Abstract
We report here the solid phase synthesis of RNA and DNA oligonucleotides containing the 2′-selenium functionality for X-ray crystallography using multiwavelength anomalous dispersion. We have synthesized the novel 2′-methylseleno cytidine phosphoramidite and improved the accessibility of the 2′-methylseleno uridine phosphoramidite for the synthesis of many selenium-derivatized DNAs and RNAs in large scales. The yields of coupling these Se-nucleoside phosphoramidites into DNA or RNA oligonucleotides were over 99% when 5-(benzylmercapto)-1H-tetrazole was used as the coupling reagent. The UV melting study of A-form dsDNAs indicated that the 2′-selenium derivatization had no effect on the stability of the duplexes with the 3′-endo sugar pucker. Thus, the stems of functional RNA molecules with the same 3′-endo sugar pucker appear to be the ideal sites for the selenium derivatization with 2′-Se-C and 2′-Se-U. Crystallization of the selenium-derivatized oligonucleotides is also reported here. The results demonstrate that this 2′-selenium functionality is suitable for RNA and A-form DNA derivatization in X-ray crystallography.
INTRODUCTION
Nucleic acids play a variety of important roles in biological systems, including the transfer and regulation of genetic information, and the RNA catalytic functions found in vivo in protein synthesis and in the life cycle of some viruses (1,2). RNAs and DNAs with catalytic and binding functions have also been identified via in vitro selection (3). Furthermore, the recent discovery of noncoding small RNAs in diverse organisms has enormously expanded the repertoire of functions of nucleic acids (4,5). This vast array of biologically active RNAs and DNAs has promoted a new front of research in the field of structural analysis to elucidate their three-dimensional (3D) structure and function relation ships. To date, in combination with synchrotron radiation, multiwavelength anomalous dispersion (MAD) and new methodologies in introducing anomalously scattering atoms into molecules, X-ray crystallography has become the most powerful and widely used technique for the 3D structure determination of proteins, DNAs and RNAs (6–9). The area of protein X-ray crystallography has been revolutionized by replacing methionine with selenomethionine (10,11). The use of selenomethionyl proteins has proved to be excellent for MAD phasing by the exploitation of the unique absorption K edge of selenium (12.6578 keV, 0.9795 Å), which is readily accessible by synchrotron radiation (7,11).
On the other hand, DNA and RNA X-ray structure determination has been more challenging due to inherent difficulties in creating suitable heavy atom derivatives for MAD phasing. Currently, there are three conventional methods for creating heavy atom derivatives for nucleic acids: (i) soaking and co-crystallization with heavy metal ions; (ii) RNA derivatization via selenomethionyl proteins; (iii) derivatization with halogens. Soaking and co-crystallization have not been very successful in nucleic acid X-ray crystallography, as opposed to the scenario in proteins, probably because of nonspecific binding of heavy metal ions and backbone cleavage by the ions. The derivatization of RNAs using the selenomethionyl protein U1A has been recently explored by Doudna’s research group. Although it has led to the successful elucidation of two ribozyme structures by X-ray crystallography (12,13), this method is labor-intensive. In addition, the utilization of selenomethionyl proteins that bind intrinsically to RNAs has facilitated the elucidation of a few other RNA crystal structures (14,15). Similarly, halogens (bromine or iodine) have been pursued as potential X-ray scattering centers in nucleic acids. The phosphoramidites of 5-halogen-uridine, 5-halogen-2′-deoxyuridine (a mimic of thymidine) and 5-halogen-2′-deoxycytidine have been synthesized for the halogen derivatization of DNA and RNA oligonucleotides via solid phase synthesis (16). However, there are two major problems associated with halogen derivatives. First, since the chemical incorporation of halogens is primarily limited to the 5-position of pyrimidine nucleosides, this lack of multiple choices of positions may limit derivatization freedom to avoid base-stacking disruptions, other structural perturbations, and possible crystallizability problems. Secondly, recent reports have indicated that the light sensitivity of the halogen derivatives can lead to their decomposition after long-time exposure to X-ray (17,18).
