Abstract
Bacteriophage ATP-based packaging motors translocate DNA into a pre-formed prohead through a dodecameric portal ring channel to high density. We investigated portal–terminase docking interactions at specifically localized residues within a terminase-interaction region (aa279–316) in the phage T4 portal protein gp20 equated to the clip domain of the SPP1 portal crystal structure by 3D modeling. Within this region, three residues allowed A to C mutations whereas three others did not, consistent with informatics analyses showing the tolerated residues are not strongly conserved evolutionarily. About 7.5 nm was calculated by FCS-FRET studies employing maleimide Alexa488 dye labeled A316C proheads and gp17 CT-ReAsH supporting previous work docking the C-terminal end of the T4 terminase (gp17) closer to the N-terminal GFP-labeled portal (gp20) than the N-terminal end of the terminase. Such a terminase–portal orientation fits better to a proposed “DNA crunching” compression packaging motor and to portal determined DNA headful cutting.
Keywords: T4 terminase, T4 portal clip region, FCS-FRET, Phyre2
Introduction
Nucleic acid translocation into a pre-formed prohead or procapsid of icosahedral double-stranded DNA and RNA bacteriophages and herpesviruses is a conserved capsid assembly mechanism (Kainov et al., 2006). Many large dsDNA viruses employ high force-generating, powerful ATP driven molecular motors to package their genomes into proheads (Smith et al., 2001). DNA packaging in the dsDNA viruses and herpesviruses involves multiple steps beginning with recognition of concatemeric viral DNA, cutting of DNA to generate an end, translocation of DNA into the preformed capsid, condensation to a near crystalline density inside the capsid, and headful cutting of DNA to terminate DNA packaging. The essential components of the packasome motor appear to share many structural and mechanistic features among intensively studied double stranded DNA phages T4, lambda, P22, SPP1, φ29, as well as others (Johnson and Chiu, 2007). In most tailed dsDNA bacteriophages translocation is driven by a two-component motor consisting of a dodecameric portal situated at a unique prohead vertex that is docked during packaging with a multimeric terminase-ATPase (Black, 1989; Casjens, 2011); in phage φ29 the packaging ATPase docks on a pRNA connected to the portal or connector (Schwartz et al., 2013). It is now generally accepted that packaging operates by a linear rather than a rotary motor mechanism (Baumann et al., 2006; Hugel et al., 2007). Two specific ATP-driven linear motor mechanisms have been proposed for phage T4 DNA packaging: an electrostatic model (Sun et al., 2008), and a “DNA crunching” or compression model (Oram et al., 2008; Ray et al., 2010).
DNA packaging of replicated concatemeric T4 DNA in vivo is dependent upon the small terminase subunit gp16 whereas the small subunit is not required for packaging linear DNA in vitro. A number of lines of evidence suggest that maturation of the T4 concatemer is gauged by synapsis of two pac site DNAs by the gp16 that leads to packaging initiation (Black, 1995; Lin and Black, 1998; Lin et al., 1997). Following gp16 synapsis of two pac site DNA segments, DNA is handed off to the large terminase-ATPase protein. The large terminase protein gp17 makes the first cut to generate a free end for packaging. The terminase–DNA complex then docks the free end of DNA to the empty prohead by interacting with the portal protein gp20, which is situated at the special fivefold vertex of the capsid (Lin et al., 1999). Using ATP hydrolysis as the driving force, the terminase protein translocates DNA through the portal channel into the capsid (Baumann and Black, 2003; Kondabagil et al., 2006). Once a headful volume of DNA is translocated, gp17 makes a cut to terminate packaging and dissociate from the packaged prohead. The large terminase protein gp17 is a 70 kDa protein. It has an N-terminal ATPase domain (aa 1–360) which fuels the DNA translocation and a C-terminal nuclease domain (aa 360–577) which cleaves the DNA before and after packaging (Kanamaru et al., 2004). A high-resolution structure of a truncated phage T4 large terminase subunit gp17 and a low resolution structure (34 Å) of the terminase on the prohead have been determined (Sun et al., 2008).
The capsid portal protein is central to the morphogenesis and structure of all dsDNA phages as well as other viruses. The portal complex is located at a specialized vertex and acts as a connector between the head and the tail. Importantly, the portal vertex provides the docking site for the terminase during DNA translocation and also acts as a headful packaging gauge by controlling the length of the DNA packaged (Casjens et al., 1992; Tavares et al., 1992). Structural studies of several phage portals (φ29, SPP1, P22, and T7) reveal dodecamers with an open channel through which DNA passes and highly conserved architecture (Kang et al., 2008; Lebedev et al., 2007; Simpson et al., 2001; Tang et al., 2011). The overall structure and symmetry of the portal protein is strictly conserved among the bacteriophages and herpesviruses. The T4 portal was the first shown to be a dodecameric turbine shaped structure 14 nm long and 7 nm in diameter with a cylinder with a hollow central channel of 3–4 nm through which DNA is pumped to high density (~500 mg/ml) within the prohead (100 nm long and 75 nm wide) (Driedonks et al., 1981). [For phage T4 packaging component models, see Figs. 5 and 6].
Fig. 5.
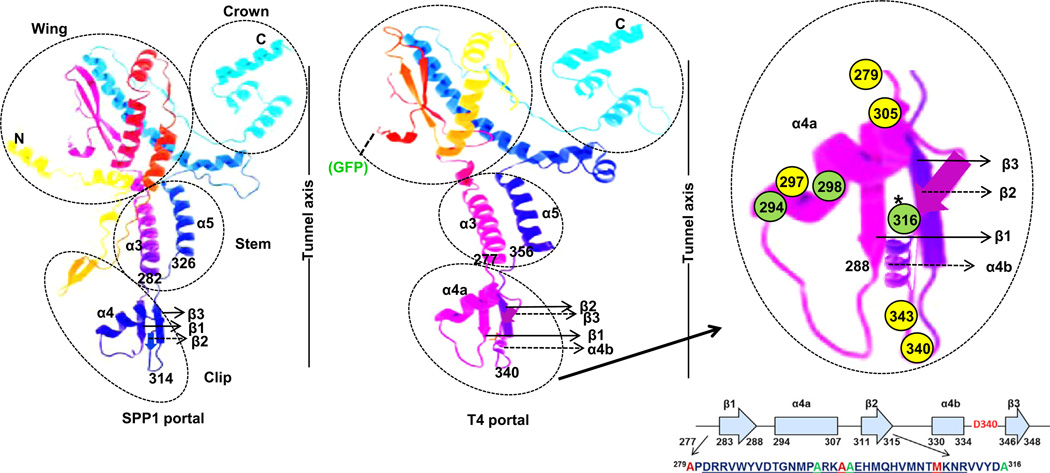
Tertiary structure prediction of T4 portal using the Phyre2 program. Model is based on template (PDBID-2jes) (SPP1 portal). 296 residues (56% of sequence) have been modeled with 78.7% confidence by the single highest scoring template. Of amino acids assigned in the clip region, those in yellow circles could not be substituted by cysteine (279, 297 and 305) or with any other amino acids (340, 343), amino acids in green circles could be substituted with cysteine. FRET transfer between CT-ReAsH terminase and Alexa 488 labeled proheads was observed with A316C residue modification. Secondary structures that are predicted by the Phyre2 program are shown by solid arrows whereas those predicted by PSIPRED analysis and not by the Phyre2 program are shown by dashed arrows. The Crown, Wing, Stem, and Clip regions of the SPP1 portal crystal structure are shown in relation to the DNA tunnel axis. The position of the N-terminal GFP in the predicted T4 portal is shown.
Fig. 6.
