ova gene by PCR (Fig. 4D, Lane 1). However, OT-I T cells did not induce loss of the gene encoding gp100 (Fig. 4D, Lane 1). Similarly, co-culture of B16ova cells with Pmel T cells induced loss of neither ova nor gp100 genes (Fig. 4D, Lane 2). Pmel T cells also failed to induce loss of the gp100 gene from B16 tumor cells even when the target tumor cells were pre-loaded with the KVPRNQDWL (hgp10025–33) peptide (Fig. 4D, lanes 5&6), a treatment which we have shown further activates the T cells and significantly increases the ability of Pmel cells to kill B16 targets (data not shown). As expected, OT-I T cells did not kill (ova negative) B16 tumor cells in vitro by chromium release assays or secrete IFN-γ upon co-culture (data not shown), and neither did they induce loss of the gp100 gene from co-cultured B16 cells (Fig. 4D, Lane 4). The differences between the effects of OT-I T cells on gene loss of ova, and Pmel on the loss of gp100, are discussed in more detail in the Discussion below.
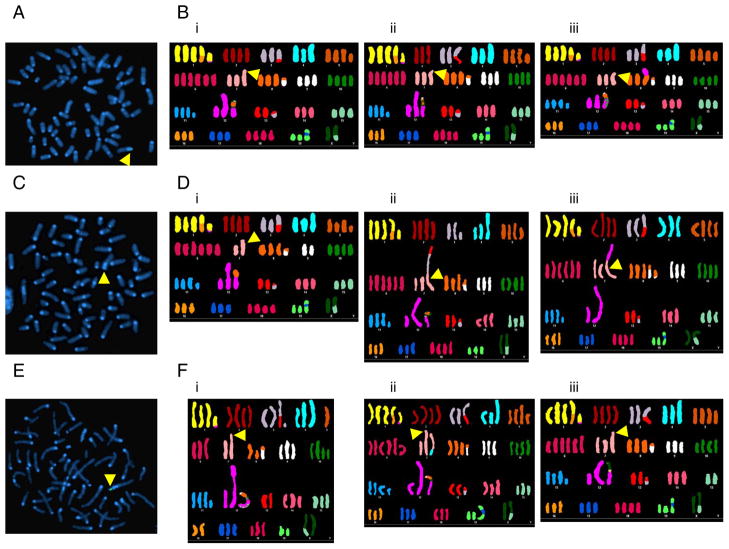
Based on the rapid loss of ova both in vitro and in vivo, we hypothesized that the direct interaction of antigen specific T cells with target tumor cells was sufficient to induce ova gene loss. To test this we generated a population of B16ova derived from a single cell clone and compared karyotypes of these cells before, and after, co-culture with OT-I T cells. Each cell in the input population had a single ova insertion (Fig. 5A). Spectral karyotyping (SKY) showed that this insertion was located on an abnormal chromosome 7 (Fig. 5B). Prior to analyzing the input population, the clone was grown in vitro (in G418) for approximately six months. During that time, the clone diverged enough so that none of the 5 karyotypes analyzed were identical, indicating ongoing genomic instability. However, the diversity in these karyotypes was limited to a single unique translocation per cell (for example, Fig. 5Biii) and loss of the fourth copy of chromosome 2 in some cells (for example, Fig. 5Bii).
Figure 5. B16ova cells have increased karyotypic variation following OT-I co-culture.
A, B16ova derived from a single-cell clone contained a single ova insert detectable by FISH. Bi–Biii, Karyotypes of 3 individual cells of untreated B16ova. Arrows indicate the location of the ova insert on an abnormal chromosome 7. C–F, The same single-cell clone shown in A and B was subjected to a single round of OT-I co-culture and analyzed by FISH and SKY. Some cells were no longer ova positive by FISH (C) but still contained the abnormal chromosome (Di–Diii). Other cells retained the ova insert (E) and had other chromosomal losses and translocations (Fi–Fiii).
