Abstract
Single-nucleotide polymorphisms (SNPs) in double-stranded DNA (dsDNA) have been straightforwardly genotyped by matrix-assisted laser desorption/ ionization time-of-flight mass spectrometry (MALDI-TOF MS). Peptide nucleic acid (PNA), a DNA analog, was used as a probe molecule. In its presence, genomic dsDNA was first treated with exonuclease III and then with nuclease S1. By these one-pot reactions, single-stranded DNA fragments including the SNP sites were formed in situ. These fragments were directly analyzed by MALDI-TOF MS, and the identity of the DNA base at the SNP site was determined in terms of mass number. By using two or more PNA probes simultaneously, multiplex analysis was also successful. Various genotypes of apolipoprotein E gene (ε2/ε2, ε3/ε3, ε4/ε4, ε2/ε3 and ε3/ε4) were identified from dsDNA obtained by PCR from corresponding patients.
INTRODUCTION
Among variations in human genomes, SNPs are the most common and well-recognized ones and are strongly related to physiological properties of individuals such as responses to medicines, dangers of hereditary diseases and others (1–3). Thus, high-throughput, accurate and low-cost genotyping of SNPs is crucially important (4,5). In order to fulfill all these requirements, the following three factors are necessary: (i) sample DNA must be easily and economically obtainable, (ii) a number of SNP sites in specimens should be simultaneously analyzable under the same conditions and (iii) alteration of nucleobase, if any, must be easily, precisely and straightforwardly detectable. Although many proposals have been previously made, none of them is completely satisfactory and a number of attempts for further improvements are being made in many laboratories (6–9).
One of the most critical problems is factor (i), since most previous genotyping methods [e.g. hybridization with allele-specific probes (10,11), oligonucleotide ligation (12,13), single-nucleotide primer extension (14) and enzymatic cleavage (15,16)] employ single-stranded DNA (ssDNA) as the specimen. Double-stranded DNA (dsDNA), which is more easily obtainable either from natural sources or by PCR, cannot be directly used. Thus dsDNA is usually first denatured at high temperatures, cooled in a controlled manner and then charged to the probes (17–19). Even with this procedure, however, the amount of ssDNA available for analysis is limited. Another method for the preparation of ssDNA involves the removal of the unnecessary counterpart strand from dsDNA using insoluble support matrices bearing the corresponding complementary oligonucleotides (20,21). Alternatively, ssDNA must be prepared by asymmetric PCR from the target portion of the patient’s DNA (22,23). However, this type of PCR is much less efficient in DNA amplification than standard PCR methods. Therefore direct detection of SNPs in dsDNA, if possible, is much more desirable.
Factor (ii) is also critical, especially when the genotyping takes advantage of the difference in stability between fully-matched and single-base mismatched duplexes. This difference is evident only near the melting temperatures of these duplexes, so the analysis temperature must be very carefully chosen (24,25). For simultaneous analysis of many SNP sites, however, this choice is difficult, as often experienced in microarray systems. As a solution to factor (iii), matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) has often been used recently (26–28). Here, any alteration of nucleobases in genes is precisely detectable in terms of a change in the mass number of a DNA fragment. Its rapidness and small sample consumption are appropriate to high-throughput analysis. However, it is not easy to prepare short DNA fragments for MS analysis (<20mer) directly from the designated site in genomic dsDNA (29–32).
In this article, we present a new method which straightforwardly genotypes SNPs in dsDNA by MALDI-TOF MS. Genomic dsDNA (amplified by standard PCR when necessary) is treated with exonuclease III and then with nuclease S1 in the presence of PNA [a DNA analog in which the phosphate backbone of DNA is replaced by a peptide backbone (33–36)]. This PNA probe protects designated portions of the DNA specimen from enzymatic digestion. As a result, ssDNA fragments of the desired length are prepared by a one-pot reaction. By analyzing these short ssDNA fragments by MALDI-TOF MS, SNPs are conclusively genotyped. All the enzymatic digestions are carried out under conditions where all the fully matched duplexes and mismatch-involving ones are completely formed, and thus the analysis can be achieved at any temperature below the melting temperatures of these duplexes. By using two or more PNA probes simultaneously, multiplex analysis is also successful. Apparently, all the factors (i)–(iii) are fulfilled. As an example of a practical application of the present method, various genotypes of the apoE gene (ε2/ε2, ε3/ε3, ε4/ε4, ε2/ε3 and ε3/ε4) (37,38) are scored. These isoforms change the amino acids 112 (nucleobase 3937: T/C variation) and 158 (nucleobase 4075: C/T variation) (DDBJ/EMBL/GenBank accession number AF261279), which are associated with lipid disorder (39) and Alzheimer’s disease (40,41).
MATERIALS AND METHODS
Synthesis of PNA and primer DNA
The PNA additives were manually synthesized and characterized according to the literature (35,36). A lysine or glycine residue was attached to their C-termini (see Table 1). The DNA primers were prepared on an Applied Biosystems 394 DNA/RNA synthesizer. These PNAs and DNAs were purified by high-quality prepacked column (Kanto Chemical Co., Tokyo) and characterized by mass analysis. Their purities were >99% according to HPLC. The concentrations of DNA and PNA in the enzymatic reactions were determined from the absorbance at 260 nm on a Jasco V-530 UV/vis spectrophotometer.