In an effort to increase the repertoire of anomalously scattering atoms for MAD phasing in nucleic acid X-ray crystallography, Huang, Egli and co-workers have undertaken chemical and enzymatic approaches to covalently introduce selenium into DNAs and RNAs (19–23). We report here the derivatization of biological function-related DNAs and RNAs using our novel 2′-methylseleno cytidine phosphoramidite and the previously reported 2′-methylseleno uridine phosphoramidite (20). We also present major improvements in the synthesis of 2′-methylseleno uridine phosphoramidite as well as new advances in the highly efficient synthesis of Se-DNA and Se-RNA oligonucleotides using 5-(benzylmercapto)-1H-tetrazole (5-BMT) as the coupling reagent. Crystallization of some selenium-derivatized oligonucleotides is also described.
MATERIALS AND METHODS
Synthesis of the 2′-selenium-derivatized uridine and cytidine phosphoramidites
2,2′-O-anhydro-1-(β-d-arabinofuranosyl)-uracil (2). Uridine (50 g, 201 mmol) and diphenyl carbonate (48 g, 224 mmol) were placed in a round flask, and N,N-dimethylformamide (50 ml) was added. The slurry was heated in an oil bath at 100°C. Dry sodium bicarbonate (400 mg) was then added, and a watch glass was used to cover the flask. The reaction mixture was heated at 130–140°C for 1 h while being monitored on thin layer chromatography (TLC) (methanol/methylene chloride, 2:8). After completion, the reaction was cooled to room temperature, filtered, and washed by methanol three times (each time 15 ml). The white powdered product was dried on high vacuum overnight (37.9 g, 82%; wt 226). 1H-NMR (d6-DMSO) δ (p.p.m.): 3.09–3.21 (m, 2H, H-5′), 3.95–4.11 (m, 1H, H-4′), 4.25–4.41 (m, 1H, H-3′), 4.91 (m, HO), 5.10 (d, J = 6.5 Hz, 1H, H-2′), 5.81 (m, HO), 5.96 (d, J = 7.5 Hz, 1H, H-5), 6.22 (d, J = 5.7 Hz, 1H, H-1′), 7.78 (d, J = 7.5 Hz, 1H, H-6).
2,2′-anhydro-1-[2′-deoxy-5′-O-(4,4-dimethoxytrity1)-β-d-arabinofuranosyl]-uracil (3). Dimethoxytrityl chloride (18.6 g, 55.0 mmol) and 2,2′-O-anhydrouridine (12.4 g, 54.9 mmol) were placed in a 250 ml round flask and dried on high vacuum for half an hour before addition of dry pyridine (110 ml, 0.5 M). The reaction was stirred for 3 h at room temperature and monitored by TLC (methanol/methylene chloride = 7.5%). After the reaction was complete, MeOH (10 ml) was added to quench the reaction, and the mixture was stirred for 10 min. NaCl (50 ml, sat.) and ethyl acetate (EtOAc, 100 ml) were then added to the flask. The organic phase was removed and the aqueous phase was extracted with EtOAc (3 × 100 ml). The combined organic phase was dried over anhydrous MgSO4 for 30 min, the salt was filtrated, and the organic solvents were evaporated under reduced pressure. The residue was purified on a silica gel column (equilibrated with EtOAc) and eluted with a methanol/EtOAc gradient [EtOAc, and CH3OH in EtOAc (0–10%)]. The fractions containing the product were combined, evaporated and dried on high vacuum overnight to yield a white foamy product (78% yield; 22.7 g, wt 528). UV (in acetonitrile), λmax: 233.8, 281.2 nm. IR (KBr): 3400 (br), 3030, 2920, 2850, 1670, 1520, 1510, 1490, 1460, 1260, 1190, 1095, 1055, 820, 770, 705, 580 cm–1. 