Cells which survived co-culture with OT-I T cells were allowed to recover for 23 days and analyzed for their karyotypes. Following co-culture, the number, and extent, of genomic variations increased significantly. All cells still contained the abnormal chromosome 7. However, this chromosome had lost the ova-containing portion in some cells (Fig. 5C, 6D). In other cells, the ova-containing chromosome remained intact (Fig. 5E, 6F). Most cells had changes to 2 or 3 chromosome pairs (Table I). Notably, the greatest number of changes (translocations or losses involving 10 of the 20 chromosome pairs) was observed in a cell which retained ova (Fig. 5Fi). To quantify karyotypic diversity, we scored each cell by counting the number of chromosome pairs which differed from their most common presentation. Table I lists the score for each karyotype. By this analysis, significantly greater karyotypic diversity was detected following co-culture than in the input population (p<0.05), suggesting that co-culture with OT-I T cells induced a significantly increased diversity of karyotypes within the surviving population compared to that which existed in the pre-co-culture population.
Table I.
Karyotypic variations in each cell analyzed
| Number of karyotypic variationsa | |
|---|---|
| Input | 0, 1, 1, 1, 2 |
| After co-culture | 1, 1, 2, 2, 3, 3, 3, 3, 10 |
A variation is counted for each chromosome pair which differs from the most common presentation of that pair among the 14 cells analyzed. Variations include translocations, loss of chromosomes, gain of chromosomes, and loss of the ova FISH signal.
Discussion
The development of tumor immunotherapies has led to the discovery of a wide array of mechanisms which tumors can employ to escape these therapies. We show here, for the first time to our knowledge, that a potent T cell response can induce tumor escape through the promotion of enhanced genomic instability. Although OT-I T cells transiently protected mice against the growth of B16ova tumors, they also concomitantly drove evolution of the surviving tumor cells into a new phenotype which allowed their escape from T cell-mediated therapy. In OT-I mice, escape tumors completely lost the ova gene. In contrast, tumors recovered from WT mice receiving OT-I T cells usually retained some ova DNA. Coupled with gene loss, escape tumors had significantly diminished immune infiltrates and MHC class I expression, suggesting that multiple escape mechanisms were operative in vivo.
We confirmed that the ova gene was stably inserted into the B16ova cells by cytogenetic analysis and was present in all 120 separate cells which were analyzed. Significantly, all of the cells which lost the ova gene in our studies also lost resistance to G418 selection and the associated neor gene. Since the ova and neor genes are closely genetically linked in the original plasmid used to generate G418r ova+ B16ova cells, the observation of the concomitant loss of boh G418 resistance and ova DNA strongly suggests that escape tumors did not emerge through the selection of pre-existing ova-negative cells, since any such cells would not have survived in G418-containing medium in vitro. These observations clearly suggest that ova loss was the result of an evolution of the tumor cell genotype in response to OT-I T cell activity. In this respect, a single round of co-culture of B16ova cells with OT-I T cells was sufficient to reduce significantly the ability of fresh OT-I T cells to respond to stimulation with B16ova (Fig. 4A). This result could be explained by one of two models in which the OT-I T cells either simply selected spontaneous ova-loss cells from the cultures or actively induced gene loss in some target cells, which subsequently survived and expanded. In the selection model, we would predict that the surviving ova-negative cells would contain an increasing level of homogeneity between their karyotypes following co-culture compared to the input population before co-culture. In contrast, the induction model would predict the emergence of cells with increased levels of diversity of both ova gene content, and karyotypic phenotypes, induced by the mutagenic activity of the OT-I T cells. Therefore, to assess the significance of co-culture with OT-I T cells in vitro on the genomic stability of B16ova cells, we measured the diversity of karyotypes between the pre-co-culture, and post-co-culture, populations. As expected, both populations had diversity of karyotypes within them (Table 1). However, the diversity of karyotypes in the pre-co-culture population was significantly more limited (restricted to a single unique translocation per cell and loss of the fourth copy of chromosome 2 in some cells) compared to the diversity observed in the post co-culture population (which involved changes ranging from 2 or 3 chromosome pairs up to 10 of the 20 chromosome pairs) (p<0.05) (Table 1). In addition, the low diversity of karyotypes in the pre-co-culture population was derived from a single cell clone over a period of 6 months in culture. In contrast, the increased diversity of karyotypes in the post-co-culture population evolved over a considerably shorter period of 3 weeks. Therefore, although it is likely that our sample sizes failed to detect the full range of karyotypic diversity in either population, these data indicate that co-culture with OT-I T cells induced a significantly increased, and more rapidly evolving, diversity of karyotypes within the surviving tumor cell population compared to that which existed in the pre-co-culture population. Finally, similar to other tumors30,31, it is clear that untreated B16ova cells are already somewhat genomically unstable. Therefore, it may also be possible that the mutagenic activity of OT-I T cells targets only a subset of cells within the B16ova population and that it is these cells which selectively survive co-culture (a model involving both induction and selection of ova loss variants).
