Abstract
Synapse formation in the developing brain depends on the coordinated activity of synaptogenic proteins, some which have been implicated in a number of neurodevelopmental disorders. Here, we show that the sushi repeat-containing domain protein X-linked 2 (SRPX2) gene encodes a protein that promotes synaptogenesis in the cerebral cortex. In humans, SRPX2 is an epilepsy- and language-associated gene that is a target of the foxhead box protein P2 (FoxP2) transcription factor. We also show that FoxP2 modulates synapse formation through regulating SRPX2 levels, and that SRPX2 reduction impairs development of ultrasonic vocalization in mice. Our results suggest FoxP2 modulates the development of neural circuits through regulating synaptogenesis and that SRPX2 is a synaptogenic factor that plays a role in the pathogenesis of language disorders.
Synapse formation is an essential process during brain development that is coordinated by many membrane and secreted proteins (1–5). Proper development of neural circuitry is required for brain function, and mutations in synaptogenic genes have been linked to many cognitive diseases, including autism, schizophrenia and mental retardation (6–8). While a number of proteins have been shown to modulate synaptogenesis, no single gene knockout has been shown to completely ablate the formation of any major class of synapses, suggesting that the brain may utilize many proteins to regulate this process. To search for synaptogenic factors, we embarked on a high throughput overexpression screen for human genes encoding membrane and secreted proteins that mediate synaptogenesis in the central nervous system. We identified SRPX2 as a secreted protein that modulates synapse density in dissociated hippocampal neurons. The SRPX2 gene is mutated in human patients suffering from rolandic (sylvian) epilepsy with associated oral and speech dyspraxia (9), and is a target of the FoxP2 gene (10), suggesting that SRPX2 may be involved in neural connectivity and language in humans. While sushi domain proteins, also known as also known as complement control protein (CCP) domain proteins, function as regulators of the immune system in vertebrates (11), they also regulate neuronal development in C.elegans (12) and Drosophila (13, 14), We therefore decided to further examine the role of SRPX2 in synapse formation.
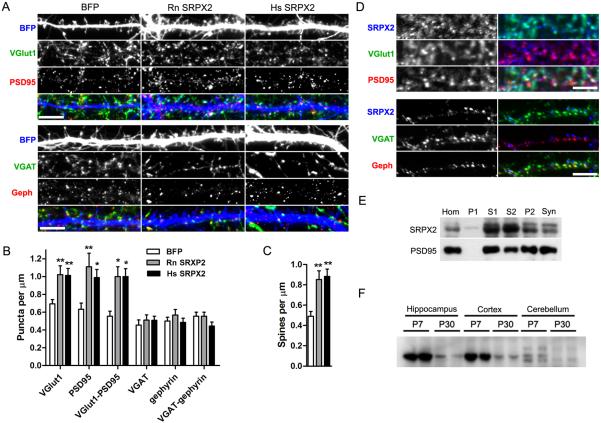
To verify that SRPX2 controls synapse density, we overexpressed rat and human SRPX2 genes in dissociated rat cortical cells. Overexpression of SRPX2 caused an increase in the density of vesicular glutamate transporter 1 (VGlut1) and PSD-95 puncta on the neurons, while leaving the density of inhibitory synaptic markers vesicular GABA transporter (VGAT) and gephyrin unchanged (Fig. 1A and 1B). Dendritic morphology was unaffected by SRPX2 overexpression (Fig. S3A). Both human and rat SRPX2 genes are capable of increasing spine density when overexpressed (Fig. 1C). Thus, SRPX2 overexpression increases the density of excitatory synapses and spines in vitro without an effect on inhibitory synapse formation.
Fig. 1. SRPX2 overexpression increases excitatory synaptic density in cultured neurons.
A, Rat cortical neurons were cotransfected with BFP and rat (Rn SRPX2) or human SRPX2 (Hs SRPX2), and stained for excitatory synapse markers VGlut1 (green) and PSD95 (red), as well as for inhibitory synaptic markers VGAT (green) and gephyrin (red). Scale bars, 5 μm. B, Quantification of synapse density shown in (A). (Means ± sem, n = 13–35, *p < 0.05, **p < 0.01). C, Quantitation of spine density shown in (A). (Means ± sem, n = 25–35, **p < 0.01). D, DIV 21 dissociated rat cortical neurons were surface-stained for SRPX2, and then permeabilized and stained for excitatory and inhibitory synaptic markers. Scale bars, 3.5 μm. E, Synaptosome preparation was immunoblotted for SRPX2 and PSD95. Fractions are crude homogenate (Hom), P1 pellet (P1), S1 supernatant (S1), S2 supernatant (S2), P2 pellet (P2), and synaptosomes (Syn). F, Western blot analysis of SRPX2 levels in the cerebral cortex, hippocampus and cerebellum of P7 and P30 mice.
