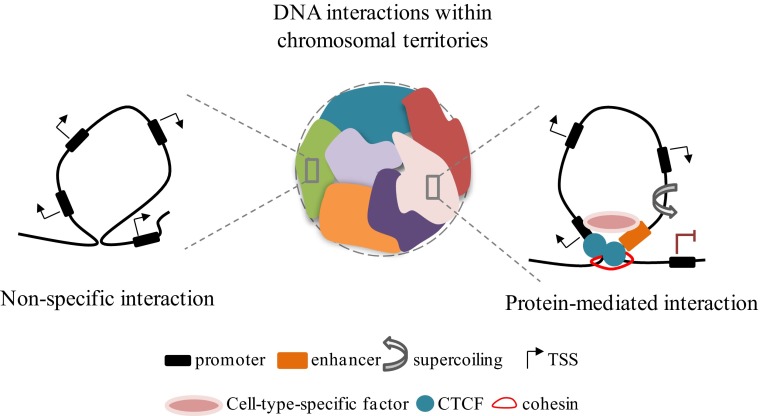
Genome function depends on its 3D organization. In the nucleus of eukaryotic cells, interphase chromosomes occupy distinct territories with respect to each other (1) (Fig. 1), and the spatial positioning of genomic segments affects ongoing DNA transactions. In recent years, the combination of advanced imaging techniques and new molecular approaches revealed the occurrence of previously underestimated long-range intrachromosomal and even interchromosomal interactions that have been linked to fundamental biological processes such as genomic imprinting (2), X-chromosome inactivation (3), and developmentally regulated programs (4). Therefore, considerable interest is growing around the study of 3D models of gene expression, focused on describing the fine-grain organization of chromatin architecture and the key players mediating and controlling long-range contacts. The development of Hi-C (high-throughput detection of chromosomal interactions) technology has markedly advanced the field and has revealed that chromosome territories are further arranged into large megabase-sized topological domains (5) that are highly conserved and stable across the cell type (6). In contrast, subregions within each domain are dynamic and may be responsible for cells' type-specific regulatory events (Fig. 1). As the boundaries between different domains dictate their spatial organization and function, understanding the contribution of factors that demarcate domain boundaries is of compelling importance. The work of Zuin et al. in PNAS explores the 3D architecture of the genome by studying the contributions of two leading players in loop organization: the CCCTC-binding factor (CTCF) and the cohesin complex (7). CTCF is a highly conserved protein with a unique structure that confers a versatile role in genome function (8). Through the 11 zinc fingers of its DNA-binding domain, CTCF binds with a broad range of sequences and coregulatory proteins and plays multiple roles in gene expression (9), including promoter repression, enhancer insulation, and, more recently, long-range interactions (6). Cohesin has been shown to co-occupy some CTCF sites and to mediate DNA looping interactions associated with gene regulation (10). Embracement of juxtaposed paired double helices, by cohesin’s ringlike shape, provides an attractive but overly simplistic mechanism to stabilize CTCF-mediated DNA loops. However, whether the two factors work independently or in concert in defining loop interactions is still conjecture. By using a combination of 4C (chromosome conformation capture coupled to sequencing), Hi-C, 3D-FISH, and ChIP-seq techniques, the authors examine changes in higher-order chromatin organization and in the transcriptome of human cells depleted of CTCF and the cohesion subunit RAD21 (depletion that destroys cohesin complex) and provide insight into the source, dynamics, and biology of long-range interactions.
Fig. 1.
CTCF and cohesin cooperate to organize the 3D structure of the mammalian genome. In the interphase nucleus, individual chromosomes occupy distinct spatial territories (Center). At the subcompartment level, chromatin is organized through megabase-sized interactions called topological domains. Chromatin looping can be the result of nonspecific interactions of the chromatin fiber packing in the crowded nucleus (Left) or of specific contacts between two loci, mediated by protein complexes and the subnuclear structure (Right). Within bounded domains, torsional stress accumulates and affects the expression of neighboring genes. In the central drawing colors represent chromosomal territories.
A general loss of short-range interactions but not of topological domains is observed on deletion of either factor. However, despite their extensive colocalization, cohesin destruction markedly affects interacting regions in the range of 100–200 kb, whereas CTCF reduction affects shorter loops (less than 100 kb), suggesting nonredundant roles during the looping process. Further, CTCF depletion leads to a gain of interactions between neighboring topological domains, which is even more pronounced when CTCF binding sites are involved. Therefore, in light of these observations, the authors propose a role for CTCF as a “genome-wide organizer” determining the specificity of interactions, and the role of cohesin as a factor facilitating intradomain loops (Fig. 1). Consistent with the idea that these two proteins contribute differently to shape chromatin architecture, RNA-seq analysis reveals only little overlap between genes differentially affected by CTCF reduction vs. by cohesin disruption. Indeed, cohesin controls gene expression through interactions between genes and their neighboring enhancers (11), whereas CTCF directly regulates the output of its genes by binding their promoters.
