Abstract
We report a method called SSH array which combines the suppression subtraction hybridization (SSH) and DNA array techniques to find species-specific DNA probes from genomic DNA (gDNA) for species identification. The method first obtains the differential gDNA fragments between two species by SSH and then hybridizes the differential gDNA fragments with arrays made of multiple whole genomes from several species to screen the unique gDNA fragments for one species. The screened unique gDNA fragments can be used as species-specific probes to differentiate the species they represent from all other species. We used five species of the genus Dendrobrium, D.aurantiacum Kerr, D.officinale Kimura et Migo, D.nobile Lindl., D.chrysotoxum Lindl. and D.fimbriatum Hook., as experimental materials to study the feasibility of the method. The results showed that the method could efficiently obtain different species-specific probes for each of the five species.
INTRODUCTION
Species identification is a growing field of interest within the study of pathogenic prokaryotes (1–3), viruses (4) and eukaryotes (5,6) due to diagnostic, taxonomic, epidemiological, medical and socio-economic importance. However, the traditional approaches for species identifications are based on morphological (7,8), anatomical (9) and chemical analyses (10), which are often affected by environmental and developmental factors during growth. To overcome these limitations, many molecular techniques were developed for species identification by genotypic patterns, including ribotyping (11,12), random amplified polymorphic DNA (RAPD) (13,14), PCR amplification of the internal transcribed spacer 16S-23S (ITS 16–23S PCR) (15), ITS-PCR-restriction fragment length polymorphism (ITS-PCR-RFLP) (16), PCR-specific identification (17) and mip gene sequencing (18). These methods provide possible tools to identify species directly based on their genomic sequences. However, none of them has been applied as a routine and reliable method for species identification, possibly due to their inconvenient detection processes or inconsistent results (11).
Some other approaches have been developed to find species-specific genomic DNA (gDNA) sequences for species identification. Corredor et al. (5) screened the species-specific probes for Debaryomyces hansenii strains by RAPD (5). Picardeau and Vincent (19) discovered a species-specific probe for Mycobacterium xenopi by PCR (19). Most DNA probes used in ribotyping are derived from highly conserved genes coding for rRNA (20,21). 16S rRNA or 23S rRNA targeted hybridization probes have been used to identify and detect some species of Lactobacillus (17,22). However, in these methods closely related species are hard to distinguish with 16S rRNA probes, as little variation of the 16S rRNA sequence exists between them (23). Limitations in the number of species-specific probes has become the bottleneck in species identification using probe-dependent techniques. There are two possible approaches to improving such methods. One is to find new species-specific probes that are able to distinguish between closely related species and another is to use multispecies-specific probes to identify the species. Therefore, it is necessary to develop new methods for the discovery of highly species-specific gDNA sequences, especially in the whole genome field.
In this paper, we describe a novel method called SSH array for screening species-specific probes from whole gDNA for species identification. The method combines suppression subtraction hybridization (SSH) and nylon membrane-based array hybridization. SSH was first used to screen out the gDNA fragments that differentiated between two species. The differential gDNA fragments were then hybridized with an array fabricated with gDNAs from all studied species to screen out the species-unique gDNA fragments for one species. The species-unique gDNA fragments could be used as species-specific probes capable of distinguishing their ‘target’ species from all other species.
Five species of the genus Dendrobrium were employed as experimental materials: D.aurantiacum Kerr, D.officinale Kimura et Migo, D.nobile Lindl., D.chrysotoxum Lindl. and D.fimbriatum Hook. These species have been used in the preparation of herbal medicine in many Asian countries for hundreds of years (24). Each species has its own specific medical effects (25). However, after processing the commercial forms of the different species of the genus Dendrobrium appear very similar to each other and are hardly distinguishable from each other or from false samples; this is a source of confusion to herbalists and to the consumers of herbal medicine (26). The accurate identification of species of the genus Dendrobrium is necessary for clinical applications. Several groups have already studied the accurate identification of species of the genus Dendrobrium by rDNA ITS (26,27). However, methods for sequencing the unique sequence in the ITS region in each experiment are costly. In this paper, we describe the way in which we have successfully screened out several highly species-specific probes from five species of the genus Dendrobrium by our newly developed method. The results also revealed that the method is highly efficient.