SRPX2 mRNA is found in neurons in multiple brain regions, including the cerebral cortex and hippocampus (9, 15). To further characterize the expression and localization of SRPX2 at the protein level, we generated an antibody against SPRX2, and used it to perform immunocytochemistry on rat cortical cultures. We verified that the antibody is specific for SRPX2, and does not recognize the closely related SRPX protein (Fig. S1). We observed that SRPX2 staining is largely colocalized with VGlut1 and PSD-95 puncta, and excluded from VGAT and gephyrin puncta (Fig. 1D), consistent with its effect on excitatory synapses. Immunoblots of synaptosome preparations showed that SRPX2 is partially retained on synaptic membranes (Fig 1E), and staining of the heterologous and neuronal cells overexpressing SRPX2 also showed large puncta of SRPX2 on the surface of cells (Fig. S2A), suggesting that SRPX2 is largely oligomerized and deposited close to the site of secretion. To investigate whether SRPX2 forms oligomers, we performed coimmunoprecipitations from culture medium of heterologous cells coexpressing SXPX2-GFP and SRPX2-Flag, and found that SRPX2 binds to itself, but not to the closely related family member SRPX (Fig. S2B). We also tested the properties of a dominant negative SRPX2 bearing a mutation associated with epilepsy and language disorders in human patients (9), and found that SRPX2-DN is secreted into the medium, and retains binding to native SRPX2 (Fig. S2B). In rat cerebral cortex lysates, SRPX2 is more highly expressed in juvenile compared to adult rats (Fig. 1F), coinciding with the period of elevated synapse formation. Thus, the spatial and temporal expression patterns of SRPX2 are consistent with a role in excitatory synaptogenesis.
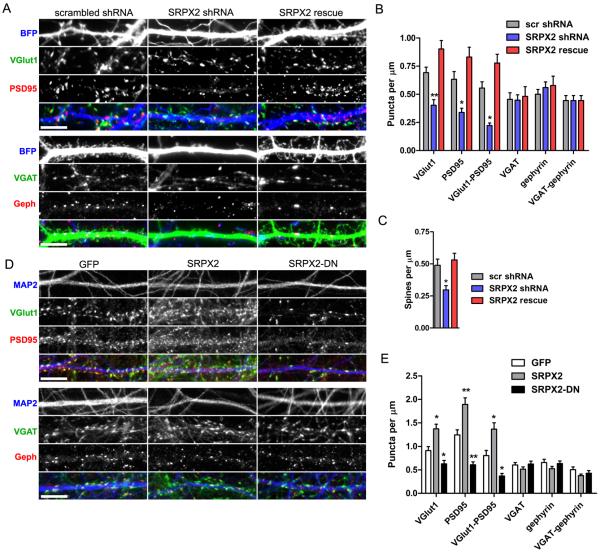
To investigate whether endogenous SRPX2 regulates synapse formation, we generated a small hairpin RNA (shRNA) that efficiently knocks down SRPX2 (Fig. S1B and S1C). When dissociated neurons were transfected with the SRPX2-directed shRNA, a reduction of excitatory synapse and spine density (Fig. 2A, 2B and 2C) was observed, without a change in inhibitory synaptic density (Fig. 2A and 2B) and dendritic morphology (Fig. S3B). To control for off-target effects of the shRNA, we also generated a rescue shRNA-resistant SRPX2 construct bearing a silent mutation, and confirmed that the rescue construct is not knocked down by the shRNA (Fig. S1D). Coexpression of the rescue construct with SRPX2-shRNA in dissociated neurons results in the reversal of the deficits in excitatory synapse and spine formation with no effect on inhibitory synapse formation (Fig. 2A, 2B and 2C). This data demonstrates that endogenous SRPX2 is critical for excitatory synapse formation. To examine if secreted SRPX2 can cause changes in synapse density, we cultured cortical neurons with conditioned medium taken from heterologous cells overexpressing GFP, SRPX2, or SRPX2-DN. We found that cortical neurons cultured with SRPX2-conditioned medium showed an increased excitatory synapse density without a change in inhibitory synapse density (Fig. 2D and 2E). Interestingly, neurons cultured with SRPX2-DN-conditioned medium showed a decrease in excitatory synapse density (Fig. 2D and 2E). All conditioned media had no effect on dendritic morphology (Fig. S2C). Taken together, these results suggest that SRPX2 expression levels can bidirectionally regulate the density of excitatory synapses in neurons, and that secreted dominant negative SRPX2 impairs excitatory synapse formation.