Table 1. The DNAs from the apoE gene and PNA probes used for the present genotypinga,b.
| Sense strands in apoE gene | |
| (1) 112 codon (Sense) | 203mer |
| apoE(S112T) (112Cys) | 5′···(3925)ATGGAGGACGTGTGCGGCCGCCTGGTG(3951)···3′ |
| apoE(S112C) (112Arg) | 5′···(3925)ATGGAGGACGTGCGCGGCCGCCTGGTG(3951)···3′ |
| PNA(S112A) | H2NCO-(Gly)-TGCACACGCC-NH2 10mer |
| PNA(S112G) | H2NCO-(Gly)-TGCACGCGCC-NH2 10mer |
| (2) 158 codon (Sense) | 106mer |
| apoE(S158T) (158Cys) | 5′···(4063)GACCTGCAGAAGTGCCTGGCAGTGTAC(4089)···3′ |
| apoE(S158C) (158Arg) | 5′···(4063)GACCTGCAGAAGCGCCTGGCAGTGTAC(4089)···3′ |
| PNA(S158G) | H2NCO-(Gly)-TCTTCGCGGA-NH2 10mer |
| Antisense strands in apoE gene | |
| (1) 158 codon (Antisense) | 203mer |
| apoE(A158A) (158Cys) | 5′···(4089)GTACACTGCCAGGCACTTCTGCAGGTC(4063)···3′ |
| apoE(A158G) (158Arg) | 5′···(4089)GTACACTGCCAGGCGCTTCTGCAGGTC(4063)···3′ |
| PNA(A158T) | H2NCO-(Lys)-GTCCGTGAAGAC-NH2 12mer |
| PNA(A158C) | H2NCO-(Lys)-GTCCGCGAAGAC-NH2 12mer |
aThe underlined part of DNA substrate is complementary with the PNA probe, and the polymorphic nucleotide is in bold.
bThe sequence data are from DDBJ/EMBL/GenBank accession no. AF261279.
Preparation of DNA samples for the genotyping
The DNA from human apoE gene was isolated from the whole blood of individuals by use of the Puregene® DNA isolation kit (Gentra Systems). First, the DNA portion containing SNP sites (from nucleobases 3900–4115) was amplified by PCR on a GeneAmp® PCR system 9700 (Perkin–Elmer–Applied Biosystems); the forward primer was (5′-GGCGCAGGCCCGGCTGGGC-3′) and the reverse primer was (5′-TCGGCGCCCTCGCGGGCC-3′). Then, the product was cloned into pGEM®-T Easy vector (Promega, USA). The genomic library was screened, and genotyped using an ABI PRISM™ 310NT genetic analyzer. These plasmids were used as the templates for the following PCR.
The forward and the reverse primers (30 pmol each), dNTP (0.2 mM each), 2.0 mM MgCl2, 1× PCR buffer and 1.25 U TaKaRa Ex Taq™ polymerase (TaKaRa Bio Inc., Tokyo) were added to 50 µl of solution containing the plasmid (20 ng). The PCR was started at 95°C for 3 min, followed by 28 cycles of 95°C for 30 s, 58°C for 30 s and 72°C for 30 s, and then finished with an additional extension step of 72°C for 3 min. The PCR products were treated with the QIAquick® PCR purification kit (QIAGEN Sciences, USA) and dissolved in water (purified by Milli-Q®, Millipore, Tokyo). The primers used for the preparation of 250, 203 and 106 bp dsDNA substrates were pApoE-U1/pGEM-D, pApoE-U1/pApoE-D and pApoE-U2/pGEM-D, respectively. Here, pApoE-U1 (5′-GGCCCGGCTGGGCGCGGAC-3′) and pApoE-U2 (5′-TCCGCGATGCCGATGAC-3′) are the forward primers, while pApoE-D (5′-CCTCGCGGGCCCCGGCCTGG-3′) and pGEM-D (5′-ATATGGTCGACCTGCAGG-3′) are the reverse primers.
Enzymatic reactions
The substrate DNA was digested by exonuclease III (TaKaRa Bio Inc., Tokyo) in 1× exonuclease III buffer at pH 8.0 (50 mM Tris–HCl, 5 mM MgCl2, 10 mM 2-mercaptoethanol) and 37°C for 5 or 10 min. Then, 1× nuclease S1 buffer pH 4.6 (30 mM sodium acetate, 1 mM zinc acetate, 5% (v/v) glycerol) and PNA additives were added (the volume ratio of the nuclease S1 buffer to the exonuclease III buffer was 1.5:1.0). Alternatively, the PNAs were added before the exonuclease III treatment. The digestion by nuclease S1 [Aspergillus oryzae (Invitrogen Life Technologies, Carlsbad, CA)] was carried out at pH 4.6 and 20°C for 10–20 min. The reaction was stopped by adding EDTA to a final concentration of 25 mM.
MALDI-TOF MS
Prior to the MS analysis, the reaction mixtures were desalted using a ZipTip® pipette tip C18 (Millipore Corporation, Bedford, MA, USA), and then mixed with MALDI matrix solution containing 3-hydroxy-2-picolinic acid and diammonium hydrogen citrate. The mass spectra were measured in negative ion mode on a Kratos Kompact MALDI-TOF mass spectrometer (unless noted otherwise) or a Bruker Daltonics® Autoflex mass spectrometer.