We do not believe that the effects described here are specific to the OT-I model. Thus, in separate experiments, we have shown that, when OT-I T cells were co-cultured with ligand negative (ova-) TC2 murine prostate tumor cells (syngeneic to C57BL/6 mice), minimal cell death was observed in vitro. These TC2 cells surviving co-culture showed very little karyotypic deviation from parental TC2 cells (not shown). In contrast, in TC2 cells co-cultured with T cells derived from mice treated with a potent anti-TC2 vaccine32, the majority of those TC2 cells surviving the co-culture showed reproducible cytogenetic changes which were not seen upon co-culture with either OT-I, or control C57BL/6, splenocytes (Kottke et al., In preparation).
The scope of our studies is limited in two principal ways. First, the targeted OVA antigen is an artificial tumor associated antigen and has no direct relevance to tumor cell survival. Therefore, B16ova cells can, presumably, lose OVA expression with no deleterious effects on tumor progression. Second, chromosomal integration of the ova plasmid may have occurred at a location relatively susceptible to breaks or rearrangements, making disruption/loss of the ova locus potentially more frequent/easy than for other genetic loci. In this respect, although we observed OT-I T cell mediated genomic loss of the ova gene from a single (haploid) chromosomal locus, we did not detect total loss of the gp100 gene by the action of Pmel T cells on B16ova or B16 tumor cells (Fig. 4D). We hypothesize that the discrepancy between these results could be for any of three principal reasons. In the first, the strength of the interaction between Pmel T cells and B16 targets is significantly weaker than that between OT-I and SIINFEKL-presenting B16ova cells. This is because the transgenic T cell receptor in Pmel cells recognizes the human gp100 peptide significantly better than the murine homologue29 and is manifested by very poor in vitro killing of B16ova, or B16, cells by Pmel T cells, compared to OT-I mediated killing of B16ova cells (ref 29 and Kaluza et al., In preparation). Thus, it may be that the signals mediating genomic instability, induced by T cell/target cell interaction, are significantly weaker in the Pmel/B16 combination, compared to those in OT-I/B16ova cultures. Second, it may be that the ova gene in B16ova cells integrated at a chromosomal site of particular fragility following plasmid transfection. Correspondingly, this site may, therefore, be acutely susceptible to rearrangement upon induction of the signals generated by T cell mediated recognition of target cells. Finally, unlike the haploid single insertion site of the ova gene, chromosomal gp100 genes are present in our tumor cells at copy numbers of at least 2 or more. Of particular significance to our studies here, gene loss of an MHC locus has been reported in escape tumors in both animal models and in human tumors25,27. Significantly, irreversible loss of an HLA class I gene, as reported by Garrido and colleagues, is often associated with coincident genetic mutation at the second allele 25, 27. Therefore, although T cell induced gene loss may be occurring in B16 cells exposed to Pmel cells, total loss of functional expression may not be detected until either more extensive T cell mediated genomic rearrangements are induced sufficient to ablate at least 2 copies of the gene, and/or alternative mutations are incorporated into the additional alleles27.
In conclusion, our results highlight a potential drawback of T cell therapies which target only a single antigen – including those using chimeric immune receptor-modified T cells1,2 – in that the interaction of tumor cells with anti-tumor T cells may actually help to generate variant tumor cells which are then capable of the evading the initial T cell therapy. This effect will be exacerbated if the selective pressure against the targeted antigen does not offer any protection against antigen-negative cells. These considerations would predict better clinical outcomes using transfer of T cells with multiple antigenic specificities. However, clinical trials involving transfer of polyclonal TIL can still result in clonal repopulation of the T cell compartments of responding patients18, suggesting that ongoing control of residual tumor cells in these patients becomes focused on a single, or a very few, antigens. In such cases, our results suggest that it would be worthwhile investigating whether tumor recurrences in those patients result, at least in part, from T cell-driven immune escape at the level of gene loss. If this proves to be the case, it will be important to combine anti-tumor T cell activity with therapies targeting other aspects of tumor growth, such as anti-angiogenic therapies33,34 or destruction of the tumor stroma35,36. We are currently testing whether such combination therapies can be used to increase the efficacy of OT-I transfer in our model by requiring tumors to develop increasingly complex, and hopefully ultimately unachievable, escape mechanisms in order to evade all components of the therapy simultaneously.
