Significance
The Mikea are the last known Malagasy population reported to be still practicing a hunter-gatherer lifestyle. Earlier writers thought the Mikea were descended from ancient forager groups who have maintained their way of life up to the present. However, our analyses show that the Mikea are not a remnant population and, to the contrary, derived from a recent admixture of two agriculturalist populations: the Bantu (from Africa) and the Austronesian (from east-Asia). Thus, it is probable that the Mikea have adopted their hunter-gatherer way of life through a recent cultural reversion.
Keywords: settlement, migration, DNA
Abstract
Linguistic and cultural evidence suggest that Madagascar was the final point of two major dispersals of Austronesian- and Bantu-speaking populations. Today, the Mikea are described as the last-known Malagasy population reported to be still practicing a hunter-gatherer lifestyle. It is unclear, however, whether the Mikea descend from a remnant population that existed before the arrival of Austronesian and Bantu agriculturalists or whether it is only their lifestyle that separates them from the other contemporary populations of South Madagascar. To address these questions we have performed a genome-wide analysis of >700,000 SNP markers on 21 Mikea, 24 Vezo, and 24 Temoro individuals, together with 50 individuals from Bajo and Lebbo populations from Indonesia. Our analyses of these data in the context of data available from other Southeast Asian and African populations reveal that all three Malagasy populations are derived from the same admixture event involving Austronesian and Bantu sources. In contrast to the fact that most of the vocabulary of the Malagasy speakers is derived from the Barito group of the Austronesian language family, we observe that only one-third of their genetic ancestry is related to the populations of the Java-Kalimantan-Sulawesi area. Because no additional ancestry components distinctive for the Mikea were found, it is likely that they have adopted their hunter-gatherer way of life through cultural reversion, and selection signals suggest a genetic adaptation to their new lifestyle.
Located 400 km of the East African coast, Madagascar has been referred to as “the single most astonishing fact of human geography” (1, 2). Despite its proximity to Africa, less than 10% of the vocabulary of the Malagasy language is from African languages (mainly Sabaki, a branch of Bantu) (3–5). In contrast, 90% of Malagasy vocabulary belongs to the Barito (6) or other subgroups of Austronesian languages of Island Southeast Asia (7–9). Although being less specific, genetic studies have generally confirmed the dual ancestry of the Malagasy population (10–14).
Low genetic differentiation of the Malagasy mtDNA and Y chromosome lineages from related lineages observed in present day Bantu and Austronesian populations support a model drawn from linguistic evidence that the Malagasy gene pool has been derived predominantly from these two dispersals of agriculturalist populations. As shown by linguistic and phylogenetic studies on cattle and crop names and their genetic diversity, both agricultural populations have apparently brought their way of life once they had settled in Madagascar (15, 16).
Although one archaeological report claims the presence of anthropic artifacts as early as 4,000 y ago (17), most research points to first human impact on the Malagasy environment around 2,400 y ago (18), which would still be before the Bantu expansion reached the East African coast (19). In addition, European traveler reports and putative archaeological artifacts support hunter-gatherers living in the south of the island until the 16th century (20–23). It has been speculated that these hunter-gatherer groups were the remnants of a pre-Bantu settlement of Madagascar (24). The cause of disappearance of the hunter-gatherers after the 16th century is unknown, but the two most likely scenarios that can be contemplated involve either a cultural shift or population replacement.
Traditions concerning the dispersal of a sedentary way of life and agriculture in the south of the island relate to the Sakalava expansion (25). These traditions recount that in the 17th century, leaders, soothsayers, and migrants from the arabo-islamized Temoro population from the southeastern coast of Madagascar colonized the southern regions of Madagascar with the intention of creating new cities and kingdoms, such as Maroserana and Andrevola (25, 26). A few decades later, new Sakalava kingdoms emerge on the southeast coast and gradually spread throughout southern Madagascar, which coincides with the disappearance of hunter-gatherer populations (25).
The survival in Madagascar of a modern hunter-gatherer population was believed to be a myth (24, 25). However, there are a variety of hunter-gatherer groups scattered across the island that have been studied and mapped since the 1920s, particularly in the Tsiribihina region (southwest Madagascar) under the names Vazimba and Beosi (27). In the Mikea forest (south of the Mangoky River, southern Madagascar) one population, the Mikea, still live as hunter-gatherers. Earlier writers thought the Mikea were descended from ancient forager groups who have maintained their way of life up to the present (24, 25, 27, 28), but most modern scholarship argues the Mikea reverted back to the forest for political or economical reasons, such as Sakalava royalty pressure or French colonization (29–31).
We address here the question of whether and to what extent the Mikea share their genetic ancestry with their neighboring Malagasy populations with a sedentary lifestyle. Specifically, we aim to detect in the Mikea patterns of genetic diversity assignable to a population that would predate Austronesian and Bantu incursions. Alternatively, we consider the scenario by which it is only their subsistence strategy that separates the Mikea from other contemporary populations in southern Madagascar.
To answer these questions, we performed a genome-wide analysis of 21 Mikea individuals, 24 individuals from a nearby Vezo population, and 24 individuals of the Temoro population, using Illumina HumanOmniExpress BeadChips, and compared the data with Southeast Asian and African populations. Based on this dataset, we have: (i) studied the genetic distance between the three Malagasy populations; (ii) tested the existence and age of admixture patterns; and (iii) tested the Mikea genome for any adaptive signal that may be associated with the hunter-gather way of life.