1H-NMR (CDCl3) δ (p.p..m.): 2.99–3.18 (m, 2H, H-5′), 3.69 (s, 6H, 2×CH3O), 4.29–4.36 (m, 1H, H-4′), 4.42–4.46 (m, 1H, H-3′), 5.20–5.25 (m, 1H, H-2′), 5.92 (d, J = 7.5 Hz, 1H, H-5), 6.07 (d, J = 5.7 Hz, 1H, H-1′), 6.98–6.79 (m, 4H, Ar-H), 7.11–7.30 (m, 10H, H-6, 9 Ar-H). 13C-NMR (CDCl3) δ (p.p.m.): 55.20 (OCH3), 62.96 (C-5′), 75.60 (C-3′), 86.16 (Ar-C), 87.64 (C-4′), 89.42 (C-2′), 90.31 (C-1′), 109.64 (C-5), 113.18, 126.91, 127.91, 129.83, 135.40, 144.39, 158.45 (Ar-C), 135.93 (C-6), 159.75 (C-2), 172.69 (C-4). FAB-HRMS: C30H29N2O7 (M+), 529.1976 (calc. 529.1974).
5′-O-(4,4-dimethoxytrity1)-2′-methylseleno-2′-deoxyuridine (4). NaBH4 (1.1 g, 6 mmol) was placed in a 250 ml round flask and suspended in dry THF (45 ml) under vigorous stirring. Dimethyl diselenide (CH3SeSeCH3, 1.99 ml, 19.9 mmol) was slowly injected, and the suspension was placed in an ice-water bath under dry argon. Anhydrous ethanol (5 ml) was added dropwise. Gas bubbles started to occur in the yellow mixture. The reaction mixture turned colorless after 60 min, and a solution of 3 (10.5 g, 19.9 mmol) in THF (20 ml) was injected to the flask. The flask for 3 was rinsed twice (2 × 5 ml) and the washings were injected to the reaction flask. The ice-water bath was removed, and the reaction was stirred under argon and monitored by TLC (5% CH3OH/CH2Cl2, product Rf = 0.35). The reaction was complete in 1 h. NaCl (sat., 50 ml) was added to the reaction, followed by dropwise addition of 20% HOAc (∼12 ml) until pH reached 7. The crude product was extracted by EtOAc (3 × 100 ml), and the combined organic layer was dried over anhydrous MgSO4, followed by filtration and solvent evaporation. The residue was purified on a silica gel column (equilibrated with hexane/CH2Cl2, 1:1) and eluted with a methanol/methylene chloride gradient [hexane/CH2Cl2, 1:1; hexane/CH2Cl2, 1:3; CH2Cl2 and CH3OH in CH2Cl2 (0.5 and 1%)] to afford the desired product 4 as a light yellow foam (92% yield; 11.4 g, wt 622). UV (in acetonitrile), λmax: 236.2, 273.4 nm. IR (KBr): 3450 (br), 3080, 3030, 2940, 1705, 1610, 1520, 1460, 1390, 1245, 1190, 1090, 1045, 850, 782, 710, 590 cm–1. 1H-NMR (CDCl3) δ: 2.08 (s, 3H, CH3Se), 3.42–3.47 (m, 2H, H-5′), 3.49–3.54 (m, 1H, H-2′), 3.78 (s, 6H, CH3O), 4.14–4.18 (m, 1H, H-4′), 4.34–4.39 (m, 1H, H-3′), 5.36 (d, J = 8.0 Hz, 1H, H-5), 6.19 (d, 1H, J = 3.3 Hz, H-1′), 6.86–6.92 (m, 4H, aromatic), 7.19–7.38 (m, 9H, aromatic), 7.77 (d, J = 7.8 Hz, 1H, H-6), 8.48 (br, 1H, NH). 13C-NMR (CDCl3) δ: 5.18 (SeCH3), 51.12 (C-2′), 55.63 (OCH3), 62.8 (C-5′), 71.90 (C-3′), 84.84 (C-4′), 87.66 (Ar-C), 87.98 (C-1′), 103.05 (C-5), 113.69, 128.43, 130.42, 135.50 (Ar-C), 139.45 (C-6), 144.36 (Ar-C), 150.55 (C-2), 158.55 (Ar-C), 163.03 (C-4). ESI-MS (positive mode): [M(80Se)+NH3]+ calc. 641, observed 641.2; [M(78Se)+NH3]+ calc. 639, observed 639.1. FAB-HRMS: C31H32N2O7Se (M+), 624.1376 (calc. 624.1374).