RESULTS AND DISCUSSION
Preparation of ssDNA samples for MS analysis from dsDNA using exonuclease III/nuclease S1/PNA systems
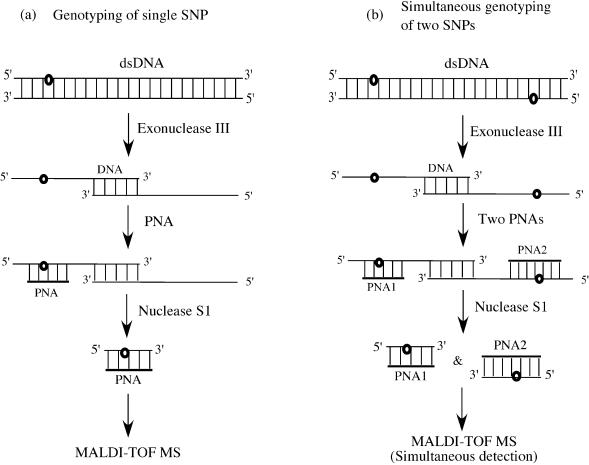
The strategy developed in this study is depicted in Scheme 1. First, the dsDNA specimen is treated with exonuclease III (42–44). This dsDNA-specific exonuclease digests both sense and antisense strands in the 3′–5′ direction, providing two ssDNAs which are bound by a short double-stranded joint (see the second row from the top). The joint region is gradually shortened and finally two ssDNAs are formed. A PNA probe, which is complementary to the DNA portion involving the SNP site, is added to these mixtures (Scheme 1a). In order to genotype two SNP sites simultaneously, two PNA probes are added (Scheme 1b). The mixture is then treated with nuclease S1. Since this ssDNA-specific endonuclease (45–47) does not hydrolyze the DNA in DNA/PNA duplexes (48,49), the corresponding portion in the DNA remains unchanged. Even when the ssDNA has a mismatch to the PNA probe, the ssDNA fragment is also obtainable (vide infra). These ssDNA fragments are directly analyzed by MALDI-TOF MS.
Scheme 1. Schematic representation of the processes for SNP detection by exonuclease III/nuclease S1/PNA systems. The small circles represent the SNP sites. (a) Genotyping of one SNP by use of single PNA probe. (b) Simultaneous genotyping of two SNPs by use of two PNA probes.
When dsDNA was directly treated with nuclease S1 in the presence of PNA probes (without the pretreatment by exonuclease III), no MS signals were detectable. There, the PNA probes could not form heteroduplexes with the dsDNA, and no DNA fragments of predetermined length were produced.
Use of the present strategy for genotyping of SNPs in dsDNA from the apoE gene
The apoE gene has three isoforms that differ at the codons 112 and 158 (ε2, cysteine/cysteine; ε3, cysteine/arginine; ε4, arginine/arginine) (see Table 2). First, the SNP site at codon 112 (nucleobase 3937) was analyzed. From the corresponding portion in the homozygote ε4/ε4 (C-allele of codon 112), dsDNA (250 bp) was prepared by PCR [its sense strand is apoE(S112C)(112Arg) in the upper part of Table 1). When this dsDNA was treated at pH 8.0 and 37°C with exonuclease III and analyzed by agarose gel electrophoresis, the band of dsDNA gradually weakened and concurrently two new bands of larger mobilities became dominant (data not presented). As presented in Scheme 1, the dsDNA was in situ converted into two ssDNAs which are bound by the joint (the band of intermediate mobility). In prolonged reactions, the joint region was gradually digested and two separate ssDNAs were formed (the band with greatest mobility). The resultant mixture was then digested by nuclease S1 at pH 4.6 and 20°C in the presence of PNA(S112G), which is complementary with A3932-G3941 in the sense strand of the substrate dsDNA.
Table 2. DNA fragments formed from apoE gene by exonuclease III/nuclease S1/PNA systems.
| SNP site | Genotype | DNA fragments | Calculated | Founda |
|---|---|---|---|---|
| 112 codon(Sense) | ε2, ε3 | p-CGTGTGCGG-OH | 2849.5 | 2849.6, 2849.8, 2848.9 |
| p-CGTGTGCGGC-OH | 3138.5 | 3138.9, 3139.1, 3139.0 | ||
| p-ACGTGTGCGGC-OH | 3451.6 | 3452.8, 3451.6 | ||
| ε4 | p-CGTGCGCGG-OH | 2834.5 | 2834.9 | |
| p-CGTGCGCGGC-OH | 3123.5 | 3123.5, 3122.6 | ||
| p-ACGTGCGCGGC-OH | 3436.6 | 3436.7, 3436.3 | ||
| 158 codon(Sense) | ε3 | p-GAAGCGCCTG-OH | 3131.5 | 3130.6 |
| p-GAAGCGCCTGG-OH | 3460.6 | 3460.5 | ||
| 158 codon(Antisense) | ε2 | p-CAGGCACTTCTG-OH | 3699.6 | 3701.0, 3699.0 |
| p-CAGGCACTTCTGC-OH | 3988.6 | 3989.1, 3988.2 | ||
| ε3, ε4 | p-CAGGCGCTTCTG-OH | 3715.6 | 3715.0, 3716.0, 3715.6 | |
| p-CAGGCGCTTCTGC-OH | 4004.6 | 4005.3, 4005.1, 4004.2 |
aThe experimental error in these values is around 2.
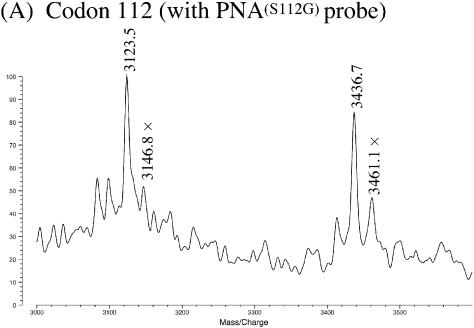
The MALDI-TOF MS of the products is presented in Figure 1A. The signal at m/z = 3436.7 corresponds to the 11mer from A3932 to C3942 (the theoretical m/z value = 3436.6; see Table 2). In this fragment, the DNA portion which is complementary with PNA(S112G) was sufficiently protected from nuclease S1, and furthermore the phosphodiester linkage next to the 3′-end of the DNA/PNA duplex was also kept intact. Another major signal at m/z = 3123.5 is for the 10mer (C3933–C3942), in which the 5′-terminal A3932 of the 11mer was removed by nuclease S1 (the theoretical m/z = 3123.5). Both of these signals were accompanied by the corresponding Na+ adducts (designated by crosses). The differences in m/z between the original fragments and their Na+ adducts are 23 as expected. By using exonuclease III/nuclease S1/PNA systems, ssDNA fragments for MS analysis were successfully obtained from the PCR products (dsDNA) of genomic DNA, and the nucleotide at position 3937 was conclusively assigned as C.