Methods
Sampling and Genotyping.
The samples analyzed in this study were collected in field seasons 2007–2009. The samples were obtained with informed consent, and were approved by Human Subjects’ Ethics Committees of the Health Ministry of Madagascar (N°036 and N°039). Buccal cells and peripheral blood were sampled. Subjects were surveyed for language affiliation, current residence, familial birthplaces, and a genealogy of four generations to establish lineage ancestry and to select unrelated individuals. The Mikea samples were collected from seven different villages (Fig. S1). From 266 individuals sampled, a total of 21 Mikea (hunter-gatherers), 24 Vezo (seminomadic fishermen), and 24 Temoro (farmers) were genotyped. Studied individuals were chosen to be representative of distinct ancestral lineages to encompass as much diversity as possible in this small community. The genotyping of 730,525 SNPs across the genome was performed using Illumina HumanOmniExpress BeadChips. All genotyped individuals passed the quality checks and had a genotype call rate higher than 98%. To remove closely related individuals, we estimated identity by descent proportions using the genome function of PLINK (32) and the King algorithm (33). We filtered the data by removing iteratively one individual from each pair until no kinship coefficient value remained higher than 0.25. Four individuals from the Mikea sample and one from the Vezo sample were removed. We also compared the data to African and Indonesian populations available from several publications and databases, such as the Human Genome Diversity Project (HGDP), HapMap3, and Pan-Asian consortia (34–39). In addition, for the purpose of comparison, Illumina HumanOmniExpress data were generated for this study from two Indonesian populations, the Lebbo (a Kayah subgroup) from Kalimantan (n = 18) and the Bajo from Sulawesi (n = 32). We merged the relevant datasets, keeping only overlapping sets of compatible SNP markers after correcting for strand consistencies; the data is available through National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) data repository (accession no. GSE53445) and available from the authors upon request.
Statistical Analyses.
To explore the distribution of pairwise genetic differences within and among populations, we computed an identity-by-state distance (1-IBS) matrix using PLINK (32). The 1-IBS statistic corresponds to the proportion of SNPs that are different between a pair of individuals.
We have estimated genome-wide 1-IBS scores for the three Malagasy populations, the two Indonesian populations, and three African populations from HAPMAP (35, 36). We used the PLINK software to remove each SNP that has an r2 value greater than 0.1 with any other SNP within a 50-SNP sliding window (advanced by 10 SNPs each time) (32). Following the pruning steps and merging, this analysis was restricted to 121,522 SNPs that were genotyped in both datasets.
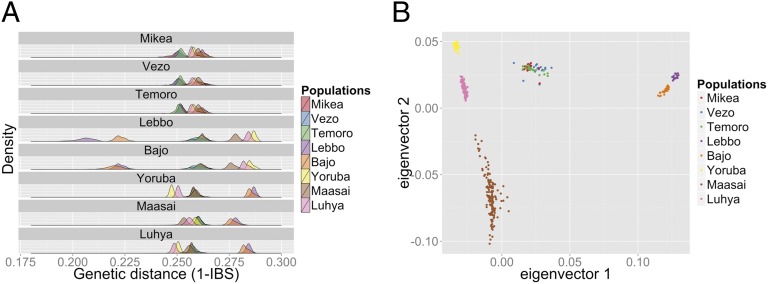
The genetic distance intrapopulation was determined by computing the mean (and SD) of the genetic distance 1-IBS separating the individuals from this population (Table S1). Similarly, the genetic distance between two populations was determined by computing genetic distance 1-IBS separating the individuals from the two populations. Based on this computation, the genetic distance between two populations was plotted using the density function from R (38). In each panel of Fig. 1A, we have represented the distance between the individuals of one population and the individuals from the other populations. In each panel we have also represented the intrapopulation distance, which is the mean of the genetic distances observed among all pairs of individuals from the given population.
Fig. 1.
Genetic variation across the Malagasy individuals compared with Indonesian and African populations. (A) Identity by structure distance intra and interpopulations using the PLINK algorithm (32). Each panel represents the distance between the individuals of one population and the individuals from the other populations. The intrapopulation distance is also represented as the genetic distance between all of the individuals from this population. (B) PCA performed using Eigensoft (40).
To explore clustering of populations and their admixture we used principle component analysis (PCA) and ADMIXTURE. The PCA analyses were performed on the same pruned dataset using the smartPCA functionality of the EIGENSOFT 4.2 software (40).
To estimate the admixture fractions from the different ancestral source populations of the three Malagasy populations, we performed a structure-like analysis using the ADMIXTURE software (41) after thinning the marker set for linkage disequilibrium.
