Abstract
Sequencing of multiple, nearly complete eukaryotic genomes creates opportunities for detecting previously unnoticed, subtle functional signals in non-coding regions. A genome-wide comparative analysis of orthologous sets of mammalian and yeast mRNAs revealed distinct patterns of evolutionary conservation at the boundaries of the untranslated regions (UTRs) and the coding region (CDS). Elevated sequence conservation was detected in ∼30 nt regions around the start codon. There seems to be a complementary relationship between sequence conservation in the ∼30 nt regions of the 5′-UTR immediately upstream of the start codon and that in the synonymous positions of the 5′-terminal 30 nt of the CDS: in mammalian mRNAs, the 5′-UTR shows a greater conservation than the CDS, whereas the opposite trend holds for yeast mRNAs. Unexpectedly, a ∼30 nt region downstream of the stop codon shows a substantially lower level of sequence conservation than the downstream portions of the 3′-UTRs. However, the sequence in this poorly conserved 30 nt portion of the 3′-UTR is non-random in that it has a higher GC content than the rest of the UTR. It is hypothesized that the elevated sequence conservation in the region immediately upstream of the start codon is related to the requirement for initiation factor binding during pre-initiation ribosomal scanning. In contrast, the poorly conserved region downstream of the stop codon could be involved in the post- termination scanning and dissociation of the ribosomes from the mRNA, which requires only the mRNA–ribosome interaction. Additionally, it was found that the choice of the stop codon in mammals, but not in yeasts, and the context in the immediate vicinity of the stop codons in both mammals and yeasts are subject to strong selection. Thus, genome-wide analysis of orthologous gene sets allows detection of previously unrecognized patterns of sequence conservation, which are likely to reflect hidden functional signals, such as ribosomal filters that could regulate translation by modulating the interaction between the mRNA and ribosomes.
INTRODUCTION
The most pronounced evolutionary conservation in genomic sequences reflects the constraints on protein structure and function. In general, coding portions of genes are much more conserved than non-coding regions. Patterns of amino acid residue conservation offer important functional clues because highly conserved amino acids are those which are crucial for protein function or folding (1,2). However, non-coding sequences, which comprise >98% of the vertebrate genomes (3,4), and, in particular, untranslated regions (UTRs) of mRNAs also contain numerous conserved stretches which, by analogy to the well established conservation-function correspondence in proteins, are also thought to comprise functional signals (5–8). Some of these signals have been studied experimentally but the role of most remains mysterious (9).
On average, 5′- and 3′-UTRs are less conserved across species than protein-coding sequences, but more conserved than untranscribed sequences (10,11). Nevertheless, highly conserved blocks have been detected in 5′-UTRs and, especially, 3′-UTRs of orthologous genes from different mammalian orders and even between mammals and birds or fish (5,6,12). In some genes, conservation of UTRs even exceeds the conservation of the corresponding coding regions (13). Generally, it is believed that conserved sequence elements in the UTRs are binding sites for proteins or antisense RNAs, which contribute to the regulation of nucleocytoplasmic transport, subcellular localization, translation and stability of mRNAs (14–16). The context around the principal functional signals, such as the start and stop codons, is thought to be an important determinant of the expression level (17).
Much of the critical action during translation takes place at the boundaries between the functional regions of the mRNA, namely: (i) 5′-UTRs and the coding sequence (CDS), (ii) the CDS and the 3′-UTR, and (iii) the 3′-UTR and the poly(A) tail. According to the scanning model of translation initiation, the eukaryotic ribosome binds to the 5′-terminal cap and starts scanning the mRNA, typically until detecting the first AUG where it initiates translation (18,19). However, the context around an AUG codon is critical for this process and determines, in large part, whether the ribosome initiates at the 5′-ultimate AUG or proceeds scanning to the next AUG (20). In mammals, the optimal context for start codon recognition is GCCRCCaugG... (21). Within this motif, the purine (R) in position –3 is the most highly conserved and functionally most important nucleotide. The initiation of translation in eukaryotes is affected by several regulatory mechanisms, which involve 5′-UTRs and include binding of regulatory proteins to specific sites within 5′-UTRs, upstream open-reading frames, and internal ribosome entry sites (22–25).
The mechanism of translation termination is understood much less thoroughly than the initiation mechanism (26). The release of the nascent polypeptide does not depend on a tRNA molecule but requires both codon-specific class-I release factors (eRFs in eukaryotes), which recognize the stop codon in mRNA, and codon-non-specific, GTP-binding class-II RFs (27,28). The context of the stop codon, in particular, the immediate downstream (+1) base, is important for termination; the modulations in the termination efficiency brought about by substitution of this nucleotide can reach an order of magnitude, which prompted the notion that the true termination signal could be a tetranucleotide (29–31). Additional complexity is introduced into the termination mechanism by the fact that there are three stop codons, for which the context effects seem to differ substantially. Thus, in yeast, UAG is most efficiently employed for termination when the next base is G, whereas, for UAA, A or U are optimal (29). The tetranucleotides that were most effective terminators have been found to be most common at the end of the coding regions in the respective genomes (29,30). However, beyond the +1 position, the 3′-UTRs have not been reported to have a distinct consensus analogous to the Kozak consensus in the 5′-UTR (31). Interestingly, the 5′-UTRs and the 3′-UTRs of eukaryotic mRNAs tend to interact through bridging protein complexes, which include certain initiation factors, polyA-binding proteins, and other subunits; this interaction adds another level of complexity to the regulation of translation initiation and termination and suggests the possibility of coordination between these processes (25,32,33).
The formation of mature eukaryotic mRNAs involves cleavage of the native transcripts and polyadenylation (34–36). The signal for transcript cleavage and polyadenylation is thought to consist of the AAUAAA hexamer, which is located 20–30 nt upstream of the cleavage-polyadenylation site in the majority, although not in all, eukaryotic mRNAs, an upstream U-rich sequence, and a downstream GU-rich sequence (34,37). However, up to 40% of human mRNAs appear to have alternative polyadenylation signals, without the AAUAAA motif (38).
