Abstract
From the muscles that control the blink of your eye to those that allow you to walk, the basic architecture of muscle is the same: muscles consist of bundles of the unit muscle cell, the muscle fiber. The unique morphology of the individual muscle fiber is dictated by the functional demands necessary to generate and withstand the forces of contraction, which in turn leads to movement. Contractile muscle fibers are elongated, syncytial cells, which interact with both the nervous and skeletal systems to govern body motion. In this review, we focus on three key cell–cell and cell–matrix contact processes, that are necessary to create this exquisitely specialized cell: cell fusion, cell elongation, and establishment of a myotendinous junction. We address these processes by highlighting recent findings from the Drosophila model system.
Introduction
The model system, Drosophila melanogaster, has been used to great effect to study fundamental issues of muscle development [1–7]. This model organism offers the cell biologist an in vivo system, coupled to a long-established genetic tradition to study muscle morphogenesis. In addition, application of genomics and varied imaging approaches makes this model a highly tractable system for the study of the cell biology of muscle.
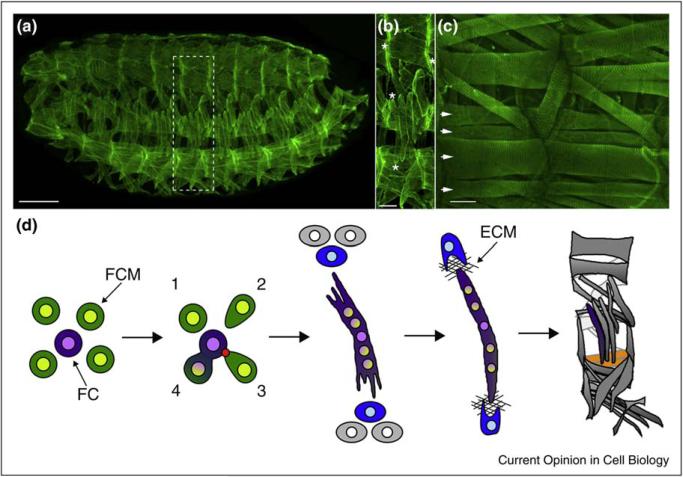
Body wall muscles in Drosophila are generated twice during the life of the fly: first, during embryogenesis to form the larval muscles [1], the process that is the main focus of this review; and subsequently during metamorphosis, in which cells set aside during embryonic myogenesis are used to generate the adult muscles [8]. In the embryo, a single fiber is considered a single muscle, whereas in the adult, multiple fibers constitute a single muscle. A similar mechanism, however, governs fiber formation in both situations: each fiber is seeded by a specialized myoblast, called a founder cell (FC), which fuses repeatedly with neighboring fusion competent myoblasts (FCMs) to generate a multi-nucleated myotube (Figure 1) [1]. Upon fusion, the newly incorporated FCM-derived nuclei adopt the transcriptional profile of the FC/myotube. By virtue of a complex developmental specification process, individual FCs/myotubes express different combinations of cell identity regulators, which endow them with unique morphological characteristics, including size (i.e. the number of fusions with FCMs), shape, and spatial orientation [9,10••].
Figure 1.
The morphogenesis of Drosophila larval body wall muscles. (a) A stage 16 Drosophila embryo labeled with antibodies against Tropomyosin (green), which reveal the segmentally repeated pattern of body wall muscles. Scale bar 50 μm. (b) A close-up of a single hemisegment from A shows that each muscle is a single fiber with unique size, shape, and attachment sites. Examples of attachment sites (*) shown for two different muscles. Scale bar 15 μm. (c) Subset of Drosophila larval muscles labeled with antibodies against Zasp (green), which labels the Z-band of the sarcomere. Scale bar 20 μm. Arrows point to the four ventral lateral (VL) muscles. (d) Schematic of steps leading to the development of a muscle fiber. FC, founder cell (purple); FCM, fusion-competent myoblast (green). First panel: the two types of myoblasts required to make a muscle. Second panel: Fusion. Numbers note FCMs that are in different states in the fusion cycle: 1—Rounded and not initiating fusion, 2—Cell shape change associated with orientation and migration to a FC; 3—Adhesion to a FC and formation of an actin focus (red) at the fusion site; 4—Pore formation and membrane vesiculation leading to cytoplasmic continuity. Panel 3: Both ends of an elongating myotube navigate towards a tendon cell (blue), positioned within the epidermis (gray cells). Panel 4: An attachment between muscle and tendon. ECM, extracellular matrix. Panel 5: The pattern of 30 muscles per abdominal hemisegment. Two VL muscles are highlighted in yellow.
Fusion is accompanied by elongation of the growing myofiber, which navigates towards tendon cells that arise in the overlying epidermis. Through interactions with these tendon cells, a stable attachment forms between muscle, epidermis, and cuticle (the exoskeleton) (Figure 1). These initial, crucial myogenic processes – fusion, elongation, and attachment – are at the heart of this review. Innervation of each muscle fiber occurs after fusion and tendon attachment, while the stereotypical arrangement of the fiber contractile apparatus (sarcomeres) appears late during muscle morphogenesis, just before hatching [1,10]. The contractile properties and inter-cellular associations provide body wall muscles with the capacity to execute and govern larval motility.
Muscle fiber as syncytial cell: focus on the actin focus
A series of recent reviews provide a comprehensive, updated description of myoblast fusion in Drosophila and in vertebrates [11•,12,13,14•]. Here we discuss recent insights to fusion, emphasizing the remaining gaps in our knowledge, with particular emphasis on the contribution of the actin-based cytoskeleton.
Myoblast fusion, as in all cases of cell–cell fusion, requires several distinct cellular behaviors [15]. Initially, FCs/myotubes and FCMs must recognize and adhere to each other. The result of recognition and adhesion is a series of cellular events that are necessary to bring the cell membranes of the two cells in close proximity to one another. Subsequently, the tethering of the plasma membranes, the formation of pores between the membranes and the expansion of these pores leads to the merging of the two myoblasts (Figure 1d).
In the Drosophila embryo, myoblast recognition and adhesion is mediated by a set of immunoglobulin (Ig) domain transmembrane proteins: Dumbfounded (Duf, a.k.a Kirre) and Roughest (Rst a.k.a IrreC), which are found primarily in FCs, and the FCM elements Sticks and stones (Sns) and Hibris (Hbs) [12–14,16•]. Fusion-related roles for vertebrate homologs of these proteins have been recently revealed [17,18], implying conservation of this system of cell recognition and adhesion. However, several significant issues pertaining to the underlying molecular mechanisms remain unclear, and require further investigation. Prominent among these are the initiation of contact between distant myoblasts (is a diffusible factor involved?), selection of fusion partners (‘first come, first serve’ vs. ‘specificity of partnering’), and generation of tightly apposed myoblast membranes, primed for fusion. Recent evidence suggests that the Ig domain recognition receptors form a ring-like structure, termed the FuRMAS [13,19•] at the fusion site. These data have been used to suggest that the recognition receptors, upon engagement, are cleared from the site of actual membrane fusion. The FuRMAS, by the nature of a ring-like structure, could also limit the site of membrane fusion to within this ring. Time-lapse imaging of tagged receptors would provide a significant step forward in verifying this model.
