Significance
In centrosomes, pericentriolar material (PCM) serves as the principle site for microtubule nucleation and anchoring. In Drosophila, the centrosomal protein spindle assembly defective-4 (Sas-4) scaffolds cytoplasmic PCM protein complexes via its N terminus and tethers them to centrioles via an unknown mechanism. By determining the crystal structure of Sas-4‘s C-terminal T complex protein 10 (TCP) domain and functional studies in Drosophila, human cells, and induced pluripotent stem cell-derived neural progenitors, we show that Sas-4 performs its tethering role via its TCP domain. Furthermore, point mutations within the TCP domain perturb PCM tethering while still allowing the protein to scaffold cytoplasmic PCM complexes. These studies provide insights into how Sas-4 proteins tether PCM complexes for the assembly of functional centrosomes.
Abstract
Pericentriolar material (PCM) recruitment to centrioles forms a key step in centrosome biogenesis. Deregulation of this process leads to centrosome aberrations causing disorders, one of which is autosomal recessive primary microcephaly (MCPH), a neurodevelopmental disorder where brain size is reduced. During PCM recruitment, the conserved centrosomal protein Sas-4/CPAP/MCPH6, known to play a role in centriole formation, acts as a scaffold for cytoplasmic PCM complexes to bind and then tethers them to centrioles to form functional centrosomes. To understand Sas-4’s tethering role, we determined the crystal structure of its T complex protein 10 (TCP) domain displaying a solvent-exposed single-layer of β-sheets fold. This unique feature of the TCP domain suggests that it could provide an “extended surface-like” platform to tether the Sas-4–PCM scaffold to a centriole. Functional studies in Drosophila, human cells, and human induced pluripotent stem cell-derived neural progenitor cells were used to test this hypothesis, where point mutations within the 9–10th β-strands (β9–10 mutants including a MCPH-associated mutation) perturbed PCM tethering while allowing Sas-4/CPAP to scaffold cytoplasmic PCM complexes. Specifically, the Sas-4 β9–10 mutants displayed perturbed interactions with Ana2, a centrosome duplication factor, and Bld-10, a centriole microtubule-binding protein, suggesting a role for the β9–10 surface in mediating protein–protein interactions for efficient Sas-4–PCM scaffold centriole tethering. Hence, we provide possible insights into how centrosomal protein defects result in human MCPH and how Sas-4 proteins act as a vehicle to tether PCM complexes to centrioles independent of its well-known role in centriole duplication.
Centrosomes consist of a pair of centrioles surrounded by a protein network of pericentriolar material (PCM), the main sites for microtubule nucleation and anchoring and thus responsible for PCM’s role as the principle microtubule-organizing centers (MTOCs) of cells (1–4). When PCM is not recruited, centrioles are unstable, and thus no functional centrosomes are generated (5, 6). Although initial proteomic studies suggested PCM to be an amorphous cloud composed of more than a 100 different proteins (7), recent superresolution microscopy of fly and human centrosomes have indicated key centrosomal proteins essential for centrosome biogenesis to be organized into distinct spatial compartments before appearing as a PCM cloud surrounding the centriole (6, 8–11).
Thus, there could be a protein providing an interface for mediating PCM tethering to a centriole, a suitable candidate of which is the conserved centrosomal protein Sas-4 (CPAP in human), forming a layer closely associated with the centriole wall and yet shown to interact with various PCM components (6, 12). Functional studies in various model organisms suggest that Sas-4 proteins are required for both centriole formation and PCM assembly (6, 12); in the absence of Sas-4, nascent centrioles form but fail to mature into centrosomes (6). Overexpression of Sas-4 in flies produces PCM-like structures (13), whereas reduced amounts of Sas-4 in worms result in centrosomes having proportionally less PCM (12). Thus, although it is clear that Sas-4 is essential for centrosome biogenesis, the mechanisms by which Sas-4 contributes to PCM assembly remains elusive.
During the course of these studies, we and others have reported that Sas-4/CPAP, a protein essential for centriole formation was found to interact with several centrosomal and PCM proteins including Cnn, Asl, D-PLP, γ-TuRC, SIL, Cep135, Cep120, and tubulin dimers (5, 6, 14–16). In Drosophila, the N-terminal domain of Sas-4 provides a scaffolding site for cytoplasmic protein complexes (hereafter referred to as Sas-4–PCM scaffold) and tethers the components of Sas-4–PCM scaffold to a centrosome matrix via its C terminus (6).
Interestingly, the C-terminal region of Sas-4 proteins contains a conserved TCP10c domain (Pfam: PF07202) (hereafter referred to as TCP for brevity) (Fig. 1A and SI Appendix, Fig. S1). An E1235V missense mutation within this domain in CPAP has been identified in patients with primary microcephaly (MCPH), resulting in a reduced interaction with STIL (Ana2 in Drosophila), a centriole duplication factor also implicated in MCPH (16–18). Accordingly, recent structural studies on CPAP-STIL complex revealed that CPAP-STIL interaction is required during centriole assembly (19, 20). The C-terminal domain of CPAP has also been shown to mediate an interaction with another MCPH protein Cep135 (Bld-10 in Drosophila) and that interaction is required for centriole assembly. Bld-10 is a core centriolar protein and is required to stabilize structural integrity of centrioles (21–23). Taken together, we therefore speculate that the TCP domain could mediate protein–protein interactions and might serve as a tethering site for Sas-4–PCM scaffold–centriole interactions.
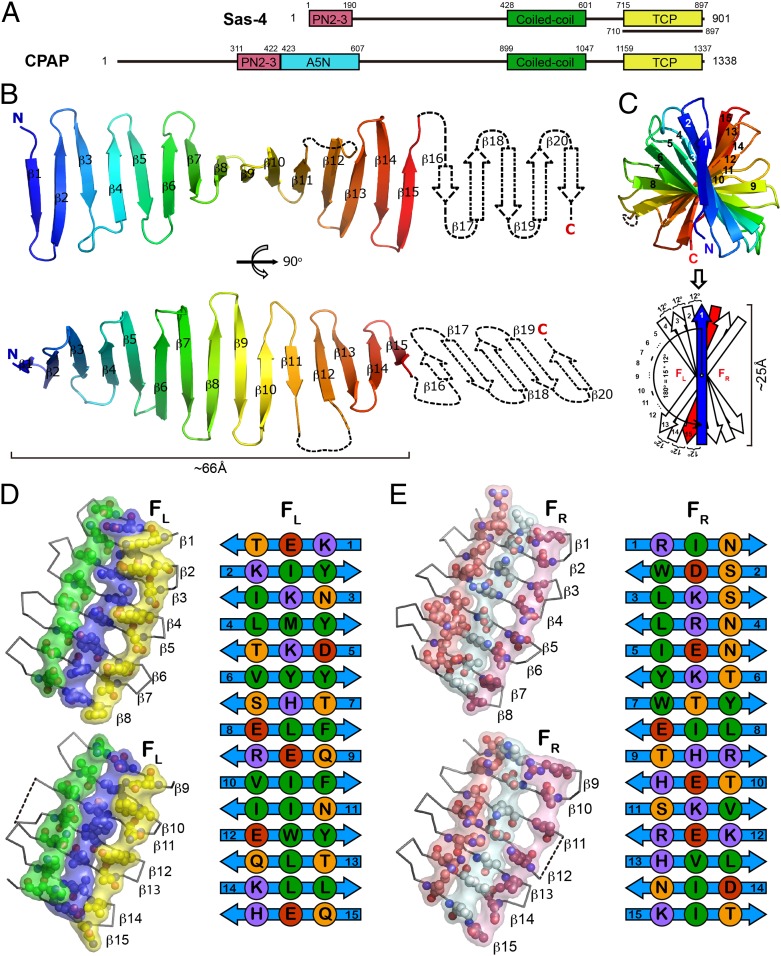
Fig. 1.