We were interested in examining the evolutionary patterns of eukaryotic mRNAs at the boundaries between functional regions by taking advantage of the availability of multiple, closely related, (nearly) complete genomes of mammals and yeasts, for which numerous pairs of probable orthologs can be reliably identified. Comparisons of mammalian and yeast genomes have been recently employed to predict numerous, previously undetected, conserved motifs, which are likely to function as transcription regulation sites (4,39,40). The boundaries between the CDS and the UTRs or between the 3′-UTR and the poly(A) tail are particularly amenable to comparative-genomic analysis because the start and stop codons provide natural phasing, which allows one to compare unrelated pairs of orthologs and potentially identify even weak signals. We demonstrate significant sequence conservation in the ∼30 nt region immediately upstream of the start codon and in the synonymous positions in ∼30 nt in the beginning of the CDS for both mammals and yeasts. In contrast, the ∼30 nt region immediately downstream of the stop codon in the 3′-UTRs is significantly less conserved than the distal part.of the UTR. However, the poorly conserved 30 nt window is enriched for GC compared with the rest of the UTR and could be involved in scanning interruption and ribosome release after termination. Additionally, we analyze the frequencies of stop codons themselves and the surrounding context and demonstrate the role of selection in their evolution. Genome-wide analysis of orthologous genes seems to allow detection of so far unrecognized, subtle functional signals in mRNAs.
MATERIALS AND METHODS
A data set of 5800 human–mouse and 3600 rat–mouse mRNA orthologous pairs with annotated 3′-UTRs was compiled from GenBank as described in the supplementary data (ftp://ftp.ncbi.nih.gov/pub/kondrashov/utr). Briefly, symmetrical best hits between proteins from the respective genomes were identified using the BLAST program (41). The nucleotide sequence alignment for identified orthologous pairs of mRNAs were produced using the OWEN program (42). For the CDS, the alignment of the nucleotide sequences was guided by the amino acid sequence alignment (43). Among the identified orthologs, 5510 human–mouse pairs of orthologs and 3234 pairs of rat–mouse orthologs had complete 3′-UTRs as determined by the presence of the AAUAAA polyadenylation signal (3′-UTRs shorter than 110 bp were excluded); 2073 human–mouse pairs and 811 rat–mouse pairs had 5′-UTRs longer than 50 bp and these were used for the analysis. The positions of 5′-UTRs and 3′-UTRs were taken from the feature tables of the respective GenBank entries.
In addition, 50 sets of orthologous mRNA sequences from representatives of four mammalian orders, Primata, Rodentia, Carnivora and Cetartiodactyla, were constructed by extracting the most similar sequences from the BLAST search outputs; whenever available, genomic synteny was analyzed to ensure the correct identification of orthologs (43). Multiple alignments of nucleotide sequences were constructed using the CLUSTALW program with default parameters (44) and edited to take into account results of pairwise comparison, which was done using the OWEN program (42). The sequences and alignments from this data set are available at ftp://ftp.ncbi.nih.gov/pub/kondrashov/utr.
Aligned sequences of orthologous genes from four yeast species, Saccharomyces cerevisiae, S.paradoxus, S.mikatae and S.bayanus were extracted from the SGD database (ftp://genome-ftp.stanford.edu/pub/yeast/data_download/sequence/fungal_genomes) (39); 2066 sets of orthologs had complete 5′-UTRs and 1441 sets had complete 3′-UTRs in all four yeasts. The alignment of start and stop codons was fixed in order to ensure the correct partitioning of 5′-UTR, the CDS and the 3′-UTR.
The degree of conservation at each nucleotide position was calculated as the number of matches over the number of pairwise alignments. The start codon, the stop codon and the polyadenylation signal provided natural coordinate systems for this analysis such that the position number was always determined as the distance from one of these signals, even if the actual alignment of orthologous sequences included gaps. Shannon entropy was calculated using the definition S = –Σ(Pilog2Pi), where Pi is the nucleotide frequency. Two alternative approaches for treating gaps (indels) in pairwise sequence alignments were employed: (i) whenever a gap was introduced in one of the sequences, the respective position was treated as a non-conserved one (a mismatch), and (ii) positions containing one or more gaps were excluded from the analysis. For multiple alignment positions containing gaps, conservation was calculated for the number of sequence pairs containing aligned nucleotides, with sequences containing gaps excluded.
RESULTS AND DISCUSSION
To investigate the constraints that might affect the evolution of the sequences at the boundaries of the functional regions of eukaryotic mRNAs, we generated profiles of sequence conservation and base composition for the 5′-UTR–CDS, the CDS–3′-UTR and 3′-UTR–poly(A) site boundaries in mammalian and yeast orthologous genes.