That the structure of topological domains remains largely intact after CTCF or cohesin depletion is intriguing and indicates that these proteins are not the sole determinants of long-range interactions. In fact, it has been recently shown that combinations of CTCF, cohesin, and mediator shape the genome at different length scales, with mediator being an additional key player in DNA looping (12). Interestingly, Phillips-Cremins et al. (12) observed that sites co-occupied by CTCF, cohesin, and mediator altogether are enriched for both constitutive and cell-specific interactions, irrespective of the cell differentiation state, a stability characteristic of topological domains. The effect of mediator depletion on topological domains and other chromatin interactions remains to be explored more fully. Of interest, CTCF occupancy depends not only on the combinatorial binding of its 11 zinc fingers that recognize specific DNA elements but also on extrinsic factors, including the DNA sequence, that either stabilize or destabilize CTCF binding (13). Evaluation of the influence of different sequence motifs on the stability of CTCF-mediated loops will help further the characterization of topological domains. Enrichment of repetitive sequences at the boundaries of topological domains elicits the exciting possibility that self-organization of DNA may also have a role in establishing the 3D structure of the genome (6, 14).
Although many efforts have been made to elucidate the principles and actors of genome folding, the chromatin and DNA dynamics of the looping that control genetic transactions are still poorly understood. From a topological point of view, when two DNA segments interact with each other, they generate a closed domain within which a process that unwinds or distorts the double helix, e.g., transcription via dynamic supercoiling, can distort the geometry of DNA and change its properties to favor formation of looped, wrapped, or melted structures that are high-energy intermediates in DNA transactions. Depending on the size and shape of the confining topological domain, the dynamic torsional stress introduced by tracking proteins such as RNA polymerase (15) can be transmitted along the helix (16), influencing the expression of neighboring loci (17). In this
The work of Zuin et al. in PNAS explores the 3D architecture of the genome by studying the contributions of two leading players in loop organization.
regard, it has been reported that DNA supercoiling is organized into large domains with specific topological features, those with a transcription permissive environment, underwound (negatively supercoiled), and less compliant overwound (positively supercoiled) regions that are more likely to be silent. Within the domains, dissipation of supercoiling is governed by topoisomerases and their breaking/resealing activity. Topoisomerases have significant affinity for the base of loop structures where two duplexes juxtapose and have been identified at tissue-specific regulatory elements such as enhancers (18). These considerations make the enzymes attractive candidates for looping regulation; therefore, further studies aiming to assess the topological state of chromatin domains and their relationship with topoisomerases would be of high relevance.
In summary, the work of Zuin et al. enriches our knowledge of the principles of genome organization and will stimulate new experiments. This unique view of chromatin loop formation indicates that CTCF is required to separate neighboring domains and to orchestrate cohesin positioning during intradomain interaction. The results here portend the illumination of a rich tapestry of interacting fibers. The work here is not yet fine grained; obtaining a high-resolution interaction map for mammalian genomes will require massive sequencing efforts. As the cost of sequencing decreases and the fine structure is revealed, the study of short-range interactions (like the ones established between promoters and enhancers or promoters and superenhancers that appear to be cell type-specific and associated with developmental or pathological processes such as cancer) will confirm and extend the known principles of gene action, and no doubt will deliver some surprises.
Footnotes
The authors declare no conflict of interest.
See companion article on page 996.
References
- 1.Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2(3):a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apostolou E, Thanos D. Virus infection induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell. 2008;134(1):85–96. doi: 10.1016/j.cell.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 3.Bacher CP, et al. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol. 2006;8(3):293–299. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- 4.Kim YJ, Cecchini KR, Kim TH. Conserved, developmentally regulated mechanism couples chromosomal looping and heterochromatin barrier activity at the homeobox gene A locus. Proc Natl Acad Sci USA. 2011;108(18):7391–7396. doi: 10.1073/pnas.1018279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuin J, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci USA. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17(9):520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 9.Phillips JE, Corces VG. CTCF: Master weaver of the genome. Cell. 2009;137(7):1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451(7180):796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 11.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467(7314):430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phillips-Cremins JE, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153(6):1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakahashi H, et al. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 2013;3(5):1678–1689. doi: 10.1016/j.celrep.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Son LS, Bacolla A, Wells RD. Sticky DNA: In vivo formation in E. coli and in vitro association of long GAA*TTC tracts to generate two independent supercoiled domains. J Mol Biol. 2006;360(2):267–284. doi: 10.1016/j.jmb.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 15.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA. 1987;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kouzine F, et al. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat Struct Mol Biol. 2013;20(3):396–403. doi: 10.1038/nsmb.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naughton C, et al. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat Struct Mol Biol. 2013;20(3):387–395. doi: 10.1038/nsmb.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baranello L, Kouzine F, Levens D. DNA Topoisomerases: Beyond the standard role. Transcription. 2013;4(5):1–6. doi: 10.4161/trns.26598. [DOI] [PubMed] [Google Scholar]