To address questions regarding the African and Southeast Asian ancestries of the three Malagasy populations, we performed two separate sets of analyses. The first analysis focused on the Southeast Asian origin, including the Malagasy and Indonesian populations (Lebbo and Bajo) from this study and all of the populations from the Pan-Asian consortium dataset speaking a language that belongs to the Austronesian family (34). We included samples from Malaysia (Malay: MY-KN, MY-TN, MY-MN; proto-Malay: MY-TM; Bidayuh: MY-BD), Taiwan (Atayal: AX-AT; Ami: AX-AM), Philippines (Ayta: PI-AE; Filipino: PI-UN, PI-UB, PI-UI; Agta: PI-AG; Iraya: PI-IR; Ati: PI-AT; Mamanwa: PI-MW; Minanubu: PI-MA), and Indonesia (Dayak: ID-DY; Toraja: ID-TR; Lembeta: ID-LE; Lamaholot: ID-LA; Manggarai: ID-RA, ID-SO; Kambera: ID-SB; Javanese: ID-JV; Sunda: ID-SU; Malay: ID-ML; Batak Karo: ID-KR; Batak Toba: ID-TB; Mentawai: ID-MT). We also included the Papuan population from the HGDP panel (39) as a known source of admixture among several extant Austronesian-speaking populations (42), and used the Yoruba population from HAPMAP (35) as an African reference group. We used the PLINK software to remove each SNP that has an r2 value greater than 0.1 with any other SNP within a 50-SNP sliding window (advanced by 10 SNPs each time) (32). Following the pruning and merging steps with the Pan-Asian panel, this analysis was based on 7,534 SNPs.
In the second analysis focusing on the potential sources of admixture from Africa and West Eurasia, we prepared a dataset including the three populations from Madagascar, African, and West Eurasian populations from several available sources, including HGDP [Biaka and Mbuti Pygmies (Central African Republic), Mandinka (Senegal), Yoruba (Nigeria), Bantu (including South-East African Bantu (Pedi, Sotto, Tswana, Zulu); Southwest African Bantu (Herero, Ovambo), San (Namibia), and French (39)], the Henn et al. data (Hadza, Khoisan and Sandawe) (38), HAPMAP data (Yoruba: YRI; Luhya: LWK; and Maasai: MKK) (35), and Behar et al. data (Saudis, Egyptians, Ethiopians, Jewish) (37). Finally, we have added the Lebbo population as a reference group representing Austronesian-speaking populations from Southeast Asia. Following the pruning steps, this analysis was based on 48,467 SNPs.
Several approaches to estimate admixture dates have been developed on the basis of exponential decay of admixture-induced linkage disequilibrium (LD) as a function of genetic distance (40, 43). Here we used the ALDER software (44) to estimate the time of admixture between Indonesian and African source population. Because the choice of the best surrogate of a source population was uncertain, we performed several admixture tests combining different African and Indonesian populations.
We have estimated the admixture age of the Papuan gene flow in several Indonesian populations as well as the Malagasy populations. Similarly, we have used the Yoruba population as a surrogate population of the Bantu gene flow and tested the admixture on several African and Malagasy populations.
We performed scans of positive selection using an integrated haplotype score (iHS) statistic (45) with tools available at http://hgdp.uchicago.edu and the HapMap b37 genetic map for calculating genetic distances between markers. The iHS statistic was calculated on a subset of sites with a minor allele frequency >0.05 and for which information about ancestral states was available from the Ensembl Variation database (46). We divided the autosomal chromosomes by 200-kb windows following the approach of Pickrell et al. (47), with a minor modification of combining windows with more than 80 SNPs into the same bin.
We performed enrichment analyses of the top 1% iHS windows using a modified algorithm of DAVID (48). We used the EASE score (49) to estimate the significance of gene ontology (GO) term associations with the 200-kb windows for which the iHS statistic had been calculated. The EASE score is a modified Fisher exact P value that penalizes (subtracts) the count of positive agreements by 1; it weights significance in favor of terms supported by more genes and, therefore, improves the family-wise true discovery rate as a whole. We use the EASE score over other multiple-test correction methods for the reasons outlined elsewhere (48). Multiple-test correction results are offered in the Supporting Information, Table S3 (50). In addition, to control for physical clustering of genes by gene families, each GO term that was associated with more than one gene in the same 200-kb window was considered only once in the estimation of enrichment score.
Results and Discussion
To test whether Mikea differ from the two other Malagasy populations in the overall pattern of genome-wide diversity, we first computed the complete matrix of pairwise identity-by-state distance in the three Malagasy populations and five reference populations from Africa and Indonesia. All three Malagasy populations were characterized by similar patterns of diversity (Fig. 1A and Table S1). Mikea did not differ significantly from the other two populations in either the interpopulation distances to other Malagasy or the reference populations.
Next, to gain better understanding about the relationships between the different populations, we performed PCA using Eigensoft (40). The first eigenvector (4% of variation explained) clearly separated the Indonesian and African populations and the second (1%) distinguished between the different African populations (Fig. 1B). All Malagasy individuals clustered tightly together at an intermediate position consistent with their admixed status. The Mikea did not appear to be genetically different from other Malagasy individuals and were no closer than the other two Malagasy populations than to either the Indonesian or the African populations (even in less-significant vectors).
ADMIXTURE Analysis.
To study in more detail the composition and structuring of genetic diversity in the Malagasy populations, we performed two distinct analyses with ADMIXTURE (41): the first focused on the Indonesian origin and the second on the African one. For both analysis with a K = 2 parameter, the results showed a clear African vs. non-African population dichotomy. These two analyses concurred in estimating the African ancestry proportion of 67% in each of the three Malagasy populations (Fig. S2 and Table S2). At increased values of K (Fig. 2 and Fig. S2) more complex patterns emerged.