5′-UTR–CDS boundary
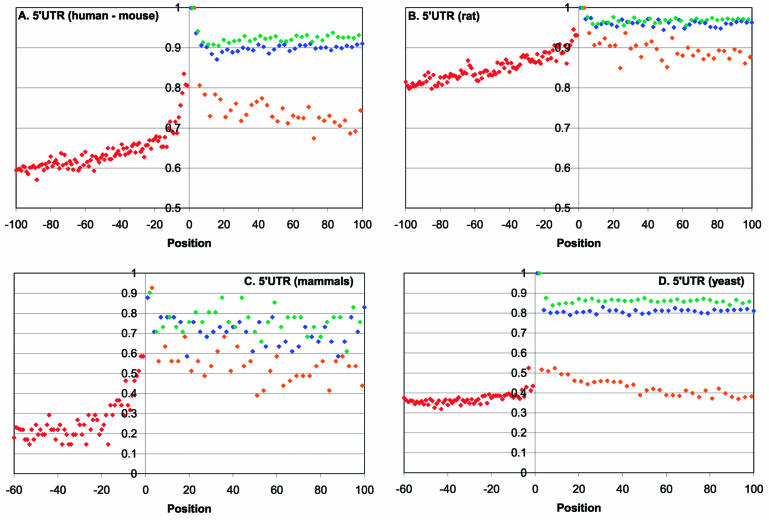
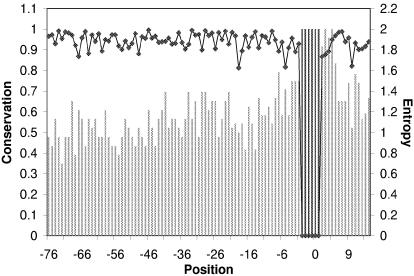
Profiles of sequence conservation at the boundaries between 5′-UTR and CDS regions in mammals and yeasts are shown on Figure 1. The portion of the 5′-UTR immediately upstream of the start codon shows substantially greater conservation than the rest of the UTR. In the human–mouse comparisons, the most pronounced conservation, ∼80% identity, is seen in the five positions of the 5′-UTRs immediately upstream of the start codon, with the peak in the critical –3 position of the Kozak consensus (Fig. 1A). In addition, non-random, statistically significant increase in conservation compared with the average level in the 5′-UTRs is seen for ∼30 nt upstream of the start codon (Fig. 1A and Table 1). Qualitatively similar results were obtained in the mouse-rate comparisons (Fig. 1B and Table 1). Further upstream into the 5′-UTR, the identity level in the human–mouse alignments drops to a plateau of ∼50% (Fig. 1A and data not shown), which is close to the neutral conservation level in human–mouse comparison (A. Y. Ogurtsov and A. S. Kondrashov, unpublished results). Examination of the conservation profile of multiple alignments of mammalian orthologs further sharpens the picture and indicates that the sequence of 5′-UTRs upstream of the position –30 is essentially random (Fig. 1C). A profile of sequence conservation for the four different yeast species shows the same peak at the 5′-UTR–CDS boundary (Fig. 1D). However, in this case, the excessive sequence conservation in the 30 nt stretch immediately upstream of the AUG, compared with the upstream portion of the UTR, albeit statistically significant, was much less pronounced than it was in mammals (Fig. 1D and Table 1).
Figure 1.
Profiles of sequence conservation around the start codons in orthologous eukaryotic mRNAs. (A) Human–mouse. (B) Rat–mouse. (C) Multiple alignments of orthologous mRNAs from four orders of mammals. (D) Multiple alignments of orthologous mRNAs from four species of yeasts. Positions from –100 to –1 correspond to 5′-UTRs and positions from 1 to 100 correspond to CDSs. Blues, first codon positions; green, second codon positions; orange, 4-fold degenerate third codon positions.
Table 1. Elevated sequence conservation in the UTR and CDS regions adjacent to the start and stop codons of eukaryotic mRNAsa.
| 5′-UTRs | 5′CDSs | 3′CDSs | 3′-UTRs | |||||
|---|---|---|---|---|---|---|---|---|
| t | P | t | P | t | P | t | P | |
| Human–mouse (pairwise alignments) | 5.923 | 5.41E–07 | 5.579 | 2.4E–06 | 2.518 | 0.0085 | –5.425 | 1.88E–06 |
| Rat–mouse (pairwise alignments) | 7.999 | 4.94E–11 | 3.19 | 0.0016 | 2.179 | 0.0184 | –4.032 | 0.0001 |
| Mammals (multiple alignments) | 5.606 | 1.4E–06 | 2.329 | 0.0133 | 0.704 | 0.2431 | –7.6 | 1.55E–10 |
| Yeast (multiple alignments) | 4.8 | 1.21E–05 | 6.975 | 3.97E–08 | 4.047 | 0.0002 | –10.327 | 9.4E–14 |
aThe table shows the results of t-test analysis for the 30 nt low conservation region in 3′-UTRs following the stop codon across species, the 30 nt high conservation region in 5′-UTRs immediately upstream of the start codon, and the 30 nt regions of the CDS adjacent to the start or stop codons. In each case, the conservation in the respective 30 nt region was compared with the conservation in the corresponding 70 nt downstream or upstream region. The P-value indicates the probability that the difference in sequence conservation is due to chance.
Predictably, the distal portions of 5′-UTRs show substantially lower conservation than non-synonymous codon positions in the CDS (Fig. 1A–D). In contrast, the conservation level in the 30 nt window upstream of the AUG is comparable with that in the synonymous (4-fold degenerate) third positions of codons in the CDS (Fig. 1A–D). Although synonymous positions are often used as a surrogate for neutral, clock-like evolution, several studies have shown that these positions evolve under weak purifying selection, presumably due, in large part, to the link between codon usage and expression level (45–48). The conservation profiles of the 5′-UTR and the CDS around the AUG showed a degree of symmetry, with the peak at the start codon (Fig. 1A–D). Apparently, the synonymous positions of several codons directly following the AUG are subject to a stronger negative selection than those in the rest of the CDS. This effect is stronger and statistically more significant in yeasts than it is in mammals (compare Fig. 1D and A–C; Table 1). Thus, there seems to be a distinct functional signal in the 5′ portion of the CDS and this signal appears to be, in a sense, complementary to the signal in the 5′-UTR: the relatively strong 5′-UTR signal in mammals is coupled with a weak signal in the CDS and, conversely, the weak UTR signal in yeasts is counterweighted by the stronger signal in the CDS.
The apparent increased conservation in the 5′-UTR immediately upstream of the AUG potentially could be explained by incorrect identification of the start codon in some of the analyzed mRNAs, which would result in an admixture of in-frame coding sequence in the 5′-UTR pool. If the frequency of such errors decreased with the length of the unannotated coding sequence, this artifact might lead to the pattern of UTR sequence conservation seen in Figure 1. However, two considerations suggest that this is not the case: (i) this type of misannotation cannot account for the conservation pattern observed in the synonymous positions in the upstream portion of the CDS, and (ii) when the analysis was repeated for only those human and mouse 5′-UTRs that did not have in-frame AUGs within 30 nt upstream of the annotated start, a pattern essentially identical to that in Figure 1 was observed (data not shown).
