Significance
High-grade serous ovarian cancers are characterized by widespread gain and loss of copy number involving large numbers of genes; however, the functional consequences of many of these changes remain unclear. To determine which of the many amplified genes exhibited tumor-promoting behavior, we developed a novel in vivo method to systematically screen potential oncogenes for tumor formation. We identified GAB2, a signaling adaptor protein, as a potent oncogene that is also significantly amplified in ovarian and breast cancer. GAB2 overexpression activates the phosphatidylinositol 3-kinase (PI3K) pathway and confers sensitivity to PI3K pathway inhibition. These results credential GAB2 as a potent oncogene in ovarian cancer and emphasize the importance of PI3K signaling in this cancer.
Keywords: functional genomics, open reading frame, ORF
Abstract
High-grade serous ovarian cancers are characterized by widespread recurrent copy number alterations. Although some regions of copy number change harbor known oncogenes and tumor suppressor genes, the genes targeted by the majority of amplified or deleted regions in ovarian cancer remain undefined. Here we systematically tested amplified genes for their ability to promote tumor formation using an in vivo multiplexed transformation assay. We identified the GRB2-associated binding protein 2 (GAB2) as a recurrently amplified gene that potently transforms immortalized ovarian and fallopian tube secretory epithelial cells. Cancer cell lines overexpressing GAB2 require GAB2 for survival and show evidence of phosphatidylinositol 3-kinase (PI3K) pathway activation, which was required for GAB2-induced transformation. Cell lines overexpressing GAB2 were as sensitive to PI3K inhibition as cell lines harboring mutant PIK3CA. Together, these observations nominate GAB2 as an ovarian cancer oncogene, identify an alternative mechanism to activate PI3K signaling, and underscore the importance of PI3K signaling in this cancer.
Several histologic subtypes are associated with unique biological behaviors collectively called “ovarian cancer,” and serous tumors represent the majority of high-grade serous ovarian epithelial (HGSOE) cancers (1). The Cancer Genome Atlas (TCGA) has performed a large-scale, multiplatform genomic profiling study of primary HGSOE cancers, and the gene expression analysis from this study and others (2) demonstrated that at least four different molecular subtypes comprise clinically defined high-grade serous and endometrioid ovarian cancer. Genomic characterization of these tumors revealed ubiquitous copy number alterations as the dominant genetic alterations in ovarian cancer and a surprisingly small number of recurrent, significant mutations (3). Although these efforts confirmed recurrent copy number alterations in well-acknowledged driver genes, such as MYC and CCNE1, in a subset of serous ovarian cancers, the identity of the driver genes resident in the majority of the 63 focal and recurrent regions of amplification remain undefined.
Functional interrogation of somatically altered genes represents a complementary approach to large-scale structural genome characterization. We and others have performed large scale, loss-of-function short hairpin RNA (shRNA) screens to identify essential cancer genes and recently reported PAX8 and ID4 as ovarian cancer dependencies (4, 5). In other cancer types, both genome-wide and targeted loss-of-function studies were used to identify novel tumor suppressors in hepatocellular carcinoma (6) and epigenetic regulators in lymphomas (7). In addition, gain-of-function, cDNA-based approaches have uncovered novel driver roles for IKBKE (8) and PAK1 (9) in breast cancer, ERBB3 in endometrial cancer (10), and FGF19 in hepatocellular cancer (11). These studies demonstrate the utility of integrating evidence from both structural and functional assays to identify genes that represent tractable therapeutic targets.
Here we have developed and implemented a multiplexed in vivo transformation assay to identify genes recurrently amplified in HGSOE cancers that suffice to induce tumorigenic growth of immortalized human cells. These observations credential GAB2 as an ovarian cancer oncogene.
Results
Amplicon-Based Pooled in Vivo Transformation Screen.
To identify recurrently amplified genes that contribute to tumorigenicity in HGSOE cancers, we initiated a systematic study in which we used genome characterization data to identify recurrent amplified genes, created a lentivirally delivered collection of ORFs, and then screened for genes that induced tumorigenicity using a multiplexed in vivo transformation assay. We queried the copy number data generated by TCGA (3) to identify 1,017 recurrently amplified genes resident in the 63 recurrently amplified regions in HGSOE cancers. Using the Center for Cancer Systems Biology (CCSB)/Broad Institute lentiviral ORF expression collection (12), we created an arrayed collection of 587 ORFs representing 455 amplified ovarian genes (Dataset S1) including AKT1 that served as a positive control.
We previously showed that human embryonic kidney cells immortalized by expression of the human catalytic subunit of telomerase (hTERT) and the SV40 Early Region (SV40 ER) (HA1E) (13, 14) (HA1E) are rendered tumorigenic by the expression of HRAS (15) or the coexpression of constitutively active alleles of MEK1-DD (HA1E-M cells) and AKT1 (myristoylated-AKT1) (8). We used this experimental model to identify IKBKE and PAK1 as breast cancer oncogenes (8, 9). Here we introduced each of the 587 ORFs into HA1E-M cells by transducing each ORF separately in arrayed format. We created 26 pools composed of ORF-expressing cell lines representing 16–24 ORFs and implanted each group s.c. in six separate replicates in immunodeficient mice (Fig. 1A). We also tested a minipool containing individually infected HA1E-M-BRAFV600E, HA1E-M-HRASV12, and HA1E-M-KRASV12 and a mixture of HA1E-M-AKT1 cells with uninfected cells at 1:24 and 15:24 ratios as well as a pool of HA1E-M-EGFP cells mixed with uninfected HA1E-M cells at a 1:24 ratio. After 120 d, 11 of 26 (42%) injected pools formed tumors, including 1 pool that formed tumors at 3 of the 6 injected sites and 4 pools that formed tumors at 2 of the 6 injected sites (Fig. 1B). In total, we recovered 21 tumors from 149 injected sites for an overall tumor incidence of 14.1%.