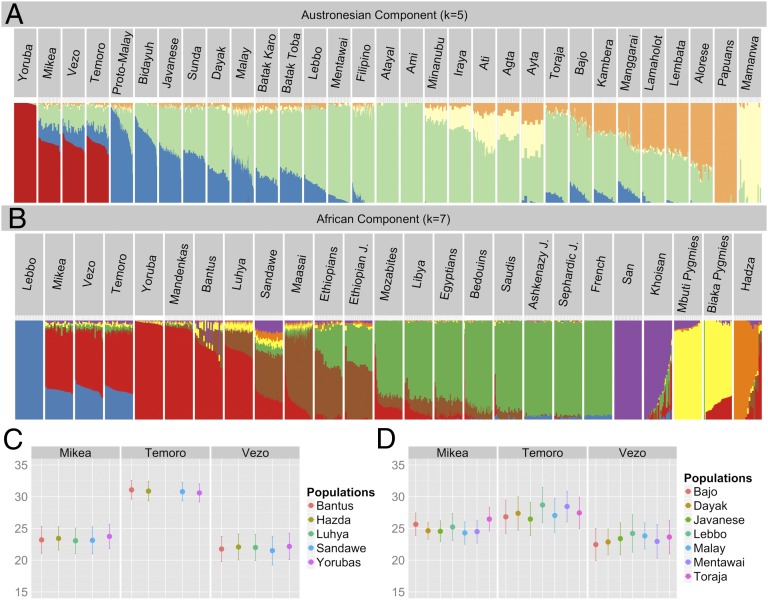
Fig. 2.
Genetic structure of three Malagasy populations. (A) ADMIXTURE analysis involving 25 Austronesian-speaking populations from Island Southeast Asia and one African and one Papuan population for reference. (B) ADMIXTURE analysis focused on African and West Eurasian populations. (C) Dates of admixture for three Malagasy populations estimated by ALDER and using the Lebbo and several African populations as sources. (D) Dates of admixture for three Malagasy populations using Yorubas and several Southeast Asian populations as sources.
In the analyses aimed at discriminating further details among the different genetic ancestry components of the Indonesian populations, cross-validation scores aimed our focus on K values 4–6 (Fig. S2). At K = 5 (Fig. 2A), the red component reflects the African ancestry and accounts, as in the K = 2 analyses, for two-thirds of the total variation in the Malagasy populations. In contrast, the pastel yellow component, predominant in the Mamanwa from the Philippines, is virtually absent among the Malagasy. The pastel orange component reflecting Papuan ancestry in several Austronesian populations from east Indonesia, such as Alorese and Lambata (42) is, similarly, almost undetectable in Malagasy. Above these three ancestry components, which were also uncovered in K = 4 analyses (Fig. S2), the K = 5 analysis revealed two additional components that together accounted for most of the Southeast Asian ancestry in Malagasy. The pastel green component, predominant in two Taiwanese groups, Atayal and Ami, characterizes one-half of the Southeast Asian ancestry proportion, whereas the blue component accounts for the other half in the Malagasy. A number of Southeast Asian populations sampled from the broad Java-Kalimantan-Sulawesi area show, as with Malagasy, an almost equal share of the pastel green and blue and a virtual absence of the Mamanwa and Papuan components. This area could, thus, be considered as a plausible source for the Southeast Asian component in all three Malagasy populations examined here.
The saturation of the cross-validation scores in the analyses focusing on the African side of admixture occurs at K = 7 (Fig. 2B and Fig. S2). The interpretation of the results is complicated because of the fact that very few populations are available from Africa and none of them are reasonably close to the putative ancestral populations of Malagasy populations (such as any Swahili populations for example). The results of these analyses should therefore be considered with caution. The red component, which represents almost 100% of the Yoruba from Nigeria, the Mandinka from Senegal, is also the predominant component in HGDP Bantu and Luhya populations and accounts for most of the African ancestry in Malagasy. Besides this West African/Bantu component, others reflected in African hunter-gatherers (Fig. 2B, yellow, purple, and orange) or East Africans and West Eurasians (Fig. 2B, brown and green) show only marginal traces in the Malagasy. The absence of significant traces of the green component suggests that genetic admixture with the European population during the colonial period and the influence of Arabic, sometimes evoked in the oral traditions and suggested by the tradition of writing in Arabic, has been minimal among the Mikea, Vezo, and Temoro.
Notably, the brown component (Fig. 2B), which is common among the populations of the East African coast, such as Maasai, is absent in the Malagasy groups considered here. This finding is not surprising considering that the Maasai speak a Nilo-Saharan language and may therefore carry ancestry components that never reached Madagascar. Unfortunately, similar data for the Swahili, who would represent the best candidate for an East African source population, is not available thus far.
