Abstract
CD1d-restricted NKT cells comprise an innate-like T cell population that exerts significant influence over early events in the developing immune response. The frequency of NKT cells is highly variable in humans and in mice, but the basis for this variability remains unclear. Here, we report a striking deficiency of Type I NKT cells in the wild-derived inbred strains, PWD/PhJ, SPRET/EiJ, and CAST/EiJ. Investigation of the underlying basis for the lack of Type I NKT cells revealed that one strain, PWD/PhJ, exhibited a significant impairment in thymocyte and splenocyte CD1d gene and protein expression. Accordingly, both thymocytes and BMDCs from PWD mice exhibited a significant impairment in the ability to present α-galactosylceramide to NKT cells. The impaired PWD CD1d gene expression was due to impaired CD1d promoter activity. Fine-mapping of the promoter activity revealed that two single nucleotide substitutions at positions -331 and -164 in the proximal promoter were each sufficient to account for the diminished PWD CD1d promoter activity. Examination of the strain distribution pattern of these polymorphisms revealed that, of 19 strains analyzed, only PWD and PWK mice possessed both CD1d promoter polymorphisms. A subsequent examination of the PWK strain revealed that it also exhibited impaired thymocyte CD1d expression and very low numbers of NKT cells. Taken together, these results provide new insight into the control of CD1d gene expression, and have implications for the evolution of CD1d and Type I NKT cells.
Introduction
CD1d-restricted NKT cells comprise an innate-like T cell subset that has been implicated in a wide variety of immune responses. NKT cells are notable for their paradoxical ability to either promote or suppress the immune response in a context-dependent manner (1). NKT cells recognize glycolipids presented by the MHC class I-like molecule CD1d (2–4). Upon activation by the prototypical glycosphingolipid ligand, α-galactosylceramide (αGalCer) NKT cells rapidly produce a wide variety of cytokines and chemokines including IFN-γ, IL-4, and TNF (5–7). Activated NKT cells can subsequently activate a number of leukocyte subsets of both the innate and adaptive arms of the immune system, including dendritic cells, NK cells, and macrophages (8–12). These observations, together with their prevalence in peripheral tissues such as the liver, lung, and colon, suggest that NKT cells play a role during the early stages of the developing immune response.
NKT cell numbers are highly variable in the human population, spanning over a 100-fold range (13, 14). This high level of variability is also observed in the mouse, which exhibits widespread strain-dependent variability in both NKT cell number and in NKT cell function (15–18). Numerous reports in both humans and mice have linked alterations in NKT cell number and function with disease susceptibility and/or pathogenesis (15, 16, 19–26). Whether this relationship is causal or correlative is still unclear. However, it remains the case that NKT cells can significantly affect the quality of the innate immune response (27, 28), and that naturally occurring variability in the NKT cell population can have a significant impact on immune outcomes.
Genetic mapping studies using inbred strains of mice have been successful in identifying loci that contribute to NKT cell development and function (29–32). However, the genetic diversity among inbred laboratory strains of mice is limited (33). Therefore, we examined the phenotype of NKT cells in the more genetically disparate wild-derived inbred strains of mice. Here, we report that each of the strains examined, PWD/PhJ, CAST/Eij, SPRETUS/EiJ, and PWK/PhJ, exhibited extremely low, almost undetectable NKT cell numbers. Using consomic PWD/PhJ mice, we identified chromosome 3 as a major contributor to the phenotype. We further demonstrate that the expression of CD1d, which is located on chromosome 3, is significantly lower in PWD/PhJ in comparison to B6 mice. Analysis of the B6 and PWD CD1d1 promoters revealed the presence of numerous polymorphisms, two of which contributed to the impaired CD1d gene expression in PWD mice. These data suggest that genetic variability in the CD1d gene itself may be an important, overlooked contributor to the widespread variability in NKT cell number and function in mice and in humans.
Materials and Methods
Mice and treatments
C57BL/6J, CAST/EiJ, SPRETUS/EiJ, PWD/PhJ, PWK/PhJ and B6.PWDchr3 mice were obtained from Jackson Laboratory (Bar Harbor, ME). All mice were housed in the specific pathogen-free facility at the University of Vermont. The alpha-galactosylceramide (Axxora Pharmaceuticals, San Diego, CA) was prepared as described (15) and was administered i.p. (100 μg/kg) in a 100 μl volume. All experiments were approved by the University of Vermont Institutional Animal Care and Use Committee.
Leukocyte isolation
Splenocytes and thymocytes were obtained by gentle pressing through nylon mesh. Red blood cells in splenocyte preparations were lysed using Gey’s solution. Intrahepatic leukocyte (IHL) isolation was performed as described (15). Briefly, anesthetized mice were perfused with PBS, after which the liver was removed, minced, and gently pressed through nylon mesh. The resulting cell suspension was washed in PBS + 2% FCS and then resuspended in isotonic 33.8% Percoll (GE Healthcare, Piscataway, NJ). After centrifugation, red blood cells were lysed using Gey’s solution, washed in PBS + 2% FCS and resuspended.
Cytokine analysis
Serum was prepared from blood collected via cardiac puncture at various times after injection. Samples were frozen at −20°C until analysis. Serum cytokines were measured by ELISA (BD Biosciences, San Jose, CA) according to the manufacturer’s instructions. For analysis of ex vivo splenocyte cytokine production, equal numbers of splenocytes from B6, B6.PWDchr3, or PWD mice were prepared 90 min after an i.p. administration of αGalCer, and incubated in complete RPMI medium for 4 hours at 37°C in a humidified 5% CO2 incubator. Supernatants were collected and cytokine secretion in the supernatant was evaluated by ELISA.
Antibodies and flow cytometry
Cells were stained at 4°C in PBS/2%FCS containing 0.1% sodium azide. TruStain FcX (Biolegend) was used in all samples prior to the addition of antibodies to block non-specific antibody binding. Antibodies used in these experiments were: anti-TCR-β H57-597), anti-CD1d (1B1), anti-IFN-γ (XMG1.2) anti-IL-4 (11B11), and anti-TNF (MP6-XT22) all from BD Biosciences, CD45 Pacific Orange (30-F11) (Caltag Laboratories). CD1d tetramer loaded with PBS57 was obtained from the NIH tetramer facility (Emory University Vaccine Center, Atlanta, GA). Thymocytes and splenocytes were gated on FSC vs. SSC. Liver samples were first gated on CD45+ cells, followed by FSC vs. SSC. The CD1d-specific 1B1 clone recognizes both CD1d1 and CD1d2 proteins (34).
To compare CD1d expression among different strains, we calculated the relative CD1d fluorescence using the following formula: [(sample CD1d median fluorescence intensity (MFI)- sample background MFI)/(Average (B6 sample MFI-B6 background MFI))]. In all cases, statistical tests using MFIs directly yielded the same results.
For intracellular cytokine staining, cells were isolated from spleen as described above and stained with antibodies to surface markers. In all experiments, cells were analyzed directly ex vivo with no cell culture or treatment with Brefeldin A or Monensin. After washing in staining buffer, cells were fixed and permeabilized using fixation/permeabilization buffer (BD Biosciences) according to the manufacturer’s instructions, followed by staining with Alexa647-conjugated anti-cytokine mAbs or isotype control mAb. Data were collected on an LSRII (BD Biosciences) and analyzed using FlowJo (TreeStar, Ashland, OR) software.
PCR, cloning, and sequencing
Full-length CD1d1 gene sequence was obtained using PCR on cDNA synthesized from splenocyte RNA from the different mouse strains. Primers used to amplify full-length CD1d were 5′CD1d: 5′-GCCAAGCTT ATGCGTACCT ACCATGC-3′ and 3′CD1d: 5′-GGCGGATCCCG GATGTCTTG ATAGGC-3′ (Eurofins MWG Operon, Huntsville, AL). PCR products were TOPO-cloned according to the manufacturer’s protocol (Invitrogen), followed by sequencing at the DNA Analysis Facility (University of Vermont, Burlington, VT). The CD1d promoter was generated using PCR amplification of genomic DNA from B6 and PWD/PhJ mice. PCR products were TOPO- cloned and sequenced. Primers used for amplification of the promoter were: CD1dFwd—5′-GCCGC AAGCTTCTCCGACTTCTGCGCTTTAC-3′, CD1dRvs—5′-GCCGCCTCGAGATCAGACTG ACACAAGTCTTC-3′.