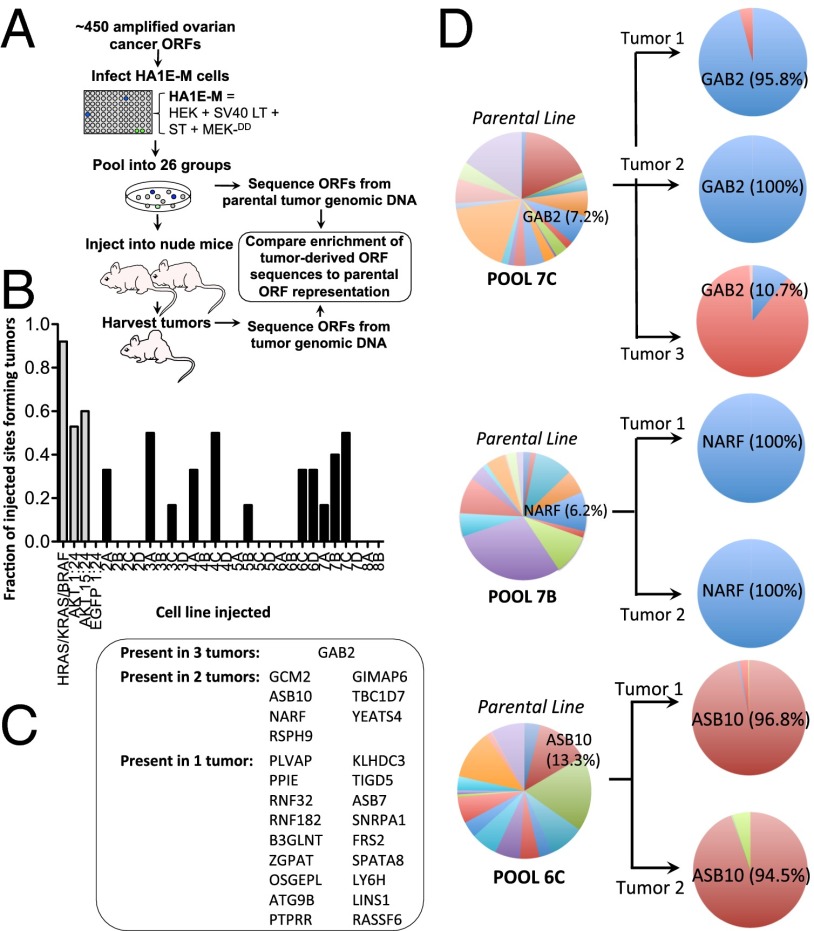
Fig. 1.
In vivo ORF transformation screen. (A) Schematic for the ORF screen. Individual ORFs were expressed in HA1E-M cells, pools created from these cells and implanted s.c. in immunodeficient mice. Genomic DNA from tumors was sequenced for ORF identification. (B) Tumor formation per ORF pool. Positive controls include a minipool containing individually infected HA1E-M-BRAFV600E, HA1E-M-HRASV12, and HA1E-M-KRASV12 and a mixture of HA1E-M-AKT1 cells with uninfected cells at 1:24 and 15:24 ratios. The negative control included HA1E-M-EGFP cells mixed with uninfected HA1E-M cells at a 1:24 ratio. (C) List of ORFs recovered in one, two, or three tumors. (D) Enrichment of ORFs from pools 7C, 7B, and 6C ORF sequence reads are shown as a percentage of the duplicated nonvector reads in either tumor or parental cell lines.
To determine the identities of the ORF sequences within each tumor, we used a massively parallel sequencing approach (SI Materials and Methods). We considered ORFs present if their sequences represented greater than 0.1% of the nonvector-aligned reads in the recovered tumor to identify weaker scoring genes in pools. Of the 455 ovarian cancer genes screened, 25 unique ovarian cancer ORF sequences were recovered. GAB2 was the only ORF identified in all three tumors that formed from its parental pool, whereas 7 ORFs were present in two tumors, and 18 ORFs were identified in a single tumor (Fig. 1C). Whereas GAB2 comprised 7.2% of the ORF sequences within the parental pool before injection, we found that GAB2 comprised nearly all of the tumor ORF sequences in two tumors and 10.7% of the ORF sequences in the third tumor from the same starting pool (Fig. 1D). Of the ORFs that scored in two tumors, we found that NARF represented 6.2% of the ORF sequences within the 7B parental pool but comprised all the sequences within the two resulting tumors from this pool. In pool 6C, ASB10 comprised 13.3% of the ORF sequences within the parental pool but 94.5% and 96.8% of the sequences within the two resulting tumors, respectively. These experiments illustrate the utility of a multiplexed in vivo approach to oncogene identification and identify several candidate oncogenes capable of transformation.
GAB2 Transforms Immortalized Ovarian and Fallopian Tube-Derived Cells.
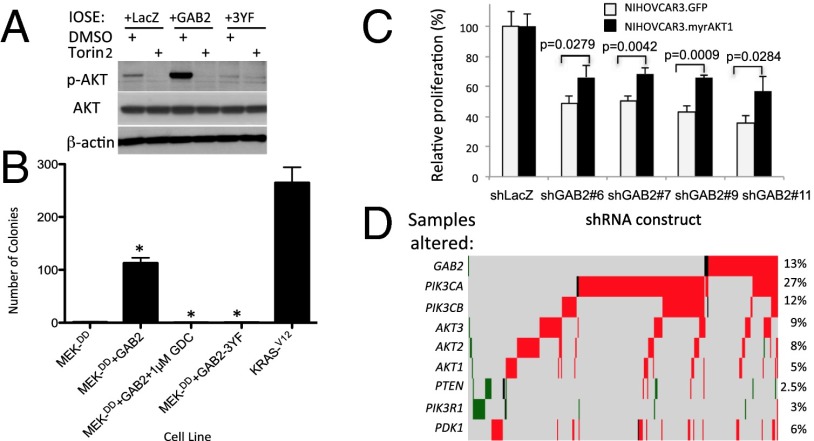
Prior work has implicated GAB2 in NeuNT-driven murine mammary tumorigenesis and migration (16, 17). However, GAB2 overexpression failed to induce the transformation of immortalized MCF10A cells (16, 17). To validate our screening results, we reintroduced GAB2 into HA1E-M cells and found that tumors formed in mice at the same rate (7 of 12, 58%) as that induced by the ovarian cancer oncogene, ID4 (5) (8 of 14, 57%). To determine whether GAB2 expression also transforms immortalized cells relevant to ovarian cancer, we introduced GAB2 into human immortalized ovarian surface epithelial (IOSE) cells expressing the SV40 ER, hTERT, and an activated MEK1-DD allele and immortalized fallopian tube secretory epithelial cells (FTSECs) expressing hTERT and the SV40 large and small T antigens (18). Expression of GAB2 induced significantly larger tumors than control or LACZ-expressing control IOSE cells when injected into immunodeficient NOD/IL2Rγc/SCID mice (Fig. 2A), and immortalized FTSEC expressing GAB2 formed significantly more anchorage-independent colonies than did control cells. FTSEC expressing both GAB2 and activated MEK1-DD formed nearly as many colonies as FTSEC expressing constitutively active KRASV12 (Fig. 2B). These observations confirmed the transforming function of GAB2 in lineage-matched, physiologically-relevant contexts in which GAB2 is expressed at high levels similar to those found in GAB2-overexpressing cell lines.