These results suggest that gene flow from Bantu-speaking populations is likely to be the main source of the present genetic background in Madagascar. Given the recent time scale of Bantu dispersal, these results suggest a recent origin of the African component in Madagascar or a complete replacement of any previous African mainland components by the Bantu. Besides the predominant red component, a minor—albeit detectable—proportion of the African ancestry in all three Malagasy populations belongs to a blend of the yellow, purple, and green components (Fig. 2). Because of the lack of many African populations, such as the Swahili, it is not possible to tell from these analyses whether this blending reflects additional gene flow or whether the ancestral Southeast African Bantu population that dispersed to Madagascar already carried these additional components. Considering the ancestry of the Mikea, regardless of their hunter-gather subsistence, the composition of their African ancestry pallet is not distinguishable from those of Vezo and Temoro.
Date of Admixture for Malagasy Populations.
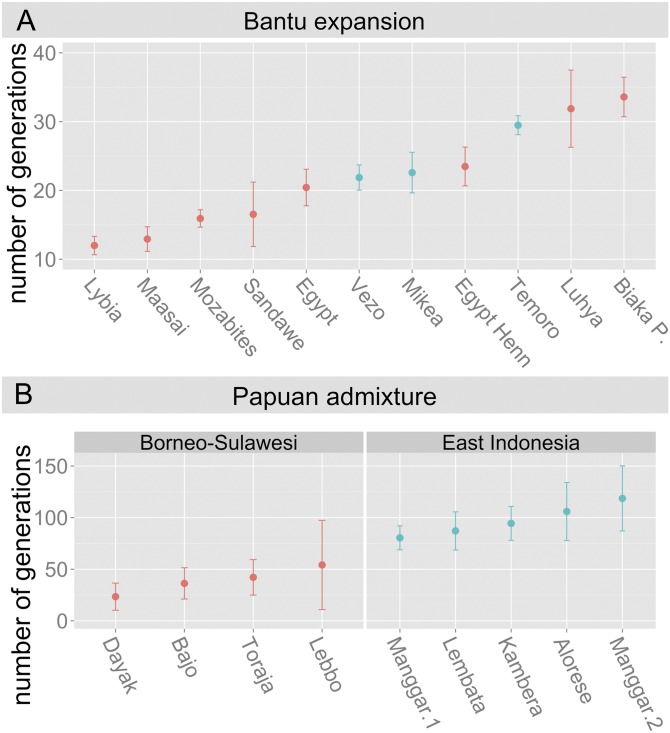
To infer the date of admixture between the two source populations (Southeast Asian and African), we have used the ALDER software. To circumvent the uncertainties about the exact ancestral source, we have performed the same admixture test using several African and Indonesian populations. For all combinations we find an admixture date between 20 and 32 generations ago (Fig. 2 B and C). These estimates, depending on an assumed generation times (20 or 25 y), date the peak of admixture between 400 and 800 y before present.
The time estimates obtained with ALDER should be taken with caution for several reasons. First, the mathematical model beyond the dating is based on a scenario of a perfect mix between the two populations, implying that since the first generation the mating has been fully random. This model is likely to be too simplistic. Second, in cases of prolonged process of admixture or multiple admixture events, the approach is designed to give higher emphasis on more recent admixture events (43). Therefore, it is not implausible that the admixture process involving the Austronesian and Bantu components could have started long before the date estimated by the method. Additionally, we should bear in mind that dating applied in this study was only on a few populations from southern Madagascar and should not be generalized to other Malagasy populations.
Taking into account all these limitations, the most conservative interpretation of our result is that the admixture between Bantu and Southeast Asian genomic components began before the 17th century and before the disappearance of hunter-gatherers, European colonization, and also before the major change of the modern societies.
Bantu and Austronesian Expansion.
To place the obtained date of admixture in Malagasy population into a broader African context, we have assessed the admixture date with the predominant West African component in several African populations as a proxy of the demographic events that may have followed the Bantu expansion in the eastern coast of Africa. In the context of these date estimates it is notable that the dates of admixture of African and Southeast Asian components we obtained for the Malagasy populations are in the same range of 20–35 generations that we obtained for the Luhya and Biaka groups, whereas the West African admixture in the Sandawe appears to be relatively younger (Fig. 3).
Fig. 3.
(A) Dates of admixture of West African component in Malagasy and several African populations (using the present Yoruba population as parental population). (B) Dates of admixture in Austronesian-speaking populations of Indonesia with the Papuan component (using the present Papuan sample as parental population).
As reported above, ADMIXTURE analyses at K = 5 detected in a number of Southeast Asian populations variable proportions of a pastel orange component that was predominant in Papuans and virtually absent in Malagasy populations. To test whether the lack of Papuan component in Malagasy populations could be further used as a clue to determine which Austronesian populations were involved as potential sources in the settlement of Madagascar, we estimated the dates of admixture between Papuan and Austronesian components in nine Austronesian-speaking populations of Indonesia. The admixture dates for the islands of East Indonesia fall in the range of 80–120 generations, which is younger than dates for the same data estimated by a different method by Xu et al. (42) (Fig. 3). Regardless of the methodological differences, the admixture dates for East Indonesia significantly predate plausible migration dates to Madagascar and thus suggest that Eastern Indonesian populations are unlikely to have contributed a significant proportion of their genes in the Malagasy settlement. However, we found that the admixture of the Sulawesi and Kalimantan populations with the Papuan genetic component is significantly more recent: between 20 and 40 generations (Fig. 3) and contemporary with the admixture dates we obtained for the Malagasy populations. Considering the fact that the Southeast Asian input to Madagascar had to precede the local admixture event with the African component, this result makes it plausible that the parental populations of the Austronesian component could come from either Java, Kalimantan, or Sulawesi, as suggested by linguistic studies (7, 9), as these source populations themselves may have admixed with Papuan sources after the settlement of Madagascar had already taken place.