Transfections
B6 and PWD CD1d were subcloned into pBabe puro IRES-EGFP (Addgene). NIH3T3 cells were transfected with using X-tremeGENE HP (Roche Applied Science) according to the manufacturer’s instructions. Cells were cultured in DMEM supplemented with 10% FCS (Atlanta Biologicals), penicillin-streptomycin, nonessential amino acids, β-mercaptoethanol, and sodium pyruvate. Stable transfectants were established by selection in puromycin (2 μg/mL). Positive transfectants were identified by flow cytometry using α-CD1d (1B1) (BioLegend). NIH3T3-CD1d/B6 and NIH3T3-CD1d/PWD cells were FACS sorted to select for transfectants with equivalent expression levels of GFP.
Antigen presentation assays
For thymocyte antigen presentation assays, B6 and PWD thymocytes (250000/well) were co-cultured with the 2C12 NKT cell hybridoma (50000/well) with or without varying amounts of α GalCer for 72 h, after which supernatants were collected for analysis by ELISA.
In other antigen presentation assays, NIH3T3 cells transfected with B6 or PWD CD1d, or bone marrow-derived dendritic cells from C57BL/6J or PWD/PhJ mice were incubated with varying amounts of αGalCer for 3 h in complete DMEM, after which they were extensively washed. Pulsed APCs were then co-cultured with the 2C12 NKT hybridoma at a ratio of 1:1 NKT: NIH3T3 cells for 24 h, after which supernatants were collected for analysis by ELISA.
Promoter analysis using luciferase reporter constructs
B6 and PWD CD1d promoters were subcloned into a luciferase reporter construct (pGL3, Promega, Madison, WI). L929 fibroblasts were transfected with equal amounts of the CD1d reporter constructs and a Renilla luciferase reporter construct (pRLTK, Promega, Madison, WI) using XtremeGene HP (Roche). Cells were cultured for 48 h at 37 °C., after which luminescence was detected using Dual-Glo (Promega) according to the manufacturer’s instructions. Data were collected using a Synergy H4 plate reader (BioTek, Winooski, VT). Individual samples were first normalized using the internal Renilla luciferase control, and then all samples were normalized to the B6 luciferase activity.
Site-directed mutagenesis
Site-directed mutagenesis was conducted according to manufacturer’s instructions (QuikChange, Stratagene). Briefly, B6 CD1d in pGL3 constructs were amplified with complementary oligonucleotides containing PWD substitutions using PfuTurbo followed by digestion with DpnI, and mutant constructs were verified by sequencing. Oligonucleotides for site-directed mutagenesis of B6 CD1d to PWD were as follows: -603-5′-GGG AGA AAA GAC ACC AGG GTA A-3′, -331-5′ GGA GTT GTA ACA GAA ATA CAA T-3′, -296(1)-5′-GGA GTT GTA ACA GAA ATA CAA T-3′, -296(2)-5′ GCT TTC GAT ATT GAG ACT TTT TGT CTC AAC CTC ATT CTT TAG-3′, -205-5′-GAA GTG GTT TGT GTA GCG GTT C-3′, -164-5′-GCC CTC AAC TAT CAG GGG GTT T-3′
qPCR
qPCR was conducted using cDNA synthesized from thymocyte mRNA. CD1d-specific primers used were: 5′CD1dqpcr: 5′-AGAAGCAAGAGAAGCCAGTG-3′ and 3′CD1dqpcr: 5′-CAGGAGAGGCAGGTGTAAGG-3′ (Eurofins MWG Operon; Huntsville, AL). These primers are predicted to bind to both CD1d1 and CD1d2 transcripts. β2m primers were used as internal controls. QPCR was performed using SYBR green for quantification. Relative expression was calculated using a standard curve.
Western Blot
Mouse thymocyte cell suspensions were lysed in lysis buffer (140 mM NaCl, 25 mM Tris-HCl, 1% Triton-X 100, protease inhibitors (Sigma)). Cell lysates were resolved using SDS-PAGE, transferred to nitrocellulose membrane, and incubated with anti-PU.1 (T-21) (Santa Cruz Biotechnology). Anti-beta actin was used as a loading control. After incubation with the appropriate secondary antibodies, the blot was visualized using ECL (Pierce).
Statistical Analysis
Statistical analysis was carried out using PRISM software (GraphPad Software, San Diego, CA). Comparisons between 2 groups were tested using a Student’s t-test. Comparisons among three or more groups were tested using one-way ANOVA, followed by the Holm-Sidak test to correct for multiple comparisons. In antigen presentation assays, the data was first fit to a 3-parameter dose response curve, and then compared using an F-test.
Results
Severe deficiency in NKT cells in genetically disparate wild-derived inbred strains
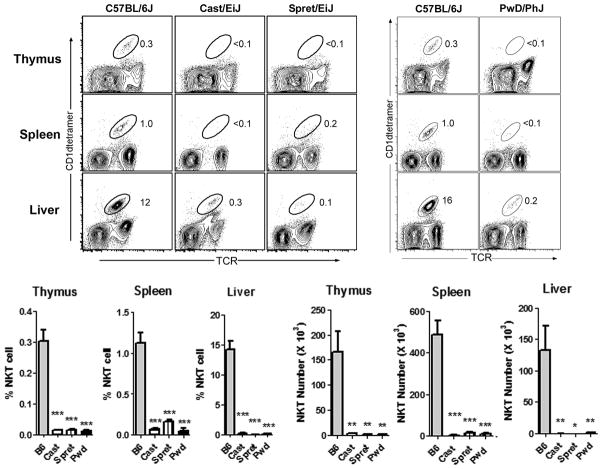
We previously reported a high level of strain-dependent variation in NKT cell number and function among commonly used inbred strains of mice (15, 16). Here, we extended this analysis by examining NKT cells in genetically disparate wild-derived inbred strains SPRET/EiJ (Mus spretus), CAST/EiJ (Mus musculus castaneus), and PWD/PhJ (Mus musculus musculus). Using CD1d tetramer staining, we found that in comparison to B6 mice, all 3 wild-derived inbred strains analyzed exhibited a striking deficiency in the frequency of NKT cells in thymus, spleen, and liver (Fig. 1). Similarly, a calculation of total NKT cell numbers in the different organs revealed a striking deficiency in NKT cell numbers in these strains (Fig. 1).
Fig. 1. Severe deficiency NKT cell numbers from divergent Mus species.
Thymus, spleen, and liver NKT cells numbers were calculated by flow cytometry after staining with anti-TCRβ and CD1dtetramer/PBS57. Representative FACS contour plots depicting the percentages of NKT cells in the different organs are shown at top. The NKT cell population is circled, and the percentages are indicated. Percentages for thymus and spleen are of total cells, while percentages for liver are of CD45+ IHLs. Cumulative percentages and numbers of NKT cells in the different organs are shown at bottom. Data represent the means ± s.d., n = 4–5 mice per strain, *p ≤ 0.01, **p ≤ 0.01, ***p ≤ 0.001.
Impaired CD1d expression in PWD/PhJ mice
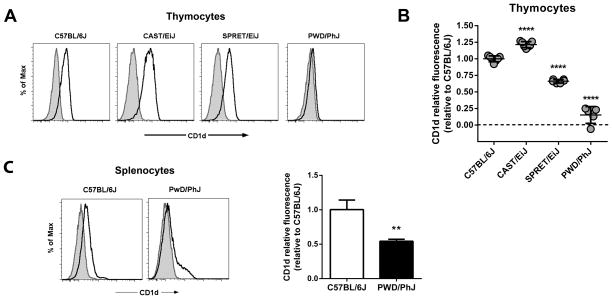
Since CD1d expression on thymocytes is necessary for NKT cell development (3), we compared thymic CD1d expression among B6, CAST, SPRET, and PWD mice. This analysis revealed a variable pattern of CD1d expression among these strains (Fig. 2A). In comparison to their B6 counterparts, SPRET thymocytes exhibited slightly lower (~63% of B6) levels of CD1d expression, while CD1d expression in CAST thymocytes was slightly (~118%) higher (Fig. 2B). Notably, however, PWD thymocytes exhibited significantly lower levels (~13%) of CD1d expression relative to their B6 counterparts (Fig. 2B). Investigation of the CD1d expression in the spleen of PWD mice revealed a similar significant decrease in CD1d expression in comparison to B6 mice indicating that the low CD1d expression was not a feature specific to thymocytes (Fig. 2C). Taken together, these observations indicated that, in comparison to commonly used laboratory inbred strains of mice, wild-derived inbred strains of different species and subspecies shared a notable deficiency in NKT cells. Moreover, the data suggested that the low number of NKT cells in PWD mice was associated with low levels of CD1d expressed on PWD thymocytes.