Another potential source of artifacts could be gaps in the alignments, which could lead to an apparent drop in the conservation level with the increase of the distance from the anchoring point (AUG). We investigated two approaches for treating gaps: (i) a gap was treated as a non-conserved position, and (ii) positions with gaps were excluded from the analysis (see Materials and Methods for details). The resulting sequence conservation profiles were not appreciably different (data not shown). Furthermore, we re-examined sequence conservation in the subset of human–mouse alignments, which did no contain gaps within 100 nt of the CDS immediately downstream of the AUG. The resulting pattern of gradually decreasing conservation level did not differ from that seen in Figure 1 (data not shown).
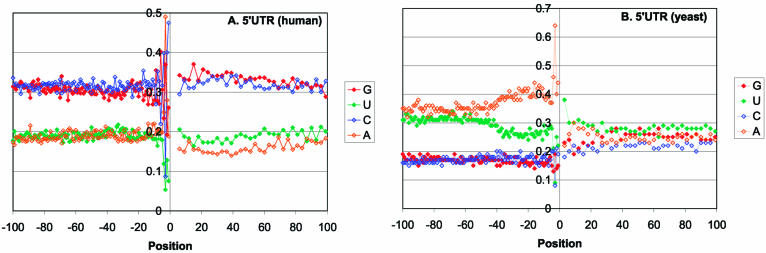
Additionally, we analyzed the nucleotide content of the 5′-UTRs. The mammalian 5′-UTRs are GC-rich, without significant strand asymmetry (Fig. 2A). In contrast, the yeast 5′-UTRs are AT-rich and show marked strand asymmetry in a ∼40 nt region upstream of the AUG, with significant enrichment for A in the sense strand (Fig. 2B; t-test: t = 10.35, P < 0.00001). Purine-pyrimidine strand asymmetry reduces the potential of the respective sequence for secondary structure formation, which could be an adaptation for ribosomal scanning in yeast mRNAs. The absence of strand asymmetry in mammalian mRNAs is likely to reflect differences between the initiation mechanisms in mammals and yeasts. Recently, significant strand asymmetry, with an excess of G+T over A+C has been reported to exist in most mammalian genes and interpreted as a result of mutational pressure caused by transcription-coupled repair (49). We did not detect this type of asymmetry in the UTRs of the analyzed mammalian genes.
Figure 2.
Profiles of nucleotide content in 5′-UTRs of eukaryotic mRNAs. (A) Human. (B) Yeast.
Stop codons and the CDS–3′-UTR boundary
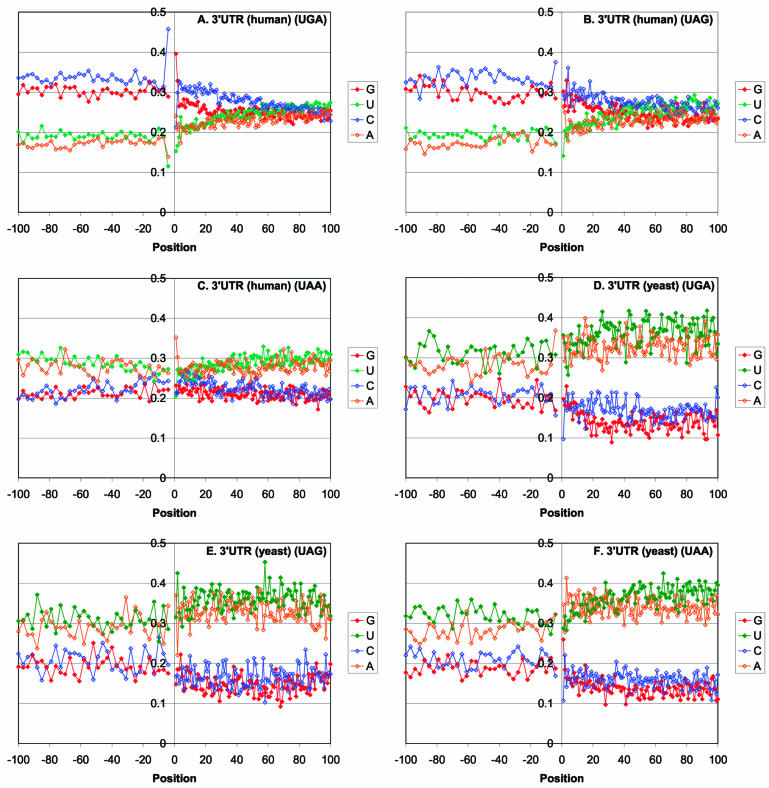
The distribution of stop codons is substantially different in mammals and in fungi. The predominant stop codon in yeasts is UAA whereas, in mammals, a large excess of UGA is observed (50) (Table 2). The dominance of a particular stop codon could be explained either by mutational or by selective pressure. A comparison of the stop codon frequencies to the frequencies of the respective triplets in sequences that appear to evolve neutrally, namely, introns of the analyzed mammalian genes and intergenic regions adjacent to the analyzed yeast genes, shows a substantially less uniform distribution among the stop codons (Table 2). The difference between the frequency of the most common stop codon and that of the respective triplet in the control non-coding sequences was statistically highly significant in both mammals and yeasts (Table 2). These observations indicate that the observed distribution of stop codons is, largely, a result of selection. The context in the immediate surroundings of the stops codons is obviously non-random and depends on the stop codon as noticed in previous studies, which analyzed smaller sets of mRNAs (51) (Fig. 3). Thus, in human genes, there is a strong preference for C in position –4 and for A in position +1 for UGA, limited preference for C in positions –4 and +3 for UAG, and notable preference for A in positions +1 and +2, but no discernible consensus in the CDS for UAA (Fig. 3A–C). In yeast genes, the context is considerably less pronounced, although the preponderance of G in position +1 after UAA codons was notable and statistically significant (Fig. 3E–F). A comparison of the frequencies of dominant tetranucleotides including the stop codon to the frequencies of the same tetranucleotides in non-coding sequences reveals major differences and indicates that, like the stop codons themselves, the adjacent positions are subject to selective pressures (Table 2).