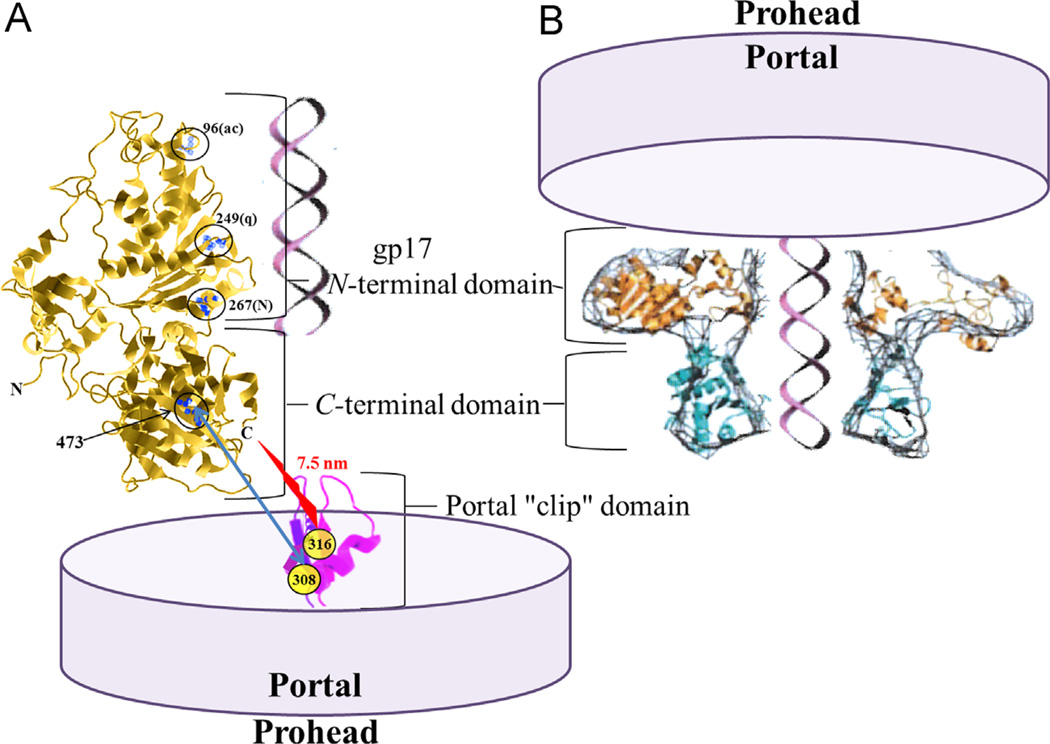
Phage T4 prohead portal gp20 binding to the terminase from (A) the C-terminal nuclease containing domain (Dixit et al., 2012) or (B) the N-terminal ATPase containing domain of the terminase (Feiss and Rao, 2012). DNA is depicted being translocated by the terminase ATP-driven motor through the portal tunnel into the prohead. In (A) the terminase intercalating dye resistance mutations at residues 96, 249, and 267 are circled, together with the circled residue 473 ts DNA translocation mutation suppressed by portal cs mutation clip domain residue 308 (blue arrow). A FRET determined distance of 7.5 nm between portal clip domain Alexa488 maleimide dye labeled residue 316 and the terminase C-terminal ReAsH dye is found in this study (red jagged arrow). Structures are not to scale, nor are the numbers of terminase or portal monomers represented in the multimeric structure constituting the two-component packaging motor complex.
Although genetic and biochemical studies were compatible with portal binding sites in gp17 in the C-terminal as well as in the N-terminal domain (Lin et al., 1999), two studies proposed that the N-terminal ATPase domain of gp17 interacts with portal protein gp20 whereas the C-terminal nuclease domain is not critical for binding (Hegde et al., 2012; Sun et al., 2008). However in our previous and present work using FCS-FRET based measurements and genetic studies we have found that although both the N-terminal and C-terminal ends of the T4 terminase are located in proximity to the portal, the C-terminal domain is closer to the portal and plays a more important role in interacting with the T4 portal during translocation (Dixit et al., 2011; Dixit et al., 2012). The N-terminal ATPase portion of the terminase is important during initiation of packaging as well as during translocation but it is unlikely to be directly apposed to the portal during translocation (see Discussion). The dynamic interactions between the motor (gp17) and the portal (gp20) are still not well understood. In this report, using bioinformatics we have predicted a tertiary structure for the exposed terminase binding or clip region of the T4 portal, also utilizing biochemical and genetic approaches we have identified critical residues in this clip region that play an important role in portal function by their role in gp17 interaction.
Results
Informatics analysis of specific residues in a central portal peptide clip region critical for portal function
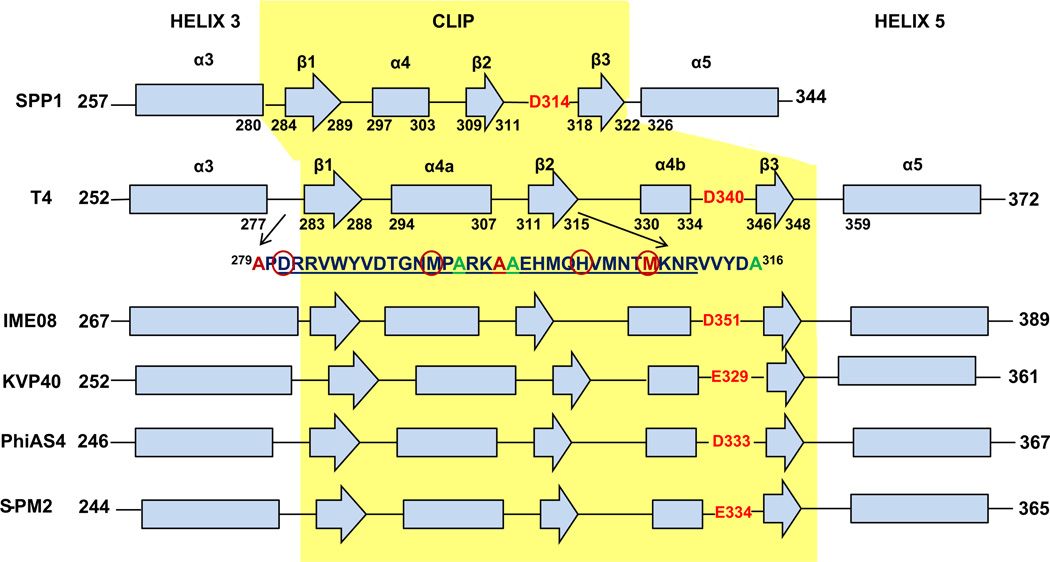
Earlier genetic, biochemical, and immunological studies established that the terminase gp17 interaction site is localized within a central D279-A316 region of the 521 residue prohead portal gp20 protein (Lin et al., 1999). We mutated six amino acid residues to cysteine within this central peptide region of gp20 to allow labeling of the portal protein with maleimide cysteine specific reactive dyes. Following site directed mutagenesis of A279C, A294C, A297C, A298C, M305C and A316C in the portal gene cloned in a plasmid followed by strong selection for recombination into the phage genome only amino acid substitutions A294C, A298C, and A316C were tolerated, allowing viable phages to be produced, as judged by phage plaque formation (Table 1). SAM (Sequence Alignment Modeling) alignment of the region incorporating the clip (residues 260–379) of 59 portal proteins (Fig. S1) homologous to T4 gp20 revealed that A279 and A297 are perfectly conserved and that M305 is strongly conserved (see Fig. 1 for representative T4-like homologs). In contrast, two residues (A294 and A298) that allowed cysteine substitution are very poorly conserved. Residues A294, A297, A298 and M305 are predicted to be in the α4a motif (aa 294–307) of the clip and the sequence of this region is overall not well conserved among various T4 like homologs (Fig. S1). Similarly, A316 could also be substituted in the phage and appears to be only moderately conserved. This is consistent with its positioning, immediately after the strongly conserved portion of the clip region in a linker region immediately following the β2 motif and prior to α4b (see later structural analysis). Additionally, residues D340 (the so-called “tunnel-loop” residue which resides at the tunnel entrance) and A343 mutated to amber codons did not apparently allow substitution by any other amino acids inserted by 13 different available amber suppressor hosts as tested (Dixit et al., 2012). It seemed unexpected that D340 could not be substituted with a glutamate residue, despite the fact that glutamate is found at this critical turn residue in some phage portals (Lebedev et al., 2007) (Fig. 1 and Fig. S1). However, inspection of this residue in homologs indicates that an aspartate in this position is apparently perfectly conserved in the phages considered to be most closely related to T4, and it is only in the more diverged T4-like phages, such as KVP40 and S-PM2 (percent identity of portal matches are 46% and 38%, respectively) that this position is held by a glutamate. Overall these results suggest that residues A279, A297, M305, D340 and A343 are critical for the T4 portal structure and function and hence do not allow substitution. However, residues A294, A298 and A316 are likely not as important for portal function and can be substituted.