MATERIALS AND METHODS
Genomic DNA preparation
The crude drugs from five commonly used Dendrobrium species was collected from Xishuangbanna in China’s Yunnan province: D.aurantiacum Kerr, D.officinale Kimura et Migo, D.nobile Lindl., D.chrysotoxum Lindl. and D.fimbriatum Hook. gDNA extraction from the fresh leaves of crude drugs were carried out in accordance with the procedure described by Sambrook et al. (28). In brief, 0.1 g of fresh leaves were powdered in liquid nitrogen and dissolved with DNA extraction buffer (100 mM Tris–HCl pH 8.0, 20 mM EDTA pH 8.0, 1.4 M NaCl, 2% CTAB, 0.2% β-mercaptoethanol). The mixture was incubated at 60°C for 2 h, and then centrifuged at 9000 r.p.m. for 10 min. The supernatant was transferred and extracted with phenol:chloroform:isoamyl alcohol (25:24:1) and chloroform:isoamyl alcohol (24:1). The gDNAs were then pelletted with cold absolute ethanol from aqueous phase and dissolved in 200 µl of TE and 2 µl of RNase A, followed by incubation at 37°C for 30 min. The gDNAs were precipitated with 100 µl of 7.5 M NH4Ac and 750 µl of 100% ethanol and washed with 70% ethanol. The gDNA pellets were air dried and dissolved in 20 µl of sterile ddH2O.
SSH and differential DNA fragment cloning
Aliquots of 1 µg of gDNA were completely digested with RsaI (Promega, USA) according to the instructions of the provider. The digestion generated shorter, blunt-ended gDNA fragments 100–500 bp in size. The digested gDNA fragments of two species randomly selected were paired as tester and driver. The SSH was done as described for the Clontech PCR-Select™ cDNA Subtraction Kit. In brief, 0.5 µg of the tester was ligated to 40 pmol of two single-stranded phosphorylated adaptors (adaptor1, 5′-CTAATACGACTCACTATAGGGC TCGAGCGGCCGCCCGGGCAGGT-3′; adaptor2R, 5′-CTA ATACGACTCACTATAGGGCAGCGTGGTCGCGGCCG AGGT-3′) (ShengYou Inc., China). The adaptor-ligated tester was then hybridized for two rounds with excessive driver. The hybridized products were first amplified with primer 1 (5′-CTAATACGACTCACTATAGGGC-3′) (ShengYou Inc., China) in a 50 µl reaction containing 2 µl of SSH products, 1× PCR buffer (20 mM Tris–HCl pH 8.4, 50 mM KCl and 1.5 mM MgCl2), 1 µM primer 1, 0.1 U/µl Taq polymerase, 200 µM dNTPs, under the following cycling conditions: filling the adaptors for 5 min at 75°C; denaturation for 2 min at 94°C; 32 cycles of 30 s at 94°C, 45 s at 66°C and 1.5 min at 72°C; a final extension for 5 min at 72°C. Aliquots of 3 µl of PCR products were then secondarily amplified in a 100 µl reaction containing 1× PCR buffer, 1 µM nested PCR primer 1 (5′-TCGAGCGGCCGCCCGGGCAGGT-3′) (ShengYou Inc., China), 1 µM nested PCR primer 2R (5′-AGCGTG GTCGCGGCCGAGGT-3′) (ShengYou Inc., China), 0.1 U/µl Taq polymerase and 200 µM dNTPs, under the following cycling conditions: 32 cycles of 33 s at 94°C, 45 s at 68°C and 1.5 min at 72°C; a final extension for 5 min at 72°C. The secondary PCR products were purified using a 3S DNA Gel purification Kit V3.1 (Shenergy Inc., Shanghai, China). Samples of 1 µg of purified PCR products were then ligated into vector pUCm-T (Shenergy Inc.) in a 10 µl ligation reaction containing 1× T4 ligation buffer (500 mM Tris–HCl, 100 mM MgCl2, 20 mM dithiothreitol and 0.5 mg/ml bovine serum albumin), 50 ng pUCm-T vector and 100 U/µl T4 DNA ligase. The ligation reaction was done at 16°C overnight. The ligated products were transformed into Escherichia coli JM109 cells and positive clones were selected as described by Sambrook et al. (28).