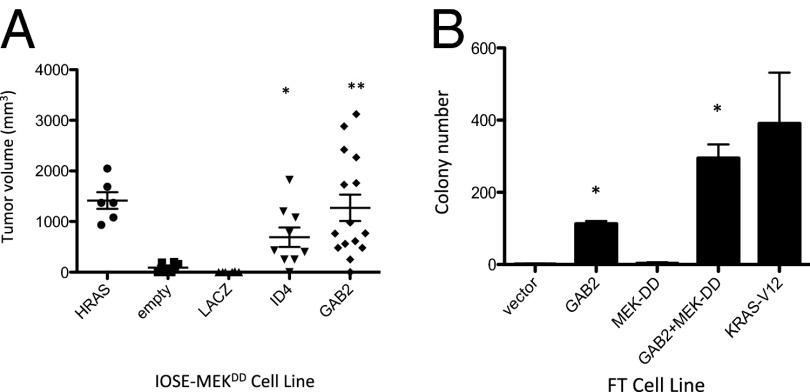
Fig. 2.
GAB2 overexpression promotes tumor formation. (A) IOSE-MEKDD cells alone or expressing HRASV12, ID4, LACZ, or GAB2 were assessed for tumor formation and tumor volume in NOD/IL2Rγc/scid mice. P values for transduced HA1E-M cell lines compared with HA1E-M “empty” cells are the following: HA1E-M-ID4, *P = 0.0286; and **HA1E-M-GAB2, P = 0.0110. (B) Immortalized FTSECs transduced with empty vector, GAB2, MEK-DD, GAB2 + MEK-DD, or HRASV12 were grown in soft agar and assessed for colony formation. P values for transduced FTSECs compared with FTSECs transduced with vector alone are *P < 0.0001 for FT-GAB2 and FT-GAB2-MEDK-DD. Error bars in A and B show standard deviation.
GAB2 Is Amplified and Overexpressed in Ovarian Cancer.
Prior work identified 11q14.1 as recurrently amplified in ovarian cancer (19), but several genes including CCND1, PAK1, and GAB2 have been nominated as potential targets of this amplification. We used the Genomic Identification of Significant Targets in Cancer Version 2.0 (GISTIC 2.0) algorithm (20) to identify recurrent copy number alterations in 562 samples within TCGA ovarian cancer dataset (3, 21). We found that GAB2 is amplified in 44% of samples. Among the samples that harbored this amplification, 24.2% demonstrated focal amplification, which reflects gain of a GAB2-containing region spanning less than 50% of the 11q arm. Furthermore, 13.7% of samples exhibited high-level gain, defined as amplification of more than 1 full copy of GAB2 (Fig. 3A). Secondly, GISTIC analysis of the 11q14.1 amplicon showed that GAB2 is located at the center of a peak that also harbors KCTD21 and USP35 that is telomeric to CCND1 and PAK1 (Fig. 3A). This region on 11q14.1 is the fourth most frequently amplified region in ovarian cancer (Fig. 3B). This observation revealed that GAB2 is a significant recurrently amplified gene in ovarian cancer. In addition, because several prior studies (16, 22) found that a subset of breast cancers harbored GAB2 amplification, we applied GISTIC analysis to the TCGA breast cancer dataset to assess the copy number status of GAB2. Across 846 samples within the TCGA breast cancer dataset, GAB2 is amplified in 24.7% of samples in which 15% harbor a focal amplification and 8.4% harbor a high-level amplification (21). This region of amplification is slightly larger than in ovarian cancer and also includes PAK1 (9). There are also a small number of lung cancers and glioblastoma that harbor GAB2 amplification. Collectively, these observations show that 11q14.1 is a major region of amplification in both ovarian and breast cancers and suggest that GAB2 is one of several targets of this region of copy number gain.
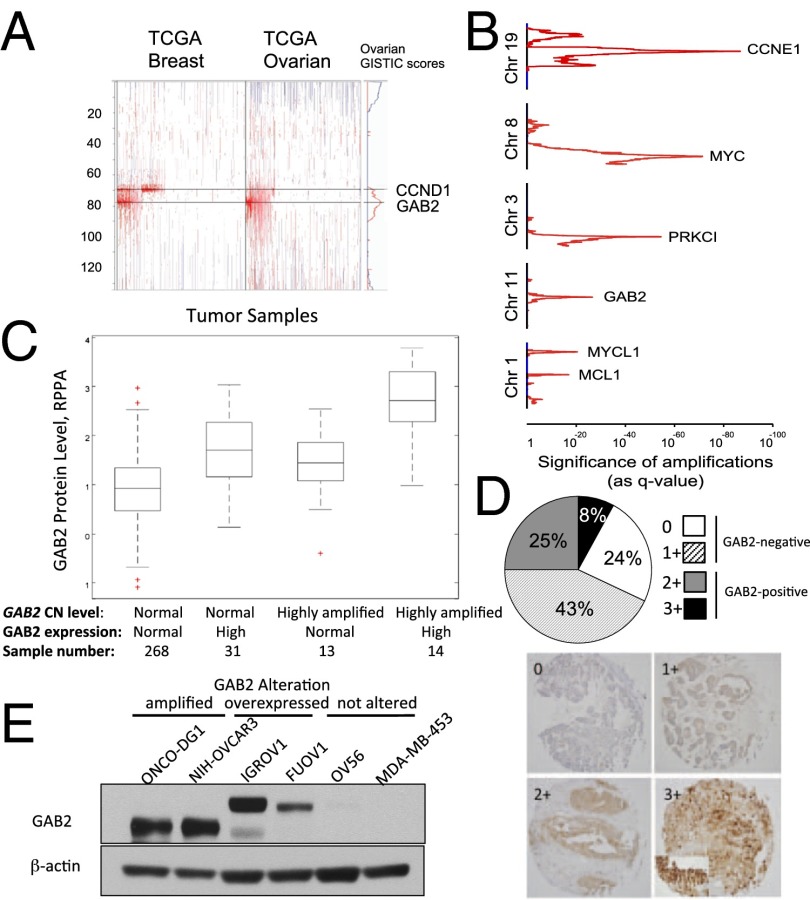
Fig. 3.
Amplification and overexpression of GAB2 in human ovarian cancers. (A) GISTIC analysis of TCGA copy number data showing GAB2 amplification on 11q14. Regions of amplification and deletion are shown in red and blue, respectively. (B) Ranking of gene peak regions of amplification in ovarian cancer by significance value analysis. (C) RPPA of TCGA ovarian cancer samples by copy number and gene expression levels. Box plots include values comprising the 25th–75th percentiles, and bars extend to extreme data values; red crosses are outliers. (D) Immunohistochemical analysis of GAB2 expression in ovarian cancer TMAs. Scores were assigned based on staining intensity: 0 (negative), 1 (weak), 2 (moderate), and 3 (strong) intensity. (E) Immunoblot analysis of GAB2 protein expression in ONCO-DG1, NIH:OVCAR3, IGROV1, FUOV1, OV56, and MDA-MB-453 cell lines.
