Abstract
The process of cancer progression involves the action of multiple proteolytic systems, among which the family of matrix metalloproteinases (MMPs) play a pivotal role. The MMPs evolved to accomplish their proteolytic tasks in multiple cellular and tissue microenvironments including lipid rafts by incorporation and deletions of specific structural domains. The membrane type-MMPs (MT-MMPs) incorporated membrane anchoring domains that display these proteases at the cell surface, and thus they are optimal pericellular proteolytic machines. Two members of the MT-MMP subfamily, MMP-17 (MT4-MMP) and MMP-25 (MT6-MMP), are anchored to the plasma membrane via a glycosyl-phosphatidyl inositol (GPI) anchor, which confers these enzymes a unique set of regulatory and functional mechanisms that separates them from the rest of the MMP family. Discovered almost a decade ago, the body of work on GPI-MT-MMPs today is still surprisingly limited when compared to other MT-MMPs. However, new evidence shows that the GPI-MT-MMPs are highly expressed in human cancer, where they are associated with progression. Accumulating biochemical and functional evidence also highlights their distinct properties. In this review, we summarize the structural, biochemical, and biological properties of GPI-MT-MMPs and present an overview of their expression and role in cancer. We further discuss the potential implications of GPI-anchoring for enzyme function. Finally, we comment on the new scientific challenges that lie ahead to better understand the function and role in cancer of these intriguing but yet unique MMPs.
Keywords: Matrix metalloproteinases, Glycosyl-phosphatidyl inositol, Membrane anchor, Proteolysis, Cancer, Lipid raft, Protease inhibitors, Plasma membrane, Extracellular matrix
1 Introduction
Modifications and alterations of the immediate microenvironment are essential for cell survival in both physiological and pathological processes. Cancer cells, like normal cells, strongly depend on their ability to influence the structure and organization of the pericellular milieu so that they can display their malignant phenotype during cancer progression and fulfill their intended biological behavior. An important characteristic of malignant cells is their ability to detach from the primary tumor and to acquire migratory and invasive properties. To accomplish these goals, cancer cells hijack various proteolytic systems, which serve to modify the pericellular milieu. The MMP family of zinc-dependent endopeptidases is one of the major protease family involved in cancer progression; and their role in regulating tumor growth, metastasis and angiogenesis has been well established [1–6]. MMPs’ role in cancer progression, however, is complex, involving both stimulatory and suppressive effects by MMPs produced by both the cancer cells and the tumor stroma [5, 7–9]. The complexity of MMPs’ biological function in cancer is compounded by their multidomain structure, diverse substrate repertoire, differential tissue expression and regulation, interacting proteins and co-factors and their inhibitor profile, just to mention a few. Furthermore, analyses of the MMP domain structure reveal that these proteases also evolved to target substrates at multiple cellular and extracellular sites. This was achieved by the deletion or addition of accessory enzyme domains, whose main function is to direct the active protease to diverse cellular and tissue microenvironments in pursuit of their favorite substrate(s). Thus, the MMP family evolved to include secreted and membrane-anchored proteases, which as a whole can process multiple substrates at many locations.
The membrane-anchored MMPs (referred to as MT-MMPs), containing an anchoring motif at the C-terminal end, are perfectly suited for pericellular proteolysis [10]. Membrane anchoring facilitates these MMPs to reach key membrane and peripheral proteins as well as closely associated extracellular matrix (ECM) components thereby playing a pivotal role during alterations of the pericellular milieu in both physiological and pathological conditions [11–13]. The anchoring domains also facilitate diffusion and translocation of the active protease on the plasma membrane to specific membrane microdomains and its recruitment to the migrating front of motile cells [14]. Finally, membrane insertion confers MT-MMPs with a unique set of regulatory mechanisms that serve to control the pool of active protease at the cell surface including endocytosis, recycling, autocatalytic processing, and ectodomain shedding, [15]. Further specialization and refinement of the MT-MMPs was achieved by the presence of two distinct anchoring systems: a transmembrane domain and a glycosyl-phosphatidyl inositol (GPI)-anchor. Thus, the MT-MMPs are divided in transmembrane MT-MMP (referred here to as TM-MT-MMPs) and GPI-anchored MT-MMPs (referred to as GPI-MT-MMPs). Since the discovery of the first TM-MT-MMP, MT1-MMP (MMP-14), more than 13 years ago [16, 17], a wealth of evidence has clearly shown that MT1-MMP as well as other TM-MT-MMPs are key players in cancer progression by promoting cell growth, migration, invasion, metastasis, and angiogenesis [10, 12, 14, 18]. A wealth of information also exists on the regulation, inhibition, and activity of TM-MT-MMPs. In contrast, the function and role of GPI-MT-MMPs in cancer remains largely elusive in spite of their identification in cancer tissues and their potential to impact cancer progression. Although there are only a limited number of studies, recent evidence suggest a unique role for GPI-MT-MMPs in cancer. Moreover, their unique structural organization has revealed distinct properties demanding new concepts in MMP regulation, substrate specificity, and inhibition. The purpose of this review is to provide a first overview of the properties, function, and regulation of the two GPI-MT-MMPs: MT4- and MT6-MMP. Although GPI-MT-MMPs have been implicated in several pathologies, our focus here is on their expression and role in cancer. We hope that this review will increase the awareness of these interesting and unique MMPs and open new avenues of research with potential clinical implications.
2 General properties of GPI-MT-MMPs
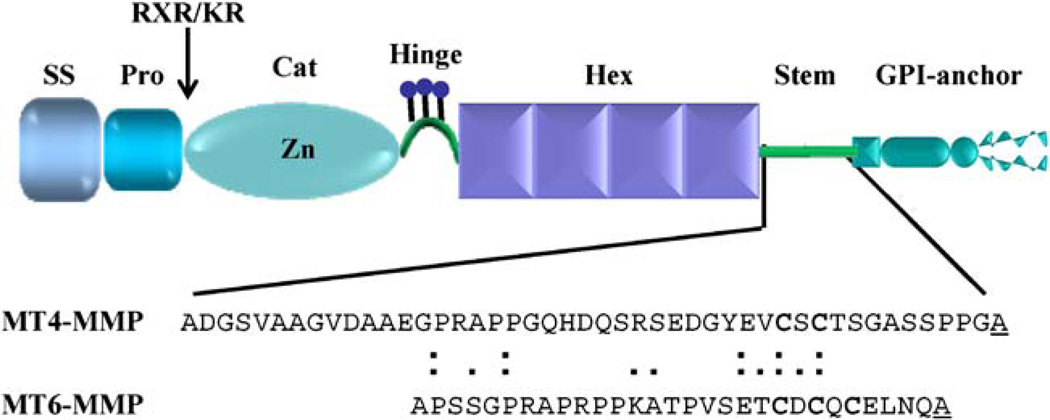
The MT-MMPs constitute a distinct set of proteases among the members of the MMP family in that they are anchored to the cell surface with their catalytic site exposed to the extracellular space. As such, the MT-MMPs are well positioned to attack many substrates located in the plasma membrane and in the immediate pericellular space [10]. The MT-MMPs subfamily is comprised of six members, four of which are tethered to the plasma membrane by a hydrophobic transmembrane domain of ~20 amino acids followed by a cytoplasmic tail, and two proteases that are tethered to the membrane via a GPI anchor. The transmembrane MT-MMPs are MT1-MMP (MMP-14) [16, 17], MT2-MMP (MMP-15) [19], MT3-MMP (MMP-16) [20] and MT5-MMP (MMP-24) [21, 22]. The GPI-MT-MMPs include MT4-MMP (MMP-17) [23, 24] and MT6-MMP (MMP-25) [25, 26]. With the exception of the anchoring region, the MT-MMPs share the same basic domain organization of most MMPs, including a signal sequence, a propeptide domain, a zinc-containing catalytic domain, a hinge region (linker 1), and a hemopexin-like domain. Both the transmembrane and the GPI-MT-MMPs also contain a RXR/KR motif at the end of the propeptide domain that serves as a recognition site for pro-convertases such as furin, which cleave the propeptide and activate the MT-MMP zymogen (Fig. 1). There are excellent reviews on the structure, function and regulation of transmembrane MT-MMPs, albeit mostly focused on MT1-MMP [10, 14, 27]. Here we will focus on the unique features of the GPI-MT-MMPs.
Fig. 1.
Structure of mature GPI-anchored MT-MMPs. SS signal-sequence, Pro prodomain, RXR/KR furin recognition motif, Cat catalytic domain, Hex hemopexin-like domain. The amino acid sequence of MT4- and MT6-MMP stem region (linker 2) is shown in detail. Cysteine residues in the stem region are indicated in bold. The homology between the stem regions of MT4- and MT6-MMP is indicated by black squares. The putative transamidase cleavage site, ω, is underlined
2.1 Identification and unique structural characteristics
MT4-MMP was first cloned from a human breast carcinoma cDNA library [24], while MT6-MMP was identified from peripheral blood leukocytes [25]. Due to the high expression of MT6-MMP in leukocytes, this MMP was also termed leukolysin [25]. Primary sequence alignment of the catalytic domain of MT4- and MT6-MMP shows they are 56% identical and 77% homologous. When compared to the MT1-MMP catalytic domain, MT4-MMP possesses 37% identity (50% similarity) while MT6-MMP possesses 42% identity (58% similarity). This suggests that GPI-MT-MMPs are structurally and functionally distant to TM-MT-MMPs. The hemopexin-like domain of the GPI-MT-MMPs is followed by a hydrophilic region (~35–45 amino acids) also called stem (linker 2) region and a hydrophobic tail of ~20–23 amino acids. The C-terminal hydrophobic tail of the GPI-MT-MMPs is removed a transamidase in the endoplasmic reticulum (ER) during incorporation of the GPI anchor, a process that is a unique characteristic of GPI-anchored proteins [28]. Biochemical studies using incorporation of [3H] ethanolamine and release by phosphatidylinositol specific phospholipase C (PI-PLC) revealed that both MT4- and MT6-MMP are indeed GPI-anchored proteins [29–31].