Fig. 2. Impaired CD1d expression in PWD mice.
(A) Comparison of thymocyte CD1d cell surface expression among inbred mouse strains. Representative histograms are shown. Shaded histograms represent isotype-matched control mAb staining. Open histograms represent anti-CD1d staining. (B) Relative thymocyte CD1d fluorescence among different inbred strains relative to C57BL/6J. Relative CD1d fluorescence was calculated as specified in Materials and Methods. Data represent the mean relative fluorescence ± s.d., n = 5 – 7 mice per strain, ****p<0.0001. (C) Comparison of CD1d expression between B6 and PWD splenocytes. Representative histograms are shown at left. Shaded histograms represent isotype-matched control mAb staining. Open histograms represent anti-CD1d staining. PWD splenocyte CD1d fluorescence relative to C57BL/6J is shown at right. Data represent the mean relative fluorescence ± s.d., n = 3 mice per strain, **p<0.01.
Analysis of PWD CD1d Ag presentation
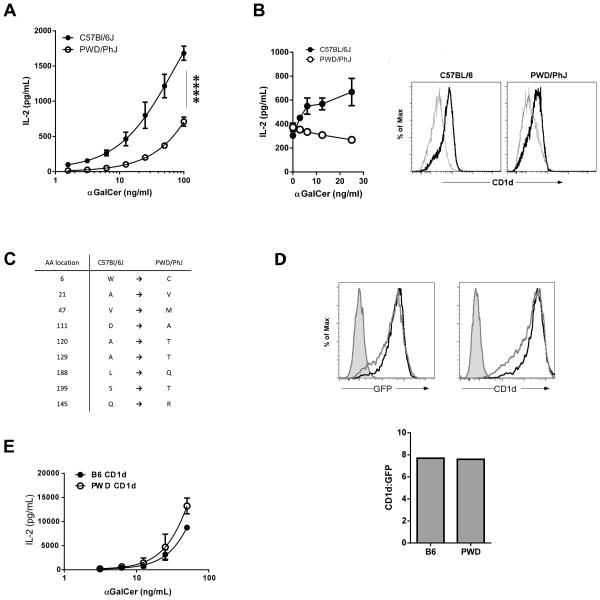
To investigate the functional significance of the low PWD thymocyte CD1d expression, we compared the ability of PWD and B6 thymocytes to stimulate NKT cells. αGalCer was added to co-cultures of an NKT cell hybridoma with either B6 or PWD thymocytes, after which NKT IL-2 production was assessed by ELISA. These data revealed that the ability of the PWD thymocytes to stimulate NKT cells was significantly impaired in comparison to their B6 counterparts (Fig. 3A). Addition of αGalCer to thymocytes alone did not result in any detectable IL-2 production (data not shown). When we compared the ability of B6 and PWD bone marrow-derived DCs (BMDCs) to present αGalCer to NKT cells, we found that PWD BMDCs were similarly impaired in their ability to stimulate NKT cells (Fig. 3B). As was observed with the thymocytes, inefficient stimulation of NKT cells by PWD BMDCs was associated with low CD1d expression (Fig. 3B).
Fig. 3. Impaired antigen presentation by PWD CD1d.
(A) Comparison of B6 and PWD thymocyte antigen presentation. B6 and PWD thymocytes were cultured with the 2C12 NKT hybridoma and varying amounts of αGalCer, after which NKT IL-2 secretion into the supernatant was assessed by ELISA. Data represent the mean IL-2 ± s.d., and are representative of two separate experiments. Incubation of B6 or PWD thymocytes with αGalCer alone did not result in the secretion of detectable levels of IL-2 (data not shown). **** p < 0.0001. (B) Comparison of B6 and PWD BMDC antigen presentation. (Left) BMDCs generated from B6 or PWD bone marrow were pulsed with varying amounts of αGalCer, after which they were co-cultured with the 2C12 NKT hybridoma. NKT IL-2 secretion into the supernatant was assessed by ELISA. Data represent the mean IL-2 ± s.d. (Right) Comparison of CD1d expression between B6 and PWD BMDCs. Grey histograms represent isotype-matched control mAb staining. Black histograms represent anti-CD1d staining. (C) Location of non-synonymous substitutions in PWD CD1d. Position 1 indicates the methionine in the signal sequence. (D) Comparison of B6 and PWD CD1d expression in NIH3T3 transfectants. B6 and PWD CD1d-IRES-GFP constructs were stably transfected in NIH 3T3 cells. Cells were sorted based on GFP expression to equalize expression. Sorted cells were stained with anti-CD1d, and expression was assessed using flow cytometry. (Left) Shaded histograms represent control staining. The black histograms represent B6 GFP expression or CD1d staining, while the gray histograms represent PWD GFP expression or CD1d staining. (Right) Comparison of the ratios of CD1d:GFP expression of B6 and PWD CD1d transfectants. (E) Evaluation of PWD CD1d polymorphisms on antigen presentation. NIH3T3 CD1d transfectants, expressing equal amounts of B6 or PWD CD1d, were pulsed with varying amounts of αGalCer, after which they were co-cultured with the 2C12 NKT hybridoma. NKT IL-2 secretion into the supernatant was assessed by ELISA. Data represent the mean IL-2 ± s.d.
It has been previously reported that amino acid differences in SPRET CD1d result in a diminished ability to present antigen to CD1d-restricted T cells (35). Comparison of the PWD and B6 CD1d gene sequences revealed numerous non-synonymous substitutions, some of which were identical to those described in SPRET CD1d, and some that were unique to PWD (Fig. 3C). To exclude the possibility that amino acid changes in the epitope recognized by the anti-CD1d mAb was impairing our ability to detect CD1d expression in PWD mice, we subcloned B6 and PWD CD1d into an IRES-GFP expression plasmid and stably transfected these constructs into CD1d-negative NIH3T3 cells. After expression was normalized between the two cell lines by FACS-sorting based on GFP, we found that there was no difference in the recognition of the B6 and PWD CD1d proteins by anti-CD1d antibody (Fig. 3D). Therefore, the amino acid differences between B6 and PWD CD1d did not affect the anti-CD1d (1B1) epitope, nor did they affect CD1d expression at the cell surface.
We next investigated whether these amino acid differences altered TCR recognition of CD1d, which could impact NKT cell development and contribute to the low NKT cell numbers in PWD mice. We compared the ability of the sorted B6 and PWD CD1d proteins expressed in 3T3 cells to present αGalCer to NKT cells. We observed no difference in the response of the 2C12 NKT hybridoma to these CD1d proteins, indicating that each was similarly recognized by NKT TCR (Fig. 3E). Thus, while numerous amino acid differences did exist between B6 and PWD CD1d proteins, they did not appear to affect the NKT TCR recognition of CD1d, nor the ability of CD1d to present glycolipid antigen. Taken together, these data suggested that low expression, not amino acid substitutions, of CD1d in PWD mice conferred a functional deficit in the ability to stimulate NKT cells, and that low CD1d expression on PWD thymocytes contributed to the impaired NKT cell development in these mice.