Table 1.
Functional and non-functional cysteine substitutions at different alanine residues close to the gp17 interaction site in the clip region of the gp20 prohead portal.
| Cysteine mutations | Functional mutations |
|---|---|
| A279C | − |
| A294C | + |
| A297C | − |
| A298C | + |
| M305C | − |
| A316C | + |
Fig. 1.
Clustalw2 alignment of T4-like phages’ portal clip regions. Amino acids in red could not be substituted in the phage and are conserved in T4-like phages. Amino acids in blue could be substituted and are not conserved in T4-like phages. The codons for the amino acids in pink were replaced by amber codons but could not be substituted by any other amino acid. Note D340 is the negatively charged residue at the tunnel entrance (see also Supplementary Fig. 1).
Specific labeling of the T4 portal clip region
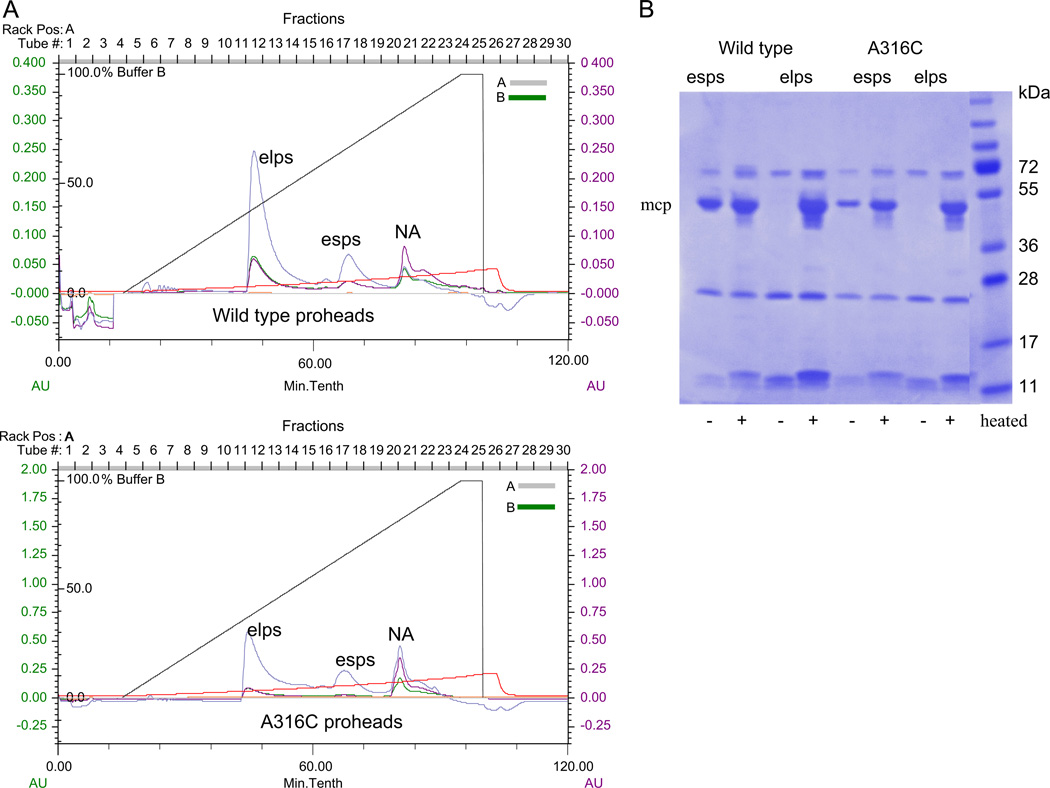
To label specifically the portal at the previously predicted gp17 interaction site, functional gp20 mutants A294C, A298C, and A316C were considered for cysteine specificmaleimide dye labeling studies. By Typhoon image analysis the A294C modified proheads showed no portal protein labeling with Alexa 488 maleimide dye (data not shown). Since a predicted clip structure suggested that the A298C modification is very close to residue 294 (~5 Å) and in a similar alpha-helical environment, labeling of this position was not tried. However Alexa 488 maleimide dye labeling of A316C modified proheads did succeed in showing specific labeling of the portal protein. Purification profiles of the wild type and A316C portal proheads produced as previously described by infection with a portal and terminase defective multiple amber mutant (see Methods, and Baumann et al., 2006) were comparable in showing the presence of a major empty large prohead (elps) peak and a minor empty small prohead (esps) peak (Fig. 2A and B). When the Alexa 488 labeled wild type and A316C proheads were observed for fluorescence on an agarose gel, both of them appeared to be comparably labeled due to uptake of dye into additional prohead proteins such as the major capsid protein gp23* and/or gp24* vertex protein (Fig. 3A). Whether this is due to labeling of the single cysteine residue in gp23* and/or labeling of the three cysteine residues in gp24* or to less specific chemical side reaction additions to these proteins that are much more abundant in the prohead (930 and 55 copies) than the gp20 portal protein (12 copies) is uncertain. The non-portal directed labeling of the proheads could not be avoided despite trying different times, temperatures, and different prohead to dye ratios for maleimide labeling (data not shown). But when the Typhoon image and SDS-PAGE profiles of the labeled wild type and A316C proheads were compared, specific labeling of the gp20 protein was observed only for the mutated gp20 (Fig. 3A). Evidently the three cysteines already present in the wild type gp20 protein are much less reactive than the SDM residue introduced into the clip region. However as shown in the next section non-specific labeling of the proheads did not interfere with our interaction experiments as wild type proheads did not show in several experiments any detectable FRET measured with CT-ReAsH terminase.
Fig. 2.
Purification and SDS-PAGE profiles of the T4 wild type and portal gp20 A316C mutant proheads. (A) Wild type and gp20 A316C proheads produced predominantly mature empty large proheads (elps) with a smaller amount of empty small proheads (esps) as seen by the linear salt gradient (straight brown line) purification profiles on FPLC-DEAE columns. The third peak in the chromatogram is nucleic acid (NA). Cyan, green, and purple are the 214, 280, and 260 nm traces respectively. (B) elps and esps are distinguished by release of the major capsid protein (mcp) gp23 into the gel by not heating (−) or heating (+) prohead samples prior to 10% SDS-PAGE electrophoresis followed by Coomassie staining.