The stem region of MT4- and MT6-MMP (residues A528-A573 in MT4-MMP and residues A509-A539 in MT6-MMP) exhibits unique features when compared to the stem of transmembrane MT-MMPs. The stem of MT4- and MT6-MMPs contains two and three cysteine residues, respectively, of unknown function (Fig. 1). Since cysteine residues are known sites for a variety of post-translational modifications including disulfide bond formation, palmitoylation, or prenylation, these cysteine residues may play an important role in regulation of protease function. For instance, cysteine residues may play a role in formation of homophilic interactions. We have recently shown that MT6-MMP [32] is found in reduction-sensitive high-molecular weight forms, which may represent dimeric and/or oligomeric forms of the protease. In the case of MT6-MMP, the protease is found at the cell surface as a major ~120-kDa species and minor ~180-kDa form that can be released from the membrane by PI-PLC [32]. The ~120 kDa form is also detected with natural MT6-MMP in extracts of human neutrophils [32]. Under reducing conditions, MT6-MMP is detected as a 57-kDa form, strongly suggesting that the ~120-kDa species represents a dimeric form of the enzyme, possibly held by a disulfide bond formed by cysteine residues at the stem region. Preliminary site-directed mutagenesis studies from our laboratory suggest that Cys532 in the stem region plays a key role in MT6-MMP homodimerization (A. Sohail, H. Zhao, Q. Sun and R. Fridman, manuscript in preparation). Furthermore, this Cys mutant exhibits enhanced enzyme degradation suggesting that homodimerization is important for protease stability. In TM-MT-MMPs, homophilic interactions have been reported with MT1-MMP, a membrane-tethered collagenase [11] and a major activator of pro-MMP-2 [16, 17]. MT1-MMP homodimerization has been suggested to play a role in collagenolysis and pro-MMP-2 activation [14]. However, the nature of the interactions and the domains involved in MT1-MMP dimeric/oligomeric forms remain unclear as both non-covalent [33, 34] and covalent [35, 36] interactions have been reported. Furthermore, as opposed to MT6-MMP, the high molecular weight forms of MT1-MMP are not readily detected by SDS-PAGE, suggesting a significant structural difference between transmembrane and GPI-anchored MMPs in regards to this unique supramolecular organization. The importance of MT6-MMP homophilic interactions for enzyme function remains to be elucidated, but it is tempting to speculate that these interactions may regulate diverse aspect of MT6-MMP including subcellular distribution, stability, endocytosis, catalysis, TIMP inhibition, and substrate specificity, just to mention a few. Recent studies have shown that oligomerization plays a key role in determining apical sorting and stabilization into rafts of GPI-anchored proteins [37, 38]. Consistent with these findings, MT6-MMP oligomers are also targeted to the lipid rafts [32]. However, the basal vs. apical sorting of MT6-MMP and its relevance for enzyme function remain to be determined.
2.2 Biosynthesis, trafficking, cellular distribution, and endocytosis
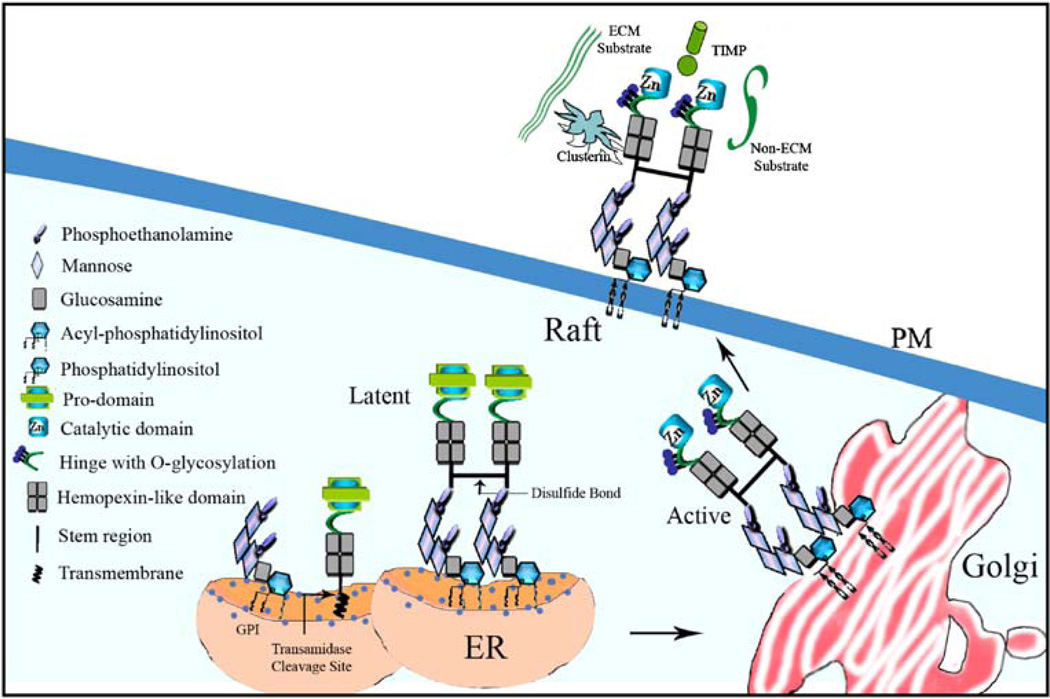
The detailed biosynthetic pathway of GPI-MT-MMPs has not yet been investigated. However, MT4- and MT6-MMP are likely to follow the biosynthetic pathway of typical GPI-anchored proteins. For a detailed review on the synthesis of GPI-anchored proteins see [28]. It is well established that GPI-anchored proteins are initially synthesized as transmembrane protein precursors. The hydrophobic tail of the precursor is cleaved by a transamidase during translation, and the protein is then linked to a pre-existing GPI-anchor on the ER membrane [28, 39] (Fig. 2). Currently, the transamidase cleavage site in MT4- and MT6-MMP is unknown. However, the C-terminal region of MT4- and MT6-MMP contain a putative transamidase cleavage site, referred to as ω, at Ala573 in MT4-MMP and Ala539 in MT6-MMP, as predicted by big-PI Predictor software [40–42]. In a typical GPI anchor, phosphatidylinositol is glycosidically linked to an inositol ring of a non-acetylated glucosamine moiety that has three mannose residues attached to it. The terminal mannose is linked to phosphoethanolamine by a phosphodiester bond. The GPI-anchor is attached to the carboxy-terminal of the protein by an amide linkage to the amino group of phosphoethanolamine [43]. Variations in the GPI structure include the attachment of galactosyl side chains and additional phosphoethanolamine or mannosyl moieties to the first mannose, which depend both on the organism and cell type. Immediately after transfer to the protein, the GPI moiety goes through a deacylation process [44] followed by the replacement of unsaturated fatty chain in phosphatidylinositol with two saturated fatty acids chains. The fatty acid remodeling, an essential step for integration of GPI-anchored proteins into lipid rafts, most likely occurs in the Trans Golgi network [45]. In the Trans Golgi, GPI proteins also undergo post-translational modifications such as glycosylation. Our preliminary studies using an O-glycosylation inhibitor suggest that MT6-MMP is O-glycosylated (Q. Sun and R. Fridman, unpublished data).
Fig. 2.
Hypothetical biosynthetic pathway of MT6-MMP. Pro-MT6-MMP is synthesized as a transmembrane precursor protein. This precursor protein is cleaved by a transamidase at the ω site releasing the carboxyl-terminal transmembrane domain and incorporating a preexisting GPI anchor in the endoplasmic reticulum (ER). Also in the ER, MT6-MMP undergoes homodimerization by formation of disulfide bonds between cysteine residues at the stem region (our unpublished data). At the Golgi network, MT6-MMP is activated by a furin-like pro-convertase and O-glycosylated (our unpublished data). In addition, unsaturated fatty chains in phosphatidylinositol are replaced with saturated fatty acids chains and MT6-MMP associates with lipid raft [32]. At the cell surface, MT6-MMP interacts with TIMPs and/or clusterin for regulation of enzymatic activity. Both ECM and non-ECM substrate are hydrolyzed by MT6-MMP at the cell surface. MT4-MMP biosynthetic pathway is likely to be similar to that of MT6-MMP
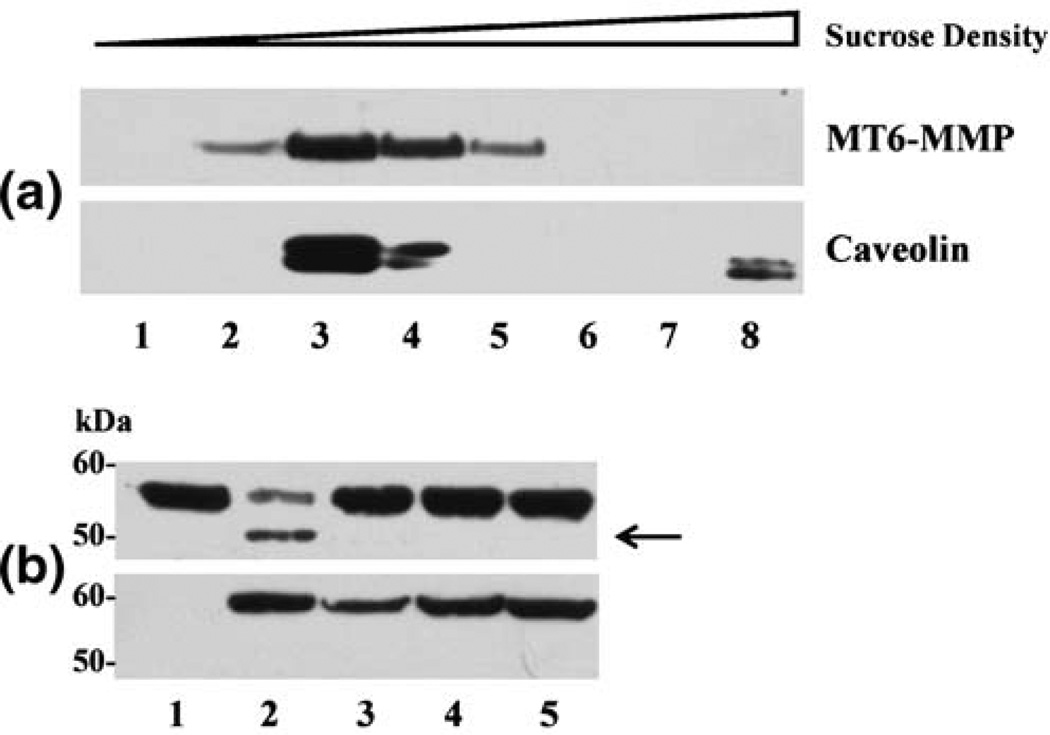
The GPI-anchor confers the protein that is attached to it with important biological functions including localization to lipid raft microdomains, sensitivity to phospholipases, apical sorting, and a role in signal transduction. We have recently shown that successive detergent fractionation of cells expressing MT6-MMP results in the distribution of MT6-MMP into the Triton X-100-insoluble fractions, which are considered to be lipid rafts [32]. This distribution was further confirmed here by sucrose density gradient centrifugation as depicted in Fig. 3(a). These data show that MT6-MMP is enriched in light buoyant density fractions that also contain the raft-associated protein Caveolin-1. These results are consistent with a previous proteomic study that identified MT6-MMP as a major component of the lipid rafts in human neutrophils [46]. Lipid rafts have received considerable attention because they are thought to be involved in many biological functions, in particular, signal transduction [47], endocytosis [48], and apical sorting of GPI-anchored proteins [49]. The presence of MT6-MMP, and likely MT4-MMP, in lipid rafts may confer GPI-MT-MMPs with the unique ability to interact with and degrade specific components of the raft microenvironment. This fundamental cellular distribution needs to be considered in future degradomic studies aimed at defining the substrate profile of GPI-MT-MMPs by proteomic approaches [50].