Figure 1.
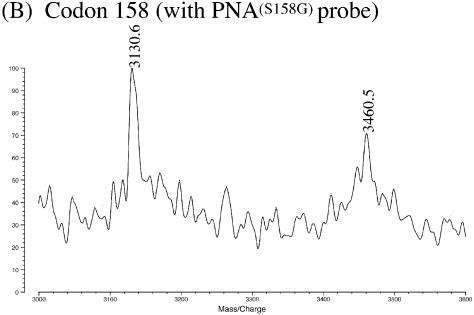
The MALDI-TOF MS spectra for genotyping of SNP in apoE gene by using exonuclease III/nuclease S1/PNA systems. (A) Analysis of dsDNA from apoE 4 (250 bp) at codon 112 using PNA(S112G). Reaction conditions: [apoE DNA] = 3 µM, [PNA(S112G)] = 5 µM, [exonuclease III] = 16.5 units/µl at pH 8.0 and 37°C for 10 min, and [nuclease S1] = 4.5 units/µl at pH 4.6 and 20°C for 15 min. The small peaks next to the major peaks (designated by crosses) correspond to the adduct with the Na+ ion. Mass spectra were recorded in a negative ion mode. (B) Analysis of dsDNA from apoE 3 (106 bp) at codon 158 using PNA(S158G).
In Figure 1B, the codon 158 in apoE (another SNP site) was analyzed. From the homozygote ε3/ε3 (the C-allele), dsDNA (106 bp) was prepared and digested consecutively by exonuclease III and nuclease S1. The PNA(S158G) probe is complementary with A4070–T4079 in the sense strand for the C-allele (Table 1). The SNP position is the nucleotide 4075 (C/T). As described above for Figure 1A, the 10mer fragment (G4071–G4080; observed m/z = 3130.6 and theoretical m/z = 3131.5) and the 11mer fragment (G4071–G4081; observed m/z = 3460.5 and theoretical m/z = 3460.6) were efficiently formed. The nucleotide at the SNP site 4075 was assigned as C. Direct and complete genotyping of PCR products (dsDNA) by MALDI-TOF MS has been further substantiated.
Simultaneous genotyping of homozygotes involving two SNP sites
Two (or more) SNP sites, which are located nearby in a gene, can also be genotyped by one-pot reactions. After the digestion of dsDNA by exonuclease III, both the sense strand upstream and the antisense strand downstream of a gene are made single-stranded (see Scheme 1). Therefore two SNP sites in these portions can be easily genotyped by using two PNA probes bound to these sites. Of course, more SNP sites are analyzable by increasing the number of PNA probes.
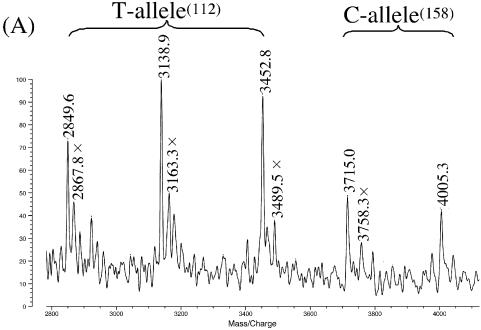
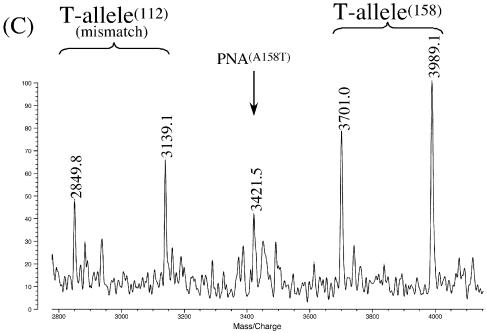
In Figure 2A, dsDNA (203 bp: the PCR product from G3906 to G4108 in the homozygote ε3/ε3) was genotyped by using two PNA probes. This dsDNA contains two SNP sites for both codon 112 (nucleotide 3937) and codon 158 (nucleotide 4075). One of the two PNA probes (PNA(S112A): 10mer) is complementary with A3932–G3941 in the sense strand, and the other probe (PNA(A158C): 12mer) is complementary with G4069–C4080 in the antisense strand. The signal at m/z = 3715.0 is for the 12mer (G4069–C4080) from the antisense strand for the C-allele at codon 158 (theoretical m/z = 3715.6; see Table 2). At m/z = 4005.3, the 13mer fragment (C4068–C4080) is also detected (theoretical m/z = 4004.6). Three major peaks on the left-hand side of Figure 2A correspond to fragments from the portion involving codon 112 in the T-allele. The signals at m/z values of 2849.6, 3138.9 and 3452.8 refer to the 9mer (C3933–G3941), 10mer (C3933–C3942) and 11mer (A3932–C3942), respectively. All the results are fully consistent with the genotype of the specimen (ε3/ε3).
Figure 2.
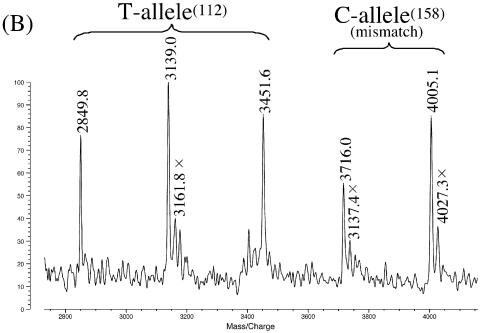
Simultaneous genotyping of two SNP sites in homozygous apoE genes by using exonuclease III, nuclease S1 and two PNA probes: (A) ε3/ε3 by PNA(S112A) and PNA(A158C); (B) ε3/ε3 by PNA(S112A) and PNA(A158T) (T–G mismatch); (C) ε2/ε2 by PNA(S112G) (T–G mismatch) and PNA(A158T). Note that mass signals are clearly observable even when there exists a mismatch between the DNA specimen and the PNA probe (see text for details). Reaction conditions: [apoE DNA (203 bp)] = 3.4 µM, [PNA probe] = 5.0 µM, [exonuclease III] = 14 units/µl at pH 8.0 and 37°C for 10 min, and [nuclease S1] = 6 units/µl at pH 4.6 and 20°C for 12 min. Spectra were recorded in a negative ion mode. The small peaks next to the major peaks (designated by crosses) correspond to the adducts with Na+ or K+ ions.