Impaired CD1d expression and low NKT cell number in B6.PWD chr3 consomic mice
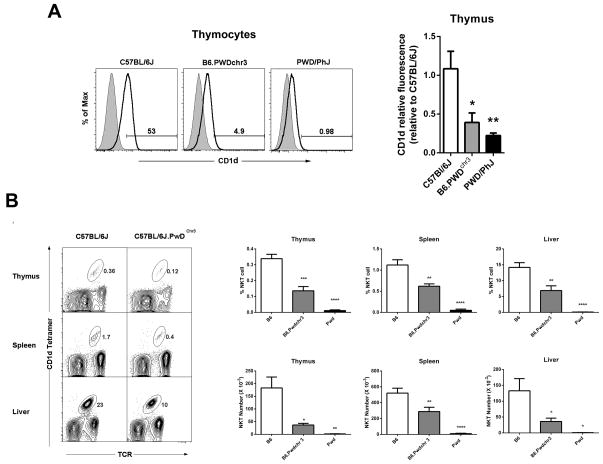
To investigate whether the low CD1d expression in PWD mice might be due to a CD1d gene-intrinsic difference, we examined CD1d expression in B6.PWDchr3 consomic mice. These are C57BL/6J mice that possess the entire PWD chromosome 3 (on which the CD1d gene is located). A comparison of thymic CD1d expression among B6.PWDchr3, B6, and PWD parental strains revealed that B6.PWDchr3 and PWD mice both expressed similar levels of CD1d, and that both were significantly lower than the B6 parental strain (Fig. 4A). These data suggested that the decreased NKT cell numbers in PWD mice was due to a gene on chromosome 3, most likely CD1d.
Fig. 4. Characterization of CD1d expression and NKT cell numbers in B6.PwDchr3 consomic mice.
(A) Comparison of CD1d expression among the B6.PWDchr3 consomic strain and the B6 and PWD parental strains. Representative thymocyte histograms are shown at left. Shaded histograms represent unstained controls, while open histograms represent anti-CD1d staining. The percent positive cells are indicated. Relative thymocyte and splenocyte CD1d fluorescence is shown at right. Relative CD1d fluorescence was calculated as specified in Materials and Methods. Data represent the mean relative fluorescence ± s.d., n = 3 – 4 mice per strain, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001. (B) Representative FACS contour plots depicting the percentages of NKT cells in different organs are shown at left. The NKT cell population is circled, and the percentages are indicated. Cumulative percentages and numbers of NKT cells in the organs of different strains are shown at right. Data represent the means ± s.e.m., n = 5–7 mice per strain, *p ≤ 0.01, **p ≤ 0.01, ***p ≤ 0.001.
To investigate whether the deficient CD1d expression observed in B6.PWDchr3 was sufficient to account for the deficiency in NKT cell numbers in the parental PWD strain, we compared the percentage of NKT cell numbers among B6, B6.PWDchr3, and PWD/PhJ strains in different organs. This analysis revealed that the percentage of NKT cells in B6.PWDchr3 mice was significantly lower in all organs studied in comparison to the B6 parental strain (Fig. 4B). Calculation of NKT cell numbers revealed a decrease in thymus, spleen, and liver in B6.PWDchr3 mice (Fig. 4B) suggesting that deficient CD1d expression was an important contributor to the low NKT cell numbers in PWD mice. We note, however, that B6.PWDchr3 did have significantly higher numbers of NKT cells than PWD/PhJ parental strain mice (Fig. 4B) suggesting that loci on other chromosomes are contributing to the low NKT cell numbers in PWD mice.
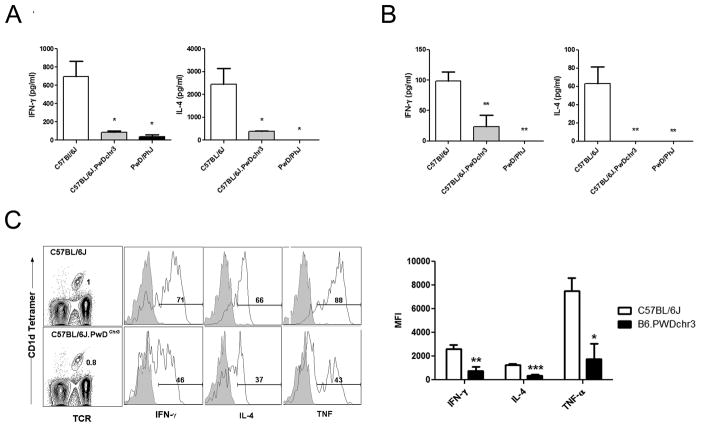
Next, we assessed whether the decreased CD1d expression and decreased NKT cell numbers in B6.PWDchr3 mice was sufficient to result in a functional impairment. A comparison of serum IFN-γ and IL-4 after administration of αGalCer i.p. revealed significantly decreased serum cytokine production in B6.PWDchr3 compared to B6 mice (Fig. 5A). Similarly, ex vivo analysis of splenocyte cytokine production after in vivo challenge with αGalCer revealed lower IFN-γ and undetectable amounts of IL-4 in B6.PWDchr3 mice compared to B6 counterparts (Fig. 5B). Since these results depend on both CD1d expression and NKT cell numbers, we assessed NKT cell cytokine production on a per cell basis using intracellular cytokine staining. Mice were administered αGalCer i.p. and 90 min later splenocytes were harvested and immediately stained for intracellular cytokine production without the use of Brefeldin A or monensin (Fig. 5C). These data revealed significantly lower IFN-γ, IL-4, and TNF production in B6.PWDchr3 mice compared to the B6 parental strain (Fig. 5C). Therefore, introgression of the CD1d-containing PWD chromsome 3 onto the B6 background resulted in impaired thymocyte CD1d expression, low NKT cell numbers, and poor responsiveness to αGalCer challenge.
Fig. 5. Impaired response to αGalCer in B6.PwDchr3 consomic mice.
(A) Comparison of serum cytokine levels among strains 90 min after i.p. αGalCer administration. (B) Comparison of ex vivo splenocyte cytokine production 90 min. after i.p. αGalCer administration. (C) Representative FACS data depicting intracellular cytokine staining by spleen NKT cells after i.p. αGalCer administration. Histograms depict intracellular cytokine staining of gated NKT cells. Shaded histograms represent isotype-matched control mAb staining, while open histograms represent the anti-cytokine staining. The percentage of NKT cells positive for cytokine production is indicated. Intracellular cytokine production in PWD NKT cells could not be determined because of the low number of cells. Cumulative MFIs of NKT cell intracellular cytokine staining is shown at right. Data represent the mean MFI ± s.d., n = 3–4 mice per strain, *p ≤ 0.01, **p ≤ 0.01, ***p ≤ 0.001.
Impaired CD1d gene expression in PWD mice
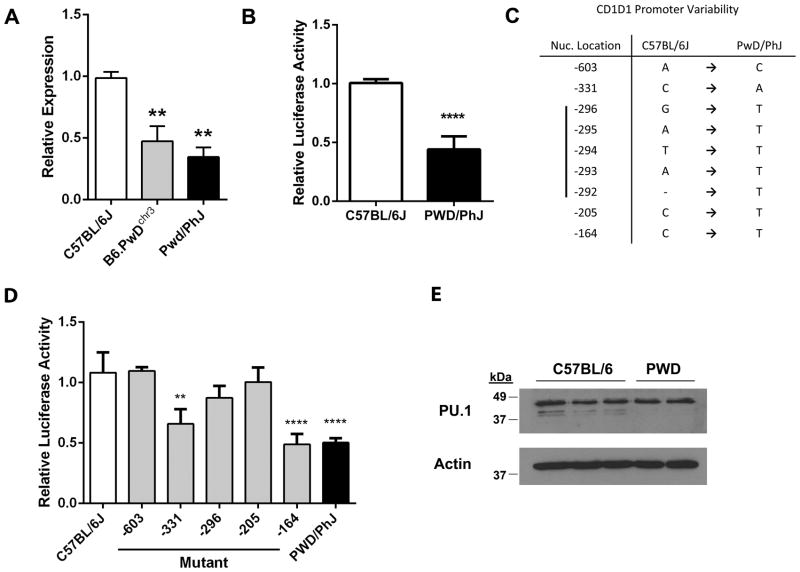
Since we could detect no obvious functional differences between B6 and PWD CD1d proteins, we investigated whether the impaired PWD CD1d expression was due to a difference in CD1d gene expression. A comparison of CD1d gene expression among B6, B6.PWDchr3, and PWD thymocytes using qPCR revealed that thymocyte CD1d gene expression was indeed significantly lower in both the B6.PWDchr3 and PWD strains in comparison to the parental B6 strain (Fig. 6A). Therefore, introgression of the PWD chromosome 3 onto the B6 background was sufficient to result in significantly lower cell surface CD1d expression, and lower protein expression was associated with impaired CD1d gene expression.