Acknowledgments
This work was supported by NIH grant CA130878. Mayo Clinic Cancer Center is supported in part by an NCI Cancer Center Support Grant 5P30 CA15083-36.
We thank the Mayo Clinic Cancer Center for the use of the Cytogenetics Shared Resource, which provided homebrew FISH probe production services, FISH analysis, and Mouse Spectral Karyotyping services. We thank Toni Higgins for expert secretarial assistance.
Abbreviations used in this paper
- FISH
fluorescence in situ hybridization
- SKY
spectral karyotyping
- TIL
tumor infiltrating lymphocytes
References
- 1.Ertl HC, Zaia J, Rosenberg SA, June CH, Dotti G, Kahn J, Cooper LJ, Corrigan-Curay J, Strome SE. Considerations for the clinical application of chimeric antigen receptor T cells: observations from a recombinant DNA Advisory Committee Symposium held June 15, 2010. Cancer Res. 2011;71:3175–81. doi: 10.1158/0008-5472.CAN-10-4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med. 2011 Aug 10; doi: 10.1056/NEJMoa1103849. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenberg SA. Cell transfer immunotherapy for metastatic solid cancer-what clinicians need to know. Nat Rev Clin Oncol. 2011 Aug 2; doi: 10.1038/nrclinonc.2011.116. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenberg SA, Restifo NP, Yang JC, Morgan RA, Dudley ME. Adoptive cell transfer: a clinical path to effective cancer immunotherapy. Nat Rev Cancer. 2008;8:299–308. doi: 10.1038/nrc2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dudley ME, Yang JC, Sherry R, Hughes MS, Royal R, Kammula U, Robbins PF, Huang J, Citrin DE, Leitman SF, Wunderlich J, Restifo NP, et al. Adoptive cell therapy for patients with metastatic melanoma: evaluation of intensive myeloablative chemoradiation preparative regimens. J Clin Oncol. 2008;26:5233–9. doi: 10.1200/JCO.2008.16.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morgan RA, Dudley ME, Rosenberg SA. Adoptive cell therapy: genetic modification to redirect effector cell specificity. Cancer J. 2010;16:336–41. doi: 10.1097/PPO.0b013e3181eb3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khammari A, Labarriere N, Vignard V, Nguyen J-M, Pandolfino M-C, Knol AC, Quereux G, Saiagh S, Brocard A, Jotereau F, Dreno B. Treatment of Metastatic Melanoma with Autologous Melan-A/Mart-1-Specific Cytotoxic T Lymphocyte Clones. J Invest Dermatol. 2009;129:2835–42. doi: 10.1038/jid.2009.144. [DOI] [PubMed] [Google Scholar]
- 8.Besser MJ, Shapira-Frommer R, Treves AJ, Zippel D, Itzhaki O, Schallmach E, Kubi A, Shalmon B, Hardan I, Catane R, Segal E, Markel G, et al. Minimally cultured or selected autologous tumor-infiltrating lymphocytes after a lympho-depleting chemotherapy regimen in metastatic melanoma patients. J Immunother. 2009;32:415–23. doi: 10.1097/CJI.0b013e31819c8bda. [DOI] [PubMed] [Google Scholar]
- 9.Kovacs-Solyom F, Blasko A, Fajka-Boja R, Katona RL, Vegh L, Novak J, Szebeni GJ, Krenacs L, Uher F, Tubak V, Kiss R, Monostori E. Mechanism of tumor cell-induced T-cell apoptosis mediated by galectin-1. Immunol Lett. 2010;127:108–18. doi: 10.1016/j.imlet.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Biswas S, Biswas K, Richmond A, Ko J, Ghosh S, Simmons M, Rayman P, Rini B, Gill I, Tannenbaum CS, Finke JH. Elevated levels of select gangliosides in T cells from renal cell carcinoma patients is associated with T cell dysfunction. J Immunol. 2009;183:5050–8. doi: 10.4049/jimmunol.0900259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramsay AG, Clear AJ, Kelly G, Fatah R, Matthews J, Macdougall F, Lister TA, Lee AM, Calaminici M, Gribben JG. Follicular lymphoma cells induce T-cell immunologic synapse dysfunction that can be repaired with lenalidomide: implications for the tumor microenvironment and immunotherapy. Blood. 2009;114:4713–20. doi: 10.1182/blood-2009-04-217687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lepique AP, Daghastanli KR, Cuccovia IM, Villa LL. HPV16 tumor associated macrophages suppress antitumor T cell responses. Clin Cancer Res. 2009;15:4391–400. doi: 10.1158/1078-0432.CCR-09-0489. [DOI] [PubMed] [Google Scholar]