Downstream of the receptors, genetic, cell, and biochemical approaches have implicated a set of actin regulatory proteins as crucial for myoblast fusion. Specifically, mutations in genes including Rac, Myoblast city (Dock 180), Kette (Nap1), Scar (WAVE), Vrp1 (Solitary; D-WIP), WASp, and Arp2/3, lead to defects in myoblast fusion [11•,12,13,14•]. While clarification is needed as to how particular actin regulatory proteins are recruited specifically to the recognition receptors and the fusion site [20,21•,22,23,24••,25••,26••,27,28,29••,30], it is clear that one crucial result of their function is a branched actin structure, termed the actin focus [19•,25••,29••]. Time-lapse imaging of Drosophila myoblasts proved conclusively that this structure marks the fusion site and revealed the dynamics of this structure. The actin focus is on average short-lived (11.9 min, range 5.7–29 min), measuring 2 μm2 in size (range 0.7–4.5 μm2); this structure disappears as cytoplasmic continuity between the fusing cells is achieved (Figure 2). Genetic analysis reveals that actin focus formation requires recognition receptor function for its formation [29••]. Consistent with these genetic experiments is the observation that the FuRMAS surrounds the actin focus [13,19•]. While double labeling experiments using actin and membrane reporter constructs suggest that the actin focus can be found on both sides of the fusion event (myotube and FCM), the distribution of actin within the focus may be biased to one cell or the other [25••,29••] (B. Richardson, I. Bothe and MKB unpub.). Whether this indicates the existence of different actin structures or reveals novel aspects of actin focus maturation remains to be investigated.
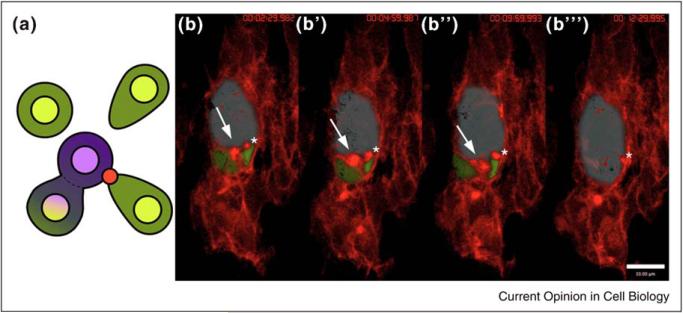
Figure 2.
The site of myoblast fusion is marked by an actin focus. (a) Schematic of myoblast fusion showing fusion between a myotube (purple) and fusion competent myoblasts (green). Note shape changes in FCMs and actin focus (red dot). (b–b′′′) Stills from time-lapse imaging reveal the progress of fusion events. Time stamps: (b) 00:02:29.982; (b′) 00:04:59.987; (b′′) 00:09:59.993; (b′′′) 00:12:29.995. An actin focus is present at each fusion site. Myotube false colored blue; FCMs false colored green. Actin (red) revealed by moesin-mcherry expressed specifically in the muscle mesoderm (using twist-GAL4). Stage 14 embryo, extended focus view of 10, 0.5 μm z-slices, frame rate is 2.30 min. Elapsed time shown upper right. Arrow points to one actin focus, asterisk to a second. Two others are also present below the false-colored myotube.
With genetics implicating SCAR/WAVE and WASp regulation of Arp2/3 as crucial to myoblast fusion, a simple model would be that these actin regulatory pathways are required for actin focus formation. However, contrary to expectation, analysis of single and double mutants between different pathway members reveals persistent and even enlarged actin foci at fusion sites [24] (B. Richardson and MKB unpub). The genetic data indicate, therefore, that these pathways are not required for formation of the foci, but may ut may influence actin rearrangements leading to dissolution of these structures. While some debate continues, these data also illustrate that the proteins required for formation of the actin focus remain to be identified. Additional Arp2/3 regulators have been uncovered in other systems [31], providing new genes to investigate for this key role in actin regulation during myoblast fusion.
An essential question arising from these studies is the function of the actin focus at the fusion site. Time-lapse imaging correlates formation and, importantly, removal of the actin focus, with cytoplasmic continuity and ultimately cell–cell fusion. With the exception of the recognition receptors and a protein implicated in receptor recycling, mutants of which do not form actin foci, all other known fusion gene mutants display actin foci that perdure at the fusion site. Of these, some mutants have enlarged foci and others have wild-type sized foci [14,29••,32]. Testing is needed to determine whether the fusion block in any of these mutants is due to the inability of the cell to remove the focus specifically, or whether the long-lived focus is just a consequence of other events gone awry. Nevertheless, several models for the cellular function of the actin focus have been put forward. These include targeting vesicles to the site of fusion [25••], supplying scaffolding to maintain cellular integrity while fusion pores are created [29••], and/or providing a force for fusion pore enlargement [26]. To provide the necessary data to evaluate these models, a combination of approaches are needed: identification of new genes and mechanisms via genetic and biochemical experiments; improved imaging, employing time-lapse videography and transmission electron microscopy; determination of composition and dynamics of cell membranes and membrane-bound vesicles associated with the fusion site; and finally, detailed description and analysis of fusion pore formation and expansion. While the Drosophila system has provided new views of a prominent and highly regulated form of cell–cell fusion, much remains to be investigated, particularly with regard to the cell-biological mechanism underlying the fusion process.
Directing myotubes to their epidermal targets
In parallel to their growth via fusion with FCMs, myotubes in the Drosophila embryo elongate towards the epidermis and attach to tendon cells at both ends (Figure 3). The highly stereotypic pattern of muscle–tendon attachments implies that regulatory mechanisms are at play, ensuring specificity and proper execution of the myotube targeting process. It is well-established that tendon cells produce spatial cues influencing the direction towards which myotubes extend. The muscles, in turn, seek out their attachment sites by sending out filopodia-like extensions that survey their environment for these instructive cues [1,33].
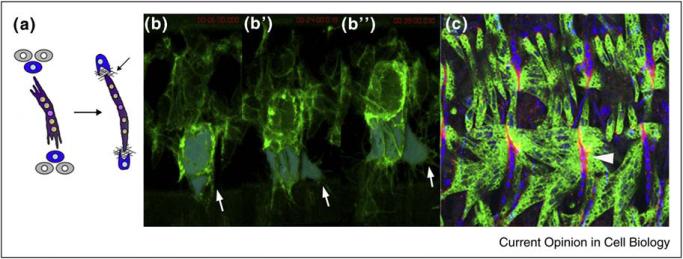
Figure 3.