Fig. 6. Decreased CD1d gene expression in PWD/PhJ is associated with SNPs in the CD1d promoter.
(A) Comparison of CD1d gene expression among age-matched, sex-matched B6, PWD, and B6.PwDchr3 strain thymocytes using qPCR. Beta-2 microglobulin was used as an endogenous control. Relative expression was calculated using a standard curve. All data are normalized to C57BL/6J mice. Data represent the means ± s.d., n = 2–3 mice per strain, **p ≤ 0.01, and are representative of two separate experiments. (B) Comparison of B6 and PWD CD1d promoter activity. The proximal ~700 bp of the B6 and PWD CD1d promoters were transfected into mouse L929 cells, after which activity was assessed using a luciferase reporter assay. The data represent the means ± s.e.m. and are the combined data of two separate experiments, ****p < 0.0001. (C) Location of nucleotide differences between B6 and PWD CD1d promoters. Positions are indicated relative to the A (+1) in the start codon. (D) Relative contribution of the PWD CD1d promoter SNPs to impaired promoter activity. Site-directed mutagenesis was used to create successive B6 to PWD mutations. The exception was the -296 to -292 region of CD1d which was mutated in its entirety (denoted as -296). B6 and PWD promoters were subcloned into a luciferase reporter construct and transiently transfected into murine L929 cells. Data represent the means ± s.d., n = 6 measurements, **p ≤ 0.01, ****p ≤ 0.0001 denote significant differences as compared to the C57BL/6J wild-type promoter, and are representative of two separate experiments. (E) Comparison of PU.1 expression between B6 and PWD thymocytes. PU.1 protein expression was assessed in thymocyte preparations from B6 (n=3) and PWD (n=2) mice, using Western blot. Each lane represents a thymocyte preparation from a single mouse. Actin expression was measured for use as a loading control.
Since decreased CD1d gene expression in PWD/PhJ mice was intrinsic to chromosome 3, we compared B6 and PWD CD1d promoter activity using a luciferase reporter assay. We PCR-amplified a ~700 bp CD1d promoter fragment from the genomic DNA of both strains, which encompassed the three previously described transcription start sites (36). A comparison of the normalized luminescence from B6 and PWD CD1d promoters transfected into mouse L929 cells revealed a significant impairment in the ability of the PWD promoter to drive CD1d transcription (Fig. 6B). Analysis of this promoter fragment sequence revealed 5 areas of difference, including 4 single nucleotide substitutions and a single 5 bp region from -296 to -292 (Fig. 6C).
To investigate the contribution of each of the five differences to the impaired PWD CD1d promoter activity, we used site-directed mutagenesis to systematically mutate the B6 nucleotides to their PWD counterparts. The exception was the -296 to -292 region, where the entire region was mutated as a single unit. Using a luciferase reporter assay, we found that mutation of two sites, -331 (rs30844068) and -164 (rs237235766), resulted in a significant impairment in the B6 promoter activity (Fig 6D). There was no significant difference between the luciferase activity of these two mutants and that of the PWD promoter suggesting that these two single nucleotide substitutions were each sufficient to account for the impaired PWD promoter activity.
Previous work has shown that members of the Ets-1 family of transcription factors, Elf-1 and PU.1, can regulate mouse CD1d gene expression (36). Therefore, we assessed the likelihood that these transcription factors played a role in the low PWD gene expression. A comparison of the CD1d promoter Ets-1 family consensus binding site sequence between B6 and PWD revealed no nucleotide differences. Although Elf-1-deficient mice exhibited no impairment in NKT cell development (36), the role of PU.1 in NKT cell development is still unclear. To rule out the possibility that differences in PU.1 protein expression accounted for the difference in CD1d gene expression between the two strains, we compared thymocyte PU.1 protein expression using Western blot. This analysis revealed no difference in thymocyte PU.1 expression between the B6 and PWD mice (Fig. 6E), suggesting that Ets-1 family transcription factors are not responsible for the low PWD CD1d gene expression. Taken together, these data indicate that novel polymorphisms in the CD1d promoter are significant contributors to the impaired CD1d expression and the associated NKT cell deficiency observed in wild-derived inbred PWD/PhJ mice.
Predictive value of CD1d promoter polymorphisms
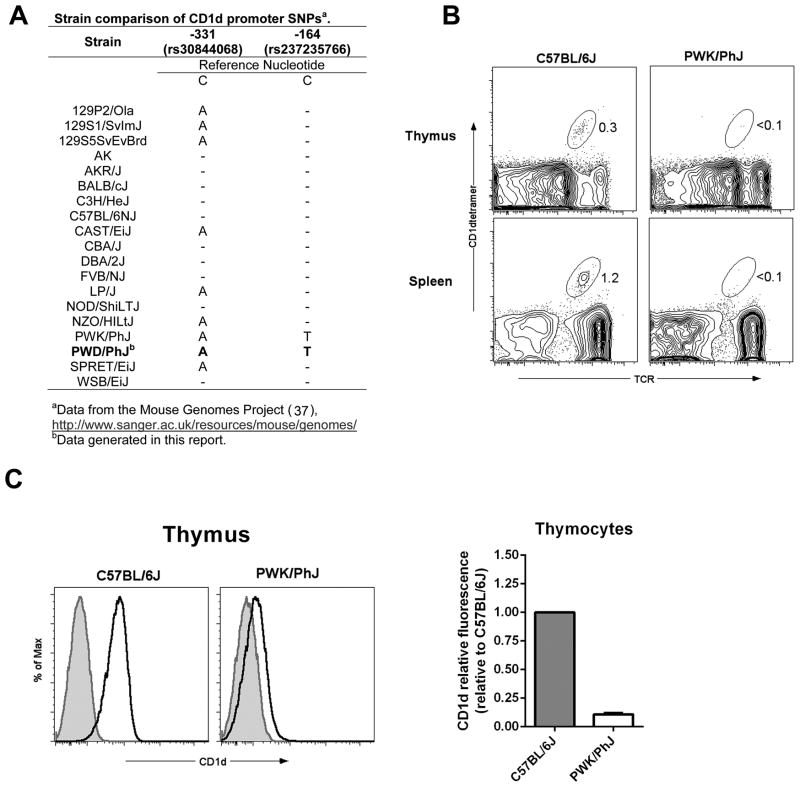
To investigate the prevalence of the -331 and -164 CD1d promoter SNPs, we conducted a database search (Mouse Genomes Project (37)) to identify the strain distribution pattern of these SNPs. This analysis revealed that of the 19 strains analyzed (including PWD/PhJ data from this report), 9 strains possessed the -331(C/A) nucleotide substitution, including CAST/EiJ and SPRET/EiJ (Fig. 7A). Since CAST/EiJ exhibits no impairment in CD1d expression, we conclude that the -331(C/A) polymorphism is not sufficient to result in impaired CD1d gene expression. In contrast, only 2 strains, PWD/PhJ and PWK/PhJ, possessed the -164(C/T) substitution (Fig. 7A), suggesting that the -164(C/T) nucleotide substitution was associated with the low CD1d expression in the PWD/PhJ mice.
Fig. 7. Association of promoter SNPs with impaired CD1d expression and low NKT cell numbers in PWK/PhJ mice.
(A) Strain distribution pattern of the -331 (C/A) and -164 (C/T) nucleotide substitutions in the CD1d1 gene. (B) Comparison of thymic and spleen NKT cell frequencies between B6 and PWK/PhJ mice. Representative FACS contour plots depicting the percentages of NKT cells in the different organs are shown. The NKT cell population is circled, and the percentages are indicated. (C) Comparison of thymocyte CD1d expression between B6 and PWK/PhJ mice. Representative histograms are shown at left. Shaded histograms represent isotype-matched control mAb staining. Open histograms represent anti-CD1d staining. Relative thymocyte CD1d fluorescence in PWK/PhJ relative to C57BL/6J is shown at right. Relative CD1d fluorescence was calculated as specified in Materials and Methods. Data represent the mean relative fluorescence ± s.d., n = 2 – 3 mice per strain.