Table 2. Frequencies of stop codons and nucleotides in the positions adjacent to stop codons in orthologous genes.
| Human | Yeast | |||||
|---|---|---|---|---|---|---|
| Stop codons | Introns | Significance of difference | Stop codons | Intergenic regions | Significance of difference | |
| UGA | 0.50 | 0.37 | 1.0E–72 | 0.29 | 0.28 | 0.38 |
| cUGA | 0.37 | 0.23 | 1.5E–141 | ND | ND | ND |
| gUGA | 0.29 | 0.22 | 1.5E–141 | ND | ND | ND |
| UGAg | 0.40 | 0.29 | 1.19E–43 | ND | ND | ND |
| UAG | 0.23 | 0.28 | 1.0E–72 | 0.24 | 0.24 | 0.84 |
| cUAG | 0.37 | 0.23 | 4.13E–47 | ND | ND | ND |
| gUAG | 0.28 | 0.22 | 4.13E–47 | ND | ND | ND |
| UAA | 0.27 | 0.35 | 1.0E–72 | 0.47 | 0.49 | 0.33 |
| UAAg | ND | ND | ND | 0.26 | 0.15 | 3.6E–9 |
Figure 3.
Profile of nucleotide content in 3′-UTRs of eukaryotic mRNAs. (A) Human mRNAs with UGA stop codon conserved in murine orthologs. (B) Human mRNAs with UAG stop codon conserved in murine orthologs. (C) Human mRNAs with UAA stop codon conserved in murine orthologs. (D) Yeast mRNAs with UGA stop codon conserved in four species. (E) Yeast mRNAs with UAG stop codon conserved in four species. (F) Yeast mRNAs with UAA stop codon conserved in four species.
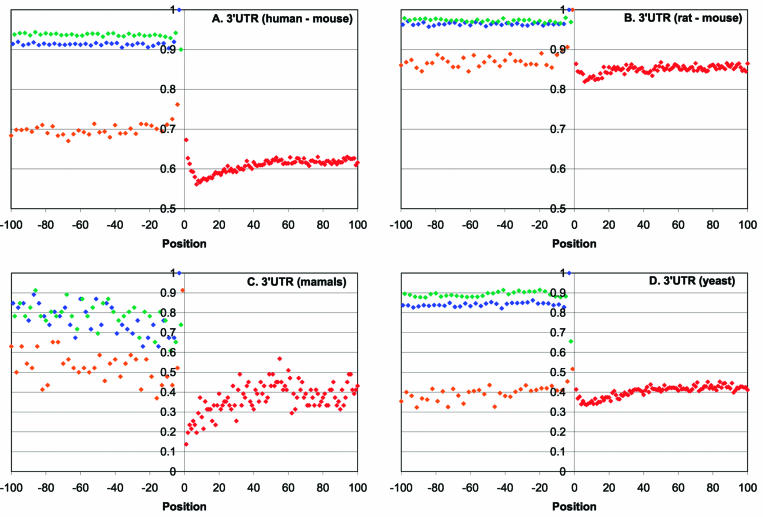
The profiles of sequence conservation at the boundaries between CDS and 3′-UTR regions showed increased conservation in the immediate vicinity of the stop codon (positions –6 to +3), which is followed by a window of low conservation in positions +5 to +35, and by a downstream region of relatively high conservation (Fig. 4). The effect was more pronounced in mammalian genes (Fig. 4A–C) than in yeast genes (Fig. 4D) but, in all cases, the difference between the ∼30 nt poorly conserved window and the downstream portion of the UTR was statistically highly significant (Table 1). To rule out the possibility that the observed increase in sequence conservation beyond 30 nt from the stop codon was due to the effect of adjacent polyadenylation signals, all 3′-UTRs <110 nt were excluded from the analyzed set. The entire 100 nt region of the 3′-UTRs downstream of the stop codon is less conserved than the synonymous codon positions in the adjacent 3′-terminal portion of the CDS (Fig. 4). Nevertheless, the conservation level in this region was slightly above the random base line, with the low-conservation window nearly reaching the 50% neutral level (A.Y. Ogurtsov and A.S. Kondrashov, unpublished observations).
Figure 4.
Profiles of sequence conservation around the stop codons in orthologous eukaryotic mRNAs. (A) Human–mouse. (B) Rat–mouse. (C) Multiple alignments of orthologous mRNAs from four orders of mammals. (D) Multiple alignments of orthologous mRNAs from four species of yeasts. Positions from –100 to –1 correspond to 5′-UTRs and positions from 1 to 100 correspond to CDSs. Empty diamonds denote the 4-fold degenerate third codon positions in CDSs.
Despite the low level of sequence conservation in the 30 nt window downstream of the stop codon, there is a statistically significant GC preference in this region of mammalian mRNAs, particularly after UGA and UAG codons (Fig. 3A–C and Table 3); in yeasts, the increase in GC content is much less pronounced and not statistically significant (Fig. 3D–F). Furthermore, in mammals, there is a statistically significant positive correlation between the composition of the poorly conserved 30 nt window in the 3′-UTR and the synonymous positions in the CDS. Specifically, the C content of the synonymous sites correlated stronger with the C content of the poorly conserved 30 nt window than with that of the entire 3′-UTRs (r = 0.47 and r = 0.40, respectively; P < 0.00001). It has been reported that expression level of vertebrate as well as plant genes positively correlated with the GC-content of the synonymous positions of the CDS (52–55). The correlation between the GC-content of the CDS and the poorly conserved 30 nt window downstream of the stop codon detected here suggests that both factors could concordantly affect the level of gene expression. None of these effects are seen in yeasts, where the UTRs are AT-rich, in accord with the genomic nucleotide composition. However, there is some increase in conservation in the synonymous positions at the 3′ end of the CDS in yeast (t = 5.307, P < 0.0005; positions –30 to –1 compared with the preceding 70 nt), an effect that is not seen in mammals.