Elongation and attachment of the myotube to tendon cells. (a) Schematic of muscle elongation and attachment to tendon cells. (b–b′′) Stills from a time-lapse movie of a late stage 15 embryo, expressing a membrane marker in the muscle mesoderm (Twist-Gal4 > UAS-plcγPH::GFP), and using extended focus of 28, 0.5 μm z-slices, frame rate, 3 min. Time Stamps: (b) 00:06:00; (b′) 00:24:00; (b′′) 00:39:00. Ventral myotubes are false-colored blue to highlight elongation. Arrow in each frame points out filopodial extensions. Elapsed time shown in upper right of each frame (red). (c) Stage 16 embryo showing mature attachments of muscles to tendon cells (arrowhead). Tendon cell nuclei (labeled using anti-Stripe) are in blue, while muscles (labeled with anti-Myosin Heavy Chain) are in green. ECM of the myotendinous junction is visualized using anti-Thrombospondin (red). Image provided by Arul Subramanian and Talila Volk.
Perhaps the sole bona fide identification of a tendon-derived guidance signal has been described in the ventral-longitudinal (VL) muscles, a repeating set of four parallel fibers that stretch between epidermal attachments sites at the adjacent embryonic segment border. Segment-border tendon cells secrete the ECM glycoprotein Slit, which directs myotube targeting by binding to the Roundabout (Robo) receptor, expressed at the edges of developing VL muscle fibers [34•]. Therefore, although the Slit–Robo ligand–receptor interaction is best known as the basis for repulsion of both axons and muscle fibers away from the ventral midline of the Drosophila embryo [35], it also functions as an attractive cue, guiding myotubes to specific epidermal target sites. Mutations in which properly fused myotubes extend randomly and fail to connect to their epidermal target sites have recently uncovered a second myotube targeting mechanism, apparently acting in parallel to the Slit/Robo pathway [36••,37•,38••,39••]. Importantly, mutant myotubes of this class are capable of forming stable attachments, but do so with incorrect tendon target cells, or even with adjacent muscles, implying specific impairment of target recognition capacity, while other aspects of myotube differentiation remain unaffected. A key player that has emerged from this group is D-Grip, the sole fly homolog of the Grip (Glutamate receptor interacting protein) adaptor protein family. Grip proteins harbor multiple PDZ domains, and have been functionally implicated in various aspects of synaptic protein trafficking and localization [40–42]. In Drosophila embryos, D-Grip is specifically expressed in developing muscles, and the D-Grip protein accumulates within endosomes at the ends of extending myofibers [39].
While D-Grip probably acts as a mediator of intracellular trafficking, it is becoming apparent that this multi-PDZ-domain adaptor serves a key myogenic role as a common platform for multiple muscle factors contributing to myotube targeting. Prominent among these is Kon-tiki (Kon; a.k.a Perdido), a transmembrane protein and potential receptor mediating recognition of tendon-cell guidance signals by growing embryonic myotubes [36••,38••]. Kon associates with D-Grip through a C-terminal PDZ binding domain, localizes to the ends of extending VL myotubes, and is required for D-Grip localization to these sites. Kon contributes to additional aspects of muscle–tendon attachment. One of these is to regulate the extent of motile exploratory activity by myotube ends [38••]. Interestingly, a similar function has been recently proposed for the tendon cell transmembrane protein LRT [43•], which can physically associate with Robo receptors. LRT may therefore restrict myotube migratory activity by signaling through Robo, or indirectly, by competing with Slit for Robo receptor binding, thereby stifling its stimulatory effect on myotube dynamics [43,44]. Such utilization of both muscle and tendon-based programs underscores the importance of setting limits on guidance-seeking mechanisms, once initial target recognition is achieved.
An additional member of the D-Grip-based guidance machinery is the Ig superfamily transmembrane protein Echinoid (Ed) [45•]. Structurally related to the vertebrate Nectins, Ed functions as a homotypic cell adhesion molecule, mediating aspects of Drosophila neurogenesis and epithelial morphogenesis [46–48]. Similar to Kon, Ed associates with D-Grip PDZ domains via its extreme C-terminal region. Furthermore, Ed colocalizes with D-Grip at the ends of VL muscles, and the two elements display strong genetic interactions with respect to VL myotube extension defects [45]. Targeting and guidance errors rather than defective adhesion are the main characteristic of embryos in which the Ed/D-Grip system is impaired, implying that Ed and D-Grip influence myotube behavior by affecting cortical and cytoplasmic features of these cells. Significantly, Ed affects the outcome of cell–cell interactions through communication with cortical micro-filaments [48–50], and therefore serves as a good candidate for a link between the D-Grip platform and the actin-based cytoskeleton.
This series of studies has therefore revealed a molecular complex, positioned at the tips of growing muscle fibers, which integrates a number of inputs aimed at achieving proper myotube targeting. Remarkably, a highly similar Grip-based platform mediates glia–neuron interactions during mammalian CNS development [51]. In this instance, the PDZ domains of Grip bring together several glial cell-surface proteins, including NG2, a mammalian homolog of Kon [52,53], which is thought to interact with a neuronal receptor. Cortical complexes of this type therefore represent a conserved machinery for instructive communication between distinct cell types, leading to functional maturation of a differentiating tissue.
While involvement of the Grip-based complex in myotube targeting implies functional reliance on the actin-based cytoskeleton, mutations resulting in defective muscle targeting have now revealed that growing myotubes also require properly polarized microtubule (MT) arrays, in order to reach their correct attachment sites [37•]. Key elements involved in this system are Tumbleweed (Tum) and Pavarotti (Pav), established components of the cytokinesis-mediating complex centralspindlin [54–56]. In Drosophila embryonic muscles, Tum and Pav are jointly responsible for localization of non-centrosomal, γ-tubulin-based microtubule organizing centers to the vicinity of myotube nuclei. These nuclei commonly cluster in the center of growing multi-nucleated fibers [37•]. The resulting microtubule array that forms extends from the myotube interior towards the periphery, and is polarized, with the microtubule plus-ends pointing towards the extending tips of the growing fibers. A similarly oriented microtubule array, nucleated from non-centrosomal organizing centers, forms in cultured mammalian myotubes [57], suggesting that reorganization of the microtubule cytoskeleton in this fashion reflects a conserved requirement during muscle differentiation. From a functional standpoint, the axial microtubule array may serve as a structural framework enabling elongation of myotube ends [37•,57], or alternatively, may provide polarized tracks for trafficking and localization of crucial guidance and targeting signals that ensure proper selection of myotube attachment sites.