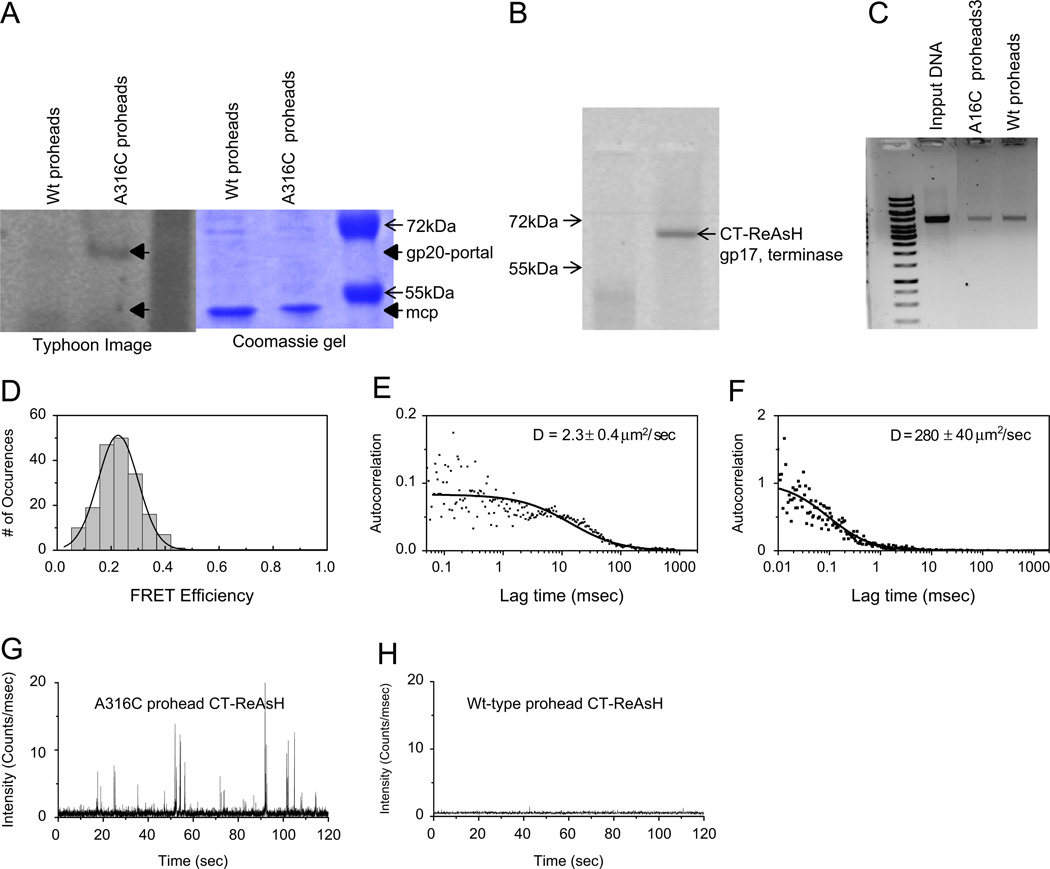
Fig. 3.
FRET properties of Alexa Fluor 488 C5-maleimide dye labeled wild type and portal A316C proheads. (A) Typhoon image and Coomassie stained image of dye labeled wild type and A316C proheads, with gp20-portal and gp23* major capsid protein (mcp) positions indicated. (B) Typhoon image of specific tetra-cysteine peptide binding ReAsH dye labeled CT-terminase, without (left lane) and with (right lane) added dye. (C) Nuclease protection assay gel showing packaged DNA in wild type and A316C proheads. (D) FRET-FCS analysis as described in methods of the Alexa 488 maleimide labeled A316C proheads and CT-gp17 labeled with ReAsH dyes revealing a C-terminal terminase to portal clip distance of 7.5 nm. (E) Diffusion coefficient of the A316C proheads (~2 µm2/s) acceptor channel from the correlated emissions. (F) FRET-FCS analysis of the Alexa 488 maleimide labeled Wt proheads and CT-gp17 labeled with ReAsH dyes showing a diffusion coefficient of ~280 µm2/s in the acceptor channel from less correlated emissions; (G) acceptor channel counts for the A316C proheads with CT-ReAsH; (H) acceptor channel counts for the wild type proheads with CT-ReAsH.
C-terminal domain of the terminase interacts with packaging competent dye labeled A316C portal proheads
Alexa 488 maleimide dye labeled A316C portal proheads were competent for DNA packaging albeit less so than control wild type labeled proheads when assayed with CT-ReAsH terminase by nuclease assay (Fig. 3B and C). Packaging mixtures containing either wild type (as control) or mutant Alexa 488 C5 maleimide labeled A316C proheads along with the CT-ReAsH gp17 and other packaging components were used in the FCS-FRET assays. A FRET value of more than 20% was observed with the A316C mutant (Fig. 3D). The FCS determined diffusion coefficient measured in the FRET acceptor channel was ~2 µm2/s, comparable with our previous measurements of the prohead, and showing that the FRET is due to transfer from the ReAsH dye labeled terminase anchored to the more slowly diffusing A316C mutant portal prohead (Dixit et al., 2011). In contrast the wild type control proheads, although active in packaging, showed these emissions were essentially uncorrelated and likely due to background, yielding an apparent diffusion coefficient of ~280 µm2/s, with much lower fluorescence in the acceptor channel (compare Fig. 3G and H). From the FRET measurement of the A316C mutant, the donor-to-acceptor distance (r) is estimated to be 7.5 nm, given a Förster critical distance of 6.2 nm (calculated from fluorescence quantum yield and spectral overlap) (Huang et al., 2009). From the measured FRET distribution (Fig. 3D), and given that the effective FRET cutoff value for this dye pair is ~10 nm, the 7.5 nm distance measured between the two fluorophores apparently represents the closest possible approach of terminase and portal monomers to each other in the complex of terminase pentamer bound to portal dodecamer. Other expected positionings of the two dyes in the multimers should be too distant to contribute to the observed FRET. In our earlier studies CT-ReAsH gp17 showed a FRET of ~50% with gp20-GFP fusion (Dixit et al., 2011). The exact spatial position of this gp20-GFP portal fusion in the prohead is unknown, but the GFP is external to the prohead since it can be removed with protease (Baumann et al., 2006) and presumably protrudes from the portal wing close to the portal clip region of the prohead.
Predicted clip region structure of the portal
Although there is low sequence homology and large variation in subunit size among portal proteins of different phages (from 36 kDa in φ29 to 84 kDa in P22), the portal complexes of dsDNA bacteriophages and herpesviruses are architecturally conserved and share a dodecameric ringlike structure as is evident in their crystallography and cryoEM structures. Consistent with this level of structural conservation, we found that secondary structure predictions using PSIPRED (McGuffin et al., 2000) of various T4-like phages’ (SPP1, IME08, PhiAS4, KVP40 and S-PM2) portal proteins show quite similar secondary structure elements (Fig. 4). Our predictive structure also revealed the T4 clip secondary structure elements similar to those in the SPP1 clip region (Lebedev et al., 2007), hence we assigned them the same nomenclature. However, instead of the single seven residue α helix 4 in SPP1, there are two α helices which we named α4a and α4b, interrupted by the short (5 residues) β2 sheet, with α4a much longer (14 residues) than α4b (5 residues). Although essentially the same structural elements are present in the two portals, the T4 portal clip has considerably more mass (58 residues) than the SPP1 clip (31 residues) from the β1 start to the tunnel loop residue. Such structural differences could account for phage T4 portal clip cs mutations that allow normal prohead assembly but block initiation of DNA packaging at low temperature (see Fig. 4) whereas such temperature mutations are apparently not found in phage SPP1 (Isidro et al., 2004); i.e. the phage T4 clip might be more structurally mobile. The negatively charged “tunnel loop residue” at the entrance into the prohead interior (highlighted in red in Fig. 4) is found in all portal sequences for which X-ray structural information is available. The cone-shaped portal consists of a wide “crown” inside the capsid, an angular “stem” formed by long helices, two per subunit, and a narrow α,β domain “stalk” that protrudes outside the capsid (Lebedev et al., 2007). We have predicted a tertiary structure for the clip region of the T4 portal based exclusively on the SPP1 portal crystal structure (PDBID-2jes) and the Phyre2 program (Kelley and Sternberg, 2009) adjusted to accommodate the PSIPRED results (Fig. 5). Based on our secondary structure predictions and biochemical and mutagenic experiments we show predicted assigned positions for the residues that were selected for mutations. Locations of the residues that could not be substituted in the portal are shown in yellow circles and those that could be substituted are shown in green circles (Fig. 5). We observed FRET between CT-ReAsH terminase and portal labeled at the A316 residue position indicated with the green circle marked with an asterisk.