Effects of mismatch between DNA specimen and PNA probe on the genotyping
It is noteworthy that DNA fragments for MS analysis can be successfully obtained even when there is a one-base mismatch between the DNA specimen and PNA probe. In Figure 2B, for example, the PNA(A158T) probe (used in place of PNA(A158C) in Fig. 2A) has a T–G mismatch to the antisense strand in the C-allele(158) (ε3/ε3). Even under these conditions, both of the signals at m/z values of 3716.0 and 4005.1 (corresponding to the theoretical values 3715.6 and 4004.6) were clearly detectable. Similar results were obtained when two SNP sites in another homozygote ε2/ε2 were simultaneously genotyped using PNA(S112G) and PNA(A158T) (Fig. 2C). The first probe PNA(S112G) has a T–G mismatch to the sense strand. It is concluded that either a fully matched or single-base mismatched PNA probe provides sufficient amounts of ssDNA fragments for precise genotyping. These results are quite significant from a practical viewpoint, since the genotype of patients is of course unknown before the analysis. Even in these cases, genotyping is successfully achievable by using a PNA probe that is complementary with either of the two alleles. Regardless of whether the DNA specimen is completely complementary with this PNA probe or has a mismatch to the probe, sufficient amounts of ssDNA fragments for MS analysis are obtained.
When there were two mismatches between the target site in the DNA specimen and PNA probe (10mer, 12mer or 14mer), however, the DNA was almost completely digested by the enzymes and hardly any ssDNA fragments for MS analysis were obtained (data not presented). As expected, no ssDNA fragments were formed when the PNA probe is not complementary with any portion in the DNA specimen. The fidelity of the present genotyping has been confirmed.
Genotyping of heterozygotes involving two SNP sites
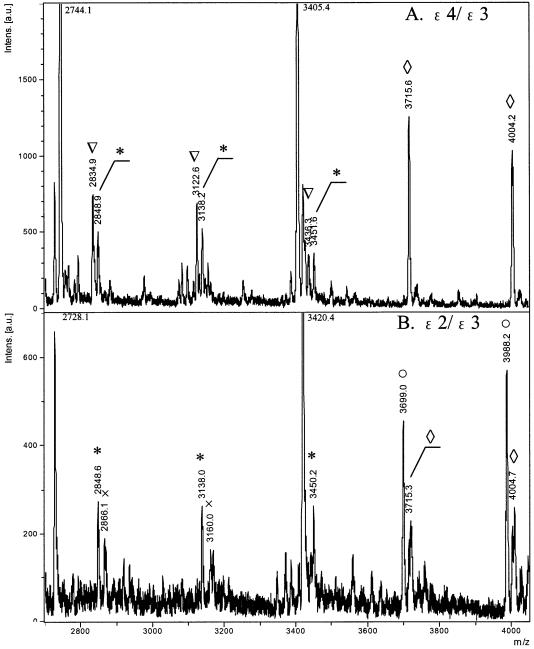
By using two PNA probes (PNA(S112G) and PNA(A158C)), two SNPs in the heterozygotes ε3/ε4 were simultaneously genotyped (see Fig. 3A). The 10mer probe PNA(S112G) is complementary with the sense strand from ε4, but has a mismatch to that of ε3. On the other hand, the 12mer probe PNA(A158C) is complementary with the antisense strands from both ε3 and ε4. The six major mass signals, marked with triangles and asterisks, are associated with codon 112. The three signals at m/z values of 2848.9, 3138.2 and 3451.6 (asterisks) are for the 9mer, 10mer and 11mer from the T-allele(112) in the isoform ε3 (Table 2). Although the PNA(S112G) probe has a mismatch to the sense strand DNA from ε3, the corresponding ssDNA fragments were efficiently obtained. The fragments from the C-allele(112) in ε4 are observed at m/z values of 2834.9, 3122.6 and 3436.3 (triangles). In the region for codon 158, only signals from the C-allele [3715.6 for the 12mer and 4004.2 for the 13mer (diamonds)] are detectable (both isoforms ε3 and ε4 are in the C-allele at the position 158). The mass signals from the PNA probes (m/z = 2744.1 and 3405.4) can serve as internal standards for accurate mass-calibration of the signals.
Figure 3.
Simultaneous genotyping of two SNP sites in heterozygous apoE genes by using exonuclease III, nuclease S1 and two PNA probes: (A) ε4/ε3 by PNA(S112G) and PNA(A158C) probes; (B) ε2/ε3 by PNA(S112A) and PNA(A158T) probes. The signals from codon 112 are shown by either triangles (from the C-allele(112)) or asterisks (from the T-allele(112)), whereas the signals from codon 158 are shown by either diamonds (from the C-allele(158)) or open circles (from the T-allele(158)). The signals at m/z values of 2728.1, 2744.1, 3405.4 and 3420.4 are from the PNA probes (PNA(S112A), PNA(S112G), PNA(A158C) and PNA(A158T), respectively). Reaction conditions: [dsDNA (203 bp)] = 2.5 µM, [PNA probes] = 2.5 µM, [exonuclease III] = 5.0 units/µl at pH 8.0 and 37°C for 5 min and [nuclease S1] = 3.0 units/µl at pH 4.6 and 20°C for 12 min. Mass spectra were recorded in a negative ion mode on a Bruker Daltonics® Autoflex mass spectrometer.