Fig. 3.
Lipid raft localization and activity of MT6-MMP. (a) Colon cancer HCT-116 cells expressing human recombinant MT6-MMP [32] were homogenized in 1% Triton X-100 containing Protease Inhibitor (Roche) mix and the homogenate was adjusted to 35% sucrose. The homogenate was placed at the bottom of a centrifuge tube and overlaid with two 4 ml layers of 30 and 5% sucrose. The samples were centrifuged at 100,000×g for 18 h at 4°C. Eight fractions (1 ml each) were collected sequentially from the top of the gradient and equal volume of each fractions were resolved by reducing 10% SDS/PAGE followed by immunoblot analyses using anti-human MT6-MMP monoclonal antibodies and a polyclonal antibody to human Caveolin, as a lipid raft marker. MT6-MMP, 57 kDa; Caveolin, 22 kDa. (b) Crude plasma membranes were isolated from HCT-116 cells transfected with empty vector (lane 1) or with human recombinant MT6-MMP (lanes 2– 4) or with a catalytically inactive E/A MT6-MMP mutant (lane 5) were incubated (18 h, 37°C in a 50 µl reaction volume with Proteinase Inhibitor cocktail without EDTA) with α1-PI in the absence (lanes 1, 2 and 5) or presence of either 200 nM recombinant human TIMP-1 (lane 3) or recombinant human TIMP-2 (lane 4). Equal amounts from each sample were mixed with Laemmli SDS-sample buffer and resolved by reducing 10% SDS–PAGE followed by immunoblot analysis with antibodies against human α1-PI (upper panel) and human MT6-MMP (lower panel) Arrow in (b) indicates the cleaved species of α1-PI
GPI-anchored proteins also undergo raft-mediated endocytosis, which serves to recycle these proteins to the plasma membrane or to target the proteins to lysosomes for degradation [48, 51]. Although the pathway involved in the endocytosis of GPI-MT-MMPs needs to be elucidated, initial results from our laboratory indicate that MT6-MMP is endocytosed and recycled back to cell surface in MT6-MMP transfected colon cancer cells (J.-A. Cho and R. Fridman, unpublished results). These studies suggest that GPI-MT-MMP activity at the cell surface is also regulated by endocytosis and recycling, as reported for MT1-MMP [14, 52].
2.3 Inhibition of GPI-MT-MMPs
The members of the MMP family are specifically inhibited by tissue inhibitors of metalloproteinases (TIMPs), a family of four proteins (TIMP-1, −2, −3, and −4) that bind to the catalytic domain of the active protease terminating catalysis. For comprehensive reviews on the structure and function of TIMPs, the readers are directed to: [53–57]. Like all MMPs, the enzymatic activity of MT-MMPs is also inhibited by TIMPs. However, structural and functional studies revealed that MT-MMPs exhibit unique interactions with TIMPs. The TM-MT-MMPs are highly sensitive to inhibition by TIMP-2, TIMP-3, and TIMP-4, which behave as high-affinity, slow-binding, reversible inhibitors of these proteases. Interestingly, TIMP-1 is a very weak inhibitor of TM-MT-MMPs, and thus under physiological conditions TM-MT-MMPs can be regarded as resistant to TIMP-1. The presence of a threonine residue at position 98 has been found to be responsible for the lack of activity of TIMP-1 against TM-MT-MMPs [58]. When the catalytic domains of GPI-MT-MMPs were examined for TIMP selectivity, it was found that both MT4- [59–61] and MT6-MMP [32, 62, 63] were efficiently inhibited by TIMP-1, TIMP-2, and TIMP-3. Thus, GPI- and TM-MT-MMPs exhibit a different TIMP inhibition profile. We showed that TIMP-1 is a more effective inhibitor of MT6-MMP than TIMP-2 (Ki values of 0.2 and 2 nM, respectively). Moreover, MT6-MMP is less sensitive to TIMP-2 inhibition than MT1- and MT3-MMP, mainly because of a slower association rate (an order of magnitude) of TIMP-2 to MT6-MMP [32]. In this regard, when we compared the association of exogenously administered TIMP-2 to cells expressing either MT6-MMP or MT1-MMP, we found that TIMP-2 could be readily detected only in MT1-MMP-expressing cells, consistent with the slow association of TIMP-2 to MT6-MMP. Interestingly, we also found that TIMP-1 failed to associate with MT6-MMP expressing cells [32]. PI-PLC-released MT6-MMP also did not form a co-precipitable complex with TIMPs. The presence of a GPI-anchor, and consequently localization in lipid rafts, was not the reason for the lack of TIMP association with MT6-MMP because a GPI-anchored MT1-MMP chimera readily bound TIMP-2 [32]. These data revealed a significant difference between MT1-MMP and MT6-MMP in their ability to form stable complexes with TIMPs when displayed at the cell surface. Therefore, we examined whether MT6-MMP activity is inhibited by TIMPs when the enzyme is present on the plasma membrane. As shown in Fig. 3(b), MT6-MMP dependent cleavage of α1-proteinase inhibitor (α1-PI) is inhibited by both TIMP-1 and TIMP-2, demonstrating the sensitivity of membrane-anchored MT6-MMP to TIMPs (Fig. 3(b)). Thus, although MT6-MMP/TIMPs complexes on the cell surface are not readily detected [32], MT6-MMP activity is sensitive to TIMP inhibition. Future studies will need to address in more detail the interactions between GPI-MT-MMPs and TIMPs in the lipid raft microenvironment.
In addition to TIMPs, a previous study showed that GPI-MT-MMPs can be inhibited by clusterin [63]. Clusterin (also known as apolipoprotein J) is a multifunctional and ubiquitously expressed protein that is involved in many physiological and pathological processes including cancer progression. Two isoforms of clusterin exist: a secreted 75– 80-kDa heterodimer present in many physiological fluids, and a truncated cytosolic/nuclear form of ~49 kDa. Clusterin plays a role in regulation of apoptosis, DNA repair, and cellular responses to stress conditions, just to mention a few [64]. The association of clusterin with MT6-MMP was identified when a recombinant soluble form of MT6-MMP, comprised of the catalytic domain and the hemopexin-like domain, co-purified with clusterin as a stable complex from the conditioned media [63]. These studies showed that clusterin binds to the hemopexin-like domain of MT6-MMP, resulting in inhibition of enzymatic activity. Clusterin inhibition of MT6-MMP activity was significantly weaker than that observed with TIMP-1 and TIMP-2 and was specific for MT6-MMP, since neither MMP-2 nor MT1-MMP were inhibited by or formed a complex with clusterin. However, a soluble MT4-MMP also formed a complex with clusterin, suggesting that this interaction is unique for the GPI-MT-MMPs. The MT6-MMP/clusterin complex was also identified with a membrane-anchored recombinant MT6-MMP and with natural MT6-MMP in human neutrophils [63]. In contrast to these findings, the studies of Sun et al. [32] showed no interactions of MT6-MMP with clusterin in colon cancer cells expressing recombinant MT6-MMP in spite of endogenous clusterin expression in these cells. These findings indicated that the high-molecular weight form of MT6-MMP (~120 kDa) detected in these cells is not a complex of the protease with clusterin but is consistent with homophilic MT6-MMP interactions [32]. At present, more studies are required to define the relevance of the interactions of GPI-MT-MMPs with clusterin and their significance in the regulation of their enzymatic activity.
2.4 Shedding of GPI-MT-MMPs
It is well established that GPI-anchored proteins can be released from the cells in membrane vesicles, referred to as exosomes [65, 66], resulting in the release of the protein with an intact GPI. Interestingly, the released GPI protein can be transferred to other cell types in a paracrine manner where it elicits biological effects [65, 67]. Previous studies have shown that both MT4- [29, 68] and MT6-MMP [30 and A. Sohail and R. Fridman, unpublished results] are shed in different cell types. Although the precise shedding mechanism of GPI-MT-MMPs remains to be elucidated, data with MT4- and MT6-MMP indicate that the molecular mass of the soluble enzyme is very similar to their membrane-anchored counterpart [29, 30], suggesting the possibility that they maintain the GPI anchor. Thus, it is tempting to speculate that GPI-MT-MMPs may also be shed via exosomes, as reported with other GPI-anchored proteins [69]. However, cleavage of the GPI anchor by a phospholipase-mediated cleavage cannot be ruled out. Indeed, shedding of MT4- and MT6-MMP was insensitive to various metalloproteinase inhibitors [29, 30]. Thus, both proteolytic and non-proteolytic shedding mechanisms may regulate the function and localization of GPI-MT-MMPs.
2.5 GPI-MT-MMP substrates
2.5.1 ECM proteins
A few studies reported the catalytic activities of GPI-MT-MMPs (Table 1). The emerging view from these studies is complex, and so far there is no clear consensus on the precise substrate repertoire of GPI-MT-MMPs (Table 1). It should be noted that most of the studies designed to define the substrate specificity of GPI-MT-MMPs utilized isolated recombinant catalytic domains expressed in bacteria [30, 59, 60, 62, 70], which cannot the address the influence of membrane insertion and other key enzyme domains on catalytic activity. In other studies, substrate identification was established by co-expression of the enzyme with a potential substrate in mammalian cells [59, 70, 71]. Nevertheless, the data so far suggest that GPI-MT-MMPs are multifunctional enzymes capable of cleaving ECM and non-ECM proteins. However, there are significant differences between MT4- and MT6-MMP in their substrate preferences. Whereas MT4-MMP exhibits minimal or no activity against many ECM proteins, MT6-MMP hydrolyzes a variety of ECM components. Specifically, soluble MT4-MMP was found to be active only against gelatin, fibrin, and fibrinogen [59–61]. However, expression of MT4-MMP failed to confer cells with the ability to invade a fibrin gel in vitro [72]. MT6-MMP, on the other hand, exhibits activity against gelatin, collagen IV, fibronectin and fibrin [30, 62]. In addition, MT6-MMP was shown to hydrolyze chondroitin and dermatan sulfate proteoglycans but showed no activity against laminin and collagen type I, II, and III [30]. The limited ECM degrading activity of the GPI-MT-MMPs is in accordance with their reported inability to support the in vitro invasion of cells through either Matrigel coated filters [32, 68] or three-dimensional fibrin gels [72]. In addition, neither MT4-MMP nor MT6-MMP played a role in invasion of basement membranes in vitro [12]. Although GPI-MT-MMPs are expressed in cancer cells [26, 32, 68, 73], these data highlight a significant functional difference among MT-MMPs in cancer cell behavior.