Table 3. Statistical significance of the differences in GC content between the 30 nt low conservation window and the rest of 3′-UTRsa.
| A | ||
|---|---|---|
| t | P | |
| Human UGA | 11.823 | 7.63E–16 |
| Human UAG | 9.471 | 2.19E–12 |
| Human UAA | 7.128 | 1.71E–09 |
| Yeast UGA | 4.278 | 5.01E–05 |
| Yeast UAG | 0.674 | 0.251452 |
| Yeast UAA | 4.299 | 4.53E–05 |
aThe comparison was between positions 1–30 and 31–100 of the UTRs.
Eukaryotic ribosomes terminate translation with the stop codon positioned in the ribosomal A site (28). However, the post-termination fate of the ribosomes and the mechanism of ribosome release are not thoroughly understood. Detailed analysis of the translation termination of short open-reading frames occurring in the 5′-UTRs of certain yeast genes, such as GCN4 and YAP1/2, provides some insights into these mechanisms (56–58). It has been shown that, after termination, ribosomes tend to resume scanning but their propensity to do so is context dependent, with AT-rich sequences downstream of the stop codon favoring scanning and GC-rich sequences promoting release. To the extent that these observations can be extrapolated to eukaryotic mRNAs in general, it seems likely that the function of the poorly conserved 30 nt region downstream of the stop codon is to interrupt post-termination ribosomal scanning. According to this hypothesis, this region is poorly conserved because it is covered by the ribosome during scanning and does not contain binding sites for regulatory proteins or RNAs. Thus, the sequence of the 30 nt region appears to be unimportant but the GC enrichment might facilitate dissociation of the ribosomes from the mRNA. In contrast, the higher level of conservation in the downstream part of the 3′-UTR suggests that, in many mRNAs, this portion contains sequence-specific functional signals.
3′-UTR–polyadenylation signal
The boundary between 3′-UTRs and poly(A) contains upstream pyrimidine-rich elements, the AAUAAA motif, and the downstream GU-rich elements; this region aligned well in the majority of the human–mouse orthologous pairs. The sequences of mammalian mRNAs around AAUAAA show elevated sequence conservation, particularly in the 10–20 nt stretch downstream of the signal (Fig. 5). This conservation differed from that in the adjacent regions at a statistically significant level (χ2 = 51.52, P < 0.001). In yeasts, the polyadenylation signal is not nearly as pronounced as it is in mammals and increased conservation around this signal was barely detectable (data not shown).
Figure 5.
Sequence conservation profile around the polyadenylation signal in orthologous eukaryotic mRNAs.
CONCLUSIONS
The advantage of genome comparisons as an approach to discovering previously undetected functional signals in sequences is 2-fold: (i) evolutionary conservation is a strong predictor of functional importance, and (ii) thanks to the large number of data points, even subtle signals can be discovered with an adequate methodology. In the analysis presented here, we attempted to integrate these two advantages and uncover such signals by compiling alignments of numerous orthologous mRNAs and assessing the average level of conservation for individual positions. This approach works best for regions at the boundaries of the functional domains of mRNAs because the principal signals, i.e. the start and stop codons and the polyadenylation motif, establish the frame for the comparison and the effect of potential misalignment is minimal for these regions.
The results of the genome-wide comparison of orthologous mRNAs in mammals and fungi support the evolutionary conservation of the context in the immediate vicinity of the principal signals, namely, the start and stop codons, and the polyadenylation site. These well recognized context elements include the Kozak consensus around the start codon, the +1 base following the stop codon, which is critical for the efficiency of termination, and the GU elements downstream of the polyadenylation AAUAAA motif. In addition, however, the present analysis revealed previously unnoticed, weaker signals that spread over longer regions of mRNAs. In the case of the start codon and the polyadenylation site, sequence conservation centers at the principal signal and drops off ∼30 nt away from it. Therefore, it seems unexpected that the stop codon and the three conserved positions immediately downstream of it are followed by a region of markedly low conservation. This observation is all the more paradoxical because both the region upstream of the start codon and the one downstream of the stop codon are thought to be the parts of mRNA that are scanned by ribosomes. However, the former sequence is highly conserved compared with the upstream portion of the 5′-UTR, whereas the latter sequence is a poorly conserved part of the 3′-UTR. The cause of the difference could be that ribosomal scanning that precedes initiation is highly dependent on binding of initiation factors to the proximal portion of the 5′-UTR (59), whereas post-termination scanning involves only the interaction between the proximal part of the 3′-UTR and the ribosome itself. Conceptually, the highly conserved region of the 5′-UTR preceding the start codon and the poorly conserved region of the 3′-UTR downstream of the stop codon could be considered ribosomal filters (60), i.e. sequence elements that modulate the interaction between the mRNA and the ribosome, and thereby contribute to the regulation of translation. These predictions made through genome-wide analysis of sequence conservation of eukaryotic mRNAs are amenable to direct experimental testing.
Acknowledgments
ACKNOWLEDGEMENT
We thank Alex Kondrashov for helpful discussions.