To determine the relationship between GAB2 amplification, gene expression, and protein expression, we evaluated ovarian cancer samples in the TCGA for which there was available data on GAB2 protein levels by reverse phase protein array (RPPA). Of 326 analyzed samples, 58 of 326 (18%) displayed elevated protein expression and harbored elevated gene expression, gene amplification, or both characteristics (Fig. 3C). Of these high-expressing samples, 14 demonstrated the highest level of expression and were defined by both high GAB2 gene amplification and elevated levels of GAB2 mRNA. Having documented GAB2 overexpression by RPPA, we performed an independent analysis of protein expression via immunohistochemical analysis of GAB2 expression in tissue microarrays (TMAs) generated from patients with high-grade serous ovarian adenocarcinoma. Of 132 tumors assessed, we found 89 of 132 (67%) to be GAB2-negative with staining scores of 0 or 1+ (Fig. 3D). However, 43 of 132 (33%) samples were GAB2-positive (scores 2+ and 3+), with expression predominantly noted in the cytoplasm. These findings paralleled what we observed using RPPA. Finally, using the copy number and expression data in the Cancer Cell Line Encyclopedia (23), we identified ovarian cancer cell lines in which GAB2 was amplified and overexpressed (ONCODG1 and NIH:OVCAR3) and not amplified but overexpressed (IGROV1 and FUOV1). In contrast, the OV56 ovarian and MDA-MB-453 breast cancer cell lines expressed low levels of GAB2 (Fig. 3E). The lower molecular weight GAB2 seen in ONCODG1 and NIH:OVCAR3 likely represents a smaller isoform of GAB2 that lacks the first exon of the GAB2 gene and is likely translated from an internal methionine of the second exon (Fig. S1). Together, these results show that GAB2 is a significant target of amplification in human ovarian cancer and is both amplified and also overexpressed in human cancer cell lines and primary tumor samples.
Ovarian Cancer Cell Dependency on GAB2.
The finding that GAB2 induced tumorigenicity and is amplified and overexpressed in a subset of ovarian cancers suggested that GAB2 was one target of the 11q14 amplicon. We investigated whether cell lines that harbor GAB2 amplification and/or overexpression required GAB2 expression for proliferation to establish that GAB2 is one target of the amplicon on chromosome 11q. We used a panel of ovarian cancer cell lines that overexpress GAB2 (NIH:OVCAR3, FUOV-1, and IGROV1) and representative ovarian (OV56) and breast (MDA-MB-453) cancer cell lines with low GAB2 expression and expressed two independent shRNAs targeting GAB2 or a control shRNA construct targeting LacZ (shLACZ, Fig. 4A). Cell lines that overexpress GAB2 exhibited significantly decreased proliferation compared with cells expressing the shRNA targeting LACZ (Fig. 4B). In contrast, suppression of representative cell lines with low GAB2 expression failed to inhibit cell proliferation. Although these observations are representative of a range of cell lines tested, we also identified a subset of cell lines with lower GAB2 expression that were sensitive to GAB2 suppression (Fig. S2 A and B), likely due to the involvement of GAB2 in other cancer-relevant signaling pathways such as those driven by receptor tyrosine kinases. These observations support the notion that GAB2 represents a dependency in GAB2-altered ovarian cancer cell lines.
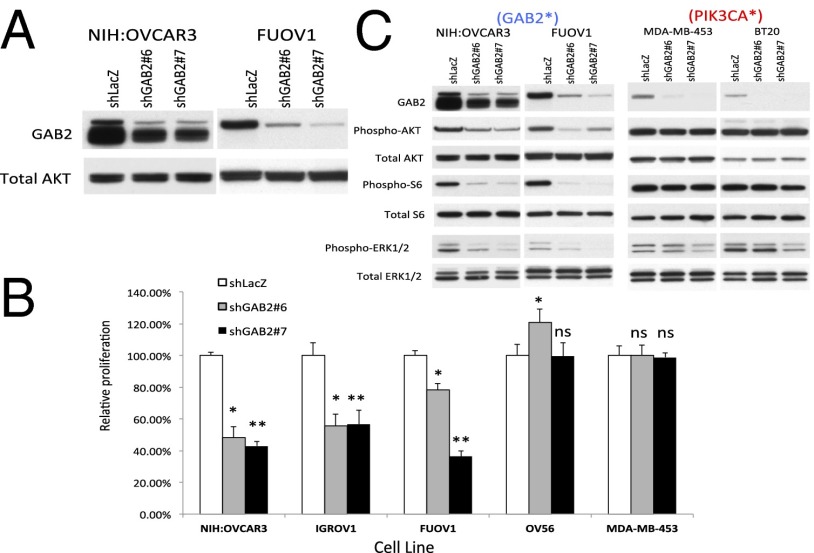
Fig. 4.
Effects of suppressing GAB2 expression. (A) Suppression of GAB2 by two independent GAB2-specific shRNAs, shGAB2 #6 and shGAB2 #7, in ovarian cancer cell lines NIH:OVCAR3 and FUOV1. (B) Effects of suppressing GAB2 expression on cell proliferation in a panel of ovarian, lung, and breast cancer cell lines that include high-expressing (NIH:OVCAR3, FUOV1, IGROV1, COV-362, NCI-H1435) and low-expressing (MDA-MB-453) cell lines. Statistically significant P values for cells expressing shGAB2 #6 (*) and shGAB2 #7 (**) compared with shLacZ control for each cell line are the following: NIH:OVCAR3, *P < 0.0001 and **P < 0.0001; IGROV1, *P < 0.0001 and **P < 0.0001; FUOV1, *P < 0.05 and **P < 0.0001; OV-56, shGAB2 #6 P < 0.0001 and shGAB2 #7 P = 0.8617; MD-MB-453, shGAB2 #6 P = 0.9847 and shGAB2 #7 P = 0.708 calculated by unpaired t test. (C) MAPK/ERK and PI3K pathway status following suppression of GAB2. GAB2*, GAB2 overexpressing; ns, not significant; PIK3CA*, PIK3CA mutant.
Signaling Pathways Activated by GAB2.