Table 1.
Substrates, inhibitors and expression in cancer tissues of GPI-anchored MT-MMPs
| MT4-MMP | MT6-MMP | ||
|---|---|---|---|
| Substrates | Gelatin | Yes [59, 61] | Yes [62] |
| Fibrinogen | Yes [59] | Yes [62] | |
| Fibrin | Yes [59] | Yes [62] | |
| Fibronectin | No [59] | Yes [30, 62] | |
| Collagen IV | No [59] | Yes [62] | |
| pro-TNFα | Ye s [59] | ND | |
| α2-macroglobulin | Yes [59] | No [101] | |
| Progelatinase-A | Yes [61, 91] | Yes [26, 71, 91] | |
| No [59, 60, 68] | No [31, 32] | ||
| Progelatinase-B | No [59] | No [32, 62] | |
| ADAMTS-4 (aggrecanase-1) | Yes [93, 94] | ND | |
| Low density lipoprotein receptor related protein | Yes [75] | ND | |
| Chondroitin sulfate proteoglycan | ND | Yes [30] | |
| Dermatan sulfate proteoglycan | ND | Yes [30] | |
| α1 proteinase inhibitor | ND | Yes [30, 70] | |
| Urokinase plasminogen activator receptor | ND | Yes [76] | |
| Galectin-3 | ND | Yes (our unpublished results) | |
| Inhibitors | TIMP-1 | Yes [59–61, 93, 102] | Yes [32, 62, 63] |
| TIMP-2 | Yes [59–61] | Yes [32, 62, 63] | |
| TIMP-3 | Yes [59, 102] | Yes [62, our unpublished results] | |
| TIMP-4 | ND | Yes (our unpublished results) | |
| Clusterin | Yes [63] | Yes [63] | |
| Ye s [32]a | |||
| Expression in cancer tissues | Anplastic astrocytomas, glioblastomas | Nob [97] | Yes [26, 97] |
| Breast carcinoma | Yes [24, 68] | ||
| Colon cancer | Yes [26, 32] | ||
| Urothelial cancer | Yes [98] | ||
| Prostate cancer | Yes [99] |
ND not determined
No complex formation detected in HCT-116 colon cancer cells transfected with human MT6-MMP
Expression decreased compared to normal brain
2.5.2 Non-ECM proteins
GPI-MT-MMPs have been shown to cleave several non-ECM proteins. For instance, MT4-MMP was found to possess ADAM (a disintegrin and metalloprotease)-17-like activity in that it can act as a sheddase of tumor necrosis factor (TNF)-α when co-transfected with pro-TNF-α in Cos-7 cells [59]. However, macrophages isolated from wild type or MT4-MMP null mice exhibited a similar extent of TNF-α in the medium when stimulated with lipopolysaccharide [74]. Thus, at least in macrophages, MT4-MMP does not appear to be a major TNF-α sheddase. The MT4-MMP catalytic domain also cleaves α2-macroglobulin [59], suggesting that this cleavage may play a role in the control of proteolytic activity because α2-macroglobulin is a major protease inhibitor. The catalytic domain of MT4-MMP also cleaves the low density lipoprotein receptor related protein (LPR) [75], which is known to play a key role in the clearance of multiple ligands at the cell surface. The cleavage of LRP is not exclusive to MT4-MMP and is also mediated by MT1-, MT2-, and MT3-MMP [75]. MT6-MMP can also cleave α1-PI, a potent inhibitor of serine proteases [70 and Fig. 3(b)]. α1-PI cleavage by MT6-MMP results in loss of its inhibitory activity, suggesting that expression of MT6-MMP in leukocytes may aid the inflammatory response by facilitating the action of serine proteases [70]. We also found that crude plasma membranes of colon cancer cells transfected with human MT6-MMP can process α1-PI in a pattern similar to that reported by Nie and Pei [70], as shown in Fig. 3(b).
MT6-MMP, like other MMPs, also releases the D1 domain of urokinase plasminogen activator receptor (uPAR) by cleaving at the linker region connecting domains 1 and 2 of uPAR [76]. Our laboratory has also found that MT6-MMP can cleave galectin-3 (A. Sohail, Q. Sun and R. Fridman, unpublished data), a 31-kDa carbohydrate-binding protein known to play a key role in inflammation, apoptosis, and cancer progression [77]. Cleavage of galectin-3 by MT6-MMP is similar to that reported with gelatinases and MT1-MMP, which generate a 21-kDa degradation product [78–80].
2.5.3 Zymogens as substrates of GPI-MT-MMPs: Role of MT4- and MT6-MMP in pro-MMP-2 and pro-MMP-9 activation
It is well established that many proteolytic enzymes are produced in a latent zymogenic form devoid of enzymatic activity. To achieve catalytic competence, the zymogen needs to be activated in a process that usually involves a specific cleavage of a portion of the enzyme, which allows the exposure of the catalytic domain [81]. MMPs, including MT-MMPs, are known to be involved in the activation of zymogens, in particular activation of pro-MMPs. In this regard, the discovery of MT1-MMP was the result of studies designed to identify the surface activator of pro-MMP-2 [17]. Later studies confirmed the importance of MT1-MMP in the physiological activation of pro-MMP-2 [11]. MMP-2 (also known as gelatinase A), together with its closely homologue MMP-9 (gelatinase B), has long been recognized as a key protease in cancer progression and tumor angiogenesis [4, 5, 82, 83]. Thus, MT1-MMP together with MMP-2 constitute a potent proteolytic axis capable of accomplishing the hydrolysis of multiple substrates at the tumor cell periphery including collagen I and collagen IV, which helps cancer cells to invade basement membranes and navigate through the interstitial collagen-rich matrix [84–86].
Biochemically, the activation of pro-MMP-2 by MT1-MMP is initiated by the recruitment of the tissue inhibitor of metalloproteinase (TIMP)-2, which binds to the active site of MT1-MMP via the amino terminal region of the inhibitor. The MT1-MMP/TIMP-2 complex then recruits pro-MMP-2, which binds to the C-terminal region of TIMP-2. This ternary complex facilitates the activation of pro-MMP-2 by a neighboring TIMP-2-free MT-MMP [17, 87]. Thus, TIMP-2 significantly enhances the rate of pro-MMP-2 activation at the cell surface by clustering MT1-MMP with its proteolytic target. TIMP-2 also enhances the activation of pro-MMP-2 by MT3-MMP [88] but has no effect on MT2-MMP-dependent pro-MMP-2 activation [89]. While MT5-MMP activates pro-MMP-2 [21], the role of TIMP-2 in this process remains unknown. The process of pro-MMP-2 activation involves two sequential cleavage sites at the Asn37–Leu38 and Asn80–Tyr81 peptide bonds, leading to the formation of an intermediate form followed by the fully active species [90]. MT1-MMP only cleaves at the Asn37–Leu38 site in the prodomain, while the second cleavage is an autocatalytic event [90]. Cell surface association of pro-MMP-2 enhances the efficiency of the reaction by promoting the second autocatalytic step.
The ability of TIMP-2 to efficiently inhibit both MT4- [59, 61] and MT6-MMP [62] raised the possibility that GPI-MT-MMPs are also involved in pro-MMP-2 activation in a mechanism analogous to that of MT1-MMP. This possibility was examined in a number of studies using various approaches and cellular systems [26, 31, 32, 59–61, 68, 71, 91]. In the case of MT4-MMP, most studies agree that this protease is not an activator of pro-MMP-2 [59, 60, 68]. Moreover, exposure of MDA-MB-231 breast carcinoma cells transfected to express MT4-MMP to concanavalin A, which is known to induce MT1-MMP-dependent activation of pro-MMP-2, had no effect on pro-MMP-2 activation [68]. Our studies also support the inability of MT4-MMP to activate pro-MMP-2 (H. Zhao and R. Fridman, unpublished data). In contrast, Wang et al. [61], using conditioned media from HT1080 cells as a source of pro-MMP-2 and an MT4-MMP catalytic domain, showed generation of the intermediate form of MMP-2. However, when using conditioned media as a source of pro-MMP-2, it is difficult to determine whether the effects observed are a direct consequence of the interaction between pro-MMP-2 and the proposed “activator”.
In the case of MT6-MMP, Velasco et al. [26] showed that co-transfection of pro-MMP-2 with MT6-MMP in Cos-7 cells resulted in activation of pro-MMP-2. Similar results were obtained by Nie and Pei [71] by co-transfecting the cDNAs of both proteins in MDCK cells. However, in both studies the efficiency of MT6-MMP was significantly lower than that of MT1-MMP or MT5-MMP When a catalytic domain of MT6-MMP was incubated with pro-MMP-2, the zymogen was fully activated [71]. Interestingly, the MT6-MMP catalytic domain was found capable of cleaving pro-MMP-2 at both the Asn37–Leu38 and Asn80 –Tyr81 sites as opposed to MT1-MMP, suggesting a unique interaction between MT6-MMP and pro-MMP-2 [71]. Contrary to these findings, Kojima et al. [31] found that expression of a FLAG-tagged MT6-MMP in Cos-1 cells did not activate exogenously administered pro-MMP-2. Likewise, Miyamori et al. [91] showed that co-transfection of pro-MMP-2 cDNA with either MT4- or MT6-MMP cDNAs did not result in activation. However, when claudin-5, a tight junction protein, was co-expressed in the cells, a modest activation of pro-MMP-2 was observed with both MT4- and MT6-MMP [91]. Although the precise mechanism by which claudin-5 stimulates pro-MMP-2 activation is unclear, the effect is not specific since both GPI-MT-MMP- and TM-MT-MMP-dependent pro-MMP-2 activation is stimulated [91]. Nevertheless, these studies suggest that GPI-MT-MMP-mediated pro-MMP-2 activation, in contrast to that accomplished by TM-MT-MMPs, may require stimulation by cooperating factors.
Extensive studies from our laboratory showed that MT6-MMP is unable to activate pro-MMP-2 under various experimental conditions including in the presence of various TIMP-2 concentrations, a factor that was not previously examined [32]. We also co-expressed pro-MMP-2 and MT6-MMP in mammalian cells, as performed by Nie and Pei [71], and found no pro-MMP-2 activation. Moreover, PI-PLC-released MT6-MMP was inactive against pro-MMP-2. Based on these negative findings [31, 32] and the modest activation reported in co-transfection systems [26, 71], we posit that MT6-MMP is unlikely to behave as an activator of pro-MMP-2 under physiological conditions. Furthermore, under conditions of MT1-MMP expression in cancer cells, pro-MMP-2 activation by GPI-MT-MMPs would not be expected to be significant.