REFERENCES
- 1.Li W.H. (1997) Molecular Evolution. Sinauer, Sunderland, MA. [Google Scholar]
- 2.Koonin E.V. and Galperin,M.Y. (2002) Sequence—Evolution—Function. Computational Approaches in Comparative Genomics. Kluwer Academic Publishers, New York, NY. [PubMed] [Google Scholar]
- 3.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 4.Waterston R.H., Lindblad-Toh,K., Birney,E., Rogers,J., Abril,J.F., Agarwal,P., Agarwala,R., Ainscough,R., Alexandersson,M., An,P. et al. (2002) Initial sequencing and comparative analysis of the mouse genome. Nature, 420, 520–562. [DOI] [PubMed] [Google Scholar]
- 5.Duret L., Dorkeld,F. and Gautier,C. (1993) Strong conservation of non-coding sequences during vertebrates evolution: potential involvement in post-transcriptional regulation of gene expression. Nucleic Acids Res., 21, 2315–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lipman D.J. (1997) Making (anti)sense of non-coding sequence conservation. Nucleic Acids Res., 25, 3580–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shabalina S.A. and Kondrashov,A.S. (1999) Pattern of selective constraint in C. elegans and C. briggsae genomes. Genet. Res., 74, 23–30. [DOI] [PubMed] [Google Scholar]
- 8.Dermitzakis E.T., Reymond,A., Scamuffa,N., Ucla,C., Kirkness,E., Rossier,C. and Antonarakis,S.E. (2003) Evolutionary discrimination of mammalian conserved non-genic sequences (CNGs). Science, 302, 1033–1035. [DOI] [PubMed] [Google Scholar]
- 9.Dieterich C., Wang,H., Rateitschak,K., Luz,H. and Vingron,M. (2003) CORG: a database for COmparative Regulatory Genomics. Nucleic Acids Res., 31, 55–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pesole G., Mignone,F., Gissi,C., Grillo,G., Licciulli,F. and Liuni,S. (2001) Structural and functional features of eukaryotic mRNA untranslated regions. Gene, 276, 73–81. [DOI] [PubMed] [Google Scholar]
- 11.Larizza A., Makalowski,W., Pesole,G. and Saccone,C. (2002) Evolutionary dynamics of mammalian mRNA untranslated regions by comparative analysis of orthologous human, artiodactyl and rodent gene pairs. Comput. Chem., 26, 479–490. [DOI] [PubMed] [Google Scholar]
- 12.Duret L. and Bucher,P. (1997) Searching for regulatory elements in human noncoding sequences. Curr. Opin. Struct. Biol., 7, 399–406. [DOI] [PubMed] [Google Scholar]
- 13.Spicher A., Guicherit,O.M., Duret,L., Aslanian,A., Sanjines,E.M., Denko,N.C., Giaccia,A.J. and Blau,H.M. (1998) Highly conserved RNA sequences that are sensors of environmental stress. Mol. Cell. Biol., 18, 7371–7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mignone F., Gissi,C., Liuni,S. and Pesole,G. (2002) Untranslated regions of mRNAs. Genome Biol., 3, REVIEWS0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grzybowska E.A., Wilczynska,A. and Siedlecki,J.A. (2001) Regulatory functions of 3′UTRs. Biochem. Biophys. Res. Commun., 288, 291–295. [DOI] [PubMed] [Google Scholar]
- 16.Bashirullah A., Cooperstock,R.L. and Lipshitz,H.D. (1998) RNA localization in development. Annu. Rev. Biochem., 67, 335–394. [DOI] [PubMed] [Google Scholar]
- 17.Kochetov A.V., Sarai,A., Vorob’ev,D.G. and Kolchanov,N.A. (2002) [The context organization of functional regions in yeast genes with high-level expression]. Mol. Biol. (Mosk.), 36, 1026–1034. [PubMed] [Google Scholar]
- 18.Kozak M. (1989) The scanning model for translation: an update. J. Cell Biol., 108, 229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pestova T.V., Kolupaeva,V.G., Lomakin,I.B., Pilipenko,E.V., Shatsky,I.N., Agol,V.I. and Hellen,C.U. (2001) Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl Acad. Sci. USA, 98, 7029–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozak M. (1999) Initiation of translation in prokaryotes and eukaryotes. Gene, 234, 187–208. [DOI] [PubMed] [Google Scholar]
- 21.Kozak M. (2002) Pushing the limits of the scanning mechanism for initiation of translation. Gene, 299, 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klausner R.D., Rouault,T.A. and Harford,J.B. (1993) Regulating the fate of mRNA: the control of cellular iron metabolism. Cell, 72, 19–28. [DOI] [PubMed] [Google Scholar]
- 23.Morris D.R. and Geballe,A.P. (2000) Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol., 20, 8635–8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hellen C.U. and Sarnow,P. (2001) Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev., 15, 1593–1612. [DOI] [PubMed] [Google Scholar]
- 25.Mazumder B., Seshadri,V. and Fox,P.L. (2003) Translational control by the 3′-UTR: the ends specify the means. Trends Biochem. Sci., 28, 91–98. [DOI] [PubMed] [Google Scholar]
- 26.Bertram G., Innes,S., Minella,O., Richardson,J. and Stansfield,I. (2001) Endless possibilities: translation termination and stop codon recognition. Microbiology, 147, 255–269. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura Y. and Ito,K. (2003) Making sense of mimic in translation termination. Trends Biochem. Sci., 28, 99–105. [DOI] [PubMed] [Google Scholar]
- 28.Kisselev L., Ehrenberg,M. and Frolova,L. (2003) Termination of translation: interplay of mRNA, rRNAs and release factors? EMBO J., 22, 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonetti B., Fu,L., Moon,J. and Bedwell,D.M. (1995) The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J. Mol. Biol., 251, 334–345. [DOI] [PubMed] [Google Scholar]
- 30.McCaughan K.K., Brown,C.M., Dalphin,M.E., Berry,M.J. and Tate,W.P. (1995) Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc. Natl Acad. Sci. USA, 92, 5431–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozawa Y., Hanaoka,S., Saito,R., Washio,T., Nakano,S., Shinagawa,A., Itoh,M., Shibata,K., Carninci,P., Konno,H. et al. (2002) Comprehensive sequence analysis of translation termination sites in various eukaryotes. Gene, 300, 79–87. [DOI] [PubMed] [Google Scholar]