Making a strong myotendinous junction
Contact between myotubes and tendon cells is closely followed by establishment of a myotendinous junction, so that muscle and epidermis maintain a strong physical connection that will be able to withstand the considerable forces imposed on it once muscle contractions initiate [58]. Integrins play a major, conserved role in this process, and indeed, the Drosophila embryonic myotendinous junction now serves as a prominent example and setting for study of integrin-based adhesion between distinct cell types [59]. Both muscle and tendon cells express αβ integrin heterodimers, which fortify cell attachment to the junction by serving as transmembrane links between the ECM and the internal actin cytoskeleton. These heterodimers are composed of a common β subunit (βPS) and distinct α subunits—αPS1 for the tendon cell integrin and αPS2 in muscles. Genetic disruption of the muscle αPS2βPS integrin does not interfere with construction, elongation, and attachment of muscle fibers. However, once the muscles of such mutant embryos begin to contract, they disconnect from their attachment sites and retract into ball-shaped structures [60–63]. This classic ‘myospheroid’ phenotype now serves as a diagnostic tool for identification of additional elements contributing to formation and consolidation of myotendinous junctions.
Several ECM components act as ligands for the Drosophila αPS2βPS integrin in the context of the myotendinous junction, including the Laminin α-chain protein Wing Blister [64], and Tiggrin [65,66]. Recent studies have now identified Drosophila Thrombospondin (Tsp) as a major αPS2βPS integrin ligand at muscle attachment sites [67••,68••]. Vertebrate Thrombospondins comprise a family of multimeric ECM glycoproteins, which have been implicated in diverse functional settings, such as cell aggregation and attachment, angiogenesis and synaptogenesis [69–71]. Drosophila Tsp is produced and secreted by tendon cells at stages corresponding to establishment of muscle–tendon contacts. The muscle detachment phenotypes of Tsp mutant embryos, genetic interactions of Tsp with integrin encoding genes, and biochemical binding assays, combine to identify Tsp as a ligand for αPS2βPS. The integrin-binding capacity and localization patterns of Tsp suggest a dynamic sequence [67••,68••], in which Tsp first contributes to the initial association of myotube ends with the tendon cell and its ECM. Subsequently, muscle–tendon interactions set in motion a general program of tendon cell differentiation [72], leading to increased expression and secretion of Tsp, which in turn fortifies the integrin-mediated myotube attachment to the myotendinous junction.
Further insight to the regulation of Tsp activity comes from the study of Slowmotion (Slow), a new player in myotendinous junction construction [73•]. Slow, the Drosophila homolog of vertebrate EGFL7 [74,75], is secreted from tendon cells in parallel to Tsp, and attenuates the Tsp-αPS2βPS integrin interaction by forming a complex with Tsp. Streamlining of muscle integrin activity in this manner turns out to be crucial for proper morphogenesis of the myotendinous junction into a structure capable of withstanding the wear-and-tear of intense larval muscle activity [73].
Progress in the understanding of integrin–ECM interactions in the context of myotendinous junction formation has been matched by refined characterization of the manner by which integrins are linked to cytoplasmic and cytoskeletal components. Thus the Drosophila homolog of ZASP (Z-band alternatively spliced PDZ-motif protein), a member of the PDZ-LIM protein family [76,77], acts as an adapter that strengthens the integrin–actin connection following its initial establishment via Talin, a ubiquitous mediator of integrin-based processes [78••]. Zasp performs this role repeatedly during myogenesis, initially to bolster the myotendinous junction, and later to mediate integrin-dependent assembly of muscle fiber sarcomeres [78].
A second adaptor of this type is Wech [79••], a well-conserved member of the growing, multi-domain TRIM protein family, the functional attributes of which are only beginning to emerge [80,81,82]. At Drosophila myotendinous junctions, Wech acts within tendon cells and myotubes to bridge a key interaction between Talin, which associates with the βPS integrin subunit, and the Integrin-linked Kinase (ILK) complex, a crucial mediator of the integrin-microfilament link [83]. This specific adaptor capacity of Wech appears to be conserved in mammals [79], suggesting that a fundamental aspect of integrin-mediated adhesion has now been uncovered.
Conclusions and perspectives
In this review we have focused on a restricted set of myogenic processes, to demonstrate the power of the Drosophila embryo as a model system for studying muscle cell properties and behaviors. A wide variety of issues, such as cell size, shape, polarity, migration, and adhesion can all be addressed using this versatile system. The relative simplicity and segmentally repeated nature of the embryonic musculature, amenable to study via classical and modern genetic approaches coupled to imaging and biochemical analysis, provide a unique, in vivo experimental setting. The recent introduction of comprehensive RNAi-based genetic methods to the Drosophila field [84,85••], along with the application of large-scale genomic techniques [36••,86–88] now promises to significantly enhance these studies. For example, the capacity to control RNAi expression in a temporal and tissue-specific manner helps overcome a variety of obstacles confronting more conventional genetic approaches. Importantly, this tool now enables comprehensive exploration of adult fly myogenesis, and we expect that considerable insight to issues of muscle cell biology will emerge from this ‘new frontier’ in the years to come.