The role of GPI-MT-MMPs in the activation of pro-MMP-9 was also investigated [32, 59, 62]. First, the primary sequence similarity of MT6-MMP with stromelysin-1 (MMP-3), a known pro-MMP-9 activator, suggested a possible role of MT6-MMP in pro-MMP-9 activation. Second, the fact that GPI-MT-MMPs are inhibited by TIMP-1 raised the question whether pro-MMP-9 activation can be promoted by GPI-MT-MMP/TIMP-1 complexes, analogous to the activation of pro-MMP-2 by MT1-MMP in the presence of TIMP-2. However, the data have shown that MT6-MMP is not a pro-MMP-9 activator regardless of the presence of TIMP-1 [32, 62]. Likewise, MT4-MMP cannot activate pro-MMP-9 [59, and unpublished data from our laboratory]. Taken together, the overwhelming evidence strongly suggests that GPI-MT-MMPs did not evolve to accomplish pro-gelatinase activation at the cell surface.
2.5.4 Other zymogen substrate
Studies with ADAMTS (a disintegrin and metalloproteinase with thrombospondin-like motif)-4, a metalloprotease with ability to cleave proteoglycans such as aggrecan [92], have shown that MT4-MMP plays a role in ADAMTS-4 activation. Thus, MT4-MMP may indirectly contribute to ADAMTS-4-mediated degradation of cartilage in normal and pathological processes [93, 94]. In summary, although only a limited number of GPI-MT-MMP substrates have been identified, it appears that the substrate profile of GPI-MT-MMPs is very different from that exhibited by other MMPs, and in light of their membrane and extracellular distribution, may involve targets in both lipid rafts and non-lipid rafts.
3 Expression and role of GPI-MT-MMPs in cancer
Since their discovery, it became evident that the mRNA of GPI-MT-MMPs is highly expressed in various human cancers. Only recently, however, data have emerged showing protein expression and localization of MT4- [68] and MT6-MMP [32] in breast and colon cancer, respectively. Here, we will summarize the current information on the expression and role of GPI-MT-MMPs in human cancer tissues (Table 1).
3.1 MT4-MMP
MT4-MMP was originally cloned from a human breast carcinoma cDNA library [24] and consistently its mRNA was found to be expressed in breast cancer tissues [24, 68]. A study by Chabottaux et al. [68] examined the expression of MT4-MMP mRNA and protein in 63 breast adenocarcinomas and 21 samples of normal breast tissue. These investigators found no significant differences in MT4-MMP mRNA. However, immunohistochemistry studies revealed higher expression of MT4-MMP in cancer cells, while normal breast epithelial cells showed variable and overall moderate staining. MT4-MMP protein was particularly elevated in metastatic lymph nodes where both tumor and inflammatory cells displayed strong staining [68]. This is consistent with the reported expression of MT4-MMP in leukocytes [24, 95]. Overexpression of recombinant MT4-MMP in the breast cancer cell line MDA-MB-231 enhanced subcutaneous tumor growth when inoculated in RAG-1 immunodeficient mice [68]. Moreover, the MT4-MMP expressing MDA-MB-231 s.c. tumors developed lung metastases in 100% of the mice. Thus, MT4-MMP promotes both tumor growth and metastasis. Interestingly, functional studies failed to reveal an effect of MT4-MMP overexpression on in vitro cell migration and invasion, pro-MMP-2 activation, VEGF production, and angiogenesis [68]. These findings are similar to those obtained with MT6-MMP in colon cancer cells, in which expression of the protease did not confer a migratory and/or invasive phenotype or ability to activate pro-MMP-2 [32]. Together, these results suggest a unique role of GPI-MT-MMP in promotion of cancer progression.
While human breast cancer tissues express MT4-MMP [24, 68] where it appears to play a role in progression, the expression and function of MT4-MMP in mouse mammary cancinoma are unclear. An in situ hybridization study showed that MT4-MMP mRNA was not expressed in mouse mammary adenocarcinomas of transgenic mice expressing the polyoma virus T antigen under control of the MMTV promoter [96]. In fact, MT4-MMP mRNA was not expressed even in normal mammary glands regardless of their developmental stage [96]. The significance of these observations is unclear and need to be reproduced in other models of mammary cancer in mice before a definite conclusion can be made about the role of MT4-MMP in mouse mammary cancer development.
MT4-MMP mRNA expression was also examined in human gliomas [97] and was found to decrease with advancing tumor grade. In fact, MT4-MMP mRNA was highly expressed in normal brain tissue [97]. Although these studies involved a limited number of patients, the data suggest that the two GPI-MT-MMPs are differentially regulated in brain cancer. Furthermore, the studies in brain and breast tumors also suggest that the association of MT4-MMP expression with tumor progression may be cancer type specific.
3.2 MT6-MMP
Elevated MT6-MMP mRNA expression was found in several human cancers including brain (anaplastic astrocytomas and glioblastomas) [26, 97], colon [26], urothelial [98], and prostate [99] cancers. In gliomas, expression of MT6-MMP mRNA was suggested to contribute to disease progression [97]. The discovery of MT6-MMP mRNA expression in colon cancer [26] led to the first immunohistochemical study of MT6-MMP expression in samples of invasive colon cancer [32]. This study showed that while MT6-MMP protein was absent from normal colonic epithelium it was highly expressed in all invasive adenocarcinomas in the majority of the cases examined (50/60). In areas of in situ dysplasia, representing early stages of progression, staining for MT6-MMP was very weak albeit evident when compared to adjacent non-dysplastic epithelium. In contrast, invasive cancer cells showed high levels of MT6-MMP protein. In all the cases studied, MT6-MMP expression levels were independent of tumor differentiation and there was no association between intensity of staining and invasion of cancer cells into lymphovascular spaces [32]. Furthermore, other factors used to define tumor grade including architectural organization, cell polarity, mitotic rate, and extent of necrosis, as well as clinical stage, were not associated with differences in MT6-MMP expression. MT6-MMP staining in the tumor stroma was almost absent with the exception of strong expression in inflammatory-like cells [32], consistent with the known expression of MT6-MMP in leukocytes [25]. The high expression of MT6-MMP protein in the colon cancer cells is striking considering that many MMPs are known to be produced in the tumor stroma. However, the contribution of inflammatory cells, which produce MT6-MMP [25, 26], may also be critical in colon cancer.
Functional studies with colon cancer cell lines (HCT-116 and HT-29) overexpressing MT6-MMP showed that MT6-MMP expression correlated with enhanced tumor growth after subcutaneous inoculation in athymic mice [32]. Distant metastases to lungs or liver were not detected in MT6-MMP tumor bearing mice. Histopathological examination of the MT6-MMP subcutaneous tumors revealed an infiltrative tumor interface with a markedly desmoplastic stroma, suggesting that MT6-MMP promotes local tumor invasion. Whether MT6-MMP contributes to metastatic dissemination of colon cancer cells in mice needs to be investigated in relevant orthotopic models. However, in vitro studies showed no effects of MT6-MMP overexpression on tumor cell migration and invasion, and thus the precise role of MT6-MMP in colon cancer remains elusive. Although more studies are required to unveil the roles of GPI-MT-MMPs in cancer, the data so far suggest that these proteases influence cancer progression by mechanisms that are different from the TM-MT-MMPs. First, GPI-MT-MMPs do not act as progelatinase activators; second, their ECM degradation profile appears to be very limited; third, GPI-MT-MMPs do not promote tumor cell migration and invasion; and fourth, their inhibition profile appears unique.
4 Concluding remarks and future challenges
It is becoming evident that the GPI-MT-MMPs represent a distinctive subset of membrane-anchored MMPs that exhibit unique structural and functional characteristics, which separate them from the rest of the MMP family. Due to their unique characteristics and expression pattern, elucidating their regulation and role in malignant processes remains a major challenge and will require implementing a different set of biochemical and functional strategies from those applied to the study of most MMPs. The evidence so far strongly suggests that GPI-MT-MMPs are involved in cancer progression, where they may function both in cancer and inflammatory cells. More studies are required to define the pattern of GPI-MT-MMPs expression in human cancers and their association with progression. Furthermore, mouse models of cancer are needed to unveil the impact of expression and downregulation of GPI-MT-MMPs in the cancer cells and in the tumor stroma on tumor development, growth, and metastasis. Assessment of GPI-MT-MMP-mediated proteolysis and function in cancer will require establishing quantitative activity-based and functional biological assays specific for GPI-MT-MMPs. Evaluation of GPI-MT-MMP specific activity in complex biological systems and how it affects cell behavior will be aided by identification of their unique substrates, likely in the raft environment, and generation of specific synthetic inhibitors. Additional studies are needed to elucidate the inhibition of GPI-MT-MMPs by TIMPs and by other potential inhibitors such as clusterin [63] and possibly RECK, a GPI-anchored protein with MMP inhibitory activity [100]. The regulation of GPI-MT-MMPs is still not fully understood, and future studies need to address their biosynthetic pathway, endocytosis, recycling, shedding, and the function and significance of dimerization/ oligomerization. The challenges are significant but surely worthwhile because they will shed light on these very unique MMPs. This is also an important task in light of the disappointments of clinical trials with MMP inhibitors and the lessons learned from these attempts. Today, it is evident that effective targeting of relevant MMP activity in cancer patients will demand a thorough understanding of the biology, function, expression, and substrate profile of all members of the MMP family and their precise roles in cancer progression.
Acknowledgments
This work is supported by NIH/National Cancer Institute Grant CA-61986 (to R.F.). The authors thank Ms. Sumayya Anjum for preparation of illustration in Fig. 2.
Contributor Information
Anjum Sohail, Department of Pathology, School of Medicine, and Proteases and Cancer Program, Barbara Ann Karmanos Cancer Institute, Wayne State University, 540 E. Canfield Ave., Detroit, MI 48201, USA.
Qing Sun, Department of Pathology, School of Medicine, and Proteases and Cancer Program, Barbara Ann Karmanos Cancer Institute, Wayne State University, 540 E. Canfield Ave., Detroit, MI 48201, USA.
Huiren Zhao, Department of Pathology, School of Medicine, and Proteases and Cancer Program, Barbara Ann Karmanos Cancer Institute, Wayne State University, 540 E. Canfield Ave., Detroit, MI 48201, USA.
M. Margarida Bernardo, Department of Pathology, School of Medicine, and Proteases and Cancer Program, Barbara Ann Karmanos Cancer Institute, Wayne State University, 540 E. Canfield Ave., Detroit, MI 48201, USA.
Jin-Ah Cho, Department of Pathology, School of Medicine, and Proteases and Cancer Program, Barbara Ann Karmanos Cancer Institute, Wayne State University, 540 E. Canfield Ave., Detroit, MI 48201, USA.
Rafael Fridman, Email: rfridman@med.wayne.edu, Department of Pathology, School of Medicine, and Proteases and Cancer Program, Barbara Ann Karmanos Cancer Institute, Wayne State University, 540 E. Canfield Ave., Detroit, MI 48201, USA.