3′-O-(2-cyanoethyl-N,N-diisopropylphosphoramidite)-5′-O- (4,4-dimethoxytrity1)-2′-methylseleno-2′-deoxyuridine (5). The starting material 4 (10 g, 16.1 mmol) was placed in a 250 ml round flask and dried on high vacuum. Dry CH2Cl2 (80 ml, final conc. 0.2 M), N,N-diisopropylethylamine (8.4 ml, 48.3 mmol) and 2-cyanoethyl N,N-diisopropylchloro-phosphoramidite (80 ml, 32.2 mmol) were then added sequentially. The reaction mixture was stirred at 0°C in an ice-water bath under dry nitrogen for 30 min and the ice bath was then removed. The mixture was further stirred for 2 h at room temperature. Reaction completion was indicated by TLC [5% CH3OH/CH2Cl2, product Rf = 0.37]. The reaction mixture was then quenched with NaHCO3 (20 ml, sat.), stirred for 15 min, and extracted with CH2Cl2 (3 × 100 ml). The combined organic layer was washed with NaCl (100 ml, sat.) and dried over anhydrous MgSO4 for 15 min, followed by filtration and solvent evaporation. The crude product was re-dissolved in CH2Cl2 (20 ml), and this solution was added dropwise to petroleum ether (1000 ml) under vigorous stirring; a white precipitate was formed. The petroleum ether solution was decanted carefully (sometimes filtration was necessary). The crude product was then loaded into a silica gel column that was equilibrated with 20% EtOAc/hexane containing 0.5% triethylamine. The column was eluded with an increasing step-wise gradient of EtOAc/hexane in the presence of 0.5% triethylamine (20, 30, 40, 50, and 60%, 400 ml each). The pooled fractions containing the pure compound were combined and evaporated under reduced pressure, and re-dissolved in 20 ml of CH2Cl2. This solution was precipitated again in petroleum ether as indicated above. The precipitate was re-dissolved in CH2Cl2, transferred into a small round flask, evaporated, and dried on high vacuum overnight to yield a white foamy product (12 g, 91%). UV (in acetonitrile), λmax: 236.6, 267.8 nm. IR (KBr): 3450 (br), 3070, 3030, 2930, 2850, 1715, 1610, 1505, 1480, 1390, 1250, 1180, 1090, 1045, 785, 715, 580 cm–1. 1H-NMR (CDCl3) δ: 1.05–1.37 (m, 24H, 8x CH3-ipr), 2.04 (s, 6H, 2x CH3CO), 2.09 (2x s, 6H, 2x CH3Se), 2.42 and 2.68 (2x t, J = 7.5 Hz, 4H, 2x CH2-CN), 3.42–3.75 (m, 12H, 4x CH-ipr, CH2-CH2-CN, 2x H-2′, 2x 2H-5′), 3.84 (s, 12H, 4x CH3O), 3.90–4.05 (m, CH2-CH2-CN), 4.22 and 4.29 (2x m, 2H, H-4′), 4.63–4.75 (m, 2H, H-3′), 5.27 and 5.35 (2x d, J = 9.6 Hz, 2H, 2x H-5), 6.38 (d, 2H, J = 8.4 Hz, 2x H-1′), 6.84–6.95 (d, J = 8.7 Hz, 8H, aromatic), 7.26–7.48 (m, 18H, aromatic), 8.34 (d, J = 9.8 Hz, 2H, H-6), 9.23 (br, 2H, NH). 13C-NMR (CDCl3) δ: 4.78 (SeCH3), 19.14, 19.67, 20.87, 24.83, 24.46, 43.32, 43.48, 46.32, 47.56, 51.47 and 51.84 (C-2′), 55.61 (OCH3), 57.05, 58.38, 62.51 (C-5′), 73.35 and 73.66 (C-3′), 84.56 (C-4′), 87.35 (Ar-C), 88.12 (C-1′), 103.35 (C-5), 113.42, 117.28, 127.21, 127.94, 128.36, 130.19, 130.25, 135.17, 135.38 (Ar-C), 139.46 (C-6), 144.18 (Ar-C), 150.39 (C-2), 158.81 (Ar-C), 163.19 (C-4). 31P-NMR (CDCl3) δ: 148.68, 149.12. HRMS (MALDI-FTMS): C40H50N4O8PSe (M+H+), 825.2538 (calc. 825.2532).