The results of genotyping of another heterozygote, ε2/ε3, are presented in Figure 3B. Of the two probes used, PNA(S112A) is complementary with the sense strands from both ε2 and ε3. However, PNA(A158T) has a mismatch to the antisense strand from ε3. The three signals at 2848.6, 3138.0 and 3450.2 (asterisks) are from the T-allele in codon 112 (both ε2 and ε3 are in the T-allele). Two pairs of signals in the region for codon 158 [3699.0 and 3988.2 (open circles); 3715.3 and 4004.7 (diamonds)] correspond to the T-allele from ε2 and the C-allele from ε3, respectively. In a single experiment using two PNA probes (one for codon 112 and another for codon 158), the heterozygotes of apoE gene have been completely genotyped.
Reaction conditions for the enzymatic treatments
Exonuclease III is specific to the hydrolysis of dsDNA. Accordingly, the ssDNA portions formed by pretreatment with this enzyme are hardly digested even in prolonged reactions (42). The reaction conditions were not rigorously chosen.
On the other hand, conditions for the digestion by nuclease S1 should be carefully controlled. In the present study, 0.8–1.8 units were used for 1 pmol of dsDNA, and the reaction was performed at 20°C for 10–20 min. Under these conditions, predetermined ssDNA fragments were successfully obtained. At higher reaction temperatures with larger amounts of the enzyme (e.g. 2.0 units at 25°C for 15 min), however, even the DNA in the DNA/PNA duplex was partially digested. This undesired digestion was efficient and small ssDNA fragments were hardly obtained when there existed a mismatch between the sample ssDNA and the PNA probe (50).
The digestion by nuclease S1 is successful even in the presence of exonuclease III. Furthermore, PNA probes show no significant inhibitory effects on the DNA digestion by exonuclease III. These results indicate that the present methods are applicable to microarray technology where PNA probes are immobilized to solid supports. By simply adding exonuclease III and nuclease S1 consecutively to these array systems, the desired DNA fragments of predetermined length for MS analysis should be easily prepared.
CONCLUSIONS
By combining two enzymes (exonuclease III and nuclease S1) and PNA probes, SNPs in dsDNA substrates have been directly, precisely and rapidly genotyped. The substrates are partially digested by these enzymes consecutively in the presence of PNA probes, and the resultant ssDNA fragments are analyzed by MALDI-TOF MS. Since dsDNA as PCR products can be directly employed for the analysis, time-consuming preparation of ssDNA specimens is unnecessary. Two (or more) SNP sites in a gene (or those in two genes which are adjacent to each other in genomic DNA) are simultaneously analyzable in a one-pot reaction. Quantitative analysis of multiple SNP sites should be facilitated. Furthermore, the present analysis requires no specific reaction conditions for the genotyping, since all the enzymatic treatments are achieved under conditions where the PNA probes are completely bound to either fully matched or single-base mismatched DNA. Thus, any analysis temperature below the melting temperatures of these duplexes is available, providing a significant advantage for high-throughput genotyping.
Importantly, short DNA fragments for MALDI-TOF MS are successfully obtained, even when there exists a mismatch between the DNA specimen and PNA probe. Thus the design of PNA probes for clinical genotyping requires no information on the genotype of a patient. Instead, we can simply use PNA that is complementary with either of the alleles. Whether the probe is complementary with the target genome or has a mismatch, conclusive MALDI-TOF MS is accomplished. Successful genotyping of dsDNA is undoubtedly advantageous for a variety of applications.
It is noteworthy that DNA specimens are almost completely digested and no MS signals are provided when they have two or more mismatches to the PNA probes. Thus the possibility that a mass signal from an unknown portion in a huge DNA induces critical errors in genotyping is highly unlikely. The lengths of PNA probes can be freely chosen, and ssDNA fragments of predetermined length are successfully obtained. Thus overlap of mass signals, if any, can be minimized. It should be possible to reduce the number of the fragments formed by the digestion with nuclease S1 in the presence of PNA by appropriate chemical modification of PNA [e.g. attachment of acridine to the PNA probe (51)].
Acknowledgments
ACKNOWLEDGEMENTS
We thank Professor Hiroyuki Aburatani of the University of Tokyo for valuable comments and also for a generous supply of human genomic DNA specimens. This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, Culture and Technology, Japan. The support of PROBRAIN is also acknowledged.