Therefore, to further explore the linkage between the -164(C/T) substitution in CD1d with low thymocyte CD1d expression and impaired NKT cell development, we examined PWK/PhJ mice. This analysis revealed that, like the other wild-derived inbred strains we analyzed, PWK/PhJ exhibited a striking deficiency in the frequency of NKT cells in the thymus and spleen (Fig. 7B). Examination of CD1d expression in thymus of PWK/PhJ mice revealed a similar impairment in CD4+CD8+ thymocyte CD1d expression (~10% of the level in C57BL/6J) as was seen in PWD/PhJ mice (Fig. 7C). Thus, of the 19 strains examined, the 2 strains of mice that possessed a -164 (C/T) nucleotide substitution in the CD1d gene exhibited striking decreases in thymocyte CD1d expression and NKT cell deficiency.
Discussion
We and others have demonstrated that there is a high level of variability in NKT cell number and function among common inbred mouse strains (13, 15, 16, 24, 29–31, 35). A previous investigation of genetic factors that regulate NKT cell frequencies in common inbred mouse strains identified two genetic loci, NKT1 and NKT2, as major regulators of thymic NKT cell numbers (31). Our investigation of NKT cell frequencies in more genetically disparate wild-derived inbred mouse strains confirmed a previous report that out of 38 inbred strains evaluated, the 4 strains with the fewest NKT cells were the wild-derived inbred strains SPRET/EiJ, CAST/EiJ, PWD/PhJ, and MSM/Ms (18).
In the present study, we focused on the wild-derived PWD strain because of its unusually low CD1d expression and because of the availability of a panel of B6.PWD consomic mice that would allow us to more easily map loci that may regulate NKT cell number and function. Our finding that the predicted amino acid sequence of the PWD CD1d protein differed from B6 at a number of positions was similar to that of a previous examining CD1d in CAST/EiJ and SPRET/Eij mice (35). In that study, it was demonstrated that introgression of the SPRETUS CD1d locus onto the B6 background resulted in a decrease in thymic NKT cell number. However, unlike PWD mice, in which CD1d expression was diminished in both thymus and spleen, B6.SPRETCD1d mice had normal CD1d expression. Rather, it was determined that amino acid changes in the SPRETUS CD1d protein resulted in less efficient NKT cell development (35). Interestingly, although some of the amino acid changes were identical between SPRETUS and PWD CD1d, we found no evidence of impaired presentation of αGalCer to an NKT cell hybridoma. The reason for this discrepancy may simply reflect the unique combination of amino acid changes present in PWD, but not SPRETUS, CD1d. It also remains possible that screening of additional NKT cell hybridomas could reveal a difference in the antigen presentation function of PWD CD1d. Nevertheless, the contribution of such differences would be expected to be relatively small in face of the striking impairment of CD1d protein expression in the PWD thymus reported here.
Although NKT cells fail to develop in mice lacking CD1d (38), NKT numbers have been reported to be comparable in CD1d +/− mice, in which there is a 50% reduction in CD1d expression (39). Although our data suggest that the ~80–90% reduction in CD1d protein expression levels observed in PWD and PWK thymocytes may have a significant impact on NKT cell development, the minimal number of thymocyte CD1d molecules needed to support NKT cell development is not known. Since a previous report found that CD1d protein expression is closely correlated with CD1d transcription (36), it is likely that the decreased CD1d gene expression due to the promoter polymorphisms identified here are sufficient to account for the impaired CD1d protein expression in PWD mice. We note, however, that additional loci likely contribute to the low NKT cell numbers, since B6.PWDchr3 mice possessed significantly higher numbers of NKT cells than wild-type PWD mice. Formal demonstration that the PWD CD1d allele contributes to impaired NKT cell development will require a knock-in of the PWD CD1d allele onto the B6 background. In the absence of these data, our findings demonstrating that impaired CD1d protein expression, CD1d gene expression, and thymic NKT cell numbers all map to chromosome 3 which contains the CD1d gene, support a model in which allele-specific variation in CD1d gene expression is negatively affecting NKT cell selection in the thymus.
Our examination of the PWD CD1d promoter revealed two distinct sites, -331 and -164, that were each independently capable of regulating B6 CD1d promoter activity. Analysis of these sites using transcription factor binding site prediction algorithms did not reveal the presence of viable candidate transcription factors that could explain the decrease in CD1d gene expression. Although the -331 site in the B6 CD1d promoter contained a possible binding site for Runx1, which has been demonstrated to be critical for NKT cell development (40), the deletion of Runx1 does not affect CD1d gene expression (40). Although both sites appeared similar in their ability to drive CD1d expression, examination of the strain distribution pattern of these polymorphisms and of CD1d expression in PWK mice suggests that the -164 site is most closely associated with poor thymocyte CD1d expression. Still, since both PWD and PWK mice possess substitutions at both -331 and -164, it remains possible that both substitutions are required to generate the phenotype in vivo. Interestingly, while structural variants of human CD1d do not exist at any appreciable frequency, CD1d alleles that are defined by nucleotide substitutions in the promoter have been described, and the frequency of these alleles differs significantly among ethnic populations (41). At present, it is unknown whether these substitutions affect the transcriptional activity of human CD1d.
NKT TCR - CD1d interactions exhibit a fair degree of evolutionary conservation (42). The observation, therefore, that wild-derived mouse strains representing different species (Mus spretus; SPRET/EiJ) and subspecies (Mus musculus musculus; PWD/PhJ and PWK/PhJ, and Mus musculus castaneus; CAST/EiJ) share a relative deficiency in the numbers of Type I NKT cells in comparison to standard laboratory strains (Mus musculus domesticus) is perhaps surprising. However, when one considers that i) the percentage of NKT cells in humans is low compared to that of mice, ii) a recent report demonstrating that rats also have very low numbers of Type I NKT cells (43), and iii) the possibility that dogs and ruminants may have very low numbers of Type I NKT cells due to their defective/altered CD1d genes, the possibility that it is the standard laboratory inbred strains of mice (e.g., C57BL/6) that are unusual in having abnormally high numbers of Type I NKT cells needs to be considered. It is important to note, however, that Type I NKT cells represent only a subset of CD1d-restricted T cells. Some of the wild-derived strains, that have few Type I NKT cells and normal expression of CD1d, may possess other CD1d-restricted T cell subsets that are not efficient at recognizing αGalCer in the context of CD1d, but nonetheless serve similar functions.
In this context, it is the observation that thymocyte CD1d expression is so low in PWD and PWK mice that is perhaps the most surprising, since it may be presumed that the development of all CD1d-restricted T cells, not just Type I NKT cells, will be affected in these strains. Although comparison of the CD1d coding sequences among a number of mammals indicates a relatively high level of evolutionary conservation, recent data suggest the CD1d gene is not fully functional in all species. For example, one recent report indicates that the CD1d gene in dogs lacks exons 2 and 3, and is therefore predicted to be non-functional (44). It has also been reported that many ruminants share a mutation that disrupts the normal CD1d start codon (45–47). Although the resulting CD1d protein is expressed, it appears incapable of presenting αGalCer to human NKT cells, although it can present glycolipid ligands with shorter hydrophobic tails (45). Last, CD1d in the African elephant is predicted to lack the cytoplasmic YXXZ motif, which has been demonstrated to be critical for internalization and trafficking of CD1d through the endosomal pathway (46, 48). Importantly, these species are known to also possess Group 1 CD1 orthologues whose function could potentially substitute for that of CD1d. In contrast, rodents are known to possess only Group 2 CD1 genes. One possibility is that PWD and PWK strains possess compensatory increases in other innate-like T cells subsets that don’t rely on CD1d (e.g., γδ T cells). However, further study is needed to address this issue.
Accumulating data have suggested that CD1d-restricted T cells play critical roles in the immune response. Our data indicate that wild-derived inbred strains of mice of different species and subspecies share a striking deficiency in Type I CD1d-restricted T cells, highlighting the variability in this unusual T cell subset. Even among the four wild-derived strains studied here, we could identify a plausible cause for the impaired Type I NKT cell development only in PWD and PWK. The mechanisms underlying the NKT cell deficiency in CAST and SPRETUS are unknown. The identification of nucleotide substitutions in the PWD and PWK CD1d promoters that are associated with restricted CD1d gene expression and with impaired thymocyte CD1d protein expression and NKT cell development suggest that CD1d promoter polymorphisms may be an overlooked contributor to the widespread variability in NKT cell development and function.