Preparation of DIG-labeled differential DNA fragments
The DIG-labeled differential DNA fragments were prepared by PCR reaction with DIG-nested PCR primer 1 in 96-well PCR plates. The amplifications were performed using 0.5 µl of saturated bacterial culture, 1 µM DIG-nested PCR primer 1, 1 µM nested PCR primer 2R, 0.1 U/µl Taq polymerase (Shenergy Inc.) and 200 µM dNTPs in 50 µl total volume of PCR buffer in each well, under the following cycling conditions: denaturation for 2 min at 94°C; 32 cycles of 35 s at 94°C, 45 s at 68°C, 1.5 min at 72°C; a final extension for 5 min at 72°C. The PCR products were precipitated with 5.0 µl of 3 M sodium acetate (pH 5.2) and 100 µl of absolute ethanol. After drying under vacuum, the DNA pellets were resuspended in 20 µl of sterilized water. The DIG-labeled differential DNA fragments were used as candidate probes for screening species-unique DNA fragments.
Fabrication of membrane-based gDNA arrays
An aliquot of 1 µg of RsaI-digested gDNA of each of the five species was denatured by mixing with the same volume of denaturation solution (0.5 N NaOH, 1.5 M NaCl) at room temperature for 30 min. The denatured gDNAs were then spotted on 20 × 25 mm positively charged nylon membranes (Roche Diagnostics, Mannheim, Germany), followed by baking at 100°C for 30 min.
Screening of the species-specific DNA fragments
The spotted array was prehybridized with 1× DIG Easy Hyb (Roche Diagnostics) in a hybridization oven (Robbins Scientific Corp., USA) for 4 h at 60°C. Samples of 10 µl (50 ng) of DIG-labeled candidate probe was denatured for 10 min at 98°C and mixed with 1× DIG Easy Hyb (1.5 ml) warmed to 60°C. The mixture was incubated with nylon membranes bearing gDNA arrays overnight at 62°C in the hybridization oven. After hybridization, the nylon membranes were washed twice with 50 ml of 2× SSC/0.1% SDS at room temperature, each for 10 min, and twice with 50 ml of 0.5× SSC/0.1% SDS at 62°C, each for 10 min, respectively. The chemiluminescent detection of hybridized nylon membranes with gDNA arrays was performed as described in the Instruction Manual of the DIG Gel Shift Kit (Roche Diagnostics).
RESULTS AND DISCUSSION
The efficiency of the PCR products of SSH and the positive clones
As the efficiency of cloning and screening using this method was unknown, as many as 192 (two 96-well plates) white color clones were selected for each subtracted species. To eliminate false positive clones as far as possible, the DIG-nested primer 1 and nested primer 2R were used instead of (sequencing/reverse) primer M13/pUC to amplify the selected clones, so that only the recombinant clones were amplified by PCR. The ratios of positive clones are shown in Table 1. As can be seen from Table 1, the ratios of positive clones of the five subtracted species varied from 78.1 to 86.4%. The average ratio of positive clones of the five subtracted species reached 82.3%. This verifies that the RsaI-digested gDNAs fragments are suitable for cloning into vector pUCm-T and are incorporated with high efficiency.