5′-O-(4,4-dimethoxytrity1)-2′-methylseleno-2′-deoxycytosine (9). Phosphorus oxychloride (1.1 ml, 12 mmol) was added to a solution of 1,2,4-triazole (3.32 g, 48 mmol) in dry acetonitrile (40 ml) under argon, and the reaction was stirred for 1 h at room temperature. Dry triethylamine (13.4 ml, 96 mmol) was added and the reaction was stirred for another hour. The suspension was then filtered directly into a round flask containing compound 4 (2.49 g, 4 mmol) reacted with 1-(trimethylsilyl)imidazole (1.17 ml, 8 mmol) in dry acetonitrile (20 ml) under argon. The reaction mixture was allowed to run for 2 h. The formation of N4-triazolide 4a was revealed by a fluorescent spot on TLC (7.5% MeOH in CH2Cl2) that moved slightly slower than the silylated intermediate. After the reaction was complete, concentrated ammonia aqueous solution (20 ml) was then injected to the solution and stirred. After 15 min, the mixture was concentrated to a solution of ∼10 ml. Concentrated ammonia aqueous solution (30 ml) and dioxane (30 ml) were then added to the flask, and the solution was stirred overnight (monitored on TLC, 5% MeOH in CH2Cl2). The reaction mixture was then evaporated to ∼20 ml before extraction with ethyl acetate (3 × 100 ml). The combined organic layer was washed with saturated NaCl (100 ml) and dried over MgSO4 (s) before evaporation. The crude product was purified on silica gel column equilibrated with hexane/CH2Cl2, 1:1. The column was eluted with hexane/CH2Cl2, 1:1, 1:3, and pure CH2Cl2, and then with a methanol/CH2Cl2 gradient (0.5, 1, 2, 3 and 4% methanol in CH2Cl2) to afford product 9 (2.13 g, Fw 622.6) as a white foam (86% yield). 1H-NMR (CDCl3) δ: 1.2 (br, 1H, OH), 1.99 (s, 3H, CH3Se), 3.38 and 3.46 (2x dd, J = 1.8, 9.6 Hz, 2H, H-5′), 3.51 (dd, J = 4.5, 4.8 Hz, 1H, H-2′), 3.68 (s, 6H, CH3O), 4.13–4.17 (m, 1H, H-4′), 4.44–4.47 (m, 1H, H-3′), 5.40 (d, J = 7.5 Hz, 1H, H-5), 6.32 (d, 1H, J = 5.9 Hz, H-1′), 6.76–6.82 (d, J = 5.9 Hz, 4H, aromatic), 7.09–7.31 (m, 9H, aromatic), 7.87 (d, J = 7.5 Hz, 1H, H-6), 12.1 (br, 2H, NH2). 13C-NMR (CDCl3) δ: 5.25 (SeCH3), 51.56 (C-2′), 56.23 (OCH3), 64.50 (C-5′), 72.04 (C-3′), 85.52 (C-4′), 86.97 (Ar-C), 88.76 (C-1′), 96.10 (C-5), 114.23, 128.84, 129.08, 129.60, 130.46, 135.73, 136.95, 142.23 (C-6), 144.05, 158.40 (Ar-C), 160.40 (C-2), 167.26 (C-4). HRMS (MALDI-FTMS): C31H33N3O6Se [M+Na]+: 646.1429 (calc. 646.1427).