- 32.Tarun S.Z. Jr and Sachs,A.B. (1996) Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J., 15, 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 33.Wells S.E., Hillner,P.E., Vale,R.D. and Sachs,A.B. (1998) Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell, 2, 135–140. [DOI] [PubMed] [Google Scholar]
- 34.Zhao J., Hyman,L. and Moore,C. (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev., 63, 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shatkin A.J. and Manley,J.L. (2000) The ends of the affair: capping and polyadenylation. Nature Struct. Biol., 7, 838–842. [DOI] [PubMed] [Google Scholar]
- 36.Proudfoot N. and O’Sullivan,J. (2002) Polyadenylation: a tail of two complexes. Curr. Biol., 12, R855–R857. [DOI] [PubMed] [Google Scholar]
- 37.Proudfoot N.J. and Brownlee,G.G. (1976) 3′ non-coding region sequences in eukaryotic messenger RNA. Nature, 263, 211–214. [DOI] [PubMed] [Google Scholar]
- 38.MacDonald C.C. and Redondo,J.L. (2002) Reexamining the polyadenylation signal: were we wrong about AAUAAA? Mol. Cell. Endocrinol., 190, 1–8. [DOI] [PubMed] [Google Scholar]
- 39.Kellis M., Patterson,N., Endrizzi,M., Birren,B. and Lander,E.S. (2003) Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature, 423, 241–254. [DOI] [PubMed] [Google Scholar]
- 40.Cliften P., Sudarsanam,P., Desikan,A., Fulton,L., Fulton,B., Majors,J., Waterston,R., Cohen,B.A. and Johnston,M. (2003) Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science, 301, 71–76. [DOI] [PubMed] [Google Scholar]
- 41.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogurtsov A.Y., Roytberg,M.A., Shabalina,S.A. and Kondrashov,A.S. (2002) OWEN: aligning long collinear regions of genomes. Bioinformatics, 18, 1703–1704. [DOI] [PubMed] [Google Scholar]
- 43.Shabalina S.A., Ogurtsov,A.Y., Lipman,D.J. and Kondrashov,A.S. (2003) Patterns in interspecies similarity correlate with nucleotide composition in mammalian 3′UTRs. Nucleic Acids Res., 31, 5433–5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kimura M. (1981) Possibility of extensive neutral evolution under stabilizing selection with special reference to nonrandom usage of synonymous codons. Proc. Natl Acad. Sci. USA, 78, 5773–5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Makalowski W. and Boguski,M.S. (1998) Evolutionary parameters of the transcribed mammalian genome: an analysis of 2,820 orthologous rodent and human sequences. Proc. Natl Acad. Sci. USA, 95, 9407–9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Makalowski W. and Boguski,M.S. (1998) Synonymous and nonsynonymous substitution distances are correlated in mouse and rat genes. J. Mol. Evol., 47, 119–121. [DOI] [PubMed] [Google Scholar]
- 48.Kondrashov F.A., Rogozin,I.B., Wolf,Y.I. and Koonin,E.V. (2002) Selection in the evolution of gene duplications. Genome Biol., 3, RESEARCH0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Green P., Ewing,B., Miller,W., Thomas,P.J. and Green,E.D. (2003) Transcription-associated mutational asymmetry in mammalian evolution. Nature Genet., 33, 514–517. [DOI] [PubMed] [Google Scholar]
- 50.Jacobs G.H., Rackham,O., Stockwell,P.A., Tate,W. and Brown,C.M. (2002) Transterm: a database of mRNAs and translational control elements. Nucleic Acids Res., 30, 310–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kochetov A.V., Ischenko,I.V., Vorobiev,D.G., Kel,A.E., Babenko,V.N., Kisselev,L.L. and Kolchanov,N.A. (1998) Eukaryotic mRNAs encoding abundant and scarce proteins are statistically dissimilar in many structural features. FEBS Lett., 440, 351–355. [DOI] [PubMed] [Google Scholar]
- 52.Carels N. and Bernardi,G. (2000) Two classes of genes in plants. Genetics, 154, 1819–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Romero H., Zavala,A., Musto,H. and Bernardi,G. (2003) The influence of translational selection on codon usage in fishes from the family Cyprinidae. Gene, 317, 141–147. [DOI] [PubMed] [Google Scholar]
- 54.Konu O. and Li,M.D. (2002) Correlations between mRNA expression levels and GC contents of coding and untranslated regions of genes in rodents. J. Mol. Evol., 54, 35–41. [DOI] [PubMed] [Google Scholar]
- 55.Versteeg R., van Schaik,B.D., van Batenburg,M.F., Roos,M., Monajemi,R., Caron,H., Bussemaker,H.J. and van Kampen,A.H. (2003) The human transcriptome map reveals extremes in gene density, intron length, GC content and repeat pattern for domains of highly and weakly expressed genes. Genome Res., 13, 1998–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grant C.M. and Hinnebusch,A.G. (1994) Effect of sequence context at stop codons on efficiency of reinitiation in GCN4 translational control. Mol. Cell. Biol., 14, 606–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vilela C., Linz,B., Rodrigues-Pousada,C. and McCarthy,J.E. (1998) The yeast transcription factor genes YAP1 and YAP2 are subject to differential control at the levels of both translation and mRNA stability. Nucleic Acids Res., 26, 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hinnebusch A.G. (1997) Translational regulation of yeast GCN4. A window on factors that control initiator-trna binding to the ribosome. J. Biol. Chem., 272, 21661–21664. [DOI] [PubMed] [Google Scholar]
- 59.Pestova T.V. and Hellen,C.U. (2000) The structure and function of initiation factors in eukaryotic protein synthesis. Cell. Mol. Life Sci., 57, 651–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mauro V.P. and Edelman,G.M. (2002) The ribosome filter hypothesis. Proc. Natl Acad. Sci. USA, 99, 12031–12036. [DOI] [PMC free article] [PubMed] [Google Scholar]