Table 1. The ratios of positive clones.
| Dendrobrium sp. | Subtracted clones | Tester | Driver | Positive clones | Ratio of positive clones (%) |
|---|---|---|---|---|---|
| D.aurantiacum Kerr | 192 | D.aurantiacum Kerr | D.nobile Lindl. | 155 | 80.7 |
| D.officinale Kimura et Migo | 192 | D.officinale Kimura et Migo | D.aurantiacum Kerr | 161 | 83.8 |
| D.nobile Lindl. | 192 | D.nobile Lindl. | D.aurantiacum Kerr | 166 | 86.4 |
| D.chrysotoxum Lindl. | 192 | D.chrysotoxum Lindl. | D.fimbriatum Hook. | 158 | 82.3 |
| D.fimbriatum Hook. | 192 | D.fimbriatum Hook. | D.officinale Kimura et Migo | 150 | 78.1 |
Screening of species-specific probes by gDNA array hybridization
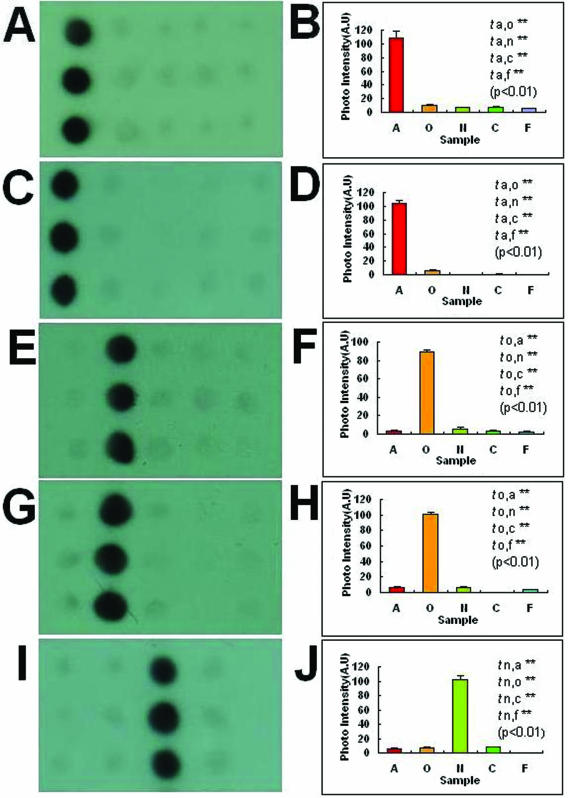
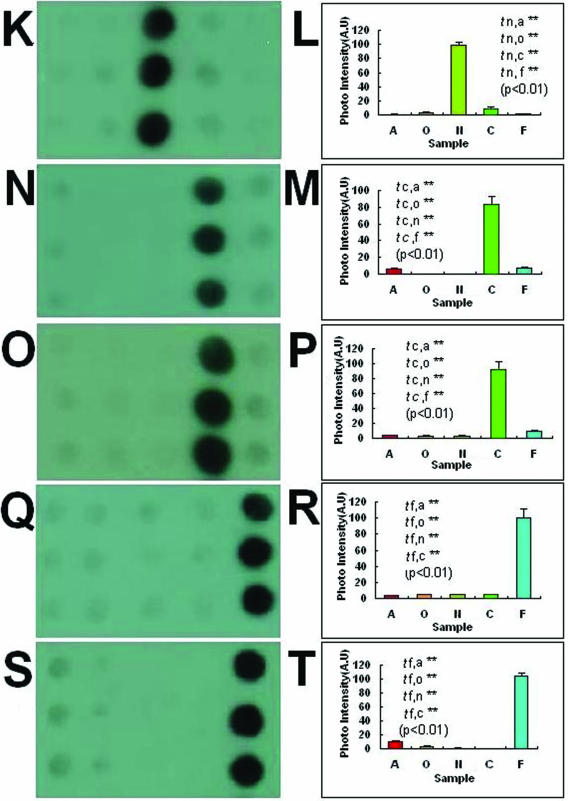
Many differential gDNA fragments were obtained through SSH and subsequent cloning. These differential gDNA fragments could be used to distinguish the tester species they represent from the corresponding driver species. However, it might not be possible to use the differential gDNA fragments to distinguish the tester species they represent from the other three species not paired as a driver. To screen out the species-specific probes for one species among the five Dendrobrium spp., the differential gDNA fragments were hybridized with gDNA arrays consisting of the five species. It is very easy to recognize those species-specific probes through the gDNA array hybridization. Two typical species-specific probes for each of the five experimental species are shown in Figure 1. Additionally, the images were analyzed by ImageJ (http://rsb.info.nih.gov/nih-image/index.html) and t-tests were performed for the average signal intensities of different samples of each image. All of the t-tests were significant (P < 0.01). All of the species-specific probes were specifically hybridized with the gDNAs they represent, but not with the other four species. The results showed that the species-specific probes we obtained have high specificity for the species they represent.
Figure 1.
Images and signal intensity plots (including t-tests) of hybridization of the species-specific probes to the gDNAs array of five species. Columns of spots on each array from left to right are D.aurantiacum Kerr, D.officinale Kimura et Migo, D.nobile Lindl., D.chrysotoxum Lindl. and D.fimbriatum Hook. D.A-SP1 (A) and D.A-SP2 (C) are D.aurantiacum Kerr-specific probes, D.O-SP1 (E) and D.O-SP2 (G) are D.officinale Kimura et Migo-specific probes, D.N-SP1 (I) and D.N-SP2 (K) are D.nobile Lindl.-specific probes, D.C-SP1 (M) and D.C-SP2 (O) are D.chrysotoxum Lindl.-specific probes and D.F-SP1 (Q) and D.F-SP2 (S) are D.fimbriatum Hook.-specific probes. (B), (D), (F), (H), (J), (L), (N), (P), (R) and (T) are signal intensities of (A), (C), (E), (G), (I), (K), (M), (O), (Q) and (S), respectively. All of the t-tests were significant (P < 0.01). A, O, N, C and F on the x-axis of the signal intensity plots represent D.aurantiacum Kerr, D.officinale Kimura et Migo, D.nobile Lindl., D.chrysotoxum Lindl. and D.fimbriatum Hook., respectively.