Crystal structure of Drosophila Sas-4–TCP domain. (A) Domain architecture of Drosophila Sas-4 and its human ortholog CPAP. The fragment used for crystallization is indicated by a black underline. (B) Cartoon view of the overall structure of Sas-4–TCP. The invisible part of β16–20 in the crystal structure is shown as dotted lines. (C) Side view of Sas-4–TCP along the longitudinal axis from the N to C termini. Twisting of the TCP β-strands is diagramed below. FL, surface left to β1; FR, surface right to β1. (D and E) Cross-strand ladder residues on FL (D) and FR (E) are shown in spheres and classified into different types by color (purple, positively charged residues; red, negatively charged residues; orange, polar residues; green, hydrophobic residues).
Although it appears that Sas-4 plays pivotal roles in centriole formation, assembling protein complexes in the cytoplasm, and tethering them to a developing centrosome, the mechanisms by which Sas-4 accomplishes its tethering role have remained unclear. In this study, we therefore investigated the structural basis of Sas-4 and show that via its conserved C-terminal TCP domain, it could provide an “extended surface-like” platform by which Sas-4 could mediate the Sas-4–PCM scaffold–centriole interaction during centrosome biogenesis.
Results
The Conserved C-Terminal TCP Domain of Sas-4 Is Required for Binding of Sas-4–PCM Scaffolds in a Centrosome.
Previously, we have shown that the N-terminal domain of Sas-4 functions as the scaffolding site for S-CAP (scaffold for cytoplasmic complexes, including Sas-4, Cnn, Asl, and D-PLP) complexes (6). Using cell-free experiments, we then demonstrated that Sas-4 could tether components of the Sas-4–PCM scaffold to the centrosomal matrix via its C-terminal region (6). To test this hypothesis in vivo, we generated transgenic Drosophila expressing Sas-4 devoid of its TCP region in sas -4s2214 null flies (Sas-4ΔTCP). We observed that the expression of Sas-4ΔTCP in sas-4s2214 null homozygote flies failed to rescue the sas-4s2214 null phenotype of uncoordination. Uncoordination is a ciliary-loss phenotype observed in centrosome mutant flies, which show unusual leg and wing coordination, including the inability to stand on their legs, as well as to have crossed legs and upward-extending wings (24) (Movie S1). Sas-4ΔTCP flies also failed to display a defined pattern of developing centrosomes in their testes and contained Sas-4ΔTCP and Asl (component of Sas-4–PCM scaffold) predominantly in the cytoplasm but not at centrosomal structures (SI Appendix, Fig. S2 A and B). However, in sas-4s2214 heterozygote flies, Sas-4ΔTCP could be targeted to endogenous centrosomes but to a lesser extend compared with sas-4s2214 homo- or heterozygotes of controls, which express full-length Sas-4 at the endogenous level (SI Appendix, Fig. S2C), an observation that is consistent with recent findings in Drosophila embryos (19). Thus, in the absence of the TCP domain, centrosomes fail to develop into functionally stable structures. The observed phenotypes could be attributable to either a failure of efficient Sas-4–PCM recruitment to developing centrosomes or inefficient tethering of the Sas-4–PCM scaffold at centrosomes.
The Sas-4–TCP Domain Adopts an Extended Single-Layer β-Sheet Structure.
To obtain mechanistic insights into how Sas-4’s TCP domain (amino acids 710–897) could play a role in tethering PCM complexes, we solved its crystal structure at a 2.7-Å resolution (Table 1). Structural determination revealed that the TCP domain adopts an extended single-layer antiparallel β-sheet fold. The observed β-folds are consistent with secondary structure predictions that estimated the presence of ∼20 β-strands across the TCP domain (Fig. 1B). However, according to the electron density, we could only trace the first 15 strands (amino acids 714–853) in the crystal structure. The loss of electron density at the β16–20 strands (amino acids 854–897) was unexpected because both secondary structure predictions and circular dichroism analysis suggested an all-β structure of this fragment (SI Appendix, Figs. S1 and S3A). Crystal packing analysis revealed that TCP β1–15 monomers are associated in a continuous “head-to-tail” mode, where β15 forms an antiparallel interaction with β1’ from an adjacent molecule (SI Appendix, Fig. S4). Mass spectrometric analysis confirmed that the missing electron density at the β16–20 strands in the crystal was not attributable to peptide cleavage (SI Appendix, Fig. S3B). We reason that β16–20 might be expelled from its original position to adapt to crystal packing, thereby being invisible in the current structure (SI Appendix, Fig. S4). It is noteworthy that β16 is predicted to be the shortest strand in length, because this might render β15–16 segments energetically subjective to breaks (SI Appendix, Fig. S1 and Fig. 1D).
Table 1.
Data collection and refinement statistics
| Crystal | SeMet-TCPSAS4, peak* | TCPSAS4, native |
| Data collection | ||
| Space group | P61 | P61 |
| Cell dimensions | ||
| a, b, c (Å) | 69.4, 69.4, 112.5 | 69.3, 69.3, 112.3 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 |
| Wavelength, Å | 0.9792 | 0.9792 |
| Resolution, Å | 50–2.9 (2.95–2.90)† | 50–2.7 (2.75–2.70)† |
| Rmerge, % | 13.1(82.8) | 9.2 (54.0) |
| I/σI | 28.4 (3.3) | 36.4 (4.2) |
| Completeness, % | 100 (100) | 100 (100) |
| Redundancy | 10.3 (10.4) | 11.2 (11.5) |
| Refinement (F > 0) | ||
| Resolution, Å | 50–2.7 | |
| No. of reflections | 8278 | |
| Rwork/Rfree, % | 24.4/27.6 | |
| No. atoms | ||
| Protein | 1,127 | |
| Ligand | 5 | |
| Water | 25 | |
| B factors, Å2 | ||
| Protein | 57.3 | |
| Ligand | 36.7 | |
| Water | 38.9 | |
| r.m.s. deviations | ||
| Bond lengths, Å | 0.002 | |
| Bond angles, ° | 0.66 |
Values in this column are based on anomalous scaling.
Values in parentheses are for highest-resolution shell.
Here, we report the experimental structure of a TCP domain; hence, the coordinates were submitted to the online Dali server for comparison with possible structural neighbors (SI Appendix, Fig. S5). The nearest structural homolog is an engineered outer-surface protein A (OspA) of Borrelia burgdorferi containing five β-hairpin repeats inserts (Protein Data Bank ID code 2FKJ) (25) with a Z score of 12.7 over 134 aligned residues. The majority of matches were assigned to the artificially engineered β-hairpins with marginal sequence identity (∼8%). Besides this, TCP shares limited similarity with other natural proteins of known structure, suggesting that it represents a fold with important physiological functions. TCP β1–15 adopts an unusually long and unbent scaffold of ∼66 Å in length and ∼25 Å in width (Fig. 1 B and C). Moreover, the TCP β-spine is also characteristic of a counterclockwise 180° twisting along the longitudinal axis over 15 consecutive strands (Fig. 1C).
Rigidity of TCP structure is likely contributed by cross-strand interactions of bulky residues, such as Tyr, Phe, Arg, Lys, Glu, and His, which may stabilize the single layer β-sheet fold by introducing cross-stand π-stacking, ion pairs, or Van der Waals contacts as previously discussed in the case of OspA (25–27) (Fig. 1 D and E).