Acknowledgments
The authors thank Cory Teuscher for helpful discussion. We thank Collette Charland and Roxana del Rio Guerra at the University of Vermont Flow Cytometry and Cell Sorting Facility for help in FACS sorting, and we thank Pamela Derickson for technical assistance. qPCR and DNA sequencing was performed at the Vermont Cancer Center DNA Analysis Facility. The CD1d tetramer was obtained from the NIH Tetramer Core Facility.
Footnotes
This work was supported by NIH R01AI067897 and P20GM103496 COBRE (J.E.B.) and AI051454 (to M.R.).
References
- 1.Godfrey DI, Berzins SP. Control points in NKT-cell development. Nat Rev Immunol. 2007;7:505–518. doi: 10.1038/nri2116. [DOI] [PubMed] [Google Scholar]
- 2.Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, Koseki H, Taniguchi M. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 3.Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med. 1995;182:2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spada FM, Koezuka Y, Porcelli SA. CD1d-restricted recognition of synthetic glycolipid antigens by human natural killer T cells. J Exp Med. 1998;188:1529–1534. doi: 10.1084/jem.188.8.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshimoto T, Paul WE. CD4pos, NK1.1pos T cells promptly produce interleukin 4 in response to in vivo challenge with anti-CD3. J Exp Med. 1994;179:1285–1295. doi: 10.1084/jem.179.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coquet JM, Chakravarti S, Kyparissoudis K, McNab FW, Pitt LA, McKenzie BS, Berzins SP, Smyth MJ, Godfrey DI. Diverse cytokine production by NKT cell subsets and identification of an IL-17-producing CD4-NK1.1- NKT cell population. Proc Natl Acad Sci U S A. 2008;105:11287–11292. doi: 10.1073/pnas.0801631105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michel ML, Mendes-da-Cruz D, Keller AC, Lochner M, Schneider E, Dy M, Eberl G, Leite-de-Moraes MC. Critical role of ROR-gammat in a new thymic pathway leading to IL-17-producing invariant NKT cell differentiation. Proc Natl Acad Sci U S A. 2008;105:19845–19850. doi: 10.1073/pnas.0806472105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carnaud C, Lee D, Donnars O, Park SH, Beavis A, Koezuka Y, Bendelac A. Cutting edge: Cross-talk between cells of the innate immune system: NKT cells rapidly activate NK cells. J Immunol. 1999;163:4647–4650. [PubMed] [Google Scholar]
- 9.Eberl G, MacDonald HR. Selective induction of NK cell proliferation and cytotoxicity by activated NKT cells. Eur J Immunol. 2000;30:985–992. doi: 10.1002/(SICI)1521-4141(200004)30:4<985::AID-IMMU985>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 10.Kitamura H, Iwakabe K, Yahata T, Nishimura S, Ohta A, Ohmi Y, Sato M, Takeda K, Okumura K, Van Kaer L, Kawano T, Taniguchi M, Nishimura T. The natural killer T (NKT) cell ligand alpha-galactosylceramide demonstrates its immunopotentiating effect by inducing interleukin (IL)- 12 production by dendritic cells and IL-12 receptor expression on NKT cells. J Exp Med. 1999;189:1121–1128. doi: 10.1084/jem.189.7.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitamura H, Ohta A, Sekimoto M, Sato M, Iwakabe K, Nakui M, Yahata T, Meng H, Koda T, Nishimura S, Kawano T, Taniguchi M, Nishimura T. alpha-galactosylceramide induces early B-cell activation through IL-4 production by NKT cells. Cell Immunol. 2000;199:37–42. doi: 10.1006/cimm.1999.1602. [DOI] [PubMed] [Google Scholar]
- 12.Sada-Ovalle I, Chiba A, Gonzales A, Brenner MB, Behar SM. Innate invariant NKT cells recognize Mycobacterium tuberculosis-infected macrophages, produce interferon-gamma, and kill intracellular bacteria. PLoS pathogens. 2008;4:e1000239. doi: 10.1371/journal.ppat.1000239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee PT, Putnam A, Benlagha K, Teyton L, Gottlieb PA, Bendelac A. Testing the NKT cell hypothesis of human IDDM pathogenesis. J Clin Invest. 2002;110:793–800. doi: 10.1172/JCI15832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berzins SP, Kyparissoudis K, Pellicci DG, Hammond KJ, Sidobre S, Baxter A, Smyth MJ, Kronenberg M, Godfrey DI. Systemic NKT cell deficiency in NOD mice is not detected in peripheral blood: implications for human studies. Immunol Cell Biol. 2004;82:247–252. doi: 10.1046/j.1440-1711.2004.01238.x. [DOI] [PubMed] [Google Scholar]
- 15.Rymarchyk SL, Lowenstein H, Mayette J, Foster SR, Damby DE, Howe IW, Aktan I, Meyer RE, Poynter ME, Boyson JE. Widespread natural variation in murine natural killer T-cell number and function. Immunology. 2008;125:331–343. doi: 10.1111/j.1365-2567.2008.02846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyson JE, NN, Nizam L, Exley MA, Strominger JL. Gestation stage-dependent mechanisms of invariant natural killer T cell-mediated pregnancy loss. PNAS. 2006;103:4580–4585. doi: 10.1073/pnas.0511025103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poulton LD, Smyth MJ, Hawke CG, Silveira P, Shepherd D, Naidenko OV, Godfrey DI, Baxter AG. Cytometric and functional analyses of NK and NKT cell deficiencies in NOD mice. Int Immunol. 2001;13:887–896. doi: 10.1093/intimm/13.7.887. [DOI] [PubMed] [Google Scholar]
- 18.Chen YG, Tsaih SW, Serreze DV. Genetic control of murine invariant natural killer T-cell development dynamically differs dependent on the examined tissue type. Genes and immunity. 2012;13:164–174. doi: 10.1038/gene.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levy O, Orange JS, Hibberd P, Steinberg S, LaRussa P, Weinberg A, Wilson SB, Shaulov A, Fleisher G, Geha RS, Bonilla FA, Exley M. Disseminated varicella infection due to the vaccine strain of varicella-zoster virus, in a patient with a novel deficiency in natural killer T cells. J Infect Dis. 2003;188:948–953. doi: 10.1086/378503. [DOI] [PubMed] [Google Scholar]
- 20.Wilson SB, Kent SC, Patton KT, Orban T, Jackson RA, Exley M, Porcelli S, Schatz DA, Atkinson MA, Balk SP, Strominger JL, Hafler DA. Extreme Th1 bias of invariant Valpha24JalphaQ T cells in type 1 diabetes. Nature. 1998;391:177–181. doi: 10.1038/34419. [DOI] [PubMed] [Google Scholar]
- 21.Kent SC, Chen Y, Clemmings SM, Viglietta V, Kenyon NS, Ricordi C, Hering B, Hafler DA. Loss of IL-4 secretion from human type 1a diabetic pancreatic draining lymph node NKT cells. J Immunol. 2005;175:4458–4464. doi: 10.4049/jimmunol.175.7.4458. [DOI] [PubMed] [Google Scholar]
- 22.Demoulins T, Gachelin G, Bequet D, Dormont D. A biased Valpha24+ T-cell repertoire leads to circulating NKT-cell defects in a multiple sclerosis patient at the onset of his disease. Immunol Lett. 2003;90:223–228. doi: 10.1016/j.imlet.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 23.Kojo S, Adachi Y, Keino H, Taniguchi M, Sumida T. Dysfunction of T cell receptor AV24AJ18+, BV11+ double-negative regulatory natural killer T cells in autoimmune diseases. Arthritis Rheum. 2001;44:1127–1138. doi: 10.1002/1529-0131(200105)44:5<1127::AID-ANR194>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 24.Aktan I, Chant A, Borg ZD, Damby DE, Leenstra PC, Lilley GW, Petty J, Suratt BT, Teuscher C, Wakeland EK, Poynter ME, Boyson JE. Slam haplotypes modulate the response to lipopolysaccharide in vivo through control of NKT cell number and function. J Immunol. 2010;185:144–156. doi: 10.4049/jimmunol.0902658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baxter AG, Kinder SJ, Hammond KJ, Scollay R, Godfrey DI. Association between alphabetaTCR+CD4-CD8- T-cell deficiency and IDDM in NOD/Lt mice. Diabetes. 1997;46:572–582. doi: 10.2337/diab.46.4.572. [DOI] [PubMed] [Google Scholar]