The efficiency of screening species-specific probes
The experiments demonstrated that the efficiency of the species-specific probes by this method is high (Table 2). Six species-specific probes for D.aurantiacum Kerr were screened out from 41 subtracted clones, and the ratio of species-specific probes reached 14.6%. Five D.officinale Kimura et Migo-specific probes were obtained from 43 subtracted clones, and the ratio of species-specific probes was 11.6%. Three D.nobile Lindl.-specific probes were found among 31 subtracted clones, and the ratio of species-specific probes reached 9.7%. Eight D.chrysotoxum Lindl.-specific probes were harvested when 62 subtracted clones were screened, and the ratio of species-specific probes was 12.9%. Three D.fimbriatum Hook.-specific probes were obtained after screening 25 subtracted clones, and the ratio of species-specific probes was 12.0%. The average ratio of species-specific probes from subtracted clones screened was up to 12.16%.
Table 2. The ratios of species-specific probes.
| Dendrobrium sp. | Subtracted clones screened | Tester | Driver | Species-specific probes | Ratio of species-specific probes (%) |
|---|---|---|---|---|---|
| D.aurantiacum Kerr | 41 | D.aurantiacum Kerr | D.nobile Lindl | 6 | 14.6 |
| D.officinale Kimura et Migo | 43 | D.officinale Kimura et Migo | D.aurantiacum Kerr | 5 | 11.6 |
| D.nobile Lindl. | 31 | D.nobile Lindl. | D.aurantiacum Kerr | 3 | 9.7 |
| D.chrysotoxum Lindl. | 62 | D.chrysotoxum Lindl | D.fimbriatum Hook. | 8 | 12.9 |
| D.fimbriatum Hook. | 25 | D.fimbriatum Hook. | D.officinale Kimura et Migo | 3 | 12.0 |
Species identification with species-specific probes
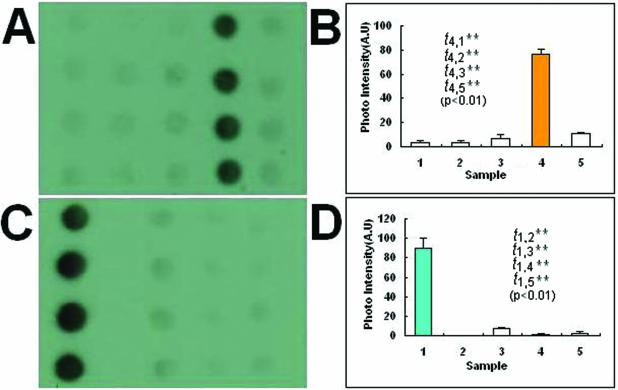
To confirm the validity of the species-specific probes, species identification experiments were carried out with hybridization of the species-specific probes to arrays consisting of the gDNAs of five Dendrobrium spp. However, in this experiment the operator did not know the origin and location of the gDNA in the array. Five DIG-labeled species-specific probes were hybridized with the unknown gDNAs array, so as to exactly find gDNA spots from the species they specifically represented. The results indicated that the species-specific probes could precisely identify the gDNAs they represented. For example, Figure 2A shows the results for probe D.O-SP1 hybridizing with an unknown gDNAs array, which revealed that the gDNAs comprising column 4 of the array should come from the species D.officinale Kimura et Migo. It was confirmed by the fabricator of the unknown gDNAs array that the identification was correct. Figure 2C shows the hybridization profile of probe D.F-SP2 to the unknown gDNAs array, which suggests that the gDNAs comprising column 1 of the array should be from the species D.fimbriatum Hook. The fabricator of the unknown gDNAs array also confirmed that. The images were also analyzed with ImageJ (and a t-test was performed for the average signal intensities of different samples of each image) (Fig. 2B and D). All of the t-tests were significant (P < 0.01).
Figure 2.