The TCP domain displays an inherent tendency to self-assemble, as illustrated by our crystal packing analysis (SI Appendix, Fig. S4). If this occurs in vivo, it may help to stabilize Sas-4–centriole tethering in a synergistic way. Another important consequence of the single-layer β-sheet fold is the exposure of the side chains of nearly every TCP residue to solvent, rendering the TCP domain an ideal platform to mediate protein–protein interactions during centrosome biogenesis. Indeed, studies in human cells have shown that the C terminus of CPAP mediates its interaction with STIL and Cep135 (Ana2 and Bld-10 in Drosophila), and this interaction is required for centriole assembly (15, 16). Interestingly, a missense mutation of E1235V within the TCP domain of CPAP (which corresponds to E792V within the β9-strand of Drosophila Sas-4) has been shown to affect CPAP-STIL interaction (16).
The β9–10 Surface of the TCP Domain Is Required to Bind Sas-4–PCM Complexes in a Centrosome.
To explore the functional role of TCP in tethering Sas-4–PCM scaffolds in a centrosome, we began by overexpressing Td-Tomato–tagged β9–10 mutant constructs with the following mutations: E792V (Sas-4–TCP-1, MCPH patient mutation), V800A/I802A (Sas-4–TCP-2, “E792-proximal, β10”), and L841K (Sas-4–TCP-3, “E792-distal, β14”) in Drosophila cells. We chose β9-strand because it harbors the MCPH patient mutation implicating its role in centrosome biogenesis. Whereas wild-type Sas-4 (control) and Sas-4–TCP-3 were normally recruited to endogenous centrosomes, both Sas-4–TCP-1 and Sas-4–TCP-2 variants displayed only moderate levels of recruitment to centrosomes with partial dispersion within the cytoplasm (SI Appendix, Fig. S6A), consistent with observations in Sas-4ΔTCP heterozygous flies (SI Appendix, Fig. S2). Cytoplasmic diffusion of the mutant proteins could be attributable to the changes in their oligomerization property. However, our filtration experiment indicated that TCP variants eluted essentially the same as the wild type, suggesting that the mutations that we explored did not alter the oligomerization property of the protein in vitro (SI Appendix, Fig. S7). These data suggest that the β9–10 surface of TCP is required for its own efficient centrosomal localization. However, the involvement of other Sas-4 regions in its centrosomal localization cannot be excluded (28).
We further overexpressed Sas-4–TCP variants in acentrosomal Drosophila cells (hereafter referred to as C131) derived from DSas-4 null homozygous mutant flies and tested their ability to restore the formation of centrosomes that could recruit PCM in a normal manner (29). Control and Sas-4–TCP-3 could completely restore centrosome formation because they recruited normal levels of Asl (SI Appendix, Fig. S6 Bi and Bii), whereas ∼70% cells expressing Sas-4–TCP-1 or Sas-4–TCP-2 variants displayed a centrosome-like foci along with cytoplasmic diffusion of Asl. Of this 70%, a fraction of centrosomal structures recruited Asl but only at moderate to weak levels (SI Appendix, Fig. S6 Biii and Biv). Electron microscopy of centrosome structures induced by TCP variants did not differ markedly from centrosomes induced by wild-type Sas-4 (SI Appendix, Fig. S6C). In summary, these observations suggest that Sas-4 is required to a certain extent for centriole formation and that in the absence of the β9–10 surface of TCP domain, the nonfunctional Sas-4 fails to efficiently target the preassembled Sas-4–PCM scaffolds in a centrosome, thus resulting in unstable centrosomes.
β9–10 Mutants Can Form Cytoplasmic Sas-4–PCM Scaffolds but Are Defective in Their Binding to a Centrosome.
To determine the specific role of β9–10 mutants in tethering Sas-4–PCM scaffolds in a centrosome, a biochemical approach was used. We began by investigating centrosomal proteins, which specifically fail to bind to β9–10 mutants. For this, we immunopurified Sas-4–containing complexes from C131 cells expressing control, TCP-1, -2, and -3 variants, respectively. Western blots showed that in addition to previously identified S-CAP components, wild-type Sas-4 also bound Ana2 and Bld-10 (Fig. 2A) (5, 6). Interestingly, the TCP-1 and -2 variants bound lesser amounts of Ana2 and Bld-10 than control and Sas-4–TCP-3. Thus, it would seem that the TCP domain is required for Ana2 and Bld-10 interaction, because a TCP domain alone failed to pull down tubulin dimers or γ-tubulin from embryonic extracts (Fig. 2B). Accordingly, compared with control or TCP3 pull downs, TCP-1 and -2 variants pulled down a reduced amount of Ana2 and Bld-10 (Fig. 2C).
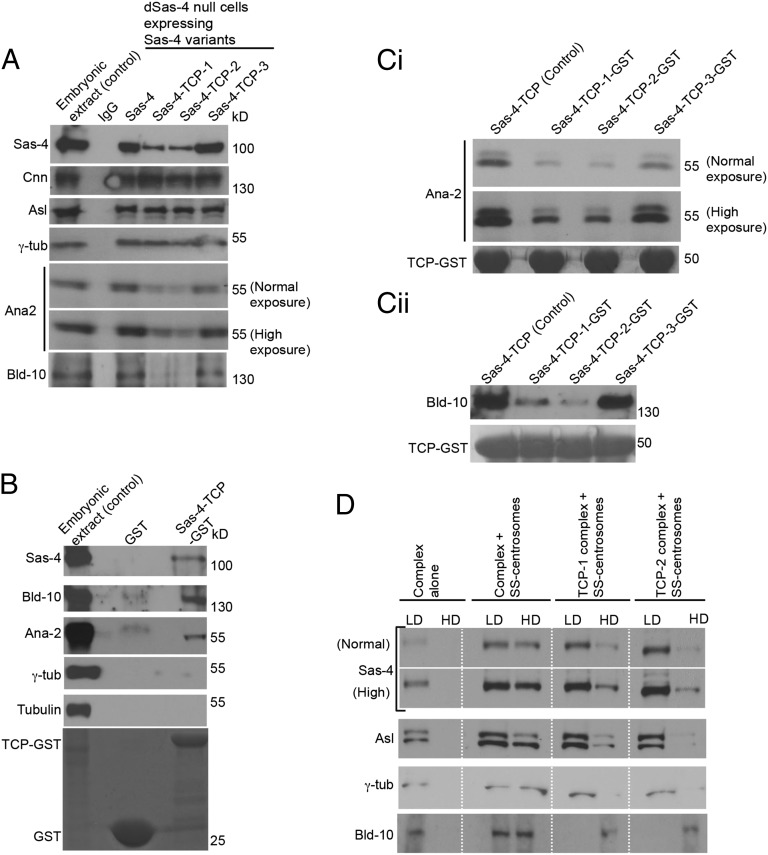
Fig. 2.
The β9–10 surface of TCP domain mediates an interaction with Bld-10 and Ana2 and is required to tether Sas-4–PCM scaffolds in a centrosome. (A) Sas-4–TCP variants can assemble cytoplasmic complexes of centrosomal proteins. Immunopurified Sas-4–TCP variants containing complexes from C131 cells reveal the presence of S-CAP components. Variants TCP-1 (E792V) and TCP-2 (V800A/I802A) specifically fail to bind Ana2 and Bld-10. Embryonic extracts were used as a positive control and mouse IgG-coated beads were used as a negative control. (B) GST pull-down assay. TCP domain alone pulls down Ana2 and Bld-10 from Drosophila embryonic extracts. Embryonic extracts were used as a positive control. (C) In a GST pull-down assay, TCP-1 (E792V) and TCP-2 (V800A/I802A) variants bind a reduced amount of Ana2 (Ci) and Bld-10 (Cii), compared with wild type or TCP-3 (L841K) variant. Both normal and high exposures are shown. (D) Cell-free binding assay of purified Sas-4–TCP variants containing complexes to stripped centrosomes by discontinuous sucrose-gradient centrifugation. Unbound complexes are found in the low-density (LD) fractions and bound complexes in the high-density (HD) fractions that contain stripped centrosomes. In the absence of stripped centrosomes (complexes alone), the components of Sas-4 complexes (Sas-4, Asl, γ-tubulin, and Bld-10) remain only in the low-density fractions. Control Sas-4 complexes but not TCP-1 (E792V)- or TCP-2 (V800A/I802A)-containing complexes sink efficiently with Bld-10–positive stripped centrosomes that are detected in the high-density fractions by Western blots. Control and TCP-1 (E792V)- or TCP-2 (V800A/I802A)-containing complexes were prepared as stated in A. Bld-10–positive stripped centrosomes were prepared as mentioned in SI Appendix, Fig. S8. TCP3 variant was not used in this experiment because this variant behaved like the wild-type control.