- 13.Movahedi K, Guilliams M, Van den Bossche J, Van den Bergh R, Gysemans C, Beschin A, De Baetselier P, Van Ginderachter JA. Identification of discrete tumor-induced myeloid-derived suppressor cell subpopulations with distinct T cell-suppressive activity. Blood. 2008;111:4233–44. doi: 10.1182/blood-2007-07-099226. [DOI] [PubMed] [Google Scholar]
- 14.Nagaraj S, Gupta K, Pisarev V, Kinarsky L, Sherman S, Kang L, Herber DL, Schneck J, Gabrilovich DI. Altered recognition of antigen is a mechanism of CD8+ T cell tolerance in cancer. Nat Med. 2007;13:828–35. doi: 10.1038/nm1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu VC, Wong LY, Jang T, Shah AH, Park I, Yang X, Zhang Q, Lonning S, Teicher BA, Lee C. Tumor evasion of the immune system by converting CD4+CD25– T cells into CD4+CD25+ T regulatory cells: role of tumor-derived TGF-beta. J Immunol. 2007;178:2883–92. doi: 10.4049/jimmunol.178.5.2883. [DOI] [PubMed] [Google Scholar]
- 16.Liu K, Caldwell SA, Greeneltch KM, Yang D, Abrams SI. CTL adoptive immunotherapy concurrently mediates tumor regression and tumor escape. J Immunol. 2006;176:3374–82. doi: 10.4049/jimmunol.176.6.3374. [DOI] [PubMed] [Google Scholar]
- 17.Jazirehi AR, Baritaki S, Koya RC, Bonavida B, Economou JS. Molecular mechanism of MART-1+/A*0201+ human melanoma resistance to specific CTL-killing despite functional tumor-CTL interaction. Cancer Res. 2011;71:1406–17. doi: 10.1158/0008-5472.CAN-10-1296. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Dudley ME, Wunderlich JR, Yang JC, Sherry RM, Topalian SL, Restifo NP, Royal RE, Kammula U, White DE, Mavroukakis SA, Rogers LJ, Gracia GJ, et al. Adoptive cell transfer therapy following non-myeloablative but lymphodepleting chemotherapy for the treatment of patients with refractory metastatic melanoma. J Clin Oncol. 2005;23:2346–57. doi: 10.1200/JCO.2005.00.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bai XF, Liu JQ, Joshi PS, Wang L, Yin L, Labanowska J, Heerema N, Zheng P, Liu Y. Different lineages of P1A-expressing cancer cells use divergent modes of immune evasion for T-cell adoptive therapy. Cancer Res. 2006;66:8241–9. doi: 10.1158/0008-5472.CAN-06-0279. [DOI] [PubMed] [Google Scholar]
- 20.Uyttenhove C, Maryanski J, Boon T. Escape of mouse mastocytoma P815 after nearly complete rejection is due to antigen-loss variants rather than immunosuppression. J Exp Med. 1983;157:1040–52. doi: 10.1084/jem.157.3.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez-Perez L, Kottke T, Diaz RM, Ahmed A, Thompson J, Chong H, Melcher A, Holmen S, Daniels G, Vile RG. Potent selection of antigen loss variants of B16 melanoma following inflammatory killing of melanocytes in vivo. Cancer Res. 2005;65:2009–17. doi: 10.1158/0008-5472.CAN-04-3216. [DOI] [PubMed] [Google Scholar]
- 22.Goldberger O, Volovitz I, Machlenkin A, Vadai E, Tzehoval E, Eisenbach L. Exuberated numbers of tumor-specific T cells result in tumor escape. Cancer Res. 2008;68:3450–7. doi: 10.1158/0008-5472.CAN-07-5006. [DOI] [PubMed] [Google Scholar]