Fig. 4.
Comparison of the SPP1 portal secondary structure (derived from the crystal structure, PDBID 2JES) over the clip and flanking regions (α3–α6) with the corresponding regions of the portal proteins of T4 and other representative T4-like phages. The secondary structures of the T4-like phages were predicted using PSI-PRED. The peptide region (amino acids 279–316) of the T4 portal with the positions of green amino acids which were mutated to cysteine and used in the studies is shown. The residues of the T4 clip (D281-R311) that have a critical role interacting with gp17 interaction are underlined. Cold sensitive (cs) portal mutation sites that block initiation of packaging at low temperature in vivo are highlighted with red circles in the clip region.
Discussion
Previous genetic, biochemical, and antibody studies established that the gp17 interaction site resides in the central D279-A316 sequence of the gp20 portal protein (Lin et al., 1999). Extending upon this work we have now demonstrated using bioinformatics that this region in gp20 equates to the clip region in the SPP1 portal crystal structure, as shown in Fig. 5. The previous analysis (Lin et al., 1999) suggested two different potential interaction regions in the terminase, at central and C-terminal locations, judging by terminase to portal and portal to terminase intergenic suppressor mutations. However by new mutant isolation and resequencing old mutations the terminase–portal clip region suppressor interactions are shown to lead to two different packaging defective phenotypes, either packaging initiation deficiency or packaging completion deficiency (Dixit et al., 2012). Terminase packaging initiation deficiency and packaging completion deficiency mutations are generally associated with the N-terminal ATPase domain and C-terminal nuclease domain respectively. Thus a csN33 (cold sensitive) portal mutation in the portal clip region (mutated residues 281 and 292) can undergo a conformational change in proheads upon temperature increase that releases a block to initiation of packaging. This was established by electron microscopy of DNA empty proheads that accumulate in vivo before temperature shift as well as by full accessibility of the unpackaged DNA to glucosylation modification measured by restriction enzyme protection of the packaged phages (Black and Silverman, 1978). This portal clip cs mutation that blocks initiation of packaging can be suppressed by a terminase temperature sensitive mutation located in the central hinge region (residue 364) located between the gp17 ATPase and nuclease domains. However, the location of this terminase mutation makes direct contact between the portal and this mutated residue in the terminase unlikely (Dixit et al., 2012) (see Fig. 6).
A number of different terminase ts mutations (residues 473, 546, 562, and 584) that are located in the C-terminal nuclease domain display a distinct interrupted translocation phenotype (partially DNA filled proheads accumulate as judged by electron microscopy and by lack of restriction enzyme protection – evidence summarized in (Lin et al., 1999)). One such ts translocation mutation at terminase residue 473 can be suppressed by a cs mutation at residue 308 in the portal clip region (Fig. 6A). Direct physical interaction of the C-terminal domain of the terminase with the portal clip region as the basis for this intergenic suppression is supported by our current work showing that a single specifically labeled C-terminal residue 610 is located near to portal residue 316 during translocation.
The position of Alexa 488 dye labeled portal residue 316C is predicted to be 2.5–5 nm from turn residue 340 of the portal clip within the tunnel axis channel. Therefore residue 610 of the terminase nuclease domain should be located 2.5–5 nm outside and above the tunnel entrance for a total 7.5 nm FRET distance. This measured distance therefore appears consistent with the C-terminal domain docking on the portal shown in Fig. 6A, but inconsistent with the structure of N-terminal ATPase domain docking shown in Fig. 6B. Although there is some uncertainly because only a truncated gp17 crystal structure is available to fix the terminase end of the FRET ruler, if the C-terminal residues of the nuclease domain are compactly organized, as is suggested by the intragenic suppressors among residues extending to nearly the C-terminal end of this nuclease domain polypeptide (Dixit et al., 2012, see Fig. 5B) then the N-terminal docking model of the cryo-EM structure shows a separation of 10–13 nm, too great for the observed FRET.
Although the N- and C-terminal positions of the T4 terminase and N-terminus of the SPP1 portal are not fixed by crystal structures, it is known that the N-terminal end of the portal is located outside the prohead in the predicted wing structure (Baumann et al., 2006) (see Fig. 5). However despite these limitations to precise structure determination, previously we have shown that the portal–terminase distance between the C-terminus of gp17 was 5.1- or 5.7-nm (Y versus linear DNA distances) whereas the N-terminus of gp17 was at a greater distance of 6.9 nm. These values were determined by FRET between the GFP-gp20 and the C-terminal versus N-terminal ReAsH-labeled terminase. Therefore our current measured donor-to-acceptor distance of 7.5 nm is consistent with our previous FRET distance measurement of the N-terminal GFP-portal containing prohead at 5.7 nm from the C-terminus of the terminase (Dixit et al., 2011). This distance is consistent with our predicted portal structure showing a ~3 nm diameter GFP projecting out from the portal wing above the portal clip tunnel axis and closer to the C-terminal nuclease domain. Overall our measurements support our determination that the C-terminal nuclease domain of the terminase is closer to the portal than the N-terminal ATPase domain, where in our current work the portal dye position is fixed to a single residue predicted to be located within the critical central tunnel axis of the prohead.
It is believed that both the λ and T3 terminase–portal binding interactions are through C-terminal residues in these non-pac site phage terminases, although their high resolution structures are unknown (summarized in (Lin et al., 1999)). Significantly, however, recent work shows that the C-terminal nuclease domain of the pac site phage SPP1 terminase with a crystal structure (Smits et al., 2009) that is comparable to the T4 nuclease C-terminal domain is directly responsible for binding SPP1 terminase to the portal; moreover mutations in this SPP1 domain are characterized that are comparable to the T4 nuclease domain ts mutations we discuss above that block DNA translocation as well as modulate activity of the ATPase (Cornilleau et al., 2013). It should be noted additionally that one potential problem with the proposed T4 N-terminal ATPase domain to portal binding as modeled by cryo-EM (see Fig. 6B) (Feiss and Rao, 2012) is that it requires a counterintuitive terminase release and flipover to cut the DNA upon receiving a headful cleavage signal from the prohead portal; otherwise the headful cut would leave a terminase size DNA length hanging outside the prohead. On the other hand, C-terminal nuclease domain binding to the portal leaves the terminase well positioned to carry out the headful cut immediately adjacent to the portal entrance channel upon receiving the full prohead portal signal.
The Phyre2 tertiary structure of the T4 portal predicts that most of the gp20 central peptide region APDRRVWYVDTGNMPARKAAEHMQHVMNTMKNRVVYDA279– 316 are buried within the clip region (Fig. 5). This predicted three dimensional structure is in good agreement with the finding that an antiserum raised against a polymerized peptide (underlined residues 287–300 above) from this region of the clip could react with the denatured portal protein but not with the native protein (Lin et al., 1999). In addition the antiserum neither immunoprecipitated the portal protein containing prohead, nor did the antiserum inhibit DNA packaging. Furthermore, a portal clip residues 287–311 sequence synthetic peptide specifically inhibited in vitro DNA packaging, and the 287–300 residue peptide fused to the N terminus of the capsid surface T4 HOC display phage also interacted with a gp17 terminase affinity column and additionally was co-immunoprecipitated by a gp17 antiserum (Lin et al., 1999). Overall results suggest that these overlapping peptides from the clip region are inaccessible to antibodies in the native portal structure within the prohead whereas they are apparently accessible to the terminase in the portal terminase translocation complex. The observed difference in antibody and terminase binding most likely reflects conformational changes in the portal clip structure upon terminase binding during translocation. Higher resolution structures of the initiation and translocation docked terminase–portal complexes are required to better understand the dynamics of this two component motor complex as well as definitively resolve the orientation and numbers of packaging components.