Images and signal intensity plots (including t-tests) of hybridization of the species-specific probes to the unknown gDNAs arrays for species identification. (A) Results of probe D.O-SP1 hybridizing with the unknown gDNAs array, suggesting that the gDNAs comprising column 4 of the array should come from the species D.officinale Kimura et Migo. (C) Image of probe D.F-SP2 hybridizing to the unknown gDNAs array, suggesting that the gDNAs comprising column 1 of the array should be from species D.fimbriatum Hook. (B and D) Signal intensities of (A) and (C), respectively. The two t-tests were significant (P < 0.01).
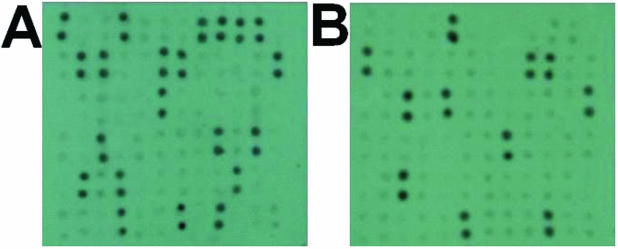
Furthermore, 72 samples of the Dendrobrium spp. were collected as fresh material from Xishuangbanna in China’s Yunnan province for a further blind trial. The 72 samples of Dendrobrium spp. contained 21 samples of D.officinale Kimura et Migo, 11 samples of D.chrysotoxum Lindl. and 40 samples of other Dendrobrium spp. DIG-labeled probes D.O-SP2 and D.C-SP2 were hybridized to a gDNA array of the 72 samples of Dendrobrium spp. The hybridization images are shown in Figure 3. Figure 3A shows that all of the 21 samples of D.officinale Kimura et Migo were correctly detected. Figure 3B shows that the 11 samples of D.chrysotoxum Lindl. were definitely detected. The identification experiments revealed that each of the species-specific probes is capable of definitely identifying the gDNA from the species it represents. These results demonstrate that the species-specific probes screened by the method presented in this paper could be reliably used to identify different Dendrobrium spp.
Figure 3.
Images of the species-specific probes hybridizing with the gDNAs array consisting of 72 Dendrobrium samples. The gDNAs of each sample were printed in double spots on the array. (A) Hybridization of probe D.O-SP2 to the array. All 21 samples of D.officinale Kimura et Migo were correctly detected. (B) Hybridization of probe D.C-SP2 to the array. Eleven samples of D.chrysotoxum Lindl. were definitely detected.
Sequence of the species-specific probes
The screening and identification experiments demonstrated that the method we present is an effective and reliable technique for isolating species-specific probes from gDNA. However, we didn’t know where these species-specific DNA fragments are located in genomic DNA and what roles they play. We presumed that these species-specific DNA fragments most probably come from the non-coding sequence, especially microsatellites. To solve these problems, two species-specific probes of each of five Dendrobrium spp. were amplified and sequenced (Shengyou Inc.) (Table 3). The sequences were then analyzed with Tandem Repeats Finder (http://c3.biomath.mssm.edu/trf.html). However, no microsatellite was found. The sequences were also analyzed with MegAlign (http://www.dnastar.com/), but the sequence similarity of the species-specific probes is very low. BLASTN 2.2.6 (http://www.ncbi.nlm.nih.gov/BLAST/) was used to analyze the sequences against GenBank holdings, but no homogeneous sequences were found in GenBank.
As in other techniques, including diversity array technology (DArT) (29) and representational difference analysis (RDA) (30), genome complexity reduction is a beneficial step in our method. However, while RDA uses multiple rounds of subtraction and amplification steps to clone unique fragments between two populations of genomic fragments and DArT initially assays unselected populations of fragments for quantitative differences in hybridization signal among input genotypes samples, SSH array screens the unique gDNA fragments for one species from many different species. Furthermore, DArT has adapted the DNA microarray platform to DNA polymorphism analysis, which is not reliant on DNA sequence information (29), but SSH array has focused on finding unique sequence fragments and using them for species identification. Although the two techniques could not be compared directly, similar potential applications of the two techniques include germplasm characterization, genetic mapping and gene tagging, molecular marker-assisted breeding and tracking genome methylation changes.