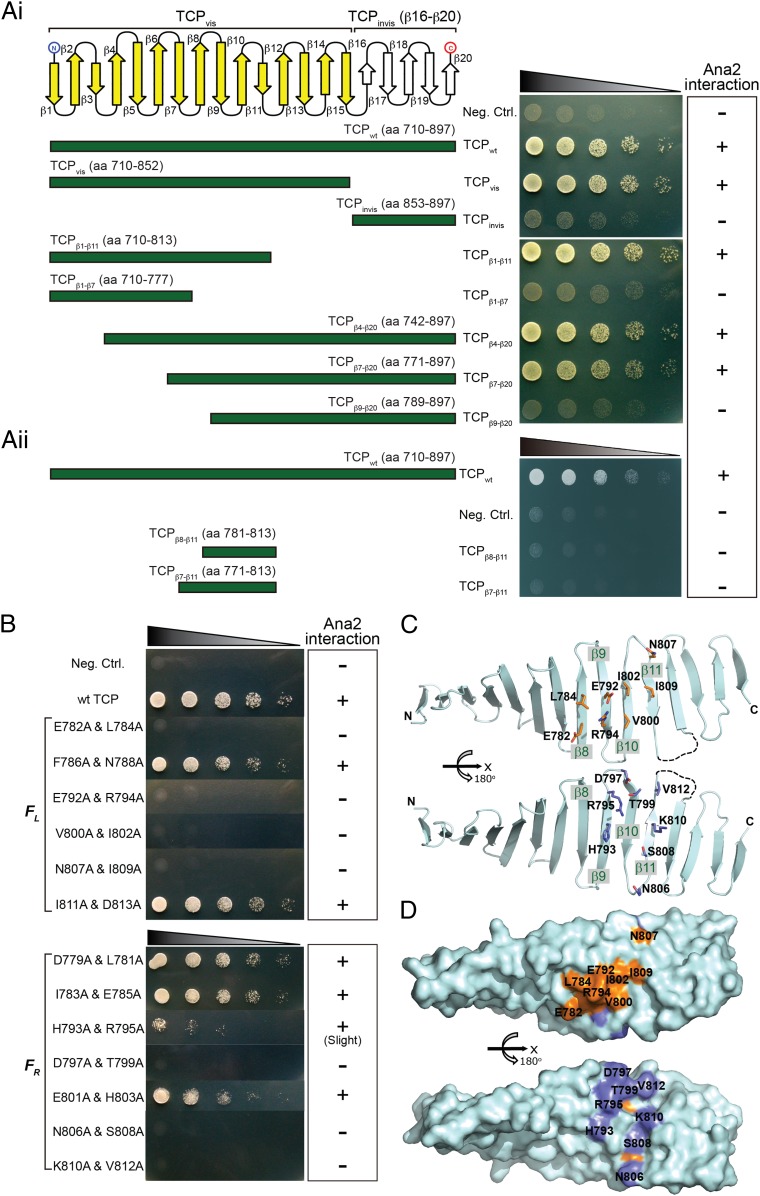
To further substantiate the TCP-Ana2 interaction, we performed yeast two-hybrid (Y2H) assays using full-length Ana2 as bait to map elements within the TCP domain responsible for this interaction. Firstly, we designed different TCP-deletion constructs within β1–20 and tested their ability to bind to Ana2 (Fig. 3Ai). Whereas the TCP β16–20 invisible region failed to interact with Ana2, the β1–15 visible region interacted with Ana2 to an extent similar to the full-length TCP domain. This suggested that β1–15 is sufficient for Ana2 binding. Whereas segments β1–11, β4–20, and β7–20 interacted with Ana2, segments β1–7 or β9–20 failed to interact with Ana2. Although we tested for sufficient residues required for Ana2 interaction, we found that β8–11 or β7–11 surface alone did not interact with Ana2 (Fig. 3Aii). From these experiments, we were able to conclude that the β8–11 fragments were critical but not sufficient for Ana2 interaction (Fig. 3 Ai and Aii).
Fig. 3.
Mapping of TCP fragment responsible for Ana2 interaction by Y2H assays. (Ai) The visible β1–15 fragment of Sas-4–TCP is highlighted in yellow. The yeast cells were dot-blotted in fivefold serial dilution for semiquantitative comparison. (Aii) β8–11 or β7–11 fragment of Sas-4–TCP is not sufficient to interact with Ana2. The yeast cells were dot-blotted in fivefold serial dilution for semiquantitative comparison. (B) Mapping of TCP surface residues required for Ana2 interaction. Residues are clustered into two categories depending which surface they reside. FL, surface left to β1; FR, surface right to β1. (Right) Surface mapping of Ana2-binding residues (orange, FL residues; blue, FR residues). (C) Positioning of key Ana2-binding residues in FL (Upper) and FR (Lower) surfaces in ribbon view. (D) A surface representation of C. Residues of FL surface are colored orange, and those of FR surface are colored in blue.
To further validate the β8–11-Ana2 interaction, we introduced alanine mutations within the β8–11 region and repeated the Y2H assay. To our surprise, except for the F786, N788, I811, D813, D779, L781, I783, or E785 mutations, none of the other mutations within the β8–11 segment retained interaction with Ana2 (Fig. 3 B–D). These experiments have therefore mapped conserved residues within the β8–11 segment of TCP that are key determinants for Ana2 binding. The finding that these key residues are clustered to both surfaces of the β8–11 sheet suggests a multivalent mode of Ana2-TCP interaction.
Finally, to test whether the β9–10 mutants play a role in binding Sas-4–PCM scaffolds in a centrosome, we adapted a cell-free binding experiment, which employs sucrose-gradient velocity sedimentation with immunopurified Sas-4 complexes and stripped centrosomes (6). First, we isolated stripped centrosomes, positive for the core-centriolar protein Bld-10 (SI Appendix, Fig. S8). We then mixed the stripped centrosomes with Sas-4–containing cytoplasmic complexes that were immunopurified from C131 cells expressing control or TCP variants (as observed in Fig. 2A) and subjected them to velocity sedimentation. We then performed Western blot analyses of the low-density fractions for unbound complexes and high-density fractions for complexes bound to stripped centrosomes. As expected, control Sas-4 complexes were found bound to Bld-10–positive stripped centrosomes in the high-density fractions. Importantly, Sas-4 complexes of TCP variants mostly existed in low-density fractions, suggesting that the β9–10 surface of TCP could mediate some aspects of Sas-4–PCM scaffold binding to a centrosome (Fig. 2D).
Taken together, our in vitro cell culture and biochemical experiments suggest that the β9–10 surface of the TCP domain can mediate Sas-4–Ana2–Bld-10 interactions and that via this interaction, Sas-4 can tether its cytoplasmic Sas-4–PCM scaffold to a centrosome. Thus, mutations in β9–10 surface can specifically perturb this interaction while still allowing Sas-4 to form its N-terminal–dependent Sas-4–PCM scaffold.
The β9–10 Surface of Sas-4’s TCP Domain Is Required for Binding of Sas-4–PCM Complexes in a Centrosome in Vivo.