Haplotype Homozygosity Analyses.
Furthermore, to assess whether the Mikea are differentiated from their neighboring populations in haplotype homozygosity signals of specific genes that might be attributable to their lifestyle, and to test whether, overall, the genome-wide estimates of admixture proportions are also reflected in the transmission rates of signals of positive selection from their parental groups, we calculated the iHS statistic for the three Malagasy populations and compared the results with those from Yoruba and two Indonesian populations. These analyses revealed (Tables S3 and S4) that, on average, a top 1% signal detected in one Malagasy population was found in 34% of the cases as well as in the top 5% empirical ranks of the two other populations. In contrast to twofold higher contributions of African ancestry, as revealed above in the general genome-wide tests, the sharing of signals of recent positive selection in the three Malagasy populations was less differentiated between the African (23%) and Indonesian (20%) sources. Interestingly, the Malagasy iHS sharing with Bajo (27.5% on average) appeared to be higher than sharing with Yoruba, whereas the sharing with Lebbo was lower than the fraction of iHS signals the Malagasy populations shared with Europeans. Only eight genomic windows had a top 1% signal in all three Malagasy populations (Tables S3 and S4). Among these genomic windows, the window containing the erythrocyte differentiation gene hemopoietic gene protein (HEMGN) was shared with the Yoruba top 1% and four windows were shared with either Bajo or Lebbo.
Of the previously ascertained genes with signatures of recent positive selection, it is worthwhile to note that the extended haplotype homozygosity signal in the 200-kb window containing the ectodysplasin A receptor (EDAR) gene, which according to previous studies (47, 51) is the topmost positive selection signal in East Asian populations, appeared among the 10 strongest signals in Vezo and also in the top 1% of Temoro and top 5% of Mikea, yet is relatively low in ranking in the top 5% signal in Bajo and absent from the top 5% signals in Lebbo. This finding may suggest either ongoing selection for the EDAR gene in Malagasy or that the homozygosity signal itself was obtained from a source population that had a higher frequency of the extended haplotype of EDAR than the two Indonesian populations considered here. Given the low genotype resolution of the Pan-Asian consortium data and lack of high-resolution genotype data from this region to date, unfortunately, scans of iHS on larger sets of populations from Southeast Asia could not be performed here. Of the top 20 iHS signals of Yoruba, 11 were observed in at least one Malagasy population in the top 1%, whereas the topmost signal at the LARGE gene, which has been associated with protection against the Lassa virus (51), was not observed in the top 5% in any of the Malagasy populations.
Analyses of enrichment of the iHS signals specific to the Mikea population are summarized in Tables S3 and S4. Compared with the other two Malagasy populations, Mikea showed a higher haplotype homozygosity in genes related to neural function (dopamine uptake, glutamate receptor signaling, and gliogenesis), adaptive immunity, striated muscle contraction, and fatty acid α-oxidation. The latter function is important in metabolism of phytanic acid, which is an important component in dairy products, ruminant animal fats, and certain fish. These results could potentially be linked to the Mikea life-style but need further biochemical and physiological investigation to confirm and characterize the putative phenotypic effect of these genes.
Conclusion
In conclusion, we found that the three Malagasy populations studied have originated recently from the admixture of Austronesian and Bantu genetic components. The Bantu gene flow represents at least 60% of the genetic background, whereas Austronesian gene flow represents around 30% and is likely coming from the Java-Kalimantan-Sulawesi area. A small fraction of the genetic background seems independent to these components; this could be a signal of either other recent genetic influences or putative remnant of an older colonization event. The admixing process began before the 17th century, which means before the hunter-gather disappearance in the south of Madagascar, before the European colonization, and before the major change of the modern societies.
We were unable to detect in Mikea unique ancestry components that would have been absent among sampled populations. These results support the hypothesis that the Mikea originated from agricultural populations and have reverted to the forest (24, 25, 27). Despite the Mikea population deriving from the same genetic admixture as Vezo and Temoro, they have reverted to a hunter-gatherer lifestyle. This reversion is one of the few observed cases that have been supported by genetic evidence and, to our knowledge, the only one reported so far for African populations (52, 53). Future genetic and anthropological work is needed on the Mikea and nearby populations to date this cultural change. In parallel, the genetic adaptation of the Mikea linked to their lifestyle suggested by the presence of selection signals need to be investigated (i.e., metabolism of phytanic acid). Future work is required on Malagasy populations, and also in Africa and Indonesia, to clarify the history of settlement of Madagascar Island.