Prior work has implicated GAB2 as a signaling intermediate in both SHP2-dependent activation of MAPK signaling (16, 17) and activation of phosphatidylinositol 3-kinase (PI3K) (24). To assess the effects of suppressing GAB2 on these signaling pathways in cell lines expressing elevated or normal levels of GAB2, we interrogated the phosphorylation levels of PI3K/AKT/mTOR pathway components AKT1 and S6 as well as the MAPK pathway component ERK. When we suppressed GAB2 with 2 independent GAB2-specific shRNAs in NIH:OVCAR3 and FUOV1 cell lines, we found decreased levels of phospho-AKT1, phospho-S6, and phospho-ERK1/2 compared with cells expressing a control shRNA, shLacZ (Fig. 4C). In contrast, suppression of GAB2 in the MDA-MB-453 and BT20 cell lines, both of which express lower levels of GAB2 and also harbor PIK3CA mutations, failed to induce changes in the observed levels of phospho-AKT1 or phospho-S6, and induced only a minimal decrease in phospho-ERK1/2 levels. Also, suppression of GAB2 in two additional low-expressing lines, OV7 and OVK18, failed to induce changes in pathway activation (Fig. S2A). These observations suggest that the PI3K/AKT1/mTOR and MAPK pathways are activated by GAB2 selectively in cell lines that overexpress this protein.
PI3K Activation in GAB2-Mediated Transformation/Dependency.
“PI3Kness” appears to be a critical feature of both type I and type II ovarian cancers (25). However, analysis of the TCGA dataset revealed that alterations in known PI3K pathway components including PTEN, PIK3CA, AKT1, and AKT2 were present in only about one-third of ovarian cancer samples (3). Although prior studies have shown that GAB2-mediated migration of melanoma (24) and ovarian cancer cells (26) can be reduced by PI3K inhibitors, we hypothesized that GAB2 amplification represents an additional mechanism of PI3K activation in ovarian cancer. We tested whether increased GAB2 expression activated AKT1. GAB2 overexpression in IOSE cells led to increased serine 473 phosphorylation of AKT1 compared with IOSE cells expressing control LacZ or the GAB2-3YF mutant protein, which impairs the ability of GAB2 to recruit p85 (Fig. 5A). These observations show that increased GAB2 levels induced AKT1 phosphorylation, likely via the recruitment of the PIK3CA complex. This phosphorylation event required the activity of the mTOR complex; treatment of IOSE cells overexpressing GAB2 with Torin2, an inhibitor of both the mTORC1 and mTORC2 complexes (27), abrogated AKT1 phosphorylation by GAB2. These data suggest that the mTOR complex is also required for the GAB2-mediated activation of AKT1. Having demonstrated that GAB2 activates AKT1 in a p85 and mTOR-dependent manner, we next tested whether GAB2-mediated AKT1 activation was necessary for the transformation of FTSEC. Compared with control FTSECs expressing only the activated MEKDD allele, FTSEC overexpressing MEKDD and GAB2 formed 90-fold more anchorage-independent colonies (Fig. 5B). We then tested the contributions of components of the PI3K pathway contributions to GAB2-mediated transformation using two approaches. First, FTSEC expressing both MEKDD and the p85-binding mutant GAB2-3YF formed significantly fewer colonies than WT GAB2-expressing cells. Secondly, FTSEC expressing MEKDD as well as WT GAB2 formed background levels of colonies when treated with the PI3K inhibitor GDC-0491 at 1 μM. In contrast, overexpression of a GAB2 mutant protein with impaired Shp2 binding failed to impair transformation in the HA1EM background. Together, these data show that PI3K pathway activation is required for GAB2-mediated transformation in a model of ovarian cancer using physiologically relevant cells of origin.
Fig. 5.
Effects of GAB2 expression on activation of the PI3K signaling. (A) mTOR-mediated phosphorylation of AKT following GAB2 expression in IOSE cell lines. (B) PI3K dependence of transformed FTSECs expressing GAB2. FTSECs overexpressing wild-type GAB2 were treated with PI3K inhibitor GDC-0941. Overexpression of the GAB2 p85 binding mutant (GAB2-3YF) was also tested. *P < 0.0001. (C) Rescue of GAB2 knockdown in NIH:OVCAR3 by overexpressing myristoylated, constitutively active ATK1. (D) Heat map of TCGA ovarian cancer samples with alterations in GAB2, PIK3CA, AKT1, or PTEN demonstrating statistically significant mutual exclusivity of GAB2 amplification and either PIK3CA amplification or PTEN loss. Red bars indicate amplification, black bars indicate mutation, and green bars indicate deletion. In C and D, error bars reflect standard deviation.
We extended these observations by exploring whether independent activation of the downstream PI3K/AKT pathway could rescue cells dependent upon GAB2 function in which GAB2 levels had been suppressed. We introduced a myristoylated, constitutively active form of AKT1 (myrAKT1) or a control vector into NIH:OVCAR3 cells and subsequently expressed a control lacZ shRNA or four independent GAB2-specific shRNAs and examined for differences in cell proliferation. Compared with GFP-expressing NIH:OVCAR3 cells in which we had suppressed GAB2, NIH:OVCAR3 cells overexpressing constitutively active myrAKT1 showed an ∼20% increase in proliferation that was statistically significant with each GAB2-specific shRNA tested (Fig. 5C). These observations suggest that GAB2 likely acts, at least in part, through AKT1.
Finally, because GAB2 activates the PI3K pathway, we investigated the pattern of genomic alterations of PI3K signaling in primary ovarian cancers. Hanrahan et al. (28) recently reported a substantial number of PI3K pathway alterations in ovarian cancer. We extended this analysis by analyzing a substantially larger number the number of samples and also incorporating GAB2 in our analysis. Specifically, we analyzed the ovarian cancer TCGA dataset to determine the incidence and type of alterations in PIK3CA, PIK3CB, GAB2, AKT1, AKT2, AKT3, PTEN, and PDK1. Of the 562 samples analyzed, we found that nearly 54% harbored significant copy number alterations or mutations in these genes. Similar to our prior analysis, 13% of samples harbored GAB2 amplifications, some of which co-occurred with amplification of other pathway components. These observations suggest that amplification of GAB2 also leads to activation of PI3K signaling.
Sensitivity of GAB2-Altered Cell Lines to PI3K Inhibition.