To specifically dissect the PCM-tethering function of Sas-4 independent of its role in centrosome formation, we generated transgenic Drosophila expressing endogenous levels of the different Sas-4–TCP variants (Sas-4–TCP-1, -2, and -3) in the sas-4s2214 null background (SI Appendix, Fig. S9A). Sas-4–TCP-3 but not Sas-4–TCP-1 or Sas-4–TCP-2 variants (hereafter referred to as TCP variants) rescued the uncoordination phenotype of sas-4s2214, but only partially (Movies S2–S4). Whereas the control and Sas-4–TCP-3 fly testes displayed similar numbers and patterns of Asl-positive centrosomes, TCP variants displayed a dramatically reduced number of Asl-positive centrosomes (SI Appendix, Fig. S9 B and C). Similar to Sas-4ΔTCP, Asl was predominantly present in the cytoplasm of spermatogonium cells, and spermatocyte cells lacked elongated centrosomes (SI Appendix, Figs. S2 and S9). These findings suggest that the β9–10 surface of TCP is critical and could mediate important aspects of Sas-4’s functions in vivo.
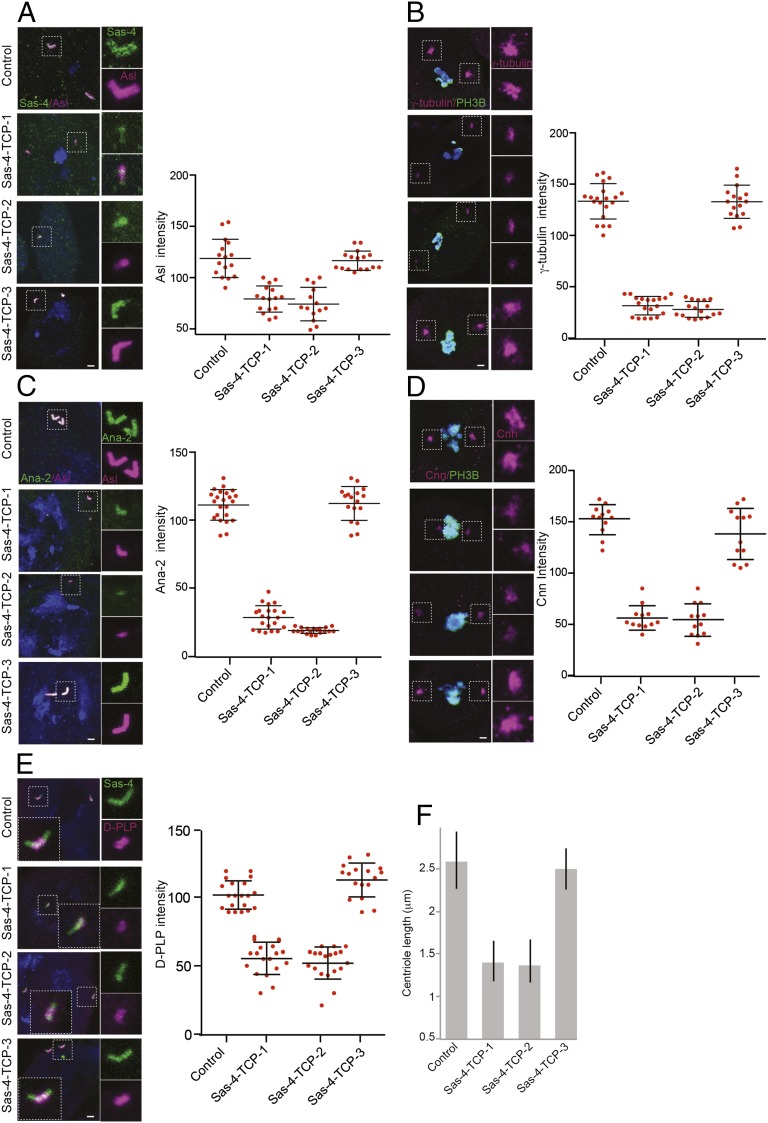
We then tested spermatogonium centrosomes for their PCM recruitment. To perform Ana2 localization studies, we generated an Ana2 monoclonal antibody (SI Appendix, Fig. S9D). Whereas all of the centrosomes in control and Sas-4–TCP-3 flies normally recruited Sas-4, Asl, γ-tubulin, Cnn, and Ana2, only a fraction of the TCP variants’ centrosomes were positive for the markers tested (Fig. 4). Notably, the TCP variants displayed only moderate to weak levels of protein recruitment at the centrosomes and exhibited a diffused cytoplasmic patterning, similar to observations for the corresponding experiments in Drosophila cells or Sas-4ΔTCP flies (Fig. 4 A–E and SI Appendix, Figs. S7 and S2). In particular, γ-tubulin, D-PLP, and Cnn recruitment in these variants was dramatically reduced (Fig. 4 B, D, and E).
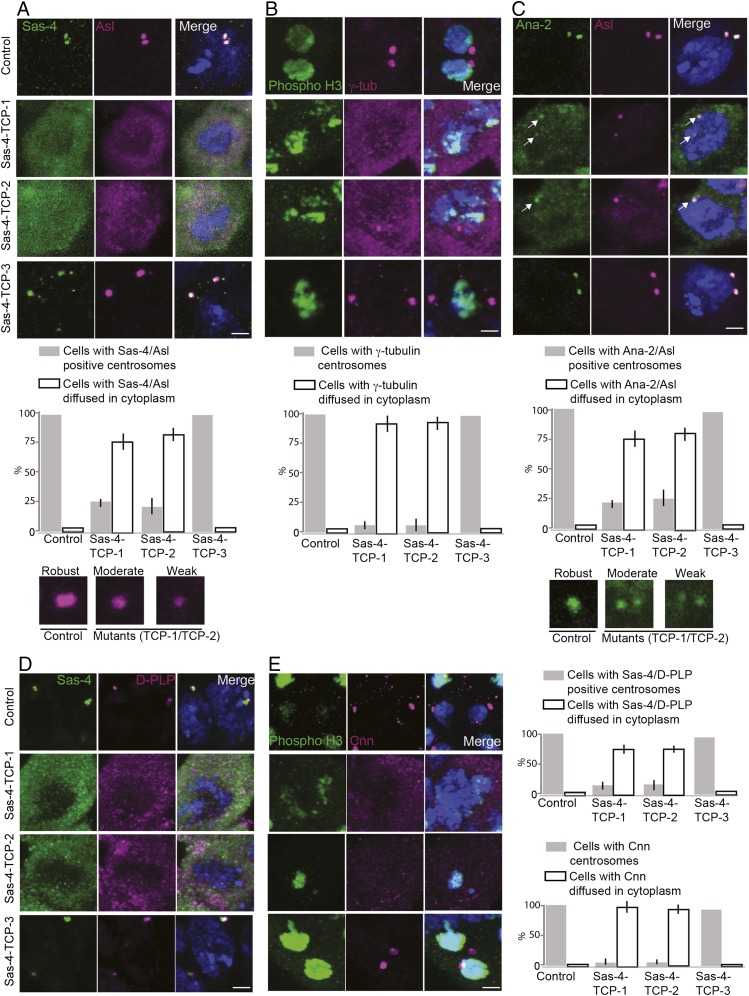
Fig. 4.
TCP domain’s β9–10 surface is essential for tethering the Sas-4–PCM scaffold in a centrosome. (A–E) Spermatogonium centrosomes of control and Sas-4–TCP-3 but not Sas-4–TCP-1 or -2 variants normally recruit Sas-4 (green), Asl (magenta), γ-tubulin (magenta), Ana2 (green), D-PLP (magenta), and Cnn (magenta); ∼75–80% of the cells expressing Sas-4–TCP-1 and -2 variants did not contain detectable centrosomes, and the components of Sas-4 complexes were diffused within the cytoplasm (corresponding graphs for A–E) The remaining cells contained centrosomes but recruited Asl, γ-tubulin, Ana2, D-PLP, or Cnn only at moderate to weak levels compared with control and Sas-4–TCP-3 (Insets shown at the bottom of A and C). The phospho-H3B antibody was used to detect dividing cells for experiments with γ-tubulin and Cnn staining. Arrows in C mark centrosomes. (Scale bar, 1 μm.)