N4-acetyl-5′-O-(4,4-dimethoxytrity1)-2′-methylseleno-2′-de oxycytosine (9a). THF (10 ml), dry triethylamine (1.11 ml, 8 mmol) and 1-(trimethylsilyl)imidazole (0.44 ml, 3 mmol) were added to a 25 ml round flask containing compound 9 (dry 1.25 g, 2 mmol) under argon at room temperature. After 15 min of stirring, a catalytic amount of N,N-dimethyl-aminopyridine (DMAP, 20 mg) and acetic anhydride (371 µl, 4 mmol) were added and the reaction was left for 2 h while being monitored by silica gel TLC (5% MeOH/CH2Cl2). After the reaction was complete, MeOH (2 ml) was added, and the mixture was stirred for another 20 min. The solvents were evaporated under reduced pressure and the resultant residue was dissolved in EtOAc (50 ml). The precipitated salt was removed by filtration and the filtrate was evaporated again under reduced pressure. This residue was dissolved in THF (10 ml), and tetrabutylammonium fluoride (4 ml, 1 M, 4 mmol) was added. The mixture was stirred for 1 h to remove the 3′-TMS group (monitored on silica gel TLC in 5% MeOH/CH2Cl2). After evaporation of THF, the crude product was dissolved in CH2Cl2 and purified on a silica gel column, equilibrated with hexane/CH2Cl2, 1:1. The column was first eluted with hexane/CH2Cl2, 1:1, 1:3, and pure CH2Cl2, and then with a methanol/CH2Cl2 gradient (0.5, 1, 2 and 3% methanol in CH2Cl2) to afford the desired product 9a (1.26 g, Fw 664.6) as a white foam (95% yield). 1H-NMR (CDCl3) δ: 2.15 (s, 3H, CH3Se), 2.23 (s, 3H, CH3CO), 3.29 (br, 1H, OH), 3.49 and 3.57 (2x dd, J = 1.9, 9.8 Hz, 2H, H-5′), 3.68 (dd, J = 4.4, 4.7 Hz, 1H, H-2′), 3.79 (s, 6H, CH3O), 4.16–4.27 (m, 1H, H-4′), 4.42–4.53 (m, 1H, H-3′), 6.34 (d, 1H, J = 6.1 Hz, H-1′), 6.81–6.95 (d, J = 6.1 Hz, 4H, aromatic), 7.19 (d, J = 7.5 Hz, 1H, H-5), 7.26–7.52 (m, 9H, aromatic), 8.32 (d, J = 7.5 Hz, 1H, H-6), 9.89 (br, 1H, NH). 13C-NMR (CDCl3) δ: 4.58 (SeCH3), 24.79 (CH3CO), 46.23 (C-2′), 55.23 (OCH3), 62.23 (C-5′), 69.92 (C-3′), 84.47 (C-4′), 87.21 (Ar-C), 90.26 (C-1′), 96.98 (C-5), 113.35, 127.17, 128.02, 128.15, 130.08, 135.20, 135.40, 144.24 (Ar-C), 144.45 (C-6), 155.37 (C-2), 158.76 (Ar-C), 162.85 (C-4), 170.75 (COCH3). HRMS (MALDI-FTMS): C33H35N3O7Se [M+Na]+: 688.1553 (calc. 688.1532).