A model in conjunction with the N-terminal ATPase portal docking model proposed that DNA packaging is driven by an electrostatic translocation force exerted on the DNA by amino acid residues in the T4 terminase nuclease domain (Sun et al., 2008). If the T4 terminase C-terminal nuclease domain in fact establishes contact with the portal the mechanism as proposed could not drive translocation. However there is no direct evidence identifying specific DNA–amino acid contacts made during translocation of DNA by the T4 or any other terminase. In fact, recent evidence favors multiple contacts at two or three sites (near residues 96, 249, 267, see Fig. 6A) between the N-terminal ATPase domain and the translocating DNA as the basis for the high motor force that is capable of “squeezing out” tight binding intercalating compounds (Dixit et al., 2012) or rupturing the phage lambda prohead (Fuller et al., 2007). Such an N-terminal ATPase domain translocation interaction fits better to the contacts known to be made between helicases and their DNA substrates (Velankar et al., 1999) as well as to the location of phage lambda terminase mutations in the ATPase domain that affect the DNA translocation motor force (Tsay et al., 2009). Moreover, given that the DnaB helicase acts also as a duplex DNA translocase, it is striking that a single stranded A form is observed to be extruded from B form DNA as visualized in a novel structure of this enzyme with its DNA substrate; additionally it is proposed that translocation on the duplex could also be on A-form (Itsathitphaisarn et al., 2012). Given comparable transient B to A form-like DNA compression that is supported by direct measurements in the T4 packaging system (Dixit et al., 2011, 2012; Ray et al., 2010) these two enzymes could well operate by quite similar molecular mechanisms.
Experimental procedures
Bacteriophage T4 strains/Escherichia coli hosts and growth conditions
T4 phage strain T4D (Doerman's wild-type strain) was used in the studies. Wild-type esps (empty small proheads) and elps (empty large proheads) were prepared by infection with T4 [13amE111 16amN66 17amA465 ΔrIIA H88] phages deficient in terminase (genes 16 and 17) and neck (gene 13) protein synthesis following procedures previously described (Black and Peng, 2006; Rao and Black, 1985); the rII mutation allows assay of phage assembly following packaging of mature T4 DNA and addition of tail components. E. coli strains used in the studies are suppressor strains CR63 for phage purification, 13 different amino acid inserting suppressor strains for multiple amino substitutions at amber mutant sites as described (Dixit et al., 2012), and nonsuppressor host E. coli BE for preparation of defective proheads following infection with terminase amber mutants. E. coli DH10B was used for cloning of plasmids pT20 (gp20 portal expression vector plasmid constructs made in plasmid pTrc99A) (Baumann et al., 2006) containing wild type gene 20 as well as those that had undergone SDM. E. coli BL21(DE3)pLysS was used for expression of recombinant proteins. Luria–Bertani (LB) medium was used for growth of cells except for phage infection experiments in which M9S/20% LB medium was used (Dixit et al., 2011). For plating bacteria carrying recombinant plasmids ampicillin was added to the media at a concentration of 100 mg/ml. The plasmid pT20 carrying the gp20 gene was the template for SDM amplifications. Cloning methodology was by routine methods including polymerase chain reaction (PCR) purification and gel extraction steps by kit specifications (Qiagen) (Dixit et al., 2012). Plating of phages was carried out on plates containing 30 ml of Bottom agar made with 10 g Bacto tryptone, 1 g Bacto yeast extract, 5 g NaCl, 0.2 g dextrose and 15 g Bacto agar/liter. Top agar contained the same ingredients as the bottom agar in the same proportions except the concentration of agar is 7 g/liter.
Site directed mutagenesis
Mutagenesis was performed using a Quickchange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's recommendations. Six cysteine substitutions (A279C, A294C, A297C, A298C, M305C and A316C) and two amber mutations D340 and A343 close to the central peptide region of gp20 were made using primers shown in the Supplementary Table S1. After verification of the mutations by sequencing (Biopolymer core sequencing facility, UMB, Baltimore) using primers listed in Supplementary Table S1, the plasmids carrying the mutated genes were used for various studies. The cells carrying the recombinant plasmid were spotted on E. coli sup+ suppressor plates. The cells were then infected with ts (temperature-sensitive) gp20 mutant N33 which were selected by growing at 42 1C to force recombination of the mutagenized gp20 gene. The progeny plaques were plated on E. coli-sup0 and E. coli-sup+ and individual plaques were screened for the presence of amber mutations on E. coli sup+. Two independent plaques for each mutant were sequenced to confirm the presence of the correct amber or cysteine mutations.
In vitro labeling of the gp17 tetra cys (CT) with ReAsH-Ethanedithiol (EDT2)
To label the terminase with a single fluorescent dye at a known position, a ReAsH-binding tetra cysteine peptide FLNCCPGCCMEP was added to the C terminus of the wild type gp17 gene in pTYB2 vector. The gp17 terminase large subunit was purified to near homogeneity from the chitin binding domain–intein fusion protein as previously described (Baumann and Black, 2003). The labeling of the purified gp17 tetra-Cys (CT) with ReAsH-EDT2 was accomplished as described previously in (Dixit et al., 2011). ReAsH fluorescence was measured on a Typhoon imager 9400 (GE Healthcare) on a 10% native polyacrylamide gel using excitation and emission wavelengths of 593 and 608 nm, respectively, as well as in a Variant Cary Eclipse fluorescence spectrophotometer (Fig. 3B).
Prohead purification and labeling
Wild-type empty large proheads (elps) were concentrated from the T4 prohead producing mutant [13amE111 16amN66 17amA465 ΔrIIA H88] infected bacteria. Prohead purification employed differential centrifugation, glycerol gradient ultracentrifugation, and FPLC-DEAE chromatography as previously described (Dixit et al., 2011). Cysteine mutant portal protein gp20 containing proheads were produced by supplying cysteine mutant gp20 in trans from an expression vector to terminase and portal deficient phage mutant infected bacteria as previously described (Baumann et al., 2006). The same purification protocol as used for wild-type proheads was followed and the FPLC-DEAE chromatographed empty large proheads (elps) which are more stable than the empty small proheads (esps) were then characterized for packaging activity by nuclease assay as previously described (Black and Peng, 2006). A 10 mM stock solution of Alexa Fluor 488 C5-maleimide (Invitrogen) was prepared in dimethyl sulfoxide (DMSO). Four nanomoles of the wild type or Cys-mutant portal expressing purified proheads were covalently labeled in the dark (60–90 min at room temperature) with 20 nmol of dye (1:5 ratio) Alexa Fluor 488 C5-maleimide dissolved in 10 mM Tris–HCl buffer. The conjugation reaction was quenched by adding final 0.5% (72 mM) β-mercaptoethanol to the reaction mixture and incubating for 10 min at room temperature. Labeled proheads were then purified using a G-25 Sephadex spin column. Diluted labeled proheads were concentrated by centrifugation at 18K for 45 min at 4 1C. The purified complex was stable for several days at 4 1C. Alexa 488 fluorescence was measured by Typhoon imager (Fig. 3A) on 0.7% agarose gel using excitation and emission wavelengths of 495 and 519 nm, respectively.