Because spermatogonium centrosomes of TCP variants lacked Sas-4–PCM scaffold components, we speculated that these centrosomes might not develop into fully functional elongated centrosomes in the later stages of development. All of the control and Sas-4–TCP-3 spermatocytes displayed normally elongated centrosomes that recruited Sas-4, Asl, Ana-2, and D-PLP (Fig. 5 A, C, and E). In contrast, only ∼30% of spermatocytes of TCP variants contained centrosomes, which were dramatically shorter, and recruited reduced levels of centrosomal proteins tested (Fig. 5 A, C, and E). This is consistent with earlier findings that Sas-4 is required for centriole elongation (30, 31). Similarly, compared with controls, the meiotic centrosomes of TCP variants recruited reduced levels of γ-tubulin and Cnn (Fig. 5 B and D).
Fig. 5.
TCP domain’s β9–10 surface is required for stabilization of developing centrosomes. Control and Sas-4–TCP-3 spermatocyte centrosomes normally elongate and recruit Sas-4, Asl, γ-tubulin, Ana2, Cnn, and D-PLP (A–E), whereas Sas-4–TCP-1 and -2 variants recruit lesser amounts of these proteins and exhibit significantly reduced centrosome lengths (graphs on the right and F). (Scale bar, 0.5 μm.)
It is possible that TCP variants fail to recruit Sas-4–PCM scaffold components because of the absence of centrioles. To test for centrioles, we stained spermatogonium and spermatocyte cells for the core centriolar markers Bld-10 and Ana-1 (6, 21, 22). Interestingly, in contrast to PCM markers, a significant number of TCP variants cells (∼50%) contained Bld-10 and Ana-1. As expected, control and Sas-4–TCP-3 centrioles recruited Sas-4 and Asl, respectively, whereas Sas-4–TCP-1 and Sas-4–TCP-2 variants failed to recruit them (SI Appendix, Fig. S10). These in vivo experiments suggest that mutations within the β9–10 surface could cause defects in PCM tethering and thus result in unstable centrosomes.
β9–10 Surface of CPAP’s TCP Domain Has a Conserved Function and Plays a Role in Tethering PCM Complexes in a Centrosome.
To examine whether the β9–10 surface of CPAP’s TCP domain (the human counterpart of Sas-4) has a conserved function in PCM recruitment, we conducted experiments in HeLa cells. CPAP harbors a conserved TCP domain related to the t-complex 10 proteins in which a missense mutation (E1235V) within this domain causes MCPH (Fig. 1) (32).
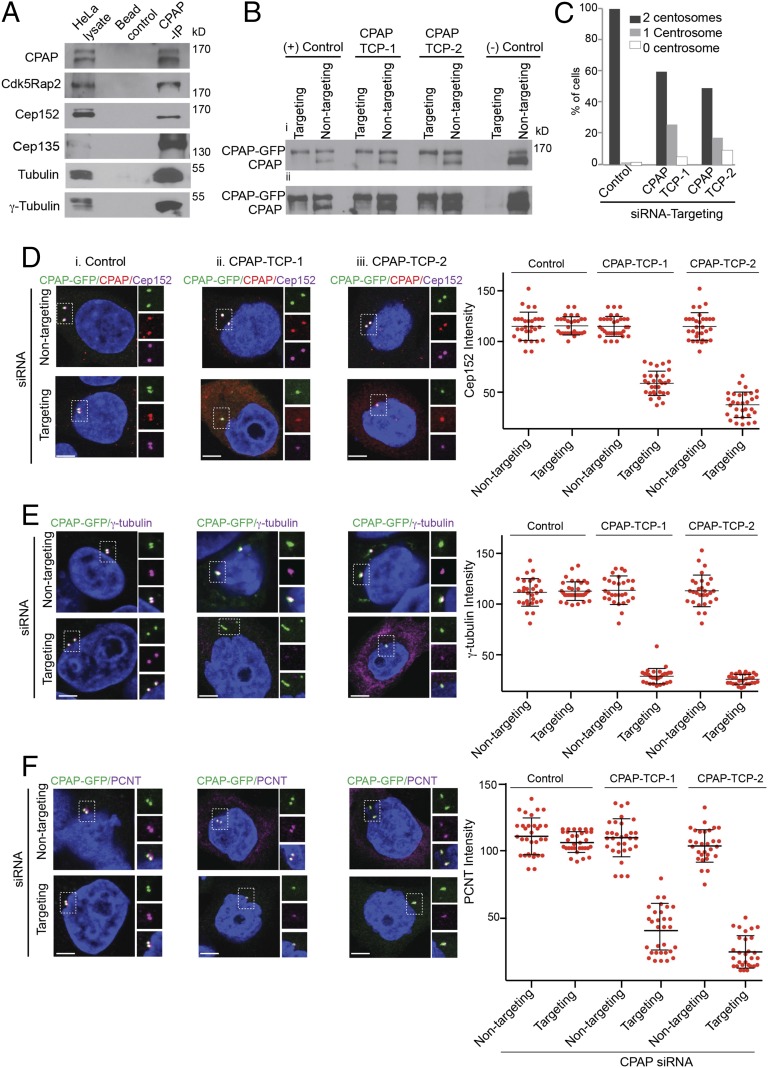
First, we generated a monoclonal CPAP antibody (SI Appendix, Fig. S11). Using this antibody, we immunoprecipitated CPAP and verified that it has binding partners similar to those of Sas-4 (Fig. 6A) (5, 6, 33, 34). We then generated analogous GFP-tagged RNAi-resistant β9−10 surface mutants of CPAP (E1235A: CPAP–TCP-1, a natural MCPH mutant; and K1243A/I1245A: CPAP–TCP-2; SI Appendix, Fig. S1), using bacterial artificial chromosome (BAC) recombineering (35, 36). In contrast to cDNA constructs, BACs contain regulatory elements required for native expression of the gene of interest (35, 36). BAC modifications including point mutations were done by homologous recombination. Finally, we established HeLa cells stably expressing CPAP-TCP variants under their own endogenous regulation. Treating cells with CPAP-targeting siRNA consistently depleted endogenous CPAP while leaving the RNAi-resistant CPAP-TCP variants unaffected (Fig. 6B). Thus, this approach allowed us to study the specific effects of β9–10 surface mutants under their endogenous regulation.
Fig. 6.
The β9–10 surface of CPAP-TCP domain has a conserved function, playing a role in tethering PCM complexes in a centrosome. (A) CPAP coimmunoprecipitates with several centrosomal proteins. HeLa lysates and empty beads were used as positive and negative controls. (B) Validation of RNAi-resistant BACs expressing CPAP-TCP variants. RNAi-resistant BACs are resistant to CPAP-targeting siRNA. In contrast, CPAP-targeting siRNA treatment completely depletes both endogenous and CPAP-GFP in cells expressing non–RNAi-resistant BACs (last lane, –control). Nontargeting siRNA (siRNA that does not target CPAP) was used as a control. Both low (Bi) and high-exposure (Bii) Western blots are shown. (C) Quantification of centrosomes in control, TCP-1, and TCP-2 variants upon treatment of cells with CPAP-targeting siRNA. (D–F) Upon treatment with CPAP-targeting siRNA (Targeting) and Control (i) but not CPAP–TCP-1 or -2 (ii and iii) variants restore the centrosome biogenesis that recruits a normal amount of centrosomal proteins (magenta) Cep152, γ-tubulin, and PCNT. CPAP is predominantly found in the cytoplasm in TCP-1 and -2 variants. CPAP–TCP-1– or CPAP–TCP-2–expressing centrosomes recruit only low to moderate levels of Cep152, γ-tubulin, and PCNT. Intensity measurements are given at the right. (Scale bar, 1 μm.)