Fig. 2. Reduction of endogenous SRPX2 and secretion of dominant negative SRPX2 decrease excitatory synaptic density in cultured neurons.
A, Rat cortical neurons were cotransfected with BFP and scrambled shRNA, SRPX2 shRNA, or SRPX2 shRNA and rescue construct (SRPX2 rescue), and then stained for excitatory or inhibitory synaptic markers. Scale bar, 5 μm. B, Quantitation of synapse density shown in (A). (Means ± sem, n = 12–35, *p < 0.05, **p < 0.01). C, Quantitation of spine density shown in (A). (Means ± sem, n = 22–35, *p < 0.05). D, Rat cortical neurons were incubated with medium conditioned by HEK293 cells overexpressing GFP, SRPX2, or SRPX2-DN, and immunostained for the dendritic marker MAP2, and excitatory or inhibitory synaptic markers. Scale bar, 5 μm. E, Quantitation of synapse density shown in (D). (Means ± sem, n = 12–21, *p < 0.05, **p < 0.01).
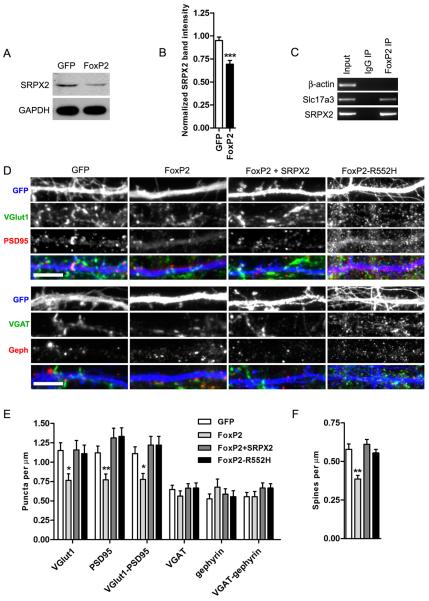
Because SRPX2 expression is repressed by FoxP2 in heterologous cells (10), we further investigated the effect of FoxP2 on synaptogenesis. To verify that SRPX2 is regulated by FoxP2 in rat cortical neurons, we electroporated dissociated neurons with FoxP2 or GFP before plating, and blotted for SRPX2 levels after 4–7 days of culture (Fig. 3A). We found that FoxP2-transfected cortical neurons contain a lower amount of SRPX2 compared to GFP-transfected neurons (Fig. 3B), suggesting that FoxP2 represses SRPX2 expression in cortical cultures. To confirm that FoxP2 directly represses the SRPX2 gene in neuronal cells, we performed chromatin immunoprecipitation (ChIP) of FoxP2 from P0 C57BL/6J mouse cortices, and found that FoxP2 binds to the promoter of the SRPX2 gene in vivo (Fig. 3C). Next, to assess FoxP2's effect on synaptic density, we overexpressed FoxP2 in a small subset of DIV 4 neurons using liposome transfection, and stained for synaptic markers 7 days after transfection. We found that neurons transfected with FoxP2 showed a lower density of excitatory synaptic markers and spines, without a change in the density of inhibitory synapses (Fig. 3D, 3E and 3F). In addition, expression of FoxP2-R552H, containing a human mutation which abolishes FoxP2 transcriptional activity, does not cause a change in synaptic density (Fig. 3E), indicating that FoxP2 transcriptional activity is required for its effects of synaptic density. To determine if FoxP2's effect on synapse formation may be due to decreases in SRPX2 expression, we coexpressed SRPX2 with FoxP2 in cortical neurons, and found that this rescued the reduction of excitatory synapses and spines caused by FoxP2 (Fig. 3D, 3E, and 3F). In addition, overexpression of FoxP2 in cortical neurons caused a decrease in the number of primary dendrites and a concomitant decrease in total dendritic length (Fig. S2D), an effect which is not rescued by SRPX2 coexpression. This suggests that FoxP2 has additional effectors which regulate dendritic morphology in cortical neurons, and that SRPX2's effect is specific to synapse formation. These results, together with data above showing that SRPX2 levels modulate synapse density bidirectionally, suggest that FoxP2 regulates synapse density through controlling SRPX2 levels.