Acknowledgements
We would like to thank members of the Baylies lab, particularly K. Dobi for critical reading, and S. Yu, I. Bothe, and T. Metzger for figures. We also thank T. Volk for kindly providing Figure 3c. We acknowledge our funding agencies: NIH (GM078318 and GM056989) and MDA to MB, ISF, and MDA to B. Shilo and EDS, and the Mary L. Ralph Fund to MB, B Shilo and EDS.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Bate M. The embryonic development of larval muscles in Drosophila. Development. 1990;110:791–804. doi: 10.1242/dev.110.3.791. [DOI] [PubMed] [Google Scholar]
- 2.Ciglar L, Furlong EE. Conservation and divergence in developmental networks: a view from Drosophila myogenesis. Curr Opin Cell Biol. 2009;21:754–760. doi: 10.1016/j.ceb.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Daczewska M, Picchio L, Jagla T, Figeac N, Jagla K. Muscle development and regeneration in normal and pathological conditions: learning from Drosophila. Curr Pharm Des. 2010;16:929–941. doi: 10.2174/138161210790883462. [DOI] [PubMed] [Google Scholar]
- 4.Figeac N, Daczewska M, Marcelle C, Jagla K. Muscle stem cells and model systems for their investigation. Dev Dyn. 2007;236:3332–3342. doi: 10.1002/dvdy.21345. [DOI] [PubMed] [Google Scholar]
- 5.Frasch M. Controls in patterning and diversification of somatic muscles during Drosophila embryogenesis. Curr Opin Genet Dev. 1999;9:522–529. doi: 10.1016/s0959-437x(99)00014-3. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen HT, Frasch M. MicroRNAs in muscle differentiation: lessons from Drosophila and beyond. Curr Opin Genet Dev. 2006;16:533–539. doi: 10.1016/j.gde.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 7.Schnorrer F, Dickson BJ. Muscle building; mechanisms of myotube guidance and attachment site selection. Dev Cell. 2004;7:9–20. doi: 10.1016/j.devcel.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 8.Bate M, Rushton E, Currie DA. Cells with persistent twist expression are the embryonic precursors of adult muscles in Drosophila. Development. 1991;113:79–89. doi: 10.1242/dev.113.1.79. [DOI] [PubMed] [Google Scholar]
- 9.Baylies MK, Michelson AM. Invertebrate myogenesis: looking back to the future of muscle development. Curr Opin Genet Dev. 2001;11:431–439. doi: 10.1016/s0959-437x(00)00214-8. [DOI] [PubMed] [Google Scholar]
- 10••.Beckett K, Baylies MK. The development of the Drosophila larval body wall muscles. Int Rev Neurobiol. 2006;75:55–70. doi: 10.1016/S0074-7742(06)75003-6. [This paper describes the spatial arrangement of the Founder cells and Fusion competent myoblasts during the 5.5 h period of myoblast fusion. An analysis of the timing of fusion is also investigated in wild-type and fusion mutants, leading to a reexamination of fusion models.] [DOI] [PubMed] [Google Scholar]
- 11•.Gildor B, Massarwa R, Shilo BZ, Schejter ED. Making muscles: Arp, two, three. Fly (Austin) 2010;4:145–148. doi: 10.4161/fly.4.2.10954. [This review focuses the discussion on the requirements of the SCAR/ WAVE and WASp actin nucleating factors in the cell behaviors required for myoblast fusion.] [DOI] [PubMed] [Google Scholar]
- 12.Haralalka S, Abmayr SM. Myoblast fusion in Drosophila. Exp Cell Res. 2010 doi: 10.1016/j.yexcr.2010.05.018. doi: 10.1016/j.yexcr.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onel SF, Renkawitz-Pohl R. FuRMAS: triggering myoblast fusion in Drosophila. Dev Dyn. 2009;238:1513–1525. doi: 10.1002/dvdy.21961. [DOI] [PubMed] [Google Scholar]
- 14•.Rochlin K, Yu S, Roy S, Baylies MK. Myoblast fusion: when it takes more to make one. Dev Biol. 2009;341:66–83. doi: 10.1016/j.ydbio.2009.10.024. [This in-depth review discusses the genes and mechanisms underlying myoblast fusion in fly, zebrafish, and mouse in light of cell-biological processes required for this process: cell recognition, migration, adhesion, vesicle trafficking, and membrane breakdown.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beckett K, Baylies MK. 3D analysis of founder cell and fusion competent myoblast arrangements outlines a new model of myoblast fusion. Dev Biol. 2007;309:113–125. doi: 10.1016/j.ydbio.2007.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Shelton C, Kocherlakota KS, Zhuang S, Abmayr SM. The immunoglobulin superfamily member Hbs functions redundantly with Sns in interactions between founder and fusion-competent myoblasts. Development. 2009;136:1159–1168. doi: 10.1242/dev.026302. [Excellent data are presented in this paper, indicating that Hbs can function redundantly with Sns to mediate cell adhesion during myoblast fusion.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sohn RL, Huang P, Kawahara G, Mitchell M, Guyon J, Kalluri R, Kunkel LM, Gussoni E. A role for nephrin, a renal protein, in vertebrate skeletal muscle cell fusion. Proc Natl Acad Sci USA. 2009;106:9274–9279. doi: 10.1073/pnas.0904398106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srinivas BP, Woo J, Leong WY, Roy S. A conserved molecular pathway mediates myoblast fusion in insects and vertebrates. Nat Genet. 2007;39:781–786. doi: 10.1038/ng2055. [DOI] [PubMed] [Google Scholar]
- 19•.Kesper DA, Stute C, Buttgereit D, Kreiskother N, Vishnu S, Fischbach KF, Renkawitz-Pohl R. Myoblast fusion in Drosophila melanogaster is mediated through a fusion-restricted myogenic-adhesive structure (FuRMAS). Dev Dyn. 2007;236:404–415. doi: 10.1002/dvdy.21035. [This paper is the first to describe the organization of the recognition and adhesion receptors in a ring-like structure, designated FuRMAS, during myoblast fusion.] [DOI] [PubMed] [Google Scholar]
- 20.Balagopalan L, Chen MH, Geisbrecht ER, Abmayr SM. The CDM superfamily protein MBC directs myoblast fusion through a mechanism that requires phosphatidylinositol 3,4,5-triphosphate binding but is independent of direct interaction with DCrk. Mol Cell Biol. 2006;26:9442–9455. doi: 10.1128/MCB.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21•.Berger S, Schafer G, Kesper DA, Holz A, Eriksson T, Palmer RH, Beck L, Klambt C, Renkawitz-Pohl R, Onel SF. WASP and SCAR have distinct roles in activating the Arp2/3 complex during myoblast fusion. J Cell Sci. 2008;121:1303–1313. doi: 10.1242/jcs.022269. [This paper identifies a new allele of Arp3. Through double mutant analysis the authors suggest a sequential requirement for actin nucleating factors and Arp3 at the site of fusion.] [DOI] [PubMed] [Google Scholar]