References
- 1.McCawley LJ, Matrisian LM. Matrix metalloproteinases: Multifunctional contributors to tumor progression. Molecular Medicine Today. 2000;6:149–156. doi: 10.1016/s1357-4310(00)01686-5. [DOI] [PubMed] [Google Scholar]
- 2.Stamenkovic I. Matrix metalloproteinases in tumor invasion and metastasis. Seminars in Cancer Biology. 2000;10:415–433. doi: 10.1006/scbi.2000.0379. [DOI] [PubMed] [Google Scholar]
- 3.Itoh Y, Nagase H. Matrix metalloproteinases in cancer. Essays in Biochemistry. 2002;38:21–36. doi: 10.1042/bse0380021. [DOI] [PubMed] [Google Scholar]
- 4.Lafleur MA, Handsley MM, Edwards DR. Metalloproteinases and their inhibitors in angiogenesis. Expert Reviews in Molecular Medicine. 2003;5:1–39. doi: 10.1017/S1462399403006628. [DOI] [PubMed] [Google Scholar]
- 5.Deryugina EI, Quigley JP. Matrix metalloproteinases and tumor metastasis. Cancer and Metastasis Reviews. 2006;25:9–34. doi: 10.1007/s10555-006-7886-9. [DOI] [PubMed] [Google Scholar]
- 6.Fingleton B. Matrix metalloproteinases: Roles in cancer and metastasis. Frontiers in Bioscience. 2006;11:479–491. doi: 10.2741/1811. [DOI] [PubMed] [Google Scholar]
- 7.Martin MD, Matrisian LM. The other side of MMPs: Protective roles in tumor progression. Cancer and Metastasis Reviews. 2007;26:717–724. doi: 10.1007/s10555-007-9089-4. [DOI] [PubMed] [Google Scholar]
- 8.Noel A, Jost M, Maquoi E. Matrix metalloproteinases at cancer tumor-host interface. Seminars in Cell & Developmental Biology. 2008;19:52–60. doi: 10.1016/j.semcdb.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 9.Jodele S, Blavier L, Yoonm JM, DeClerck YA. Modifying the soil to affect the seed: Role of stromal-derived matrix metalloproteinases in cancer progression. Cancer and Metastasis Reviews. 2006;25:35–43. doi: 10.1007/s10555-006-7887-8. [DOI] [PubMed] [Google Scholar]
- 10.Zucker S, Pei D, Cao J, Lopez-Otin C. Membrane type-matrix metalloproteinases (MT-MMP) Current Topics in Developmental Biology. 2003;54:1–74. doi: 10.1016/s0070-2153(03)54004-2. [DOI] [PubMed] [Google Scholar]
- 11.Holmbeck K, Bianco P, Yamada S, Birkedal-Hansen H. MT1-MMP: A tethered collagenase. Journal of Cellular Physiology. 2004;200:11–19. doi: 10.1002/jcp.20065. [DOI] [PubMed] [Google Scholar]
- 12.Hotary K, Li XY, Allen E, Stevens SL, Weiss SJ. A cancer cell metalloprotease triad regulates the basement membrane transmigration program. Genes & Development. 2006;20:2673–2686. doi: 10.1101/gad.1451806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbolina MV, Stack MS. Membrane type 1-matrix metalloproteinase: Substrate diversity in pericellular proteolysis. Seminars in Cell & Developmental Biology. 2008;19:24–33. doi: 10.1016/j.semcdb.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itoh Y, Seiki M. MT1-MMP: A potent modifier of pericellular microenvironment. Journal of Cellular Physiology. 2006;206:1–8. doi: 10.1002/jcp.20431. [DOI] [PubMed] [Google Scholar]
- 15.Osenkowski P, Toth M, Fridman R. Processing, shedding, and endocytosis of membrane type 1-matrix metalloproteinase (MT1-MMP) Journal of Cellular Physiology. 2004;200:2–10. doi: 10.1002/jcp.20064. [DOI] [PubMed] [Google Scholar]
- 16.Sato H, Takino T, Okada Y, Cao J, Shinagawa A, Yamamoto E, et al. A matrix metalloproteinase expressed on the surface of invasive tumour cells. Nature. 1994;370:61–65. doi: 10.1038/370061a0. [DOI] [PubMed] [Google Scholar]
- 17.Strongin AY, Collier I, Bannikov G, Marmer BL, Grant GA, Goldberg GI. Mechanism of cell surface activation of 72-kDa type IV collagenase. Isolation of the activated form of the membrane metalloprotease. Journal of Biological Chemistry. 1995;270:5331–5338. doi: 10.1074/jbc.270.10.5331. [DOI] [PubMed] [Google Scholar]
- 18.Arroyo AG, Genis L, Gonzalo P, Matias-Roman S, Pollan A, Galvez BG. Matrix metalloproteinases: New routes to the use of MT1-MMP as a therapeutic target in angiogenesis-related disease. Current Pharmaceutical Design. 2007;13:1787–1802. doi: 10.2174/138161207780831284. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka M, Sato H, Takino T, Iwata K, Inoue M, Seiki M. Isolation of a mouse MT2-MMP gene from a lung cDNA library and identification of its product. FEBS Letters. 1997;402:219–222. doi: 10.1016/s0014-5793(96)01537-2. [DOI] [PubMed] [Google Scholar]
- 20.Takino T, Sato H, Shinagawa A, Seiki M. Identification of the second membrane-type matrix metalloproteinase (MT- MMP-2) gene from a human placenta cDNA library. MT-MMPs form a unique membrane-type subclass in the MMP family. Journal of Biological Chemistry. 1995;270:23013–23020. doi: 10.1074/jbc.270.39.23013. [DOI] [PubMed] [Google Scholar]
- 21.Pei D. Identification and characterization of the fifth membrane-type matrix metalloproteinase MT5-MMP. Journal of Biological Chemistry. 1999;274:8925–8932. doi: 10.1074/jbc.274.13.8925. [DOI] [PubMed] [Google Scholar]
- 22.Llano E, Pendas AM, Freije JP, Nakano A, Knauper V, Murphy G, et al. Identification and characterization of human MT5-MMP, a new membrane-bound activator of progelatinase a overexpressed in brain tumors. Cancer Research. 1999;59:2570–2576. [PubMed] [Google Scholar]
- 23.Kajita M, Kinoh H, Ito N, Takamura A, Itoh Y, Okada A, et al. Human membrane type-4 matrix metalloproteinase (MT4-MMP) is encoded by a novel major transcript: Isolation of complementary DNA clones for human and mouse mt4-mmp transcripts. FEBS Letters. 1999;457:353–356. doi: 10.1016/s0014-5793(99)01065-0. [DOI] [PubMed] [Google Scholar]
- 24.Puente XS, Pendas AM, Llano E, Velasco G, Lopez-Otin C. Molecular cloning of a novel membrane-type matrix metalloproteinase from a human breast carcinoma. Cancer Research. 1996;56:944–949. [PubMed] [Google Scholar]
- 25.Pei D. Leukolysin/MMP25/MT6-MMP: A novel matrix metalloproteinase specifically expressed in the leukocyte lineage. Cell Research. 1999;9:291–303. doi: 10.1038/sj.cr.7290028. [DOI] [PubMed] [Google Scholar]
- 26.Velasco G, Cal S, Merlos-Suarez A, Ferrando AA, Alvarez S, Nakano A, et al. Human MT6-matrix metalloproteinase: Identification, progelatinase A activation, and expression in brain tumors. Cancer Research. 2000;60:877–882. [PubMed] [Google Scholar]
- 27.Overall CM. Matrix metalloproteinase substrate binding domains, modules and exosites. Overview and experimental strategies. Methods in Molecular Biology. 2001;151:79–120. [PubMed] [Google Scholar]
- 28.Udenfriend S, Kodukula K. How glycosylphosphatidylinositol-anchored membrane proteins are made. Annual Review in Biochemistry. 1995;64:563–591. doi: 10.1146/annurev.bi.64.070195.003023. [DOI] [PubMed] [Google Scholar]
- 29.Itoh Y, Kajita M, Kinoh H, Mori H, Okada A, Seiki M. Membrane type 4 matrix metalloproteinase (MT4-MMP, MMP-17) is a glycosylphosphatidylinositol-anchored proteinase. Journal of Biological Chemistry. 1999;274:34260–34266. doi: 10.1074/jbc.274.48.34260. [DOI] [PubMed] [Google Scholar]
- 30.Kang T, Yi J, Guo A, Wang X, Overall CM, Jiang W, et al. Subcellular distribution and cytokine- and chemokine-regulated secretion of leukolysin/MT6-MMP/MMP-25 in neutrophils. Journal of Biological Chemistry. 2001;276:21960–21968. doi: 10.1074/jbc.M007997200. [DOI] [PubMed] [Google Scholar]
- 31.Kojima S, Itoh Y, Matsumoto S, Masuho Y, Seiki M. Membrane-type 6 matrix metalloproteinase (MT6-MMP, MMP-25) is the second glycosyl-phosphatidyl inositol (GPI)-anchored MMP. FEBS Letters. 2000;480:142–146. doi: 10.1016/s0014-5793(00)01919-0. [DOI] [PubMed] [Google Scholar]
- 32.Sun Q, Weber CR, Sohail A, Bernardo MM, Toth M, Zhao H, et al. MMP25 (MT6-MMP) is highly expressed in human colon cancer, promotes tumor growth, and exhibits unique biochemical properties. Journal of Biological Chemistry. 2007;282:21998–22010. doi: 10.1074/jbc.M701737200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Itoh Y, Takamura A, Ito N, Maru Y, Sato H, Suenaga N, et al. Homophilic complex formation of MT1-MMP facilitates proMMP-2 activation on the cell surface and promotes tumor cell invasion. EMBO Journal. 2001;20:4782–4793. doi: 10.1093/emboj/20.17.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lehti K, Lohi J, Juntunen MM, Pei D, Keski-Oja J. Oligomerization through hemopexin and cytoplasmic domains regulates the activity and turnover of membrane-type 1 matrix metalloproteinase. Journal of Biological Chemistry. 2002;277:8440–8448. doi: 10.1074/jbc.M109128200. [DOI] [PubMed] [Google Scholar]
- 35.Galvez BG, Genis L, Matias-Roman S, Oblander SA, Tryggvason K, Apte SS, Arroyo AG. Membrane type 1-matrix metalloproteinase is regulated by chemokines monocyte-chemoattractant protein-1/ccl2 and interleukin-8/ CXCL8 in endothelial cells during angiogenesis. Journal of Biological Chemistry. 2005;280:1292–1298. doi: 10.1074/jbc.M408673200. [DOI] [PubMed] [Google Scholar]
- 36.Rozanov DV, Deryugina EI, Ratnikov BI, Monosov EZ, Marchenko GN, Quigley JP. Strongin AY: Mutation analysis of membrane type-1 matrix metalloproteinase (MT1-MMP). The role of the cytoplasmic tail Cys(574), the active site Glu(240), and furin cleavage motifs in oligomerization, processing, and self-proteolysis of MT1-MMP expressed in breast carcinoma cells. Journal of Biological Chemistry. 2001;276:25705–25714. doi: 10.1074/jbc.M007921200. [DOI] [PubMed] [Google Scholar]