In the control, effects of siRNA-mediated CPAP depletion could be rescued by the expression of wild-type siRNA-resistant CPAP. In contrast, siRNA-resistant TCP variants failed to rescue the effects of CPAP depletion completely indicating impaired PCM recruitment and thus centrosome duplication defects (16, 17) (Fig. 6C). In centrosome forming cells, TCP variants were still targeted to centrosomes. These results are consistent with recent reports (16, 17). However, compared with the control, there was a higher amount of TCP variants predominantly diffused within the cytoplasm, and centrosomes of cells expressing TCP variants showed only moderate levels of Cep152, γ-tubulin and PCNT recruitment (Fig. 6 D–F). A fraction of cells also displayed γ-tubulin or PCNT diffused within cytoplasm (Fig. 6 E and F).
MCPH is a neurodevelopmental disorder implicated in defective neural progenitor functions (37). To test whether the natural MCPH mutant centrosomes (E1235A: CPAP–TCP-1) have a similar PCM recruitment defect in neural-progenitor cells, we generated human induced pluripotent stem (iPS) cell-derived neural progenitor cells. First, we confirmed that these iPS cell-derived neural progenitor cells could be differentiated into various neuronal cell lineages (SI Appendix, Fig. S12). We then tested γ-tubulin, Cep152, and PCNT recruitment to the centrosomes of neural progenitor cells expressing siRNA-resistant CPAP–TCP-1. In addition to centrosome duplication defects, CPAP–TCP-1–expressing centrosomes were defective in PCM recruitment because they recruited only moderate levels of γ-tubulin, Cep152, and PCNT, an observation that is consistent with that in HeLa cells (SI Appendix, Fig. S13).
In summary, these results reflect the similar phenotypes observed in Drosophila, suggesting that the β9–10 surface of TCP domain has a conserved function and plays a role in tethering PCM complexes in a centrosome.
Discussion
In order for centrosomes to form functional organelles as the MTOCs of a cell, it must first successfully recruit PCM around centrioles. Thus, PCM assembly forms a crucial step in centrosome biogenesis. PCM recruitment begins with the formation of centrosomal protein complexes in the cytoplasm, which are then targeted to centrioles (6, 34).
MCPH is a developmental disorder caused by mutations in centrosomal proteins where brain size is severely reduced (32, 37). Interestingly, the finding that several known autosomal recessive primary MCPH proteins (MCPH1 to -8) are found in centrosomes supports an involvement of a centrosomal mechanism (6, 32, 37, 38). However, it remains unclear how these proteins interact with each other for successful recruitment to a centrosome. Recent elegant molecular studies have uncovered that these MCPH-related centrosomal proteins interact in a domain-specific manner and that these interactions are indeed required for centrosome assembly (15, 16). In particular, the C-terminal domain of CPAP harboring the TCP domain that has been reported to interact with SIL and Cep135, both of which have been implicated in MCPH (15, 16). These studies have collectively provided a molecular link by which Sas-4/CPAP proteins could mediate the formation of MCPH protein complexes during centrosome biogenesis. Thus, a mutation within the TCP domain as observed in natural MCPH patients (E1235V; Sas-4/CPAP–TCP-1 variant in this study) could perturb proper CPAP-SIL-Cep135 interactions, which might ultimately form malfunctioning protein complexes affecting the proper formation of functional centrosomes. Accordingly, our in vivo and in vitro experiments support this idea (Figs. 2–6).
In addition to this, our structural work reveals a highly solvent-exposed single-layer β-sheet fold of TCP domain. Despite the report of such a fold in an engineered OspA protein (25) (SI Appendix, Fig. S5), here, we identified a natural module of similar fold that plays an important physiological role in centrosome biogenesis. In particular, TCP adopts an unusually long and unbent β-spine conformation that presents nearly all its side chains for scaffolding potential protein–protein interactions. Indeed, our Y2H analysis revealed a cluster of residues on opposite surfaces that are essential for Ana2 interaction, suggesting that TCP could exploit its different surfaces for binding interactions. In the crystal, TCP displayed strong propensity for self-assembly (SI Appendix, Fig. S4), an intrinsic property of β-hairpin repeats (25).
Accordingly, Sas-4 proteins are also localized around the centriole wall and play a critical role in PCM organization (6, 12, 13). It is therefore conceivable that TCP may contribute to Sas-4 self-assembly around the centriole wall so as to stabilize centriole from its outer surface. Thus, by this property, TCP could mediate protein–protein interactions serving as a tethering site for Sas-4–PCM scaffold–centriole interaction. Taken together, our structural data suggest that Sas-4 proteins could perform multifaceted molecular functions, which could integrate tubulin, microtubule, and PCM binding during centrosome biogenesis.
Thus, in combination with these recently published studies, our findings in the presented work advances our knowledge on Sas-4–mediated PCM recruitment (SI Appendix, Fig. S14). Structural insights given in this work explain how the β9–10 surface of TCP domain plays a role in tethering PCM complexes during centrosome biogenesis. Our structure-based functional studies have dissected Sas-4’s role in PCM tethering in which the N-terminal domain of Sas-4 provides a scaffolding site for cytoplasmic protein complexes and the β9–10 surface of TCP serves as a tethering site at the centriole via a coordinated Sas-4–Ana2–Bld-10 interaction (Figs. 2–5 and SI Appendix, Fig. S14). When this ternary complex formation is perturbed (i.e., when β9–10 surface is mutated), the components of the Sas-4–PCM scaffold remain in cytoplasm. Our in vivo and cell-free binding assays support this idea (Figs. 2D, 4, and 5).
Although our in vivo and in vitro experiments substantiate the function of Sas-4 in PCM tethering, what remains elusive is why some of the cells in TCP variants lack centrosomal structures. One possibility is that Sas-4 complexes are temporally regulated and stabilized by the coordinated function of Sas-4–Ana2–Bld-10 interaction at the PCM-tethering site around centrioles (SI Appendix, Fig. S14). This notion is supported by studies in human cells showing that the C terminus of CPAP mediates an interaction with SIL and Cep135 (Ana2 and Bld-10 in Drosophila) and that this interaction is required for centriole assembly. This aspect is further supported by our current study, where in vivo, the Sas-4–TCP variants lack an interaction with Ana2 and Bld-10. To add to that, TCP mutants phenocopy Ana2 mutants to a certain extent because they also lack detectable centrosomes in some cells. Studies in Drosophila have further showed that Bld-10 is a centriole microtubule-binding protein and is required to stabilize structural integrity of centrioles (21–23). Taken together, it seems that a coordinated function of this ternary complex is essential for successful PCM recruitment and for generating stable centrosomes. Future structural and functional characterizations of the Sas-4–containing ternary complex may uncover this mystery.