- 22.Chen EH, Olson EN. Antisocial, an intracellular adaptor protein, is required for myoblast fusion in Drosophila. Dev Cell. 2001;1:705–715. doi: 10.1016/s1534-5807(01)00084-3. [DOI] [PubMed] [Google Scholar]
- 23.Chen EH, Pryce BA, Tzeng JA, Gonzalez GA, Olson EN. Control of myoblast fusion by a guanine nucleotide exchange factor, loner, and its effector ARF6. Cell. 2003;114:751–762. doi: 10.1016/s0092-8674(03)00720-7. [DOI] [PubMed] [Google Scholar]
- 24••.Gildor B, Massarwa R, Shilo BZ, Schejter ED. The SCAR and WASp nucleation-promoting factors act sequentially to mediate Drosophila myoblast fusion. EMBO Rep. 2009;10:1043–1050. doi: 10.1038/embor.2009.129. [This report indicates a sequential requirement of these actin nucleating factors during fusion, with SCAR required before fusion pore formation and WASp required during pore expansion. In addition, this study indicates that Scar contributes to myoblast migration by facilitating myoblast shape changes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25••.Kim S, Shilagardi K, Zhang S, Hong SN, Sens KL, Bo J, Gonzalez GA, Chen EH. A critical function for the actin cytoskeleton in targeted exocytosis of prefusion vesicles during myoblast fusion. Dev Cell. 2007;12:571–586. doi: 10.1016/j.devcel.2007.02.019. [This paper investigates the role of the solitary allele of Vrp1 in myoblast fusion, and proposes a model in which filamentous actin mediates targeting of vesicles to the fusion site.] [DOI] [PubMed] [Google Scholar]
- 26••.Massarwa R, Carmon S, Shilo BZ, Schejter ED. WIP/WASp-based actin-polymerization machinery is essential for myoblast fusion in Drosophila. Dev Cell. 2007;12:557–569. doi: 10.1016/j.devcel.2007.01.016. [This paper identifies both Vrp1 (D-WIP) and WASp as crucial for myoblast fusion and suggests that these proteins target actin branching at the fusion site, leading to the expansion of fusion pores.] [DOI] [PubMed] [Google Scholar]
- 27.Menon SD, Chia W. Drosophila rolling pebbles: a multidomain protein required for myoblast fusion that recruits D-Titin in response to the myoblast attractant Dumbfounded. Dev Cell. 2001;1:691–703. doi: 10.1016/s1534-5807(01)00075-2. [DOI] [PubMed] [Google Scholar]
- 28.Rau A, Buttgereit D, Holz A, Fetter R, Doberstein SK, Paululat A, Staudt N, Skeath J, Michelson AM, Renkawitz-Pohl R. rolling pebbles (rols) is required in Drosophila muscle precursors for recruitment of myoblasts for fusion. Development. 2001;128:5061–5073. doi: 10.1242/dev.128.24.5061. [DOI] [PubMed] [Google Scholar]
- 29••.Richardson BE, Beckett K, Nowak SJ, Baylies MK. SCAR/WAVE and Arp2/3 are crucial for cytoskeletal remodeling at the site of myoblast fusion. Development. 2007;134:4357–4367. doi: 10.1242/dev.010678. [This paper characterizes the dynamic behavior of the actin focus in wild-type and fusion mutant embryos using time-lapse videography. The paper also indicates that Scar and Arp2/3 are required for myoblast fusion in the Drosophila embryo.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schroter RH, Lier S, Holz A, Bogdan S, Klambt C, Beck L, Renkawitz-Pohl R. kette and blown fuse interact genetically during the second fusion step of myogenesis in Drosophila. Development. 2004;131:4501–4509. doi: 10.1242/dev.01309. [DOI] [PubMed] [Google Scholar]
- 31.Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richardson BE, Nowak SJ, Baylies MK. Myoblast fusion in fly and vertebrates: new genes, new processes and new perspectives. Traffic. 2008;9:1050–1059. doi: 10.1111/j.1600-0854.2008.00756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Volk T. Singling out Drosophila tendon cells: a dialogue between two distinct cell types. Trends Genet. 1999;15:448–453. doi: 10.1016/s0168-9525(99)01862-4. [DOI] [PubMed] [Google Scholar]
- 34•.Kramer SG, Kidd T, Simpson JH, Goodman CS. Switching repulsion to attraction: changing responses to slit during transition in mesoderm migration. Science. 2001;292:737–740. doi: 10.1126/science.1058766. [Identification of the Slit–Robo system as a basis for myotube attraction to tendon cells during Drosophila embryogenesis.] [DOI] [PubMed] [Google Scholar]
- 35.Kidd T, Bland KS, Goodman CS. Slit is the midline repellent for the robo receptor in Drosophila. Cell. 1999;96:785–794. doi: 10.1016/s0092-8674(00)80589-9. [DOI] [PubMed] [Google Scholar]
- 36••.Estrada B, Gisselbrecht SS, Michelson AM. The transmembrane protein Perdido interacts with Grip and integrins to mediate myotube projection and attachment in the Drosophila embryo. Development. 2007;134:4469–4478. doi: 10.1242/dev.014027. [See notes to reference [38••]..] [DOI] [PubMed] [Google Scholar]
- 37•.Guerin CM, Kramer SG. RacGAP50C directs perinuclear gamma-tubulin localization to organize the uniform microtubule array required for Drosophila myotube extension. Development. 2009;136:1411–1421. doi: 10.1242/dev.031823. [This study describes and characterizes the involvement of muscle fiber microtubule arrays in myotube guidance and extension.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38••.Schnorrer F, Kalchhauser I, Dickson BJ. The transmembrane protein Kon-tiki couples to Dgrip to mediate myotube targeting in Drosophila. Dev Cell. 2007;12:751–766. doi: 10.1016/j.devcel.2007.02.017. [This paper and the study from Estrada et al. (reference [36]) identify the transmembrane protein Kon-tiki/Perdido as an important component of the D-Grip pathway mediating myotube guidance and extension.] [DOI] [PubMed] [Google Scholar]
- 39••.Swan LE, Wichmann C, Prange U, Schmid A, Schmidt M, Schwarz T, Ponimaskin E, Madeo F, Vorbruggen G, Sigrist SJ. A glutamate receptor-interacting protein homolog organizes muscle guidance in Drosophila. Genes Dev. 2004;18:223–237. doi: 10.1101/gad.287604. [The initial discovery that D-Grip, a PDZ-domain adaptor, mediates myotube guidance and extension in Drosophila embryos.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ataman B, Ashley J, Gorczyca D, Gorczyca M, Mathew D, Wichmann C, Sigrist SJ, Budnik V. Nuclear trafficking of Drosophila Frizzled-2 during synapse development requires the PDZ protein dGRIP. Proc Natl Acad Sci USA. 2006;103:7841–7846. doi: 10.1073/pnas.0600387103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong H, O'Brien RJ, Fung ET, Lanahan AA, Worley PF, Huganir RL. GRIP: a synaptic PDZ domain-containing protein that interacts with AMPA receptors. Nature. 1997;386:279–284. doi: 10.1038/386279a0. [DOI] [PubMed] [Google Scholar]