Supplementary Material
Acknowledgments
We thank J. M. Dugoujon [Laboratoire d’Anthropologie Moléculaire et Imagerie de Synthèse (AMIS), Centre National de la Recherche Scientifique (CNRS), Université de Toulouse] for facilitating access to the Temoro population; Eric Crubézy (Laboratoire AMIS CNRS, Université de Toulouse) for helpful discussion; Phillipe Grangé, Charles Illouz, and Chandra Nuraini (Faculté des Lettres, Langues, Arts et Sciences Humaines, Université de La Rochelle, Laboratoire Centre de Recherche en Histoire Internationale et Atlantique EA, France) for facilitating access to the Bajo population; Bambang Sugiyanto (Balai Arkeologi, Banjarmasin, Indonesia), Adhi Agus Oktaviana (National Research Center for Archaeology, Jakarta, Indonesia), Antonio Guerreiro (CNRS Unité Mixte de Recherche 6571, University Aix-Marseille), and Budi Amuranto (Dinas Kebudayaan dan Pariwisata, East Kutai Sangata, Indonesia) for facilitating access to the Lebo population; and Louis Paul Randriamarolaza (Laboratoire d’Anthropologie, Faculté des Lettres et Sciences Humaines, Université d’Antananarivo, Madagascar). This research was funded by the Région Aquitaine (Grant “project MAGE”); European Research Council Starting Investigator Grant FP7-261213 (to T.K.); French ANR-12-PDOC-0037-01 (grant “GENOMIX”); the Ministry of Foreign and European Affairs [French Archaeological Mission in Borneo (MAFBO)]; l’Association Contre les Maladies Mitochondriales; the National Research Center for Archaeology (Jakarta, Indonesia), and the Laboratoire Centre de Recherche en Histoire Internationale et Atlantique Equipe d'accueil (Faculté des Lettres, Langues, Arts et Sciences Humaines; University of La Rochelle, France); and the University Haluoleo (University Haluoleo, Kendari, Indonesia).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE53445).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321860111/-/DCSupplemental.
References
- 1.Diamond J. Guns, Germs and Steel: A Short History of Everybody for the Last 13,000 Years. London: Vintage; 1998. p. 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar A. The single most astonishing fact of human geography: Indonesia’s far west colony. Indonesia. 2011;92(October):59–96. [Google Scholar]
- 3. Vérin P (2000) Madagascar (Karthala, Paris) 269 Ed.
- 4.Blench RDM. New palaeo-zoogeographical evidence for the settlement of Madagascar. Azania XLII. Journal of the British Institute in Eastern Africa. 2007;42(1):69–82. [Google Scholar]
- 5.Blench RDM. The Austronesians in Madagascar and their interaction with the Bantu of East African coast: Surveying the linguistic evidence for domestic and translocated animals. In: Brainard S, editor. Philippines Journal of Linguistics. Vol 18. Manila: SIL; 2009. pp. 18–43. [Google Scholar]
- 6.Dahl OC. Malgache et Maanjan. Une Comparaison Linguistique. Oslo: Egede-Institutet; 1951. p. 408. [Google Scholar]
- 7.Adelaar A. Towards an integrated theory about the Indonesian migrations to Madagascar. In: Peregrine P, Feldman M, editors. Ancient Human Migrations: An Interdisciplinary Approach. Salt Lake City: Univ of Utah Press; 2009. [Google Scholar]
- 8.Adelaar AK. Asian Roots of the Malagasy: A Linguistic Perspective. Leiden: Bijdragen tot de Taal-, Land- en Volkenkunde; 1995. pp. 325–356. [Google Scholar]
- 9.Beaujard MP. Les arrivées austronésiennes à Madagascar: Vagues ou continuum? (Partie 1 et 2) Études Océan Indien. 2003;35-36:59–147. [Google Scholar]
- 10.Hurles ME, Sykes BC, Jobling MA, Forster P. The dual origin of the Malagasy in Island Southeast Asia and East Africa: Evidence from maternal and paternal lineages. Am J Hum Genet. 2005;76(5):894–901. doi: 10.1086/430051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Razafindrazaka H, et al. Complete mitochondrial DNA sequences provide new insights into the Polynesian motif and the peopling of Madagascar. Eur J Hum Genet. 2010;18(5):575–581. doi: 10.1038/ejhg.2009.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soodyall H, Vigilant L, Hill AV, Stoneking M, Jenkins T. mtDNA control-region sequence variation suggests multiple independent origins of an “Asian-specific” 9-bp deletion in sub-Saharan Africans. Am J Hum Genet. 1996;58(3):595–608. [PMC free article] [PubMed] [Google Scholar]
- 13.Tofanelli S, et al. On the origins and admixture of Malagasy: New evidence from high-resolution analyses of paternal and maternal lineages. Mol Biol Evol. 2009;26(9):2109–2124. doi: 10.1093/molbev/msp120. [DOI] [PubMed] [Google Scholar]
- 14.Regueiro M, et al. Austronesian genetic signature in East African Madagascar and Polynesia. J Hum Genet. 2008;53(2):106–120. doi: 10.1007/s10038-007-0224-4. [DOI] [PubMed] [Google Scholar]
- 15.Razafindraibe H, et al. Mitochondrial DNA origin of indigenous malagasy chicken. Ann N Y Acad Sci. 2008;1149:77–79. doi: 10.1196/annals.1428.047. [DOI] [PubMed] [Google Scholar]
- 16.Walsh M. Island subsistence: Hunting, trapping and the translocation of wildlife in the western Indian Ocean, Azania. Journal of the British Institute in Eastern Africa. 2007;42(1):30. [Google Scholar]
- 17.Gommery D, et al. Les plus anciennes traces d’activités anthropiques de Madagascar sur des ossements d’hippopotames subfossiles d’Anjohibe (Province de Mahajanga) C R Palevol. 2011;10(4):271–278. [Google Scholar]