DNA packaging assays
A nuclease protection assay measured the amount of packaged DNA by agarose gel electrophoresis. After in vitro packaging for 30 min at room temperature, the proheads and packaging components were incubated at 37 1C for 30 min with pancreatic DNase, followed by release of the packaged prohead DNA by incubation at 65 1C for 30 min in the presence of EDTA, proteinase K, and 1% SDS as described previously (Dixit et al., 2011). The packaged ethidium bromide-stained DNAs were quantitated using UVP software to integrate gel DNA band densities in comparison with known quantities of DNA in a standard 1 kb DNA ladder (Fermentas). For nuclease assay of packaging following FCS measurements, the sample was removed from the microscope observation stage and processed by the nuclease assay described above to determine the packaging that had been achieved during the FCS measurements. Packaging was assayed in addition by change in the diffusion of the added dye-labeled DNA as compared with the added DNA without packaging (omitting either terminase or proheads) (Lakowicz, 2006).
FCS and FRET-FCS measurements and analyses
FCS measurements were performed using a Picoquant Micro-Time 200 confocal microscope (Picoquant microtime system coupled to an Olympus IX71 microscope). The excitation laser (λex~470 nm) was reflected by a dichroic mirror (centered at 476 nm) to a high numerical aperture (NA) oil objective (100×, NA 1.3) and focused onto the solution sample. The fluorescence signals were collected through the same objective and dichroic mirror. An additional longpass filter was used to eliminate the scattered laser light. Fluorescence responses from the donor and the acceptor molecules were separated by a beam splitter and detected by two avalanche photodiode detectors (APD) using the method of time-correlated single photon counting and the Time-Tagged Time-Resolved (TTTR) mode of the TimeHarp 200 board. High quality bandpass (Chroma) filters were used for recording donor and acceptor fluorescence in two separate detection channels. FCS measurements were performed in a constant detection volume. PicoQuant Symphotime software was used to generate the autocorrelation curves and in analyzing the FRET data. The autocorrelation curves were fitted with the following diffusion model. The autocorrelation function for n fluorescent species traversing a 3D Gaussian volume with radius ω0 and half axial z0 is:
where τ is the lag time, N is the number of molecules in the volume and fi are the fractions of the corresponding diffusion coefficients Di. (Lakowicz, 2006)
The collected single photon data was binned by 1 msec bin in each channel (donor or acceptor) which resulted in intensity-time traces and count-rate histograms. Threshold values in each channel were used to identify the single molecule bursts from the corresponding background signal level. Significant signals from adjacent intervals were binned together to get a sum photon number for each burst. Thus, fluorescence bursts were recorded simultaneously in donor and acceptor channels and FRET efficiencies were calculated using E=n A/(n A+γnD) where nD and n A are the sums of donor counts and acceptor counts for each burst, taking into account the possible difference in the detection efficiency (γ) in two separate channels (Schuler et al., 2002). The donor-to-acceptor distance (r) in terms of efficiency of energy transfer (E) and Förster Distance (R0) is given by r=R0 [1/E–1]1/6. We have used the value of R0 of 6.749 nm for the Alexa 488 (donor) and Cy3 (acceptor) pair for estimating the donor-to-acceptor distance (Elangovan et al., 2003). We calculated the value of R0 of 6.2 nm for the Alexa 488 (donor) to ReASH (acceptor) pair from the spectral properties of the donor and acceptor and the donor quantum yield. For a related portal–terminase GFP (donor) and ReASH (acceptor) pair, a comparable R0 of 5.5 nm was calculated (Lakowicz, 2006; Martin et al., 2005).
Supplementary Material
Acknowledgments
ABD, JAT and LWB were supported by NIH grant AI11676. K. R. was supported by NIH grant AI087968. A set of 13 amber E. coli suppressor strains (given in (Dixit et al., 2012)) was generously provided by J.H. Miller (UCLA). We thank Dr. S.C. Hardies for allowing access to his collection of bioinformatics software and the UTHSCSA Bioinformatics Center for assistance with computational aspects of the project.We thank Qin Dan for technical support. The authors thank Dr. J. R. Lakowicz and the Center of Fluorescence Spectroscopy for access to the facility in performing the fluorescence measurements.
Abbreviations
- ReAsH
specific tetra-cysteine peptide binding Fluorescein arsenical helix bis-EDT adduct
- Alexa 488
Alexa Fluor fluorescent dye
- FCS-FRET
Fluorescence correlation spectroscopy-Förster resonance energy transfer
- SDM
site directed mutagenesis
- ts
temperature sensitive mutation
- cs
cold sensitive mutation
Footnotes
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.virol.2013.07.011.
References
- Baumann RG, Black LW. Isolation and characterization of T4 bacteriophage gp17 terminase, a large subunit multimer with enhanced ATPase activity. J. Biol. Chem. 2003;278:4618–4627. doi: 10.1074/jbc.M208574200. [DOI] [PubMed] [Google Scholar]
- Baumann RG, Mullaney J, Black LW. Portal fusion protein constraints on function in DNA packaging of bacteriophage T4. Mol. Microbiol. 2006;61:16–32. doi: 10.1111/j.1365-2958.2006.05203.x. [DOI] [PubMed] [Google Scholar]
- Black LW. DNA packaging in dsDNA bacteriophages. Annu. Rev. Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- Black LW. DNA packaging and cutting by phage terminases: control in phage T4 by a synaptic mechanism. Bioessays. 1995;17:1025–1030. doi: 10.1002/bies.950171206. [DOI] [PubMed] [Google Scholar]
- Black LW, Peng G. Mechanistic coupling of bacteriophage T4 DNA packaging to components of the replication-dependent late transcription machinery. The J. Biol. Chem. 2006;281:25635–25643. doi: 10.1074/jbc.M602093200. [DOI] [PubMed] [Google Scholar]
- Black LW, Silverman DJ. Model for DNA packaging into bacteriophage T4 heads. J. Virol. 1978;28:643–655. doi: 10.1128/jvi.28.2.643-655.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, Wyckoff E, Hayden M, Sampson L, Eppler K, Randall S, Moreno ET, Serwer P. Bacteriophage P22 portal protein is part of the gauge that regulates packing density of intravirion DNA. J. Mol. Biol. 1992;224:1055–1074. doi: 10.1016/0022-2836(92)90469-z. [DOI] [PubMed] [Google Scholar]
- Casjens SR. The DNA-packaging nanomotor of tailed bacteriophages. Nat. Rev. Microbiol. 2011;9:647–657. doi: 10.1038/nrmicro2632. [DOI] [PubMed] [Google Scholar]
- Cornilleau C, Atmane N, Jacquet E, Smits C, Alonso JC, Tavares P, Oliveira L. The nuclease domain of the SPP1 packaging motor coordinates DNA cleavage and encapsidation. Nucleic Acids Res. 2013;41:340–354. doi: 10.1093/nar/gks974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit A, Ray K, Lakowicz JR, Black LW. Dynamics of the T4 bacteriophage DNA packasome motor: endonuclease VII resolvase release of arrested Y-DNA substrates. J. Biol. Chem. 2011;286:18878–18889. doi: 10.1074/jbc.M111.222828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit AB, Ray K, Black LW. Compression of the DNA substrate by a viral packaging motor is supported by removal of intercalating dye during translocation. Proc. Natl. Acad. Sci U.S.A. 2012;109:20419–20424. doi: 10.1073/pnas.1214318109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driedonks RA, Engel A, tenHeggeler B, van D. Gene 20 product of bacteriophage T4 its purification and structure. J. Mol. Biol. 1981;152:641–662. doi: 10.1016/0022-2836(81)90121-2. [DOI] [PubMed] [Google Scholar]