N4-acetyl-3′-O-(2-cyanoethyl-N,N-diisopropylphosphor- amidite)-5′-O-dimethoxytrityl-2′-methylseleno-2′-deoxy cytosine (10). See the synthesis of compound 5. A white foamy product resulted (Fw: 864.8; 1.28 g, 92%). 1H-NMR (CDCl3) δ: 1.04–1.35 (m, 24H, 8x CH3-ipr), 2.13 (s, 6H, 2x CH3CO), 2.16 and 2.19 (2x s, 6H, 2x CH3Se), 2.41 and 2.67 (2x t, J = 7.5 Hz, 4H, 2x CH2-CN), 3.45–3.78 (m, 12H, 4x CH-ipr, CH2-CH2-CN, 2x H-2′, 2x 2H-5′), 3.83 (s, 12H, 4x CH3O), 3.92–4.03 (m, CH2-CH2-CN), 4.36–4.41 (m, 2H, H-4′), 4.65–4.71 (m, 2H, H-3′), 6.44 (d, 2H, J = 5.1 Hz, H-1′), 6.82–6.94 (d, J = 8.7 Hz, 8H, aromatic), 7.09 and 7.14 (2× d, J = 7.5 Hz, 2H, H-5), 7.27–7.46 (m, 18H, aromatic), 8.31 and 8.35 (2× d, J = 7.5 Hz, 2H, H-6), 10.56 (br, 2H, NH). 13C-NMR (CDCl3) δ: 4.65 (SeCH3), 19.12, 19.89, 20.56, 22.94 and 22.96 (CH3CO), 24.77, 24.83, 43.34, 43.47, 45.32 and 45.38 (C-2′), 46.77, 47.64, 55.25 (OCH3), 57.95, 58.34, 61.94 (C-5′), 73.32 and 73.46 (C-3′), 84.36 (C-4′), 87.15 (Ar-C), 90.92 (C-1′), 97.22 (C-5), 113.32, 117.25, 127.23, 127.99, 128.30, 130.09, 130.23, 135.14, 135.32, 144.16 (Ar-C), 144.46 (C-6), 155.39 (C-2), 158.81 (Ar-C), 163.09 (C-4), 171.20 (COCH3). 31P-NMR (CDCl3) δ: 148.67, 149.05. HRMS (MALDI-FTMS): C42H52N5O8PSe [M+H]+: 866.2795 (calc. 866.2791).
Synthesis of 2′-Se-functionalized RNA and DNA oligonucleotides
All DNA and RNA oligonucleotides were synthesized chemically on a 1.0 or 10 µmol scale using an ABI392 DNA/RNA Synthesizer (24–26). The concentration of the Se-nucleoside phosphoramidites was identical to that of the conventional phosphoramidites (0.1 M in acetonitrile). Coupling was carried out using a 5-BMT solution (0.3 M) in acetonitrile (27). The coupling time for the Se-nucleoside phosphoramidites was 25 s. The 5′-detritylation was done using 3% trichloroacetic acid in methylene chloride. Syntheses were performed on control pore glass (CPG-500) immobilized with the appropriate nucleoside through a succinate linker (Glen Research). All the oligonucleotides were prepared with DMTr-on. In the case of RNA, the syntheses were done with nucleoside phosphoramidites containing the 2′-O-triisopropylsilyloxymethyl (2′-O-TOM) protecting groups. After synthesis, the DNA oligonucleotides were cleaved from the solid support and fully deprotected by aqueous ammonia (conc.) treatment for 14 h at 55°C. Similarly, the 2′-O-TOM protected RNA oligonucleotides were deprotected following recommendations from Glen Research with minor modifications. Briefly, 1 ml of a methylamine solution [prepared by mixing 40% aqueous methylamine with 33% ethanolic methylamine (Fluka) in a 1:1 ratio] was added to an Eppendorf tube containing the RNA resin (1 µmol), followed by incubation for 6 h at 35°C or overnight at room temperature. The supernatant was evenly transferred into two sterile 1.5 ml Eppendorf tubes. After these tubes were chilled at –20°C for 10 min, the solvents were completely evaporated on a speed vacuum. Each portion was treated with 25 equivalents of tetrabutylammonium fluoride [0.25 ml (for 0.5 µmol RNA 20mer), 1.0 M in THF; Fluka] and incubated for 6 h at 35°C. Following evaporation of THF on a speed vacuum, the two portions were combined in 1.0 ml of 1.0 M Tris–HCl buffer (RNase-free, pH 7.5), incubated with shaking overnight at 25°C, and filtrated through a 0.2 µm pore-sized filter. The 5′-DMTr deprotection of both DNA and RNA oligonucleotides was performed in a 2% trichloroacetic acid solution (from 10% w/w, 0.9 M in water) for 1.5 min, followed by neutralization to pH 7.0 with a freshly made aqueous solution of triethylamine (1.1 M) and petroleum ether extraction to remove DMTr-OH.