- 42.Hoogenraad CC, Milstein AD, Ethell IM, Henkemeyer M, Sheng M. GRIP1 controls dendrite morphogenesis by regulating EphB receptor trafficking. Nat Neurosci. 2005;8:906–915. doi: 10.1038/nn1487. [DOI] [PubMed] [Google Scholar]
- 43•.Wayburn B, Volk T. LRT, a tendon-specific leucine-rich repeat protein, promotes muscle–tendon targeting through its interaction with Robo. Development. 2009;136:3607–3615. doi: 10.1242/dev.040329. [This paper shows that LRT, expressed by tendon cells, acts to restrict migratory activity of extending myotubes by interacting with the Robo guidance receptor.] [DOI] [PubMed] [Google Scholar]
- 44.Gilsohn E, Volk T. Fine tuning cellular recognition: the function of the leucine rich repeat (LRR) trans-membrane protein. LRT, in muscle targeting to tendon cells. Cell Adhes Migr. 2010;4:368–371. doi: 10.4161/cam.4.3.11606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45•.Swan LE, Schmidt M, Schwarz T, Ponimaskin E, Prange U, Boeckers T, Thomas U, Sigrist SJ. Complex interaction of Drosophila GRIP PDZ domains and Echinoid during muscle morphogenesis. EMBO J. 2006;25:3640–3651. doi: 10.1038/sj.emboj.7601216. [This study suggests that the transmembrane protein Echinoid influences the D-Grip myotube guidance pathway, suggesting a link with microfilament organization and dynamics.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fetting JL, Spencer SA, Wolff T. The cell adhesion molecules Echinoid and Friend of Echinoid coordinate cell adhesion and cell signaling to regulate the fidelity of ommatidial rotation in the Drosophila eye. Development. 2009;136:3323–3333. doi: 10.1242/dev.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rawlins EL, Lovegrove B, Jarman AP. Echinoid facilitates Notch pathway signalling during Drosophila neurogenesis through functional interaction with Delta. Development. 2003;130:6475–6484. doi: 10.1242/dev.00882. [DOI] [PubMed] [Google Scholar]
- 48.Wei SY, Escudero LM, Yu F, Chang LH, Chen LY, Ho YH, Lin CM, Chou CS, Chia W, Modolell J, et al. Echinoid is a component of adherens junctions that cooperates with DE-Cadherin to mediate cell adhesion. Dev Cell. 2005;8:493–504. doi: 10.1016/j.devcel.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 49.Laplante C, Nilson LA. Differential expression of the adhesion molecule Echinoid drives epithelial morphogenesis in Drosophila. Development. 2006;133:3255–3264. doi: 10.1242/dev.02492. [DOI] [PubMed] [Google Scholar]
- 50.Lin HP, Chen HM, Wei SY, Chen LY, Chang LH, Sun YJ, Huang SY, Hsu JC. Cell adhesion molecule Echinoid associates with unconventional myosin VI/Jaguar motor to regulate cell morphology during dorsal closure in Drosophila. Dev Biol. 2007;311:423–433. doi: 10.1016/j.ydbio.2007.08.043. [DOI] [PubMed] [Google Scholar]
- 51.Stegmuller J, Werner H, Nave KA, Trotter J. The proteoglycan NG2 is complexed with alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors by the PDZ glutamate receptor interaction protein (GRIP) in glial progenitor cells. Implications for glial-neuronal signaling. J Biol Chem. 2003;278:3590–3598. doi: 10.1074/jbc.M210010200. [DOI] [PubMed] [Google Scholar]
- 52.Karram K, Chatterjee N, Trotter J. NG2-expressing cells in the nervous system: role of the proteoglycan in migration and glial-neuron interaction. J Anat. 2005;207:735–744. doi: 10.1111/j.1469-7580.2005.00461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishiyama A, Komitova M, Suzuki R, Zhu X. Polydendrocytes (NG2 cells): multifunctional cells with lineage plasticity. Nat Rev Neurosci. 2009;10:9–22. doi: 10.1038/nrn2495. [DOI] [PubMed] [Google Scholar]
- 54.Mishima M, Kaitna S, Glotzer M. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev Cell. 2002;2:41–54. doi: 10.1016/s1534-5807(01)00110-1. [DOI] [PubMed] [Google Scholar]
- 55.D'Avino PP. How to scaffold the contractile ring for a safe cytokinesis—lessons from Anillin-related proteins. J Cell Sci. 2009;122:1071–1079. doi: 10.1242/jcs.034785. [DOI] [PubMed] [Google Scholar]
- 56.Zavortink M, Contreras N, Addy T, Bejsovec A, Saint R. Tum/RacGAP50C provides a critical link between anaphase microtubules and the assembly of the contractile ring in Drosophila melanogaster. J Cell Sci. 2005;118:5381–5392. doi: 10.1242/jcs.02652. [DOI] [PubMed] [Google Scholar]
- 57.Musa H, Orton C, Morrison EE, Peckham M. Microtubule assembly in cultured myoblasts and myotubes following nocodazole induced microtubule depolymerisation. J Muscle Res Cell Motil. 2003;24:301–308. [PMC free article] [PubMed] [Google Scholar]
- 58.Brown NH, Gregory SL, Martin-Bermudo MD. Integrins as mediators of morphogenesis in Drosophila. Dev Biol. 2000;223:1–16. doi: 10.1006/dbio.2000.9711. [DOI] [PubMed] [Google Scholar]
- 59.Bokel C, Brown NH. Integrins in development: moving on, responding to, and sticking to the extracellular matrix. Dev Cell. 2002;3:311–321. doi: 10.1016/s1534-5807(02)00265-4. [DOI] [PubMed] [Google Scholar]
- 60.Newman SM, Jr, Wright TR. A histological and ultrastructural analysis of developmental defects produced by the mutation, lethal(1)myospheroid, in Drosophila melanogaster. Dev Biol. 1981;86:393–402. doi: 10.1016/0012-1606(81)90197-4. [DOI] [PubMed] [Google Scholar]
- 61.Leptin M, Bogaert T, Lehmann R, Wilcox M. The function of PS integrins during Drosophila embryogenesis. Cell. 1989;56:401–408. doi: 10.1016/0092-8674(89)90243-2. [DOI] [PubMed] [Google Scholar]
- 62.Brown NH. Null mutations in the alpha PS2 and beta PS integrin subunit genes have distinct phenotypes. Development. 1994;120:1221–1231. doi: 10.1242/dev.120.5.1221. [DOI] [PubMed] [Google Scholar]
- 63.Brabant MC, Brower DL. PS2 integrin requirements in Drosophila embryo and wing morphogenesis. Dev Biol. 1993;157:49–59. doi: 10.1006/dbio.1993.1111. [DOI] [PubMed] [Google Scholar]
- 64.Martin D, Zusman S, Li X, Williams EL, Khare N, DaRocha S, Chiquet-Ehrismann R, Baumgartner S. wing blister, a new Drosophila laminin alpha chain required for cell adhesion and migration during embryonic and imaginal development. J Cell Biol. 1999;145:191–201. doi: 10.1083/jcb.145.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bunch TA, Graner MW, Fessler LI, Fessler JH, Schneider KD, Kerschen A, Choy LP, Burgess BW, Brower DL. The PS2 integrin ligand tiggrin is required for proper muscle function in Drosophila. Development. 1998;125:1679–1689. doi: 10.1242/dev.125.9.1679. [DOI] [PubMed] [Google Scholar]