- 37.Paladino S, Sarnataro D, Pillich R, Tivodar S, Nitsch L, Zurzolo C. Protein oligomerization modulates raft partitioning and apical sorting of GPI-anchored proteins. Journal of Cell Biology. 2004;167:699–709. doi: 10.1083/jcb.200407094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paladino S, Sarnataro D, Tivodar S, Zurzolo C. Oligomerization is a specific requirement for apical sorting of glycosyl-phosphatidylinositol-anchored proteins but not for nonraft-associated apical proteins. Traffic. 2007;8:251–258. doi: 10.1111/j.1600-0854.2006.00522.x. [DOI] [PubMed] [Google Scholar]
- 39.Sharom FJ, Lehto MT. Glycosylphosphatidylinositol-anchored proteins: Structure, function, and cleavage by phosphatidylinositol-specific phospholipase C. Biochemistry and Cell Biology. 2002;80:535–549. doi: 10.1139/o02-146. [DOI] [PubMed] [Google Scholar]
- 40.Eisenhaber B, Bork P, Eisenhaber F. Sequence properties of GPI-anchored proteins near the omega-site: Constraints for the polypeptide binding site of the putative transamidase. Protein Engineering. 1998;11:1155–1161. doi: 10.1093/protein/11.12.1155. [DOI] [PubMed] [Google Scholar]
- 41.Eisenhaber B, Bork P, Eisenhaber F. Prediction of potential GPI-modification sites in proprotein sequences. Journal of Molecular Biology. 1999;292:741–758. doi: 10.1006/jmbi.1999.3069. [DOI] [PubMed] [Google Scholar]
- 42.Eisenhaber B, Bork P, Yuan Y, Loffler G, Eisenhaber F. Automated annotation of GPI anchor sites: Case study C. elegans. Trends in Biochemical Sciences. 2000;25:340–341. doi: 10.1016/s0968-0004(00)01601-7. [DOI] [PubMed] [Google Scholar]
- 43.Ferguson MA, Williams AF. Cell-surface anchoring of proteins via glycosyl-phosphatidylinositol structures. Annual Review of Biochemistry. 1988;57:285–320. doi: 10.1146/annurev.bi.57.070188.001441. [DOI] [PubMed] [Google Scholar]
- 44.Chen R, Walter EI, Parker G, Lapurga JP, Millan JL, Ikehara Y, et al. Mammalian glycophosphatidylinositol anchor transfer to proteins and posttransfer deacylation. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:9512–9517. doi: 10.1073/pnas.95.16.9512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maeda Y, Tashima Y, Houjou T, Fujita M, Yoko-o T, Jigami Y, et al. Fatty acid remodeling of GPI-anchored proteins is required for their raft association. Molecular Biology of the Cell. 2007;18:1497–1506. doi: 10.1091/mbc.E06-10-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nebl T, Pestonjamasp KN, Leszyk JD, Crowley JL, Oh SW, Luna EJ. Proteomic analysis of a detergent-resistant membrane skeleton from neutrophil plasma membranes. Journal of Biological Chemistry. 2002;277:43399–43409. doi: 10.1074/jbc.M205386200. [DOI] [PubMed] [Google Scholar]
- 47.Stijlemans B, Baral TN, Guilliams M, Brys L, Korf J, Drennan M, et al. A glycosylphosphatidylinositol-based treatment alleviates trypanosomiasis-associated immunopathology. Journal of Immunology. 2007;179:4003–4014. doi: 10.4049/jimmunol.179.6.4003. [DOI] [PubMed] [Google Scholar]
- 48.Rollason R, Korolchuk V, Hamilton C, Schu P, Banting G. Clathrin-mediated endocytosis of a lipid-raft-associated protein is mediated through a dual tyrosine motif. Journal of Cell Science. 2007;120:3850–3858. doi: 10.1242/jcs.003343. [DOI] [PubMed] [Google Scholar]
- 49.Paladino S, Pocard T, Catino MA, Zurzolo C. GPI-anchored proteins are directly targeted to the apical surface in fully polarized MDCK cells. Journal of Cell Biology. 2006;172:1023–1034. doi: 10.1083/jcb.200507116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Butler GS, Overall CM. Proteomic validation of protease drug targets: Pharmacoproteomics of matrix metalloproteinase inhibitor drugs using isotope-coded affinity tag labelling and tandem mass spectrometry. Current Pharmaceutical Design. 2007;13:263–270. doi: 10.2174/138161207779313524. [DOI] [PubMed] [Google Scholar]
- 51.Kumari S, Mayor S. ARF1 is directly involved in dynamin-independent endocytosis. Nature Cell Biology. 2008;10:30–41. doi: 10.1038/ncb1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang A, Lehti K, Wang X, Weiss SJ, Keski-Oja J, Pei D. Regulation of membrane-type matrix metalloproteinase 1 activity by dynamin-mediated endocytosis. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:13693–13698. doi: 10.1073/pnas.241293698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baker AH, Edwards DR, Murphy G. Metalloproteinase inhibitors: Biological actions and therapeutic opportunities. Journal of Cell Science. 2002;115:3719–3727. doi: 10.1242/jcs.00063. [DOI] [PubMed] [Google Scholar]
- 54.Brew K, Dinakarpandian D, Nagase H. Tissue inhibitors of metalloproteinases: Evolution, structure and function. Biochimica et Biophysica Acta. 2000;1477:267–283. doi: 10.1016/s0167-4838(99)00279-4. [DOI] [PubMed] [Google Scholar]
- 55.Chirco R, Liu XW, Jung KK, Kim HR. Novel functions of TIMPs in cell signaling. Cancer and Metastasis Reviews. 2006;25:99–113. doi: 10.1007/s10555-006-7893-x. [DOI] [PubMed] [Google Scholar]
- 56.Maskos K, Bode W. Structural basis of matrix metalloproteinases and tissue inhibitors of metalloproteinases. Molecular Biotechnology. 2003;25:241–266. doi: 10.1385/MB:25:3:241. [DOI] [PubMed] [Google Scholar]
- 57.Lambert E, Dasse E, Haye B, Petitfrere E. TIMPs as multifacial proteins. Critical Reviews in Oncology/Hematology. 2004;49:187–198. doi: 10.1016/j.critrevonc.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 58.Lee MH, Rapti M, Knauper V, Murphy G. Threonine 98, the pivotal residue of tissue inhibitor of metalloproteinases (TIMP)-1 in metalloproteinase recognition. Journal of Biological Chemistry. 2004;279:17562–17569. doi: 10.1074/jbc.M312589200. [DOI] [PubMed] [Google Scholar]
- 59.English WR, Puente XS, Freije JM, Knauper V, Amour A, Merryweather A, et al. Membrane type 4 matrix metalloproteinase (MMP17) has tumor necrosis factor-alpha convertase activity but does not activate pro-MMP2. Journal of Biological Chemistry. 2000;275:14046–14055. doi: 10.1074/jbc.275.19.14046. [DOI] [PubMed] [Google Scholar]
- 60.Kolkenbrock H, Essers L, Ulbrich N, Will H. Biochemical characterization of the catalytic domain of membrane-type 4 matrix metalloproteinase. Biological Chemistry. 1999;380:1103–1108. doi: 10.1515/BC.1999.137. [DOI] [PubMed] [Google Scholar]
- 61.Wang Y, Johnson AR, Ye QZ, Dyer RD. Catalytic activities and substrate specificity of the human membrane type 4 matrix metalloproteinase catalytic domain. Journal of Biological Chemistry. 1999;274:33043–33049. doi: 10.1074/jbc.274.46.33043. [DOI] [PubMed] [Google Scholar]
- 62.English WR, Velasco G, Stracke JO, Knauper V, Murphy G. Catalytic activities of membrane-type 6 matrix metalloproteinase (MMP25) FEBS Letters. 2001;491:137–142. doi: 10.1016/s0014-5793(01)02150-0. [DOI] [PubMed] [Google Scholar]
- 63.Matsuda A, Itoh Y, Koshikawa N, Akizawa T, Yana I, Seiki M. Clusterin, an abundant serum factor, is a possible negative regulator of MT6-MMP/MMP-25 produced by neutrophils. Journal of Biological Chemistry. 2003;278:36350–36357. doi: 10.1074/jbc.M301509200. [DOI] [PubMed] [Google Scholar]
- 64.Shannan B, Seifert M, Leskov K, Willis J, Boothman D, Tilgen W, et al. Challenge and promise: Roles for clusterin in pathogenesis, progression and therapy of cancer. Cell Death Differentiation. 2006;13:12–19. doi: 10.1038/sj.cdd.4401779. [DOI] [PubMed] [Google Scholar]
- 65.Rooney IA, Heuser JE, Atkinson JP. GPI-anchored complement regulatory proteins in seminal plasma. An analysis of their physical condition and the mechanisms of their binding to exogenous cells. Journal of Clinical Investigation. 1996;97:1675–1686. doi: 10.1172/JCI118594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robertson C, Booth SA, Beniac DR, Coulthart MB, Booth TF, McNicol A. Cellular prion protein is released on exosomes from activated platelets. Blood. 2006;107:3907–3911. doi: 10.1182/blood-2005-02-0802. [DOI] [PubMed] [Google Scholar]
- 67.Suzuki K, Okumura Y. GPI-linked proteins do not transfer spontaneously from erythrocytes to liposomes. New aspects of reorganization of the cell membrane. Biochemistry. 2000;39:9477–9485. doi: 10.1021/bi000113v. [DOI] [PubMed] [Google Scholar]
- 68.Chabottaux V, Sounni NE, Pennington CJ, English WR, van den Brule F, Blacher S, et al. Membrane-type 4 matrix metalloproteinase promotes breast cancer growth and metastases. Cancer Research. 2006;66:5165–5172. doi: 10.1158/0008-5472.CAN-05-3012. [DOI] [PubMed] [Google Scholar]
- 69.Lauc G, Heffer-Lauc M. Shedding and uptake of gangliosides and glycosylphosphatidylinositol-anchored proteins. Biochimica et Biophysica Acta. 2006;1760:584–602. doi: 10.1016/j.bbagen.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 70.Nie J, Pei D. Rapid inactivation of alpha-1-proteinase inhibitor by neutrophil specific leukolysin/mem-brane-type matrix metalloproteinase 6. Experimental Cell Research. 2004;296:145–150. doi: 10.1016/j.yexcr.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 71.Nie J, Pei D. Direct activation of pro-matrix metalloproteinase-2 by leukolysin/membrane-type 6 matrix metalloproteinase/matrix metalloproteinase 25 at the asn(109)-Tyr bond. Cancer Research. 2003;63:6758–6762. [PubMed] [Google Scholar]