- 23.Yee C, Thompson JA, Byrd DR, Riddell SR, Roche P, Celis E, Greenberg PD. Adoptive T cell therapy using antigen-specific CD8+ T cell clones for the treatment of patients with metastatic melanoma: in vivo persistence, migration, and antitumor effect of transferred T cells. Proc Natl Acad Sci U S A. 2002;99:16168–73. doi: 10.1073/pnas.242600099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carretero R, Romero JM, Ruiz-Cabello F, Maleno I, Rodriguez F, Camacho FM, Real LM, Garrido F, Cabrera T. Analysis of HLA class I expression in progressing and regressing metastatic melanoma lesions after immunotherapy. Immunogenetics. 2008;60:439–47. doi: 10.1007/s00251-008-0303-5. [DOI] [PubMed] [Google Scholar]
- 25.Garrido F, Cabrera T, Aptsiauri N. “Hard” and “soft” lesions underlying the HLA class I alterations in cancer cells: Implications for immunotherapy. Intl J Can. 2010;127:249–56. doi: 10.1002/ijc.25270. [DOI] [PubMed] [Google Scholar]
- 26.Torres MJ, Ruiz-Cabello F, Skoudy A, Berrozpe G, Jimenez P, Serrano A, Real FX, Garrido F. Loss of an HLA haplotype in pancreas cancer tissue and its corresponding tumor derived cell line. Tiss Antigens. 1996;47:372–81. doi: 10.1111/j.1399-0039.1996.tb02572.x. [DOI] [PubMed] [Google Scholar]
- 27.Maleno I, Aptsiauri N, Cabrera T, Gallego A, Paschen A, Lopez-Nevot MA, Garrido F. Frequent loss of heterozygosity in the b2-microglobulin region of chromosome 15 in primary human tumors. Immunogenetics. 2010;63:65–71. doi: 10.1007/s00251-010-0494-4. [DOI] [PubMed] [Google Scholar]
- 28.Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 29.Overwijk WW, Theoret MR, Finkelstein SE, Surman DR, De Jong LA, Vyth-Dreese FA, Dellemijn TA, Antony PA, Spiess PJ, Palmer DC, Heimann DM, Klebanoff CA, et al. Tumor regression and autoimmunity after reversal of a functionally tolerant state of self-reactive CD8+ T cells. J Exp Med. 2003;198:569–80. doi: 10.1084/jem.20030590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kloor M, Michel S, von Knebel Doeberitz M. Immune evasion of microsatellite unstable colorectal cancers. Int J Cancer. 2010;127:1001–10. doi: 10.1002/ijc.25283. [DOI] [PubMed] [Google Scholar]
- 31.Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, Morsberger LA, Latimer C, McLaren S, Lin ML, McBride DJ, Varela I, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–13. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kottke T, Errington F, Pulido J, Galivo F, Thompson J, Wongthida P, Diaz RM, Chong H, Ilett E, Chester J, Pandha H, Harrington K, et al. Broad antigenic coverage induced by vaccination with virus-based cDNA libraries cures established tumors. Nat Med. 2011;17:854–59. doi: 10.1038/nm.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dings RP, Vang KB, Castermans K, Popescu F, Zhang Y, A oude Egbrink MG, Mescher M, Farrar M, Griffioen AW, Mayo KH. Enhancement of T-cell mediated anti-tumor response: angiostatic adjuvant to immunotherapy against cancer. Clin Cancer Res. 2011 Jan 20; doi: 10.1158/1078-0432.CCR-10-2443. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gyorffy S, Palmer K, Podor TJ, Hitt M, Gauldie J. Combined treatment of a murine breast cancer model with type 5 adenovirus vectors expressing murine angiostatin and IL-12: A role for combined anti-angiogenesis and immunotherapy. J Immunol. 2001;166:6212–17. doi: 10.4049/jimmunol.166.10.6212. [DOI] [PubMed] [Google Scholar]
- 35.Spiotto MT, Rowley DA, Schreiber H. Bystander elimination of antigen loss variants in established tumors. Nat Med. 2004;10:294–8. doi: 10.1038/nm999. [DOI] [PubMed] [Google Scholar]
- 36.Spiotto MT, Schreiber H. Rapid destruction of the tumor microenvironment by CTLs recognizing cancer-specific antigens cross-presented by stromal cells. Cancer Immun. 2005;5:8. [PubMed] [Google Scholar]