Fig. 3. FoxP2 regulates excitatory synapse density through SRPX2.
A, Western blot analysis of SRPX2 levels in cortical neurons eletroporated with GFP or FoxP2 at plating and cultured for 4 days. B, Quantitation of SRPX2 levels shown in (A). (Means ± sem, n = 6–7, ***p < 0.001. C, ChIP assay of FoxP2 in cortices of P0 C57BL/6J mice. FoxP2 does not bind to the β-actin promoter, but does bind the promoter of the previously identified FoxP2 target gene Slc17a3(32), and the promoter of the SRPX2 gene. D, Rat cortical neurons were cotransfected with GFP and FoxP2, FoxP2 and SRPX2, or FoxP2-R552H, and immunostained for excitatory and inhibitory synaptic markers. Scale bars, 5 μm. E, Quantitation of synapse density shown in (D). (Means ± sem, n = 15–17, *p < 0.05, **p < 0.01). F, Quantitation of spine density shown in (D). (Means ± sem, n = 32–36, **p < 0.01).
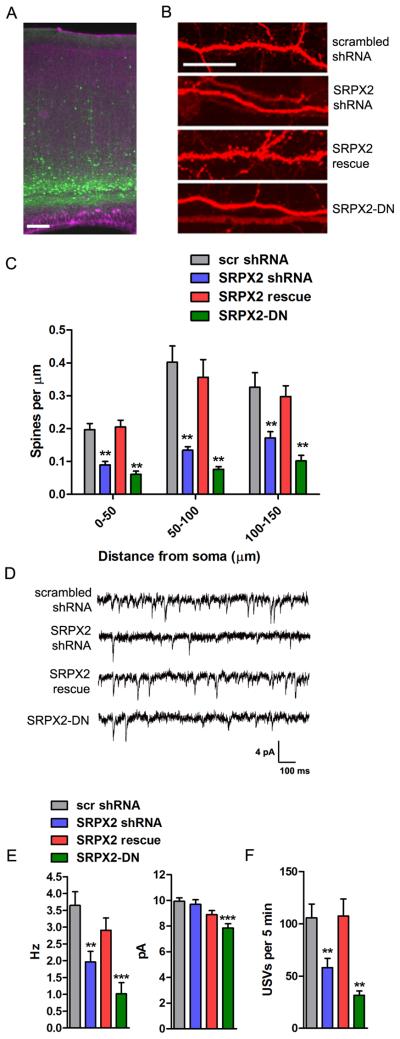
To investigate the effects of SRPX2 in vivo, we electroporated C57BL/6J mice at E12.5 in utero, targeting layers V/VI cortical neurons in a region spanning the motor and somatosensory cortices in the mouse cerebral cortex (Fig. 4A). This region of the brain was chosen because it is analogous to the language area in the human cortex, which displays cytoarchitectural defects in human patients bearing SRPX2 mutations (9). Layers V/VI neurons were chosen for their expression of the FoxP2 gene, as well as the presence of a long apical dendrite well suited for morphological assays. Neurons transfected with SRXP2-directed shRNA showed a decrease in the density of dendritic spines on the apical dendrite of layer V/VI neurons (Fig. 4B and 4C), an effect which is reversed by cotransfection of the shRNA-resistant SRPX2 construct. Neurons transfected with the dominant negative human SRPX2-DN construct also showed a decrease in dendritic spine density (Fig. 4B and 4C), indicating that SRPX2 mutations regulate apical dendritic spine density in vivo. Next, to test whether the spine density changes were reflected in electrophysiological measures of synapse density, we recorded miniature excitatory postsynaptic currents (mEPSCs) from electroporated layer V/VI neurons at P17–20 in whole-cell voltage clamp configuration. When compared against scrambled shRNA transfected control neurons, both SRPX2 shRNA transfected and SRPX2-DN transfected neurons showed a significant reduction in mEPSC frequency, while neurons cotransfected with SRPX2 shRNA and the SRPX2 rescue plasmid show no difference in frequency (Fig. 4D and 4E). In addition, there was a small decrease in the mEPSC amplitude of SRPX2-DN transfected neurons (Fig. 4E) and a small increase in the decay time constant (Fig. S4A). These results suggest that SRPX2 can regulate excitatory synapse number in the intact mouse brain.