- 72.Hotary KB, Yana I, Sabeh F, Li XY, Holmbeck K, Birkedal-Hansen H, et al. Matrix metalloproteinases (MMPs) regulate fibrin-invasive activity via MT1-MMP-dependent and -independent processes. Journal of Experimental Medicine. 2002;195:295–308. doi: 10.1084/jem.20010815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grant GM, Giambernardi TA, Grant AM, Klebe RJ. Overview of expression of matrix metalloproteinases (MMP-17, MMP-18, and MMP-20) in cultured human cells. Matrix Biology. 1999;18:145–148. doi: 10.1016/s0945-053x(99)00003-7. [DOI] [PubMed] [Google Scholar]
- 74.Rikimaru A, Komori K, Sakamoto T, Ichise H, Yoshida N, Yana I, et al. Establishment of an MT4-MMP-deficient mouse strain representing an efficient tracking system for MT4-MMP/MMP-17 expression in vivo using beta-galactosidase. Genes Cells. 2007;12:1091–1100. doi: 10.1111/j.1365-2443.2007.01110.x. [DOI] [PubMed] [Google Scholar]
- 75.Rozanov DV, Hahn-Dantona E, Strickland DK, Strongin AY. The low density lipoprotein receptor-related protein LRP is regulated by membrane type-1 matrix metalloproteinase (MT1-MMP) proteolysis in malignant cells. Journal of Biological Chemistry. 2004;279:4260–4268. doi: 10.1074/jbc.M311569200. [DOI] [PubMed] [Google Scholar]
- 76.Andolfo A, English WR, Resnati M, Murphy G, Blasi F, Sidenius N. Metalloproteases cleave the urokinase-type plasminogen activator receptor in the D1-D2 linker region and expose epitopes not present in the intact soluble receptor. Thrombosis and Haemostasis. 2002;88:298–306. [PubMed] [Google Scholar]
- 77.Dumic J, Dabelic S, Flogel M. Galectin-3: An open-ended story. Biochimica et Biophysica Acta. 2006;1760:616–635. doi: 10.1016/j.bbagen.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 78.McClung HM, Thomas SL, Osenkowski P, Toth M, Menon P, Raz A, et al. SPARC upregulates MT1-MMP expression, MMP-2 activation, and the secretion and cleavage of galectin-3 in U87MG glioma cells. Neuroscience Letters. 2007;419:172–177. doi: 10.1016/j.neulet.2007.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ochieng J, Fridman R, Nangia-Makker P, Kleiner DE, Liotta LA, Stetler-Stevenson WG, et al. Galectin-3 is a novel substrate for human matrix metalloproteinases-2 and −9. Biochemistry. 1994;33:14109–14114. doi: 10.1021/bi00251a020. [DOI] [PubMed] [Google Scholar]
- 80.Toth M, Osenkowski P, Hesek D, Brown S, Meroueh S, Sakr W, et al. Cleavage at the stem region releases an active ectodomain of the membrane type 1 matrix metalloproteinase. Biochemistry Journal. 2005;387:497–506. doi: 10.1042/BJ20041324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nagase H. Activation mechanisms of matrix metalloproteinases. Biological Chemistry. 1997;378:151–160. [PubMed] [Google Scholar]
- 82.Giannelli G, Antonaci S. Gelatinases and their inhibitors in tumor metastasis: From biological research to medical applications. Histology and Histopathology. 2002;17:339–345. doi: 10.14670/HH-17.339. [DOI] [PubMed] [Google Scholar]
- 83.Bjorklund M, Koivunen E. Gelatinase-mediated migration and invasion of cancer cells. Biochimica et Biophysica Acta. 2005;1755:37–69. doi: 10.1016/j.bbcan.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 84.Hotary KB, Allen ED, Brooks PC, Datta NS, Long MW, Weiss SJ. Membrane type I matrix metalloproteinase usurps tumor growth control imposed by the three-dimensional extracellular matrix. Cell. 2003;114:33–45. doi: 10.1016/s0092-8674(03)00513-0. [DOI] [PubMed] [Google Scholar]
- 85.Stetler-Stevenson WG, Yu AE. Proteases in invasion: Matrix metalloproteinases. Seminars in Cancer Biology. 2001;11:143–152. doi: 10.1006/scbi.2000.0365. [DOI] [PubMed] [Google Scholar]
- 86.Wolf K, Wu YI, Liu Y, Geiger J, Tam E, Overall C, et al. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nature Cell Biology. 2007;9:893–904. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]
- 87.Hernandez-Barrantes S, Bernardo M, Toth M, Fridman R. Regulation of membrane type-matrix metalloproteinases. Seminars in Cancer Biology. 2002;12:131–138. doi: 10.1006/scbi.2001.0421. [DOI] [PubMed] [Google Scholar]
- 88.Zhao H, Bernardo MM, Osenkowski P, Sohail A, Pei D, Nagase H, et al. Differential inhibition of membrane type 3 (MT3)-matrix metalloproteinase (MMP) and MT1-MMP by tissue inhibitor of metalloproteinase (TIMP)-2 and TIMP-3 rgulates pro-MMP-2 activation. Journal of Biological Chemistry. 2004;279:8592–8601. doi: 10.1074/jbc.M308708200. [DOI] [PubMed] [Google Scholar]
- 89.Morrison CJ, Butler GS, Bigg HF, Roberts CR, Soloway PD, Overall CM. Cellular activation of MMP-2 (gelatinase A) by MT2-MMP occurs via a TIMP-2-independent pathway. Journal of Biological Chemistry. 2001;276:47402–47410. doi: 10.1074/jbc.M108643200. [DOI] [PubMed] [Google Scholar]
- 90.Will H, Atkinson SJ, Butler GS, Smith B, Murphy G. The soluble catalytic domain of membrane type 1 matrix metalloproteinase cleaves the propeptide of progelatinase A and initiates autoproteolytic activation. Regulation by TIMP-2 and TIMP-3. Journal of Biological Chemistry. 1996;271:17119–17123. doi: 10.1074/jbc.271.29.17119. [DOI] [PubMed] [Google Scholar]
- 91.Miyamori H, Takino T, Kobayashi Y, Tokai H, Itoh Y, Seiki M, et al. Claudin promotes activation of promatrix metalloproteinase-2 mediated by membrane-type matrix metalloproteinases. Journal of Biological Chemistry. 2001;276:28204–28211. doi: 10.1074/jbc.M103083200. [DOI] [PubMed] [Google Scholar]
- 92.Tortorella MD, Burn TC, Pratta MA, Abbaszade I, Hollis JM, Liu R, et al. Purification and cloning of aggrecanase-1: A member of the ADAMTS family of proteins. Science. 1999;284:1664–1666. doi: 10.1126/science.284.5420.1664. [DOI] [PubMed] [Google Scholar]
- 93.Gao G, Plaas A, Thompson VP, Jin S, Zuo F, Sandy JD. ADAMTS4 (aggrecanase-1) activation on the cell surface involves C-terminal cleavage by glycosylphosphatidyl inositol-anchored membrane type 4-matrix metalloproteinase and binding of the activated proteinase to chondroitin sulfate and heparan sulfate on syndecan-1. Journal of Biological Chemistry. 2004;279:10042–10051. doi: 10.1074/jbc.M312100200. [DOI] [PubMed] [Google Scholar]
- 94.Patwari P, Gao G, Lee JH, Grodzinsky AJ, Sandy JD. Analysis of ADAMTS4 and MT4-MMP indicates that both are involved in aggrecanolysis in interleukin-1-treated bovine cartilage. Osteoarthritis Cartilage. 2005;13:269–277. doi: 10.1016/j.joca.2004.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gauthier MC, Racine C, Ferland C, Flamand N, Chakir J, Tremblay GM. Expression of membrane type-4 matrix metalloproteinase (metalloproteinase-17) by human eosinophils. International Journal of Biochemistry and Cell Biology. 2003;35:1667–1673. doi: 10.1016/s1357-2725(03)00136-5. [DOI] [PubMed] [Google Scholar]
- 96.Szabova L, Yamada SS, Birkedal-Hansen H, Holmbeck K. Expression pattern of four membrane-type matrix metalloproteinases in the normal and diseased mouse mammary gland. Journal of Cellular Physiology. 2005;205:123–132. doi: 10.1002/jcp.20385. [DOI] [PubMed] [Google Scholar]
- 97.Nuttall RK, Pennington CJ, Taplin J, Wheal A, Yong VW, Forsyth PA, et al. Elevated membrane-type matrix metalloproteinases in gliomas revealed by profiling proteases and inhibitors in human cancer cells. Molecular Cancer Research. 2003;1:333–345. [PubMed] [Google Scholar]
- 98.Wallard MJ, Pennington CJ, Veerakumarasivamm A, Burtt G, Mills IG, Warren A, et al. Comprehensive profiling and localisation of the matrix metalloproteinases in urothelial carcinoma. British Journal of Cancer. 2006;94:569–577. doi: 10.1038/sj.bjc.6602931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Riddick AC, Shukla CJ, Pennington CJ, Bass R, Nuttall RK, Hogan A, et al. Identification of degradome components associated with prostate cancer progression by expression analysis of human prostatic tissues. British Journal of Cancer. 2005;92:2171–2180. doi: 10.1038/sj.bjc.6602630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Noda M, Oh J, Takahashi R, Kondo S, Kitayama H, Takahashi C. RECK: A novel suppressor of malignancy linking oncogenic signaling to extracellular matrix remodeling. Cancer and Metastasis Reviews. 2003;22:167–175. doi: 10.1023/a:1023043315031. [DOI] [PubMed] [Google Scholar]
- 101.Tortorella MD, Arner EC, Hills R, Easton A, Korte-Sarfaty J, Fok K, et al. Alpha2-macroglobulin is a novel substrate for ADAMTS-4 and ADAMTS-5 and represents an endogenous inhibitor of these enzymes. Journal of Biological Chemistry. 2004;279:17554–17561. doi: 10.1074/jbc.M313041200. [DOI] [PubMed] [Google Scholar]
- 102.Plaisier M, Kapiteijn K, Koolwijk P, Fijten C, Hanemaaijer R, Grimbergen JM, et al. Involvement of membrane-type matrix metalloproteinases (MT-MMPs) in capillary tube formation by human endometrial microvascular endothelial cells: Role of MT3-MMP. Journal of Clinical Endocrinology and Metabolism. 2004;89:5828–5836. doi: 10.1210/jc.2004-0860. [DOI] [PubMed] [Google Scholar]