DDBJ/EMBL/GenBank accession no.+ AF261279
REFERENCES
- 1.Brookes A.J. (1999) The essence of SNPs. Gene, 234, 177–186. [DOI] [PubMed] [Google Scholar]
- 2.International SNP Map Working Group (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature, 409, 928–933. [DOI] [PubMed] [Google Scholar]
- 3.Roses A.D. (2000) Pharmacogenetics and the practice of medicine. Nature, 405, 857–865. [DOI] [PubMed] [Google Scholar]
- 4.Carlson C.S., Newman,T.L. and Nickerson,D.A. (2001) SNPing in the human genome. Curr. Opin. Chem. Biol., 5, 78–85. [DOI] [PubMed] [Google Scholar]
- 5.Kirk B.W., Feinsod,M., Favis,R., Kliman,R.M. and Barany,F. (2002) Single nucleotide polymorphism seeking long term association with complex disease. Nucleic Acids Res., 30, 3295–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Syvänen A.-C. (2001) Accessing genetic variation: genotyping single nucleotide polymorphisms. Nat. Rev. Genet., 2, 930–942. [DOI] [PubMed] [Google Scholar]
- 7.Gut I.G. (2001) Automation in genotyping of single nucleotide polymorphisms. Hum. Mutat., 17, 475–492. [DOI] [PubMed] [Google Scholar]
- 8.Landegren U., Nilsson,M. and Kwok,P.-Y. (1998) Reading bits of genetic information: methods for single-nucleotide polymorphism analysis. Genome Res., 8, 769–776. [DOI] [PubMed] [Google Scholar]
- 9.Wang D.G., Fan,J.-B., Siao,C.-J., Berno,A., Young,P., Sapolsky,R., Ghandour,G., Perkins,N., Winchester,E., Spencer,J. et al. (1998) Large-scale identification, mapping and genotyping of single-nucleotide polymorphisms in the human genome. Science, 280, 1077–1082. [DOI] [PubMed] [Google Scholar]
- 10.Griffin T.J., Tang,W. and Smith,L.M. (1997) Genetic analysis by peptide nucleic acid affinity MALDI-TOF mass spectrometry. Nat. Biotechnol., 15, 1368–1372. [DOI] [PubMed] [Google Scholar]
- 11.Prince J.A., Feuk,L., Howell,W.M., Jobs,M., Emahazion,T., Blennow,K. and Brookes,A.J. (2001) Robust and accurate single nucleotide polymorphism genotyping by dynamic allele-specific hybridization (DASH): design criteria and assay validation. Genome Res., 11, 152–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nickerson D.A., Kaiser,R., Lappin,S., Stewart,J., Hood,L. and Landegren,U. (1990) Automated DNA diagnostic using an ELISA-based oligonucleotide ligation assay. Proc. Natl Acad. Sci. USA, 87, 8923–8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X., Livak,K.J. and Kwok,P.-Y. (1998) A homogeneous, ligase-mediated DNA diagnostic test. Genome Res., 8, 549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Syvänen A.-C. (1999) From gels to chips: “minisequencing” primer extension for analysis of point mutations and single nucleotide polymorphisms. Hum. Mutat., 13, 1–10. [DOI] [PubMed] [Google Scholar]
- 15.Lyamichev V., Mast,A.L., Hall,J.G., Prudent,J.R., Kaiser,M.W., Takova,T., Kwiatkowski,R.W., Sander,T.J., de Arruda,M., Arco,D.A. et al. (1999) Polymorphism identification and quantitative detection of genomic DNA by invasive cleavage of oligonucleotide probes. Nat. Biotechnol., 17, 292–296. [DOI] [PubMed] [Google Scholar]
- 16.Mein C.A., Barratt,B.J., Dunn,M.G., Siegmund,T., Smith,A.N., Esposito,L., Nutland,S., Stevens,H.E., Wilson,A.J., Phillips,M.S. et al. (2000) Evaluation of single nucleotide polymorphism typing with invader on PCR amplicons and its automation. Genome Res., 10, 330–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo J., Bergstrom,D.E. and Barany,F. (1996) Improving the fidelity of Thermus thermophilus DNA ligase. Nucleic Acids Res., 24, 3071–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuppuswamy M.N., Hoffmann,J.W., Kasper,C.K., Spitzer,S.G., Groce,S.L. and Bajaj,S.P. (1991) Single nucleotide primer extension to detect genetic diseases: experimental application to hemophilia B (factor IX) and cystic fibrosis genes. Proc. Natl Acad. Sci. USA, 88, 1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rao K.V.N., Stevens,P.W., Hall,J.G., Lyamichev,V., Neri,B.P. and Kelso,D.M. (2003) Genotyping single nucleotide polymorphisms directly from genomic DNA by invasive cleavage reaction on microspheres. Nucleic Acids Res., 31, e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang-Baucom P. and Girard,J.E. (1997) DNA typing of human leukocyte antigen sequence polymorphisms by peptide nucleic acid probes and MALDI-TOF mass spectrometry. Anal. Chem., 69, 4894–4898. [DOI] [PubMed] [Google Scholar]
- 21.Pastinen T., Kurg,A., Metspalu,A., Peltonen,L. and Syvänen,A.-C. (1997) Minisequencing: a specific tool for DNA analysis and diagnostics on oligonucleotide arrays. Genome Res., 7, 606–614. [DOI] [PubMed] [Google Scholar]
- 22.Stoerker J., Mayo,J.D., Tetzlaff,C.N., Sarracino,D.A., Schwope,I. and Richert,C. (2000) Rapid genotyping by MALDI-monitored nuclease selection from probe libraries. Nat. Biotechnol., 18, 1213–1216. [DOI] [PubMed] [Google Scholar]
- 23.Dong S., Wang,E., Hsie,L., Cao,Y., Chen,X. and Gingeras,T.R. (2001) Flexible use of high-density oligonucleotide arrays for single-nucleotide polymorphism discovery and validation. Genome Res., 11, 1418–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross P., Hall,L., Smirnov,I. and Haff,L. (1998) High level multiplex genotyping by MALDI-TOF mass spectrometry. Nat. Biotechnol., 16, 1347–1351. [DOI] [PubMed] [Google Scholar]