Fig. 4. SRPX2 regulates excitatory synapse density and ultrasonic vocalization in vivo.
A, Low magnification micrograph of electroporated layer V/VI neurons in cortices of P21 C57BL/6J mice. B, Representative micrographs of apical dendrites of cortical neurons electroporated in utero with scrambled shRNA, SRPX2 shRNA, SRPX2 shRNA and rescue construct, or SRPX2 dominant negative construct. Scale bar, 10 μm. C, Quantitation of spine density on apical dendrite of layer V/VI neurons electroporated with various constructs. (Means ± sem, n = 16–49, **p < 0.01). D, Representative traces of mEPSC recordings from layer V/VI cortical neurons electroporated with scrambled shRNA, SRPX2 shRNA, SRPX2 shRNA and rescue construct, and SRPX2 dominant negative construct. E, Average frequency and amplitude of mEPSC recordings. (Means ± sem, n = 8–11, **p < 0.01, ***p < 0.001). F, Average number of USVs emitted in 5 minutes by P7 mouse pups electroporated with various SRPX2 constructs. (Means ± sem, n = 22–46, **p < 0.01).
Because humans who bear mutated SRPX2 alleles suffer from electrical and morphological developmental defects in the language cortex which impair language acquisition, we wanted to examine whether SRPX2 knockdown can cause a similar defect in mice. When separated from their mothers, infant mouse pups emit ultrasonic vocalizations which elicit a search-and-retrieve behavior in nearby dams (16, 17). The isolation-induced infant pup ultrasonic vocalization task has been widely used to characterize mouse models of human language, social, and arousal disorders, including mice containing FoxP2 mutations (18–22). We observed that pups electroporated with SRPX2 shRNA showed a decreased vocalization frequency compared to pups electroporated with scrambled shRNA or GFP controls (Fig. 4F and fig. S4B), and this defect is reversed by cotransfection of the rescue shRNA-resistant SRPX2 construct. Moreover, electroporation of SRPX2-DN also decreased ultrasonic vocalization frequency. Hence, we show that dysregulation of SRPX2 results in impaired ultrasonic vocalization in infant mice.
In summary, we have shown that SRPX2 is a sushi domain protein involved in synapse formation. In invertebrates, sushi domain proteins have been shown to cluster AChRs at synapses in C. elegans (12), and is localized to the nascent synaptic cleft in Drosophila (13, 14). In vertebrates, sushi domain proteins are primarily studied as regulators of the classical complement cascade (11). Our results suggest that sushi domain proteins may also play roles in regulating synaptic development and organization in vertebrates. In addition, as genes of the classical complement cascade have been shown to regulate synapse elimination (23), we speculate that SRPX2 may act through modulation of components of the complement cascade.
To date, FoxP2 is the only gene that has been shown to be involved in a human monogenic language disorder (24), although the cellular mechanisms involved remain obscure. Previous studies have suggested that FoxP2 may regulate neurite growth (25), dendritic morphology and synaptic physiology of basal ganglia neurons (26, 27). We show here that FoxP2 can regulate synaptogenesis of excitatory synapses in cortical neurons through SRXP2. While activity-regulated transcription factors have been shown to regulate synaptogenesis (28, 29), developmental synapse formation can occur in the complete absence of activity (30, 31), and it is unclear whether such synapse formation is also regulated by activity-independent transcription factors. We show here that FoxP2 is an activity-independent transcription factor that regulates synaptogenesis through SRPX2. In conclusion, our study suggests that FoxP2 can affect the development of language-related neural circuitry through regulating synaptogenesis, and that SRPX2 may be involved in the pathogenesis of language disorders.