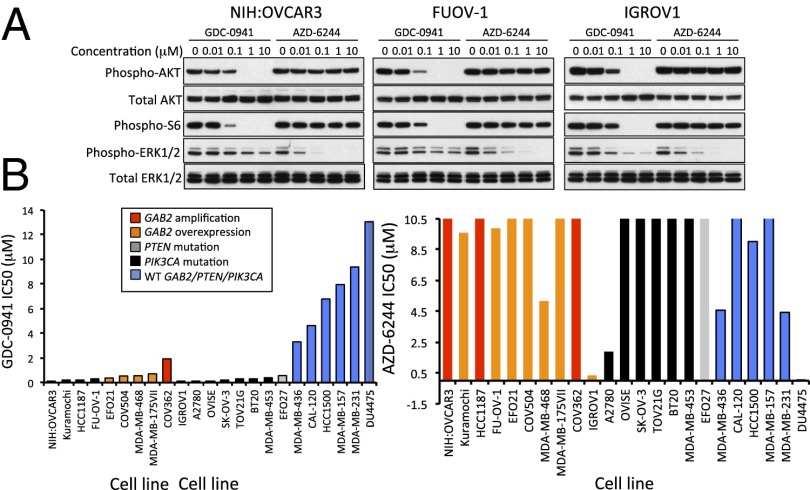
Having shown that the PI3K pathway was required for GAB2-mediated transformation and that activated AKT1 could partially rescue GAB2-dependent cell lines depleted of GAB2, we next determined whether GAB2 alterations represented a feature that correlated with enhanced sensitivity to PI3K pathway inhibition. Several studies suggest that PI3K inhibition attenuated GAB2-dependent migration phenotypes (24, 26) and pointed to the types of pathway alterations that activate AKT1 (28). However, although recent work has been directed at correlating PI3K pathway alterations with sensitivity to pathway inhibition in breast cancer (29), it is unclear whether increased GAB2 expression increases cell sensitivity to PI3K pathway inhibition in a manner similar to the sensitivity conferred by canonical mutations in PIK3CA and PTEN. We performed dose–response experiments to determine the IC50 of inhibitors against PI3K (GDC-0941) or MEK1 (AZD-6244) in a panel of 12 ovarian and 11 breast cancer cell lines 3 d posttreatment. In GAB2-overexpressing cell lines, GDC-0941 treatment abolished levels of both phospho-AKT1 and phospho-S6, and AZD-6244 treatment abolished phospho-ERK 1/2 (Fig. 6A). We observed similar effects in cell lines with low GAB2 expression (Fig. S2A). We observed that 10 cell lines that harbored amplification/overexpression of GAB2 formed a distinct subgroup from 9 cell lines that harbored mutation of PIK3CA or loss of PTEN, with the exception of IGROV1 cells that harbor both loss of PTEN and overexpression of GAB2 (Fig. 6B). Using nonlinear regression followed by comparison of the best-fit parameters, we found that the mean IC50 of GDC-0941 were comparable between the cell lines harboring amplification/overexpression of GAB2 (0.485 ± 0.553 μM) and the cell lines harboring activating mutations of PIK3CA or loss of PTEN (0.583 ± 1.032 μM). In contrast, the five cell lines with normal GAB2 or PIK3CA/PTEN exhibit significantly higher IC50 of GDC-0941 (mean ± SD = 8.3 ± 3.1 μM) compared with cell lines with alterations in GAB2 or PIK3CA/PTEN. These observations suggest that GAB2-overexpressing cell lines exhibit sensitivity to PI3K pathway inhibition that is similar to cell lines harboring other PI3K pathway alterations.
Fig. 6.
Effects of a PI3K inhibitor on cell proliferation. (A) Effect of increasing doses of the PI3K inhibitor GDC-0941 or the MEK inhibitor AZD-6244 on AKT1, ERK1/2, and S6 phosphorylation in GAB2-overexpressing cell lines NIH:OVCAR3, FUOV-1, and IGROV1. (B) Half-maximal inhibitory concentrations (IC50) for GDC-0941 (Left) and AZD-6244 (Right) in 23 ovarian and breast cancer cell lines. Genomic alterations are depicted by color as shown in Inset.
Discussion
Here we screened 455 genes amplified in ovarian cancer for the ability to transform an immortalized human cancer cell line using a pooled high-throughput in vivo approach and identified 26 genes that induced tumorigenicity. We demonstrated that GAB2 induces the transformation of ovarian and fallopian tube cells by activating PI3K signaling. Ovarian cell lines that harbor GAB2 amplifications or overexpress GAB2 are dependent on GAB2 for proliferation and exhibit sensitivity to PI3K inhibition. These observations identify GAB2 as an ovarian cancer oncogene and underscore the importance of PI3K signaling in ovarian cancer.
Multiplexed in Vivo Transformation Screens.
We used a human embryonic kidney cell line expressing the activated MEK-DD gene to facilitate a high-throughput, stringent in vivo transformation screen. This model is useful due to its rapid in vivo growth kinetics for oncogene discovery and its low background transformation rate. The HA1E-M model expresses an active MEK allele, thereby facilitating the identification of PI3K-activating genes such as GAB2. Further systematic follow-up will be necessary to validate additional candidate oncogenes identified using this system that may vary in strength of transforming capacity. We note that GAB2 also induced transformation in both ovarian and fallopian tube epithelial cells indicating that the HA1E-M model enabled the identification of ovarian cancer drivers. Additional oncogenes may be uncovered if screened in a cellular context emphasizing other signaling cascades. Although we describe a gain-of-function approach focused on oncogene discovery in ovarian cancer, this methodology can be applied to any cancer phenotype. We used a next-generation sequencing approach that allowed us both to identify ORFs within tumors and quantitate their enrichment. This approach could be applied easily to a range of scalable ORF-expressing pool sizes and phenotypic readouts using ORF collections that do not require additional barcoded cassettes. The observation that one of the three GAB2-scoring tumors displayed a lower level of GAB2 enrichment is likely due to the stochastic selection of clones that eventually form tumors at each site, owing either to a slight variation in starting pooled inoculum or interactions with the local microenvironment. In addition to GAB2, we also identified additional candidates, such as NARF and ASB10, as genes that induced tumors when overexpressed in HA1E-M cells that merit further investigation. Taken together, these studies provide further evidence that large-scale functional genomics approaches complement ongoing structural approaches to decipher genes and pathways involved in cancer pathogenesis.
Features of the 11q14 Amplicon.
GAB2 resides on the fourth most significant amplicon in high-grade serous ovarian cancer (3), the focal peak located on 11q14.1. This amplified region has been identified as a recurrent alteration in breast (9, 22) and ovarian cancer (19) as well as in metastatic melanoma (24). Across over 6,300 different TCGA samples, this region is among the 26 most amplified regions in all cancers (21). This amplicon is slightly broader in breast cancer, encompassing both GAB2 and PAK1 (9, 21) as well as the nearby region harboring CCND1. Whereas GAB2 amplification does not appear to be correlated with survival, there is a significant association with GAB2 amplification and the Her2-enriched (P = 0.042) and luminal B (P = 0.028) breast cancer subsets described by TCGA. In ovarian cancer, the 11q14.1 amplicon at is smaller and centered over GAB2. Although prior studies in ovarian cancer have highlighted the 11q14.1 amplification (3, 19), the identities of the candidate gene targets of amplification were not well clarified. These studies do not preclude the possibility that several genes on the same amplicon may cooperate to drive oncogenic programs. GAB2 may also contribute to the pathogenesis of individual ovarian cancers in which the center of the region of copy number gain does not fall on GAB2 itself but on neighboring candidate genes, such as RSF1 (30), PAK1 (9, 19), or CCND1. Also, there are a significant number of samples that harbor elevated GAB2 gene expression in the absence of amplification, supporting the idea that amplification is one of several mechanisms underlying GAB2 protein overexpression and tumorigenesis. We note that GAB2 is somatically mutated in four TCGA samples, although these amino acid changes do not affect known functional domains.