- 26.Hansen DS, Siomos MA, Buckingham L, Scalzo AA, Schofield L. Regulation of murine cerebral malaria pathogenesis by CD1d-restricted NKT cells and the natural killer complex. Immunity. 2003;18:391–402. doi: 10.1016/s1074-7613(03)00052-9. [DOI] [PubMed] [Google Scholar]
- 27.Nagarajan NA, Kronenberg M. Invariant NKT cells amplify the innate immune response to lipopolysaccharide. J Immunol. 2007;178:2706–2713. doi: 10.4049/jimmunol.178.5.2706. [DOI] [PubMed] [Google Scholar]
- 28.De Santo C, Salio M, Masri SH, Lee LY, Dong T, Speak AO, Porubsky S, Booth S, Veerapen N, Besra GS, Grone HJ, Platt FM, Zambon M, Cerundolo V. Invariant NKT cells reduce the immunosuppressive activity of influenza A virus-induced myeloid-derived suppressor cells in mice and humans. J Clin Invest. 2008;118:4036–4048. doi: 10.1172/JCI36264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esteban LM, TT, Jordan MA, Roach D, Poulton LD, Brooks A, Naidenko OV, Sidobre S, Godfrey DI, Baxter AG. Genetic control of NKT cell numbers maps to major diabetes and lupus loci. J Immunol. 2003;171:2873–2878. doi: 10.4049/jimmunol.171.6.2873. [DOI] [PubMed] [Google Scholar]
- 30.Fletcher JM, Jordan MA, Snelgrove SL, Slattery RM, Dufour FD, Kyparissoudis K, Besra GS, Godfrey DI, Baxter AG. Congenic analysis of the NKT cell control gene Nkt2 implicates the peroxisomal protein Pxmp4. J Immunol. 2008;181:3400–3412. doi: 10.4049/jimmunol.181.5.3400. [DOI] [PubMed] [Google Scholar]
- 31.Jordan MA, Fletcher JM, Pellicci D, Baxter AG. Slamf1, the NKT cell control gene Nkt1. J Immunol. 2007;178:1618–1627. doi: 10.4049/jimmunol.178.3.1618. [DOI] [PubMed] [Google Scholar]
- 32.Zhang F, Liang Z, Matsuki N, Van Kaer L, Joyce S, Wakeland EK, Aune TM. A murine locus on chromosome 18 controls NKT cell homeostasis and th cell differentiation. J Immunol. 2003;171:4613–4620. doi: 10.4049/jimmunol.171.9.4613. [DOI] [PubMed] [Google Scholar]
- 33.Yang H, Bell TA, Churchill GA, Pardo-Manuel de Villena F. On the subspecific origin of the laboratory mouse. Nat Genet. 2007;39:1100–1107. doi: 10.1038/ng2087. [DOI] [PubMed] [Google Scholar]
- 34.Monzon-Casanova E, Steiniger B, Schweigle S, Clemen H, Zdzieblo D, Starick L, Muller I, Wang CR, Rhost S, Cardell S, Pyz E, Herrmann T. CD1d expression in paneth cells and rat exocrine pancreas revealed by novel monoclonal antibodies which differentially affect NKT cell activation. PloS one. 2010:5. doi: 10.1371/journal.pone.0013089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zimmer MI, Nguyen HP, Wang B, Xu H, Colmone A, Felio K, Choi HJ, Zhou P, Alegre ML, Wang CR. Polymorphisms in CD1d affect antigen presentation and the activation of CD1d-restricted T cells. Proc Natl Acad Sci U S A. 2009;106:1909–1914. doi: 10.1073/pnas.0808476106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geng Y, Laslo P, Barton K, Wang CR. Transcriptional regulation of CD1D1 by Ets family transcription factors. J Immunol. 2005;175:1022–1029. doi: 10.4049/jimmunol.175.2.1022. [DOI] [PubMed] [Google Scholar]
- 37.Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, Furlotte NA, Eskin E, Nellaker C, Whitley H, Cleak J, Janowitz D, Hernandez-Pliego P, Edwards A, Belgard TG, Oliver PL, McIntyre RE, Bhomra A, Nicod J, Gan X, Yuan W, van der Weyden L, Steward CA, Bala S, Stalker J, Mott R, Durbin R, Jackson IJ, Czechanski A, Guerra-Assuncao JA, Donahue LR, Reinholdt LG, Payseur BA, Ponting CP, Birney E, Flint J, Adams DJ. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smiley ST, Kaplan MH, Grusby MJ. Immunoglobulin E production in the absence of interleukin-4-secreting CD1-dependent cells. Science. 1997;275:977–979. doi: 10.1126/science.275.5302.977. [DOI] [PubMed] [Google Scholar]
- 39.Lang GA, Johnson AM, Devera TS, Joshi SK, Lang ML. Reduction of CD1d expression in vivo minimally affects NKT-enhanced antibody production but boosts B-cell memory. Int Immunol. 2011;23:251–260. doi: 10.1093/intimm/dxq477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Egawa T, Eberl G, Taniuchi I, Benlagha K, Geissmann F, Hennighausen L, Bendelac A, Littman DR. Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity. 2005;22:705–716. doi: 10.1016/j.immuni.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 41.Chen QY, Jackson N, Vargas A, Chalew S, Rao J, Batzer M, Lan MS, Chang YH, Mokhashi M, Liu D. Identification of three genomic haplotypes 5′ to the human CD1D gene and their distribution in four ethnic groups. Tissue Antigens. 2003;62:442–448. doi: 10.1034/j.1399-0039.2003.00116.x. [DOI] [PubMed] [Google Scholar]
- 42.Brossay L, Chioda M, Burdin N, Koezuka Y, Casorati G, Dellabona P, Kronenberg M. CD1d-mediated recognition of an alpha-galactosylceramide by natural killer T cells is highly conserved through mammalian evolution. J Exp Med. 1998;188:1521–1528. doi: 10.1084/jem.188.8.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monzon-Casanova E, Paletta D, Starick L, Muller I, Sant’Angelo DB, Pyz E, Herrmann T. Direct identification of rat iNKT cells reveals remarkable similarities to human iNKT cells and a profound deficiency in LEW rats. Eur J Immunol. 2013;43:404–415. doi: 10.1002/eji.201242565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Looringh van Beeck FA, Leegwater PA, Herrmann T, Broere F, Rutten VP, Willemse T, Van Rhijn I. Tandem repeats modify the structure of the canine CD1D gene. Anim Genet. 2013;44:352–355. doi: 10.1111/age.12002. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen TK, Koets AP, Vordermeier M, Jervis PJ, Cox LR, Graham SP, Santema WJ, Moody DB, van Calenbergh S, Zajonc DM, Besra GS, Van Rhijn I. The bovine CD1D gene has an unusual gene structure and is expressed but cannot present alpha-galactosylceramide with a C26 fatty acid. Int Immunol. 2013;25:91–98. doi: 10.1093/intimm/dxs092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Looringh van Beeck FA, Reinink P, Hermsen R, Zajonc DM, Laven MJ, Fun A, Troskie M, Schoemaker NJ, Morar D, Lenstra JA, Vervelde L, Rutten VP, van Eden W, Van Rhijn I. Functional CD1d and/or NKT cell invariant chain transcript in horse, pig, African elephant and guinea pig, but not in ruminants. Mol Immunol. 2009;46:1424–1431. doi: 10.1016/j.molimm.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Rhijn I, Koets AP, Im JS, Piebes D, Reddington F, Besra GS, Porcelli SA, van Eden W, Rutten VP. The bovine CD1 family contains group 1 CD1 proteins, but no functional CD1d. J Immunol. 2006;176:4888–4893. doi: 10.4049/jimmunol.176.8.4888. [DOI] [PubMed] [Google Scholar]
- 48.Chiu YH, Jayawardena J, Weiss A, Lee D, Park SH, Dautry-Varsat A, Bendelac A. Distinct subsets of CD1d-restricted T cells recognize self-antigens loaded in different cellular compartments. J Exp Med. 1999;189:103–110. doi: 10.1084/jem.189.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]