- 18.Burney DA, et al. A chronology for late prehistoric Madagascar. J Hum Evol. 2004;47(1-2):25–63. doi: 10.1016/j.jhevol.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Ehret C. An African Classical Age: Eastern and Southern Africa in World History, 1000 B.C. to A.D.400. Charlottesville: Univ Press of Virginia; 1998. [Google Scholar]
- 20.Radimilahy C. Contribution à l'archéologie du Sud-ouest de Madagascar. In: Radimilahy C, Rajaonarimanana N, editors. Civilisations des Mondes Insulaires; Madagascar, îles du Canal de Mozambique, Masccareignes, Polynésie, Guyanes. Paris: Karthala; 2011. pp. 825–853. [Google Scholar]
- 21.Burney DA. Late Holocene environmental-changes in arid southwestern Madagascar. Quat Res. 1993;40(1):98–106. [Google Scholar]
- 22.Dewar RE, Wright HT. The culture history of Madagascar. J World Prehist. 1993;7(4):417–466. [Google Scholar]
- 23.Drury R. In: Madagascar: Or Robert Drury's Journal During Fifteen Years Captivity on that Island. Oliver PS, editor. London: T Fisher Unwin; 1729 [1890]. [Google Scholar]
- 24.Stiles D. The Mikea hunter-gatherers of southewestern Madagascar: Ecology and socioeconomics. Afr Study Monogr. 1998;19(3):127–148. [Google Scholar]
- 25.Lombard J. Le Royaume Sakalava du Menabe - Essai d'analyse d'un système politique à Madagascar du XVIIème au XXème siècle. Paris: Orstrom; 1988. [Google Scholar]
- 26.Fagereng E. Origine des dynasties ayant régné dans le sud et l’ouest de Madagascar. Omaly Anio. 1981;13-14(125):125–140. [Google Scholar]
- 27.Birkeli E. 1936. Les Vazimba de la Côte Ouest de Madagascar. Mémoires de l’Academie Malgache 198 (Imprimerie Officielle de la Colonie, Antananarivo)
- 28.Dina J, Hoerner JM. Etude sur les populations Mikea du sud-ouest de Madagascar. Omaly Anio. 2004;47(1–2):25–63. [Google Scholar]
- 29.Poyer L, Kelly RL. Mystification of the Mikea: Constructions of foraging identity in southwest Madagascar. J Anthropol Res. 2000;56(2):163–185. [Google Scholar]
- 30.Yount JW, Tsiazonera B, Tucker T. Constructing Mikea identity: Past or present links to forest and foraging. Ethnohistory. 2001;48(1-2):257–291. [Google Scholar]
- 31.Bram TT. The Behavioral Ecology and Economics of Variation, Risk, and Diversification Among Mikea Forager-Farmers of Madagascar. Chapel Hill: Faculty of the Department of Anthropology, Univ of North Carolina at Chapel Hill; 2001. [Google Scholar]
- 32.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manichaikul A, et al. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abdulla MA, et al. HUGO Pan-Asian SNP Consortium Indian Genome Variation Consortium Mapping human genetic diversity in Asia. Science. 2009;326(5959):1541–1545. doi: 10.1126/science.1177074. [DOI] [PubMed] [Google Scholar]
- 35.Altshuler DM, et al. International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anonymous International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Behar DM, et al. The genome-wide structure of the Jewish people. Nature. 2010;466(7303):238–242. doi: 10.1038/nature09103. [DOI] [PubMed] [Google Scholar]
- 38.Henn BM, et al. Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proc Natl Acad Sci USA. 2011;108(13):5154–5162. doi: 10.1073/pnas.1017511108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319(5866):1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 40.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2(12):e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu S, Pugach I, Stoneking M, Kayser M, Jin L. HUGO Pan-Asian SNP Consortium Genetic dating indicates that the Asian-Papuan admixture through Eastern Indonesia corresponds to the Austronesian expansion. Proc Natl Acad Sci USA. 2012;109(12):4574–4579. doi: 10.1073/pnas.1118892109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moorjani P, et al. The history of African gene flow into Southern Europeans, Levantines, and Jews. PLoS Genet. 2011;7(4):e1001373. doi: 10.1371/journal.pgen.1001373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loh PR, et al. Inferring Admixture Histories of Human Populations Using Linkage Disequilibrium. Ithaca, NY: Cornell Univ Library; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4(3):e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flicek P, et al. Ensembl 2012. Nucleic Acids Res. 2012;40(Database issue):D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pickrell JK, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19(5):826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang W, et al. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8(9):R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4(10):R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125(1-2):279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 51.Sabeti PC, et al. International HapMap Consortium Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449(7164):913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henn BM, et al. Y-chromosomal evidence of a pastoralist migration through Tanzania to southern Africa. Proc Natl Acad Sci USA. 2008;105(31):10693–10698. doi: 10.1073/pnas.0801184105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oota H, et al. Recent origin and cultural reversion of a hunter-gatherer group. PLoS Biol. 2005;3(3):e71. doi: 10.1371/journal.pbio.0030071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.