GAB2 Alterations in Cancer.
GAB2 plays critical roles in several different cancers. GAB2 is required for BCR/ABL-mediated transformation in chronic myeloid leukemia (CML) (31) and also HER2-mediated mammary carcinogenesis through ERK activation (16). Moreover, GAB2 is overexpressed in breast cancer (9, 22) and some metastatic melanomas (24) and promotes survival in breast cancer (22) and migration in both malignancies (17, 24). Several ovarian cancer cell lines also overexpress GAB2, which was linked to increased migration and the epithelial-mesenchymal transition (26). Davis et al. also demonstrated that a panel of ovarian cancer cell lines with GAB2 amplification were susceptible to siRNA-mediated GAB2 knockdown (32), consistent with our findings. Although these prior studies suggest that GAB2 expression is altered in cancer, the observations presented herein show that GAB2 is a bona fide oncogene important for both tumor initiation and maintenance.
GAB2-Mediated Signaling and the PI3K Pathway.
Recent work suggests that over 70% of ovarian cancers exhibit activation of the PI3K pathway (33), but there are likely more alterations that influence this pathway than have been identified. We identified PI3K pathway alterations in 54% of the high-grade serous ovarian cancer samples we analyzed from the TCGA dataset. PIK3CA amplification was a common event and has been seen in other cancers (34, 35). Co-occurrence of GAB2 amplification and PI3K pathway component alteration was observed in a subset of ovarian cancer samples. Co-occurrence of altered PI3K pathway components has been observed in other PI3K-altered cancer types and could potentiate PI3K-mediated signaling. Based on our findings, GAB2 overexpression represents a subset of pathway alterations that result in dysregulated PI3K signaling. Several studies have addressed PI3K/AKT pathway inhibition in ovarian cancer. In one study, a subset of ovarian cancer cell lines was sensitive to inhibition of AKT1 and AKT2, whereas those expressing AKT3 required inhibition of all three isoforms (28). In a separate study, modest clinical responses were seen in a phase-I trial incorporating mTOR inhibitors in the patients with PIK3CA mutant compared with PIK3CA wild-type cancers (36). These findings point to the importance of careful genetic annotation in tailoring pathway-specific therapies. Our findings suggest that cell lines characterized as GAB2 overexpressing are as sensitive to PI3K pathway inhibition as cell lines harboring classical PIK3CA mutations, and subsequent work has shown that additional cell lines with low GAB2 expression, OV7 and OVK18, exhibit nearly fourfold higher IC50 values in response to PI3K inhibitor treatment. These observations provide a rationale to consider inhibition of this pathway in human ovarian cancers as well as breast cancers and metastatic melanomas exhibiting appropriate pathway-specific genomic features.
Adapter Proteins as Therapeutic Targets.
In addition to its ability to activate the PI3K pathway, GAB2 can also activate the ERK pathway through Shp2. In a mammary epithelial cell model, NeuNT-driven multiacinar structure formation through GAB2 requires Shp2/ERK signaling (16). However, we failed to observe a decrease in transformation using a GAB2 mutant unable to bind Shp2 and did not find that ovarian cancer cell lines were as sensitive to MEK inhibition as they were to PI3K inhibition. Together, these observations suggest that signaling events downstream of GAB2 may be context or lineage specific. Further work will be necessary to address these possibilities.
Adapter proteins amplify receptor-initiated signaling events by recruiting downstream modular signaling proteins. Thus, genes such as GAB2 and CRKL (37) are powerful transforming oncogenes because of the number of pathways influenced by their overexpression. As was demonstrated in CRKL-overexpressing mutant EGFR lung cancer cells resistant to EGFR inhibition (37), cells overexpressing GAB2 may be resistant to inhibitors of upstream receptor tyrosine kinases, such as HER2, with which GAB2 associates. Adaptor proteins may represent a class of cancer-relevant targets that warrant further study. The observation that GAB2 copy number alterations correlate with sensitivity to PI3K pathway inhibition supports the prospective annotation of GAB2 amplification or elevated GAB2 protein expression in clinical trials of PI3K inhibitors.
Materials and Methods
Pooled in Vivo ORF Screen.
Five hundred eighty-eight ORFs representing genes recurrently amplified in the ovarian cancer TCGA dataset as well as controls were obtained from the CCSB/Broad Institute ORF collection (12). HA1E-M cells were infected in arrayed fashion, pooled, and injected into three sites each in two NCr nude mice (Taconic) per pool. Growing tumors were harvested, and ORFs were amplified from genomic DNA and recombined into common vectors, and pooled recombined plasmids were recovered from transformed bacteria and subjected to next-generation sequencing. Extended details are described in SI Materials and Methods.
Additional Materials and Methods.
Plasmids, cell lines and reagents, chemicals, immunoblotting, cell proliferation and anchorage-independence assays, TMA and immunohistochemistry, genomics/proteomics analysis, and animal injections are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank John Doench, Glenn Cowley, and Carsten Russ from the Broad Institute for helpful discussions. The following grant support is acknowledged: RC2 CA148268 (to W.C.H.), U01 CA176058 (to W.C.H.), U54 CA143798 (to R.B.) U54 CA112962 (to W.C.H.), NIH R25 (to G.P.D.), the American Brain Tumor Association (G.P.D.), a Canadian Institutes of Health Research Fellowship (to A.M.K.), a Kaleidoscope of Hope Foundation Young Investigator Research Award (to A.M.K.), U01 CA152990 (to R.D.), The Mary Kay Foundation (R.D.), The Sandy Rollman Ovarian Cancer Research Foundation (R.D.), The Robert and Debra First Fund (R.D.), The Executive Council of the Susan Smith Center for Women’s Cancers at Dana–Farber Cancer Institute (R.D.), The V Foundation for Cancer Research (H.W.C.), American Cancer Society Institutional Research Grant IRG-97-219-14 (to H.W.C.), and The Marsha Rivkin Center for Ovarian Cancer Research (H.W.C.).
Footnotes
Conflict of interest statement: R.B. and W.C.H. are consultants to Novartis as noted in this work.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311909111/-/DCSupplemental.