- Elangovan M, Wallrabe H, Chen Y, Day RN, Barroso M, Periasamy A. Characterization of one- and two-photon excitation fluorescence resonance energy transfer microscopy. Methods. 2003;29:58–73. doi: 10.1016/s1046-2023(02)00283-9. [DOI] [PubMed] [Google Scholar]
- Feiss M, Rao VB. The bacteriophage DNA packaging machine. Adv. Exp. Med. Biol. 2012;726:489–509. doi: 10.1007/978-1-4614-0980-9_22. [DOI] [PubMed] [Google Scholar]
- Fuller DN, Raymer DM, Rickgauer JP, Robertson RM, Catalano CE, Anderson DL, Grimes S, Smith DE. Measurements of single DNA molecule packaging dynamics in bacteriophage lambda reveal high forces, high motor processivity, and capsid transformations. J. Mol. Biol. 2007;373:1113–1122. doi: 10.1016/j.jmb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde S, Padilla-Sanchez V, Draper B, Rao VB. Portal-large terminase interactions of the bacteriophage T4 DNA packaging machine implicate a molecular lever mechanism for coupling ATPase to DNA translocation. J. Virol. 2012;86:4046–4057. doi: 10.1128/JVI.07197-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Rajagopalan S, Settanni G, Marsh RJ, Armoogum DA, Nicolaou N, Bain AJ, Lerner E, Haas E, Ying L, Fersht AR. Multiple conformations of full-length p53 detected with single-molecule fluorescence resonance energy transfer. Proc. Natl. Acad. SciUSA. 2009;106:20758–20763. doi: 10.1073/pnas.0909644106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugel T, Michaelis J, Hetherington CL, Jardine PJ, Grimes S, Walter JM, Falk W, Anderson DL, Bustamante C. Experimental test of connector rotation during DNA packaging into bacteriophage phi29 capsids. PLoS Biol. 2007;5:e59. doi: 10.1371/journal.pbio.0050059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isidro A, Santos MA, Henriques AO, Tavares P. The high-resolution functional map of bacteriophage SPP1 portal protein. Mol. Microbiol. 2004;51:949–962. doi: 10.1046/j.1365-2958.2003.03880.x. [DOI] [PubMed] [Google Scholar]
- Itsathitphaisarn O, Wing RA, Eliason WK, Wang J, Steitz TA. The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell. 2012;151:267–277. doi: 10.1016/j.cell.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Chiu W. DNA packaging and delivery machines in tailed bacteriophages. Curr. Opin. Struct. Biol. 2007;17:237–243. doi: 10.1016/j.sbi.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Kainov DE, Tuma R, Mancini EJ. Hexameric molecular motors: P4 packaging ATPase unravels the mechanism. Cell Mol. Life Sci. 2006;63:1095–1105. doi: 10.1007/s00018-005-5450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamaru S, Kondabagil K, Rossmann MG, Rao VB. The functional domains of bacteriophage t4 terminase. J. Biol. Chem. 2004;279:40795–40801. doi: 10.1074/jbc.M403647200. [DOI] [PubMed] [Google Scholar]
- Kang S, Poliakov A, Sexton J, Renfrow MB, Prevelige PE., Jr Probing conserved helical modules of portal complexes by mass spectrometry-based hydrogen/deuterium exchange. J. Mol. Biol. 2008;381:772–784. doi: 10.1016/j.jmb.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Kondabagil KR, Zhang Z, Rao VB. The DNA translocating ATPase of bacteriophage T4 packaging motor. J. Mol. Biol. 2006;363:786–799. doi: 10.1016/j.jmb.2006.08.054. [DOI] [PubMed] [Google Scholar]
- Lakowicz JR. Principles of Fluorescence Spectroscopy. Third ed. Springer; 2006. [Google Scholar]
- Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, Turner J, Dodson EJ, Tavares P, Antson AA. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26:1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Black LW. DNA requirements in vivo for phage T4 packaging. Virology. 1998;242:118–127. doi: 10.1006/viro.1997.9019. [DOI] [PubMed] [Google Scholar]
- Lin H, Rao VB, Black LW. Analysis of capsid portal protein and terminase functional domains: interaction sites required for DNA packaging in bacteriophage T4. J. Mol. Biol. 1999;289:249–260. doi: 10.1006/jmbi.1999.2781. [DOI] [PubMed] [Google Scholar]
- Lin H, Simon MN, Black LW. Purification and characterization of the small subunit of phage T4 terminase, gp16, required for DNA packaging. J. Biol. Chem. 1997;272:3495–3501. doi: 10.1074/jbc.272.6.3495. [DOI] [PubMed] [Google Scholar]
- Martin BR, Giepmans BN, Adams SR, Tsien RY. Mammalian cell-based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nat. Biotechnol. 2005;23:1308–1314. doi: 10.1038/nbt1136. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Oram M, Sabanayagam C, Black LW. Modulation of the packaging reaction of bacteriophage T4 terminase by DNA structure. J. Mol. Biol. 2008;381:61–72. doi: 10.1016/j.jmb.2008.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VB, Black LW. DNA packaging of bacteriophage T4 proheads in vitro. Evidence that prohead expansion is not coupled to DNA packaging. J. Mol. Biol. 1985;185:565–578. doi: 10.1016/0022-2836(85)90072-5. [DOI] [PubMed] [Google Scholar]
- Ray K, Sabanayagam CR, Lakowicz JR, Black LW. DNA crunching by a viral packaging motor: compression of a procapsid-portal stalled Y-DNA substrate. Virology. 2010;398:224–232. doi: 10.1016/j.virol.2009.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler B, Lipman EA, Eaton WA. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature. 2002;419:743–747. doi: 10.1038/nature01060. [DOI] [PubMed] [Google Scholar]
- Schwartz C, De Donatis GM, Fang H, Guo P. The ATPase of the phi29 DNA packaging motor is a member of the hexameric AAA + superfamily. Virology. 2013;443(1):20–27. doi: 10.1016/j.virol.2013.04.004. http://dx.doi.org/10.1016/j.virol.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AA, Leiman PG, Tao Y, He Y, Badasso MO, Jardine PJ, Anderson DL, Rossmann MG. Structure determination of the head-tail connector of bacteriophage phi29. Acta Crystallogr. D Biol. Crystallogr. 2001;57:1260–1269. doi: 10.1107/s0907444901010435. [DOI] [PubMed] [Google Scholar]
- Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- Smits C, Chechik M, Kovalevskiy OV, Shevtsov MB, Foster AW, Alonso JC, Antson AA. Structural basis for the nuclease activity of a bacteriophage large terminase. EMBO Rep. 2009;10:592–598. doi: 10.1038/embor.2009.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Kondabagil K, Draper B, Alam TI, Bowman VD, Zhang Z, Hegde S, Fokine A, Rossmann MG, Rao VB. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135:1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Tang J, Lander GC, Olia AS, Li R, Casjens S, Prevelige P, Jr, Cingolani G, Baker TS, Johnson JE. Peering down the barrel of a bacteriophage portal: the genome packaging and release valve in p22. Structure. 2011;19:496–502. doi: 10.1016/j.str.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares P, Santos MA, Lurz R, Morelli G, de Lencastre H, Trautner TA. Identification of a gene in Bacillus subtilis bacteriophage SPP1 determining the amount of packaged DNA. J. Mol. Biol. 1992;225:81–92. doi: 10.1016/0022-2836(92)91027-m. [DOI] [PubMed] [Google Scholar]
- Tsay JM, Sippy J, Feiss M, Smith DE. The Q motif of a viral packaging motor governs its force generation and communicates ATP recognition to DNA interaction. Proc. Natl. Acad. SciUSA. 2009;106:14355–14360. doi: 10.1073/pnas.0904364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.