- 66.Fogerty FJ, Fessler LI, Bunch TA, Yaron Y, Parker CG, Nelson RE, Brower DL, Gullberg D, Fessler JH. Tiggrin, a novel Drosophila extracellular matrix protein that functions as a ligand for Drosophila alpha PS2 beta PS integrins. Development. 1994;120:1747–1758. doi: 10.1242/dev.120.7.1747. [DOI] [PubMed] [Google Scholar]
- 67••.Subramanian A, Wayburn B, Bunch T, Volk T. Thrombospondin-mediated adhesion is essential for the formation of the myotendinous junction in Drosophila. Development. 2007;134:1269–1278. doi: 10.1242/dev.000406. [See notes to reference [68••].] [DOI] [PubMed] [Google Scholar]
- 68••.Chanana B, Graf R, Koledachkina T, Pflanz R, Vorbruggen G. AlphaPS2 integrin-mediated muscle attachment in Drosophila requires the ECM protein Thrombospondin. Mech Dev. 2007;124:463–475. doi: 10.1016/j.mod.2007.03.005. [This paper, together with reference [67••], identifies and characterizes Drosophila Thrombospondin as a tendon-cell derived integrin ligand, essential for proper formation of embryonic myotendinous junctions.] [DOI] [PubMed] [Google Scholar]
- 69.Adams JC, Lawler J. The thrombospondins. Int J Biochem Cell Biol. 2004;36:961–968. doi: 10.1016/j.biocel.2004.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Christopherson KS, Ullian EM, Stokes CC, Mullowney CE, Hell JW, Agah A, Lawler J, Mosher DF, Bornstein P, Barres BA. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120:421–433. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 71.Bentley AA, Adams JC. The evolution of thrombospondins and their ligand-binding activities. Mol Biol Evol. 2010;27:2187–2197. doi: 10.1093/molbev/msq107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yarnitzky T, Min L, Volk T. The Drosophila neuregulin homolog Vein mediates inductive interactions between myotubes and their epidermal attachment cells. Genes Dev. 1997;11:2691–2700. doi: 10.1101/gad.11.20.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73•.Gilsohn E, Volk T. Slowdown promotes muscle integrity by modulating integrin-mediated adhesion at the myotendinous junction. Development. 2010;137:785–794. doi: 10.1242/dev.043703. [Identification and characterization of a novel tendon-derived secreted element, which fortifies the larval myotendinous junction.] [DOI] [PubMed] [Google Scholar]
- 74.Bicker F, Schmidt MH. EGFL7: a new player in homeostasis of the nervous system. Cell Cycle. 2010:9. doi: 10.4161/cc.9.7.11091. [DOI] [PubMed] [Google Scholar]
- 75.Schmidt MH, Bicker F, Nikolic I, Meister J, Babuke T, Picuric S, Muller-Esterl W, Plate KH, Dikic I. Epidermal growth factor-like domain 7 (EGFL7) modulates Notch signalling and affects neural stem cell renewal. Nat Cell Biol. 2009;11:873–880. doi: 10.1038/ncb1896. [DOI] [PubMed] [Google Scholar]
- 76.Faulkner G, Pallavicini A, Formentin E, Comelli A, Ievolella C, Trevisan S, Bortoletto G, Scannapieco P, Salamon M, Mouly V, et al. ZASP: a new Z-band alternatively spliced PDZ-motif protein. J Cell Biol. 1999;146:465–475. doi: 10.1083/jcb.146.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Krcmery J, Camarata T, Kulisz A, Simon HG. Nucleocytoplasmic functions of the PDZ-LIM protein family: new insights into organ development. Bioessays. 2010;32:100–108. doi: 10.1002/bies.200900148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78••.Jani K, Schock F. Zasp is required for the assembly of functional integrin adhesion sites. J Cell Biol. 2007;179:1583–1597. doi: 10.1083/jcb.200707045. [This paper demonstrates multiple essential roles for the Drosophila ZASP homolog in mediating integrin-based adhesions.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79••.Loer B, Bauer R, Bornheim R, Grell J, Kremmer E, Kolanus W, Hoch M. The NHL-domain protein Wech is crucial for the integrin-cytoskeleton link. Nat Cell Biol. 2008;10:422–428. doi: 10.1038/ncb1704. [Identification of a novel and apparently conserved factor mediating the functional interaction of integrins with the actin cytoskeleton.] [DOI] [PubMed] [Google Scholar]
- 80.Lin YC, Hsieh LC, Kuo MW, Yu J, Kuo HH, Lo WL, Lin RJ, Yu AL, Li WH. Human TRIM71 and its nematode homologue are targets of let-7 microRNA and its zebrafish orthologue is essential for development. Mol Biol Evol. 2007;24:2525–2534. doi: 10.1093/molbev/msm195. [DOI] [PubMed] [Google Scholar]
- 81.Maller Schulman BR, Liang X, Stahlhut C, DelConte C, Stefani G, Slack FJ. The let-7 microRNA target gene, Mlin41/Trim71 is required for mouse embryonic survival and neural tube closure. Cell Cycle. 2008;7:3935–3942. doi: 10.4161/cc.7.24.7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Munir M. TRIM proteins: another class of viral victims. Sci Signal. 2010;3:jc2. doi: 10.1126/scisignal.3118jc2. [DOI] [PubMed] [Google Scholar]
- 83.Zervas CG, Gregory SL, Brown NH. Drosophila integrin-linked kinase is required at sites of integrin adhesion to link the cytoskeleton to the plasma membrane. J Cell Biol. 2001;152:1007–1018. doi: 10.1083/jcb.152.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 85••.Schnorrer F, Schonbauer C, Langer CC, Dietzl G, Novatchkova M, Schernhuber K, Fellner M, Azaryan A, Radolf M, Stark A, et al. Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature. 2010;464:287–291. doi: 10.1038/nature08799. [A tour-de-force application of whole-genome RNAi screens to the study of Drosophila myogenesis. Over 2700 genes are shown to have a role in myogenesis and are classified into functional groups.] [DOI] [PubMed] [Google Scholar]
- 86.Estrada B, Choe SE, Gisselbrecht SS, Michaud S, Raj L, Busser BW, Halfon MS, Church GM, Michelson AM. An integrated strategy for analyzing the unique developmental programs of different myoblast subtypes. PLoS Genet. 2006;2:e16. doi: 10.1371/journal.pgen.0020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sandmann T, Jensen LJ, Jakobsen JS, Karzynski MM, Eichenlaub MP, Bork P, Furlong EE. A temporal map of transcription factor activity: mef2 directly regulates target genes at all stages of muscle development. Dev Cell. 2006;10:797–807. doi: 10.1016/j.devcel.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 88.Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EE. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature. 2009;462:65–70. doi: 10.1038/nature08531. [DOI] [PubMed] [Google Scholar]