References
- 1.Soslow RA. Histologic subtypes of ovarian carcinoma: An overview. Int J Gynecol Pathol. 2008;27(2):161–174. doi: 10.1097/PGP.0b013e31815ea812. [DOI] [PubMed] [Google Scholar]
- 2.Tothill RW, et al. Australian Ovarian Cancer Study Group Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14(16):5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 3. Cancer Genome Atlas Network (2011) Integrated genomic analyses of ovarian carcinoma. Nature 474(7353):609–615. [DOI] [PMC free article] [PubMed]
- 4.Cheung HW, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci USA. 2011;108(30):12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ren Y, et al. Targeted tumor-penetrating siRNA nanocomplexes for credentialing the ovarian cancer oncogene ID4. Sci Transl Med. 2012;4(147):ra112. doi: 10.1126/scitranslmed.3003778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zender L, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135(5):852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rui L, et al. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell. 2010;18(6):590–605. doi: 10.1016/j.ccr.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boehm JS, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129(6):1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 9.Shrestha Y, et al. PAK1 is a breast cancer oncogene that coordinately activates MAPK and MET signaling. Oncogene. 2012;31(29):3397–3408. doi: 10.1038/onc.2011.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang H, et al. Whole-exome sequencing combined with functional genomics reveals novel candidate driver cancer genes in endometrial cancer. Genome Res. 2012;22(11):2120–2129. doi: 10.1101/gr.137596.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sawey ET, et al. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by Oncogenomic screening. Cancer Cell. 2011;19(3):347–358. doi: 10.1016/j.ccr.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang X, et al. A public genome-scale lentiviral expression library of human ORFs. Nat Methods. 2011;8(8):659–661. doi: 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn WC, et al. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol Cell Biol. 2002;22(7):2111–2123. doi: 10.1128/MCB.22.7.2111-2123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Counter CM, et al. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc Natl Acad Sci USA. 1998;95(25):14723–14728. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahn WC, Weinberg RA. Rules for making human tumor cells. N Engl J Med. 2002;347(20):1593–1603. doi: 10.1056/NEJMra021902. [DOI] [PubMed] [Google Scholar]
- 16.Bentires-Alj M, et al. A role for the scaffolding adapter GAB2 in breast cancer. Nat Med. 2006;12(1):114–121. doi: 10.1038/nm1341. [DOI] [PubMed] [Google Scholar]
- 17.Ke Y, et al. Role of Gab2 in mammary tumorigenesis and metastasis. Oncogene. 2007;26(34):4951–4960. doi: 10.1038/sj.onc.1210315. [DOI] [PubMed] [Google Scholar]
- 18.Karst AM, Levanon K, Drapkin R. Modeling high-grade serous ovarian carcinogenesis from the fallopian tube. Proc Natl Acad Sci USA. 2011;108(18):7547–7552. doi: 10.1073/pnas.1017300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown LA, et al. Amplification of 11q13 in ovarian carcinoma. Genes Chromosomes Cancer. 2008;47(6):481–489. doi: 10.1002/gcc.20549. [DOI] [PubMed] [Google Scholar]
- 20.Mermel CH, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. The Broad Institute Portal Team (2012) TCGA Copy Number Portal. Available at www.broadinstitute.org/tcga/home. Accessed November 20, 2012 (data freeze)
- 22.Bocanegra M, et al. Focal amplification and oncogene dependency of GAB2 in breast cancer. Oncogene. 2010;29(5):774–779. doi: 10.1038/onc.2009.364. [DOI] [PubMed] [Google Scholar]
- 23.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horst B, et al. Gab2-mediated signaling promotes melanoma metastasis. Am J Pathol. 2009;174(4):1524–1533. doi: 10.2353/ajpath.2009.080543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bast RC, Jr, Mills GB. Dissecting “PI3Kness”: The complexity of personalized therapy for ovarian cancer. Cancer Discov. 2012;2(1):16–18. doi: 10.1158/2159-8290.CD-11-0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Sheng Q, Spillman MA, Behbakht K, Gu H. Gab2 regulates the migratory behaviors and E-cadherin expression via activation of the PI3K pathway in ovarian cancer cells. Oncogene. 2012;31(20):2512–2520. doi: 10.1038/onc.2011.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu L, et al. Characterization of Torin2, an ATP-competitive inhibitor of mTOR, ATM, and ATR. Cancer Res. 2013;73(8):2574–2586. doi: 10.1158/0008-5472.CAN-12-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanrahan AJ, et al. Genomic complexity and AKT dependence in serous ovarian cancer. Cancer Discov. 2012;2(1):56–67. doi: 10.1158/2159-8290.CD-11-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Brien C, et al. Predictive biomarkers of sensitivity to the phosphatidylinositol 3′ kinase inhibitor GDC-0941 in breast cancer preclinical models. Clin Cancer Res. 2010;16(14):3670–3683. doi: 10.1158/1078-0432.CCR-09-2828. [DOI] [PubMed] [Google Scholar]
- 30.Shih IeM, et al. Amplification of a chromatin remodeling gene, Rsf-1/HBXAP, in ovarian carcinoma. Proc Natl Acad Sci USA. 2005;102(39):14004–14009. doi: 10.1073/pnas.0504195102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sattler M, et al. Critical role for Gab2 in transformation by BCR/ABL. Cancer Cell. 2002;1(5):479–492. doi: 10.1016/s1535-6108(02)00074-0. [DOI] [PubMed] [Google Scholar]
- 32.Davis SJ, et al. Functional analysis of genes in regions commonly amplified in high-grade serous and endometrioid ovarian cancer. Clin Cancer Res. 2013;19(6):1411–1421. doi: 10.1158/1078-0432.CCR-12-3433. [DOI] [PubMed] [Google Scholar]
- 33.Bast RC, Jr, Hennessy B, Mills GB. The biology of ovarian cancer: New opportunities for translation. Nat Rev Cancer. 2009;9(6):415–428. doi: 10.1038/nrc2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi J, et al. Highly frequent PIK3CA amplification is associated with poor prognosis in gastric cancer. BMC Cancer. 2012;12:50. doi: 10.1186/1471-2407-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cancer Genome Atlas Network (2012) Comprehensive molecular portraits of human breast tumours. Nature 490(7418):61–70. [DOI] [PMC free article] [PubMed]
- 36.Janku F, et al. PI3K/AKT/mTOR inhibitors in patients with breast and gynecologic malignancies harboring PIK3CA mutations. J Clin Oncol. 2012;30(8):777–782. doi: 10.1200/JCO.2011.36.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheung HW, et al. Amplification of CRKL induces transformation and epidermal growth factor receptor inhibitor resistance in human non-small cell lung cancers. Cancer Discov. 2011;1(7):608–625. doi: 10.1158/2159-8290.CD-11-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.