Supplementary Material
Acknowledgments
We thank M. Pletnikov and J. Yocum of the Behavioral Core Facility, and M. Pucak of the Multiphoton/Electrophysiology Core Facility. We also thank H. Vega, S. Yang, N. O'Sullivan, and X. Wang for technical assistance. This study was supported by National Institutes of Health grant P50MH084020 to R.L. Huganir, and grant NS050274 to the Multiphoton Core Facility.
References
- 1.Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Cell. 2000;101:657–69. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 2.Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM. Cell. 2004;119:1013–26. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biederer T, et al. Science. 2002;297:1525–31. doi: 10.1126/science.1072356. [DOI] [PubMed] [Google Scholar]
- 4.Ko J, et al. Neuron. 2006;50:233–245. doi: 10.1016/j.neuron.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 5.de Wit J, et al. Neuron. 2009;64:799–806. doi: 10.1016/j.neuron.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szatmari P, et al. Nat. Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walsh T, et al. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 8.Laumonnier F, et al. Am. J. Hum. Genet. 2004;74:552–557. doi: 10.1086/382137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roll P, et al. Hum Mol Genet. 2006;15:1195–207. doi: 10.1093/hmg/ddl035. [DOI] [PubMed] [Google Scholar]
- 10.Roll P, et al. Hum Mol Genet. 2010;19:4848–60. doi: 10.1093/hmg/ddq415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirkitadze MD, Barlow PN. Immunol. Rev. 2001;180:146–161. doi: 10.1034/j.1600-065x.2001.1800113.x. [DOI] [PubMed] [Google Scholar]
- 12.Gendrel M, Rapti G, Richmond JE, Bessereau J-L. Nature. 2009;461:992–996. doi: 10.1038/nature08430. [DOI] [PubMed] [Google Scholar]
- 13.Hoshino M, Matsuzaki F, Nabeshima Y, Hama C. Neuron. 1993;10:395–407. doi: 10.1016/0896-6273(93)90329-p. [DOI] [PubMed] [Google Scholar]
- 14.Hoshino M, Suzuki E, Nabeshima Y, Hama C. Dev. Camb. Engl. 1996;122:589–597. doi: 10.1242/dev.122.2.589. [DOI] [PubMed] [Google Scholar]
- 15.Lein ES, et al. Nature. 2007;445:168–76. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 16.Fischer J, Hammerschmidt K. Genes Brain Behav. 2011;10:17–27. doi: 10.1111/j.1601-183X.2010.00610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crawley JN. Brain Pathol. Zurich Switz. 2007;17:448–459. doi: 10.1111/j.1750-3639.2007.00096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shu W, et al. Proc Natl Acad Sci U S. 2005;102:9643–8. doi: 10.1073/pnas.0503739102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujita E, et al. Proc Natl Acad Sci U S. 2008;105:3117–22. doi: 10.1073/pnas.0712298105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groszer M, et al. Curr Biol. 2008;18:354–62. doi: 10.1016/j.cub.2008.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaub S, Groszer M, Fisher SE, Ehret G. Genes Brain Behav. 2010;9:390–401. doi: 10.1111/j.1601-183X.2010.00570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.French CA, et al. Genesis. 2007;45:440–6. doi: 10.1002/dvg.20305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stevens B, et al. Cell. 2007;131:1164–1178. doi: 10.1016/j.cell.2007.10.036. [DOI] [PubMed] [Google Scholar]
- 24.Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. Nature. 2001;413:519–23. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- 25.Vernes SC, et al. PLoS Genet. 2011;7:e1002145. doi: 10.1371/journal.pgen.1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enard W, et al. Cell. 2009;137:961–71. doi: 10.1016/j.cell.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 27.Reimers-Kipping S, Hevers W, Pääbo S, Enard W. Neuroscience. 2011;175:75–84. doi: 10.1016/j.neuroscience.2010.11.042. [DOI] [PubMed] [Google Scholar]
- 28.Flavell SW, et al. Science. 2006;311:1008–12. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- 29.Lin Y, et al. Nature. 2008;455:1198–204. doi: 10.1038/nature07319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verhage M, et al. Science. 2000;287:864–9. doi: 10.1126/science.287.5454.864. [DOI] [PubMed] [Google Scholar]
- 31.Varoqueaux F, et al. Proc Natl Acad Sci U S. 2002;99:9037–42. doi: 10.1073/pnas.122623799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vernes SC, et al. Am J Hum Genet. 2007;81:1232–50. doi: 10.1086/522238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.