- 25.Germer S., Holland,M.J. and Higuchi,R. (2000) High-throughput SNP allele-frequency determination in pooled DNA samples by kinetic PCR. Genome Res., 10, 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tost J. and Gut,I.G. (2002) Genotyping single nucleotide polymorphisms by mass spectrometry. Mass Spectrom. Rev., 21, 388–418. [DOI] [PubMed] [Google Scholar]
- 27.Lechner D., Lathrop,G.M. and Gut,I.G. (2001) Large-scale genotyping by mass spectrometry: experience, advances and obstacles. Curr. Opin. Chem. Biol., 6, 31–38. [DOI] [PubMed] [Google Scholar]
- 28.Bray M.S., Boerwinkle,E. and Doris,P.A. (2001) High-throughput multiplex SNP genotyping with MALDI-TOF mass spectrometry: practice, problems and promise. Hum. Mutat., 17, 296–304. [DOI] [PubMed] [Google Scholar]
- 29.Laken S.J., Jackson,P.E., Kinzler,K.W., Vogelstein,B., Strickland,P.T., Groopman,J.D. and Friesen,M.D. (1998) Genotyping by mass spectrometric analysis of short DNA fragments. Nat. Biotechnol., 13, 1352–1356. [DOI] [PubMed] [Google Scholar]
- 30.Griffin T.J., Hall,J.G., Prudent,J.R. and Smith,L.M. (1999) Direct genetic analysis by matrix-assisted laser desorption/ionization mass spectrometry. Proc. Natl Acad. Sci. USA, 96, 6301–6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haff L.A. and Smirnov,I.P. (1997) Single-nucleotide polymorphism identification assays using a thermostable DNA polymerase and delayed extraction MALDI-TOF mass spectrometry. Genome Res., 7, 378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jurinke C., van den Boom,D., Jacob,A., Tang,K., Wörl,R. and Köster,H. (1996) Analysis of ligase chain reaction products via matrix-assisted laser desorption/ionization time-of-flight-mass spectrometry. Anal. Biochem., 237, 174–181. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen P.E., Egholm,M., Berg,R.H. and Buchardt,O. (1991) Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science, 254, 1497–1500. [DOI] [PubMed] [Google Scholar]
- 34.Egholm M., Buchardt,O., Christensen,L., Behrens,C., Freier,S.M., Driver,D.A., Berg,R.H., Kim,S.K., Norden,B. and Nielsen,P.E. (1993) PNA hybridizes to complementary oligonucleotides obeying the Watson–Crick hydrogen-bonding rules. Nature, 365, 566–568. [DOI] [PubMed] [Google Scholar]
- 35.Hyrup B. and Nielsen,P.E. (1996) Peptide nucleic acids (PNA): synthesis, properties and potential applications. Bioorg. Med. Chem., 4, 5–23. [DOI] [PubMed] [Google Scholar]
- 36.Uhlmann E., Peyman,A., Breipohl,G. and Will,D.W. (1998) PNA: synthetic polyamide nucleic acids with unusual binding properties. Angew. Chem. Int. Ed., 37, 2796–2823. [DOI] [PubMed] [Google Scholar]
- 37.Hixson J.E. and Vernier,D.T. (1990) Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with Hha I. J. Lipid Res., 31, 545–548. [PubMed] [Google Scholar]
- 38.Weisgraber K.H. (1994) Apolipoprotein E: structure-function relationships. Adv. Protein Chem., 45, 249–302. [DOI] [PubMed] [Google Scholar]
- 39.Davignon J., Cohn,J.S., Mabile,L. and Bernier L. (1999) Apolipoprotein E and atherosclerosis: insight from animal and human studies. Clin. Chim. Acta, 286, 115–143. [DOI] [PubMed] [Google Scholar]
- 40.Meyer M.R., Tschanz,J.T., Norton,M.C., Welsh-Bohmer,K.A., Steffens,D.C., Wyse,B.W. and Breitner,J.C.S. (1998) APOE genotype predicts when—not whether—one is predisposed to develop Alzheimer disease. Nature Genet., 19, 321–322. [DOI] [PubMed] [Google Scholar]
- 41.Strittmatter W.J. and Roses,A.D. (1995) Apolipoprotein E and Alzheimer disease. Proc. Natl Acad. Sci. USA, 92, 4725–4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rogers S.G. and Weiss,B. (1980) Exonuclease III of Escherichia coli K-12, an AP endonuclease. Methods Enzymol., 65, 201–211. [DOI] [PubMed] [Google Scholar]
- 43.Roychoudhury R. and Wu,R. (1977) Novel properties of Escherichia coli exonuclease III. J. Biol. Chem., 252, 4786–4789. [PubMed] [Google Scholar]
- 44.Mol C.D., Che-FuKuo, Thayer,M.M., Cunningham,R.P. and Tainer,J.A. (1995) Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature, 374, 381–386. [DOI] [PubMed] [Google Scholar]
- 45.Shishido K. and Habuka,N. (1986) Purification of S1 nuclease to homogeneity and its chemical, physical and catalytic properties. Biochim. Biophys. Acta, 884, 215–218. [DOI] [PubMed] [Google Scholar]
- 46.Silber J.R. and Loeb,L.A. (1981) S1 nuclease does not cleave DNA at single-base mismatches. Biochim. Biophys. Acta, 656, 256–264. [DOI] [PubMed] [Google Scholar]
- 47.Desai N.A. and Shankar,V. (2003) Single-strand-specific nuclease. FEMS Microbiol. Rev., 26, 457–491. [DOI] [PubMed] [Google Scholar]
- 48.Ye S., Liang,X., Yamamoto,Y. and Komiyama,M. (2003) DNA protection by PNA from enzymatic digestion for mass-spectroscopic genotyping. Chem. Lett., 32, 10–11. [Google Scholar]
- 49.Demidov V., Frank-Kamenetskii,M.D., Egholm,M., Buchardt,O. and Nielsen,P.E. (1993) Sequence selective double strand DNA cleavage by peptide nucleic acid (PNA) targeting using nuclease S1. Nucleic Acids Res., 21, 2103–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Komiyama M., Ye,S., Liang,X., Yamamoto,Y., Tomita,T., Zhou,J.-M. and Aburatani,H. (2003) PNA for one-base differentiating protection of DNA from nuclease and its use for SNPs detection. J. Am. Chem. Soc., 125, 3758–3762. [DOI] [PubMed] [Google Scholar]
- 51.Ren B., Ye,S., Liang,X. and Komiyama M. (2003) Acridine-bearing PNA for efficient protection of designated site of DNA from nuclease S1. Chem. Lett., 32, 714–715. [Google Scholar]