In conclusion, the method presented here is demonstrated to be a new highly effective and reliable approach for screening species-specific probes from a bulky and complex genome. In comparison with other approaches to finding species-specific probes that can be used for species identification, this method has great advantages. The most important feature of the method is that it is capable of screening out many species-specific probes from the whole genome. As the genome of higher eukaryotes contains bulky genetic material, it forms a reservoir of abundant DNA molecular markers. Our experiments demonstrate that the average ratio of species-specific probes from subtracted clones screened reaches 12.16%, which means it is easy to obtain many species-specific probes for definite species authentication. The larger number of species-specific probes is very helpful for exact species identification. We are fabricating DNA arrays with the larger number of species-specific probes that we screened out, so as to provide a highly parallel platform for genetic analysis and species identification. We believe this method will lead to a revolutionary improvement in species authentication studies. Moreover, the successful identification of the origin and function of the numerous species-specific gDNA fragments will be a great challenge to biologists in the future.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by Projects 30371750 and 60121101 of National Natural Science Foundation of China and also grants 2001AA2Z2012 and 2002AA2Z2041 from the National High Tech Program.
REFERENCES
- 1.Giffel M.C., Beumer,R.R., Lijn,N.K., Wagendorp,A.F. and Rombouts,M. (1997) Discrimination between Bacillus cereus and Bacillus thuringiensis using specific DNA probes based on variable regions of 16S rRNA. FEMS Microbiol. Lett., 146, 47–51. [DOI] [PubMed] [Google Scholar]
- 2.Zhong W., Millsap,K., Bialkowska-Hobrzanska,H. and Reid,G. (1998) Differentiation of Lactobacillus species by molecular typing. Appl. Environ. Microbiol., 64, 2418–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barsotti O., Décoret,D. and Renaud,F.N.R. (2002) Identification of Streptococcus mitis group species by RFLP of the PCR-amplified 16S-23S rDNA intergenic spacer. Res. Microbiol., 153, 687–691. [DOI] [PubMed] [Google Scholar]
- 4.Shamloul A.M., Faggioli,F., Keith,J.M. and Hadidi,A. (2002) A novel multiplex RT-PCR probe capture hybridization (RT-PCR-ELISA) for simultaneous detection of six viroids in four genera: Apscaviroid, Hostuviroid, Pelamoviroid and Pospiviroid. J. Virol. Methods, 105, 115–121. [DOI] [PubMed] [Google Scholar]
- 5.Corredor M., Davila,A.M., Gaillardin,C. and Casaregola,S. (2000) DNA probes species for the yeast species Debaryomyces hansenii useful tools for rapid identification. FEMS Microbiol. Lett., 193, 171–177. [DOI] [PubMed] [Google Scholar]
- 6.Wan Q.H. and Fang,S.G. (2003) Application of species-specific polymerase chain reaction in the forensic identification of tiger species. Forensic Sci. Int., 131, 75–78. [DOI] [PubMed] [Google Scholar]
- 7.Carlsward B.S., Stern,W.L., Judd,W.S. and Lucansky,T.W. (1997) Comparative leaf anatomy and systematics in Dendrobium, sections Aporum and Rhizobium (Orchidaceae). Int. J. Plant Sci., 158, 332–342. [Google Scholar]
- 8.Stern W.L., Morris,M.W. and Judd,W.S. (1994) Anatomy of the thick leaves in Dendrobium section Rhizobium (Orchidaceae). Int. J. Plant Sci., 155, 716–729. [Google Scholar]
- 9.Namba T. and Lin,C.C. (1981) Pharmacognostical studies on the crude drugs of Orchidaceae from Taiwan (IV) on ‘Chioh-hak’. Shoyakugaku Zasshi, 35, 221–232. [Google Scholar]
- 10.Anonymous (1988) Chinese Materia Medica (Zhong Yao Zhi), 2nd Edn. People’s Health Publishing House, Beijing, Vol. 4, pp. 230–446. [Google Scholar]
- 11.Salloum G., Meugnier,H., Reyrolle,M., Grimont,F., Grimont,P.A.D., Etienne,J. and Freney,J. (2002) Identification of Legionella species by ribotyping and other molecular methods. Res. Microbiol., 153, 679–686. [DOI] [PubMed] [Google Scholar]
- 12.Regnault B., Grimont,F. and Grimont,P.A.D. (1997) Universal ribotyping method using a chemically labelled oligonucleotide probe mixture. Res. Microbiol., 148, 649–659. [DOI] [PubMed] [Google Scholar]