Materials and Methods
Transgenic flies and Sas-4 constructs were previously described but with second chromosome specific pattB-UAST vector. The TCP domain (residues 710-897) of D. melanogaster Sas-4 was cloned into the pET-28b (Novagen) vector and expressed as a N-terminal His-tagged protein E. coli BL21 (DE3) (Novagen). Protein preparation, crystallization, structure determination, molecular biology technics, transgenic flies, cell cultures, biochemical methods, and electron and light microscopy methods are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank staff members at Shanghai Synchrotron Research Facility beam line 17U for help with data collection. We thank Dr. P. Keller for CPAP and Ana2 antibody generation, Drs. T. Kaufman and T. Megraw for the Cnn antibody, Dr. M. Bettencourt for the Bld-10 antibody, Dr. T. K. Tang for Cep135 antibodies, and Dr. Nigg for Cep152 antibodies. We also thank Anand Ramani, Marit Leuschner, Andrea Ssykor, and Mai Pham for technical help and Dr. K. Nohroudi (Anatomy Institute, University of Cologne) for EM experiments. This work was supported by the Center for Molecular Medicine of the Cologne University (J.G.), Deutsche Forschungsgemeinschaft (DFG) Grant GO 2301/2-1, and the Köln Fortune Program of the Medical Faculty of the University of Cologne (E.G. and T.Š.). H.L. is supported by grants from the Tsinghua University Initiative Scientific Research Program and the Program for New Century Excellent Talents in University. T.A.-R. is supported by National Institute of General Medical Sciences Grant R01GM098394. The work of I.P. at the laboratory of A.A.H. is supported by European Community’s Seventh Framework Programme FP7/2007-2013 under Grant 241548 (MitoSys Project).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4MPZ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1317535111/-/DCSupplemental.
References
- 1.Moritz M, Braunfeld MB, Sedat JW, Alberts B, Agard DA. Microtubule nucleation by gamma-tubulin-containing rings in the centrosome. Nature. 1995;378(6557):638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- 2.Zheng Y, Wong ML, Alberts B, Mitchison T. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 1995;378(6557):578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
- 3.Oegema K, et al. Characterization of two related Drosophila gamma-tubulin complexes that differ in their ability to nucleate microtubules. J Cell Biol. 1999;144(4):721–733. doi: 10.1083/jcb.144.4.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139(4):663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 5.Gopalakrishnan J, et al. Tubulin nucleotide status controls Sas-4-dependent pericentriolar material recruitment. Nat Cell Biol. 2012;14(8):865–873. doi: 10.1038/ncb2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gopalakrishnan J, et al. Sas-4 provides a scaffold for cytoplasmic complexes and tethers them in a centrosome. Nat Commun. 2011;2:359. doi: 10.1038/ncomms1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersen JS, et al. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426(6966):570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 8.Mennella V, et al. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat Cell Biol. 2012;14(11):1159–1168. doi: 10.1038/ncb2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu J, Glover DM. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2012;2(8):120104. doi: 10.1098/rsob.120104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawo S, Hasegan M, Gupta GD, Pelletier L. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat Cell Biol. 2012;14(11):1148–1158. doi: 10.1038/ncb2591. [DOI] [PubMed] [Google Scholar]
- 11.Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open. 2012;1(10):965–976. doi: 10.1242/bio.20122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirkham M, Müller-Reichert T, Oegema K, Grill S, Hyman AA. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell. 2003;112(4):575–587. doi: 10.1016/s0092-8674(03)00117-x. [DOI] [PubMed] [Google Scholar]
- 13.Kleylein-Sohn J, et al. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13(2):190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Comartin D, et al. CEP120 and SPICE1 cooperate with CPAP in centriole elongation. Curr Biol. 2013;23(14):1360–1366. doi: 10.1016/j.cub.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Lin YC, et al. Human microcephaly protein CEP135 binds to hSAS-6 and CPAP, and is required for centriole assembly. EMBO J. 2013;32(8):1141–1154. doi: 10.1038/emboj.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang CJ, et al. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J. 2011;30(23):4790–4804. doi: 10.1038/emboj.2011.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitagawa D, et al. Spindle positioning in human cells relies on proper centriole formation and on the microcephaly proteins CPAP and STIL. J Cell Sci. 2011;124(Pt 22):3884–3893. doi: 10.1242/jcs.089888. [DOI] [PubMed] [Google Scholar]
- 18.Vulprecht J, et al. STIL is required for centriole duplication in human cells. J Cell Sci. 2012;125(Pt 5):1353–1362. doi: 10.1242/jcs.104109. [DOI] [PubMed] [Google Scholar]
- 19.Cottee MA, et al. Crystal structures of the CPAP/STIL complex reveal its role in centriole assembly and human microcephaly. ELife. 2013;2:e01071. doi: 10.7554/eLife.01071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatzopoulos GN, et al. Structural analysis of the G-box domain of the microcephaly protein CPAP suggests a role in centriole architecture. Structure. 2013;21(11):2069–2077. doi: 10.1016/j.str.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carvalho-Santos Z, et al. BLD10/CEP135 is a microtubule-associated protein that controls the formation of the flagellum central microtubule pair. Dev Cell. 2012;23(2):412–424. doi: 10.1016/j.devcel.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Mottier-Pavie V, Megraw TL. Drosophila bld10 is a centriolar protein that regulates centriole, basal body, and motile cilium assembly. Mol Biol Cell. 2009;20(10):2605–2614. doi: 10.1091/mbc.E08-11-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roque H, et al. Drosophila Cep135/Bld10 maintains proper centriole structure but is dispensable for cartwheel formation. J Cell Sci. 2012;125(Pt 23):5881–5886. doi: 10.1242/jcs.113506. [DOI] [PubMed] [Google Scholar]
- 24.Blachon S, et al. Drosophila asterless and vertebrate Cep152 Are orthologs essential for centriole duplication. Genetics. 2008;180(4):2081–2094. doi: 10.1534/genetics.108.095141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makabe K, et al. Atomic structures of peptide self-assembly mimics. Proc Natl Acad Sci USA. 2006;103(47):17753–17758. doi: 10.1073/pnas.0606690103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koide S, et al. Design of single-layer beta-sheets without a hydrophobic core. Nature. 2000;403(6768):456–460. doi: 10.1038/35000255. [DOI] [PubMed] [Google Scholar]
- 27.Pham TN, Koide A, Koide S. A stable single-layer beta-sheet without a hydrophobic core. Nat Struct Biol. 1998;5(2):115–119. doi: 10.1038/nsb0298-115. [DOI] [PubMed] [Google Scholar]
- 28.Zhao L, et al. Dimerization of CPAP orchestrates centrosome cohesion plasticity. J Biol Chem. 2010;285(4):2488–2497. doi: 10.1074/jbc.M109.042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lecland N, et al. Establishment and mitotic characterization of new Drosophila acentriolar cell lines from DSas-4 mutant. Biol Open. 2013;2(3):314–323. doi: 10.1242/bio.20133327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang CJ, Fu RH, Wu KS, Hsu WB, Tang TK. CPAP is a cell-cycle regulated protein that controls centriole length. Nat Cell Biol. 2009;11(7):825–831. doi: 10.1038/ncb1889. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt TI, et al. Control of centriole length by CPAP and CP110. Curr Biol. 2009;19(12):1005–1011. doi: 10.1016/j.cub.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 32.Bond J, et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet. 2005;37(4):353–355. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- 33.Dzhindzhev NS, et al. Asterless is a scaffold for the onset of centriole assembly. Nature. 2010;467(7316):714–718. doi: 10.1038/nature09445. [DOI] [PubMed] [Google Scholar]
- 34.Hung LY, Tang CJ, Tang TK. Protein 4.1 R-135 interacts with a novel centrosomal protein (CPAP) which is associated with the gamma-tubulin complex. Mol Cell Biol. 2000;20(20):7813–7825. doi: 10.1128/mcb.20.20.7813-7825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bird AW, et al. High-efficiency counterselection recombineering for site-directed mutagenesis in bacterial artificial chromosomes. Nat Methods. 2012;9(1):103–109. doi: 10.1038/nmeth.1803. [DOI] [PubMed] [Google Scholar]
- 36.Poser I, et al. BAC TransgeneOmics: A high-throughput method for exploration of protein function in mammals. Nat Methods. 2008;5(5):409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thornton GK, Woods CG. Primary microcephaly: Do all roads lead to Rome? Trends Genet. 2009;25(11):501–510. doi: 10.1016/j.tig.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Basto R, et al. Flies without centrioles. Cell. 2006;125(7):1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.