- 13.Bansal N.S. and McDonell,F. (1997) Identification and DNA fingerprinting of Legionella strains by randomly amplified polymorphic DNA analysis. J. Clin. Microbiol., 35, 2310–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LoPresti F., Riffard,S., Vandenesch,F. and Etienne,J. (1998) Identification of Legionella species by random amplified polymorphic DNA profiles. J. Clin. Microbiol., 36, 3193–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tannock G.W., Tilsala-Timisjarvi,A., Rodtong,S., Ng,J., Munro,K. and Alatossava,T. (1999) Identification of Lactobacillus isolates from the gastrointestinal tract, silage and yoghurt by 16S-23S rRNA gene intergenic spacer region sequence comparison. Appl. Environ. Microbiol., 65, 4264–4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amelio S.D., Mathiopoulos,K.D., Santos,C.P., Pugachev,O.N., Webb,S.C., Picanco,M. and Paggi,L. (2000) Genetic markers in ribosomal DNA for the identification of members of the genus Anisakis (Nematoda: Ascaridoidea) defined by polymerase chain reaction-based restriction fragment length polymorphism. Int. J. Parasitol., 30, 223–226. [DOI] [PubMed] [Google Scholar]
- 17.Song Y.L., Kato,N., Liu,C.X., Matsumiya,Y., Kato,H. and Watanabe,K. (2000) Rapid identification of 11 human intestinal Lactobacillus species by multiplex PCR assays using group and species-specific primers derived from the 16S-23S rRNA intergenic spacer region and its flanking 23S rRNA. FEMS Microbiol. Lett., 187, 167–173. [DOI] [PubMed] [Google Scholar]
- 18.Ratcliff R.M., Lanser,J.A., Manning,P.A. and Heuzenroeder,M.W. (1998) Sequence-based classification scheme for the genus Legionella targeting the mip gene. J. Clin. Microbiol., 36, 1560–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Picardeau M. and Vincent,V. (1995) Development of a species-specific probe for Mycobacterium xenopi. Res. Microbiol., 146, 237–243. [DOI] [PubMed] [Google Scholar]
- 20.Johansson M.L., Molin.G., Pettersson,B.U., Uhoen,M. and Ahrne,S. (1995) Charactersation and species recongnization of Lactobacillus plantarum strains by restriction fragment length polymorphism (RFLP) of the 16S rRNA gene. J. Appl. Bacteriol., 79, 536–541. [Google Scholar]
- 21.Rodrigues U.M., Aguirre,M., Facklam,R.R. and Conllins,M.D. (1991) Specific and intraspecific molecular typing of lactococci based on polymorphism of DNA encoding rRNA. J. Appl. Bacteriol., 71, 509–516. [DOI] [PubMed] [Google Scholar]
- 22.Hertel C., Ludwig,W., Obst,M., Vogel,R.F., Hammes,W. and Schleifer,K.H. (1991) 23S rRNA-targeted oligonucleotide probes for the rapid identification of meat lactobacilli. Syst. Appl. Microbiol., 14, 173–177. [Google Scholar]
- 23.Schleifer K.H., Ehrmann,M., Beimfohr,C., Brockmann,E., Ludwig,W. and Amann,R. (1995) Application of molecular methods for the classification and identification of lactic acid bacteria. Int. Dairy J., 5, 1081–1094. [Google Scholar]
- 24.Chinese Pharmacopoeia Editorial Committee (2000) Chinese Pharmacopoeia, 2000 Edn. Chemical Technology Press, Beijing, Vol 1, p. 70. [Google Scholar]
- 25.Jiangsu New Medical College (1986) Dictionary of Chinese Medicines. Shanghai Scientic and Technologic Press, Shanghai, pp. 586–592. [Google Scholar]
- 26.Ding X.Y., Xu,L.S., Wang,Z.T., Zhou,K.Y., Xu,H and Wang,Y.Q. (2002) Authentication of stems of Dendrobium officinale by rDNA ITS region sequence. Planta Med., 68, 191–192. [DOI] [PubMed] [Google Scholar]
- 27.Lau D.T., Shaw,P.C., Wang,J. and But,P.P. (2001) Authentication of medicinal Dendrobium species by the internal transcribed spacer of ribosomal DNA. Planta Med., 67, 456–460. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 29.Jaccoud D., Peng,K., Feinstein,D. and Kilian,A. (2001) Diversity array: a solid state technology for sequence information independent genotyping. Nucleic Acids Res., 29, e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lisitsyn N., Lisitsyn,N. and Wigler,M. (1993) Cloning the differences between two complex genomes. Science, 259, 946–951. [DOI] [PubMed] [Google Scholar]