HPLC analysis and purification
The DNA and RNA oligonucleotides were analyzed and purified by reverse-phase high performance liquid chromatography (RP-HPLC) both DMTr-on and DMTr-off. Purification was carried out using a 21.2 × 250 mm Zorbax, RX-C8 column at a flow rate of 10 ml/min. Buffer A consisted of 50 mM triethylammonium acetate (TEAAc, pH 7.1, RNase-free water), while buffer B contained 50% aqueous acetonitrile and 50 mM TEAAc, pH 7.1. Similarly, analysis was performed on a Zorbax SB-C18 column (4.6 × 250 mm) at a flow rate of 1.0 ml/min using the same buffer system. The DMTr-on oligonucleotides were eluted with up to 90% buffer B in 25 min in a linear gradient, while the DMTr-off oligonucleotides were eluted with up to 40% of buffer B in a linear gradient in the same period of time. The collected fractions were lyophilized; the purified compounds were re-dissolved in RNase-free water. The pH was adjusted to 7.0 after the final purification of the Se-oligonucleotides without the DMTr group.
Electrospray mass spectrometry analysis
Crude and purified oligonucleotides containing the selenium derivatization were analyzed by LC-MS using electrospray negative ion mode. The general analytical procedures for the liquid chromatography were elution (1 ml/min, 4.5 × 150 mm 300SB-C8 column) with buffer A (5 mM ammonium acetate, pH 6.5) for 2 min, and then elution with a linear gradient from buffer A to 100% buffer B (60% acetonitrile and 40% of buffer A) in 13 min.
Thermodenaturization of duplex DNAs
Solutions of the duplex DNAs (2 µM) were prepared by dissolving the DNAs in a buffer containing NaCl (90 mM), sodium phosphate (10 mM, pH 7.2), and EDTA (1 mM). The solutions were then heated to 95°C for 2 min, cooled slowly to room temperature, and stored at 5°C overnight before measurement. Prior to thermal denaturation, helium was bubbled through the samples. Denaturation curves were acquired at 254 nm at a heating rate of 0.5°C/min using an 8453 UV-Visible Spectrometer from Agilent Technologies. This system is equipped with a Peltier Temperature Controller. The data were analyzed in accordance with the convention of Puglisi and Tinoco (28).
Crystallization of the Se-derivatized oligonucleotides
The purified oligonucleotides (2 mM) containing selenium labels were first heated to 90°C for 1 min, and the samples were then allowed to slowly cool to 25°C. Crystallization conditions were first screened using the nucleic acid screening kits from Hampton Research. To minimize the amount of material used, 1 or 2 µl of the appropriate oligonucleotide solutions were typically used in each of these screens. Crystallization was carried out using the hanging drop method by vapor diffusion at 25 and 4°C.
RESULTS AND DISCUSSION
Synthesis of 2′-methylseleno uridine and cytidine phosphoramidites
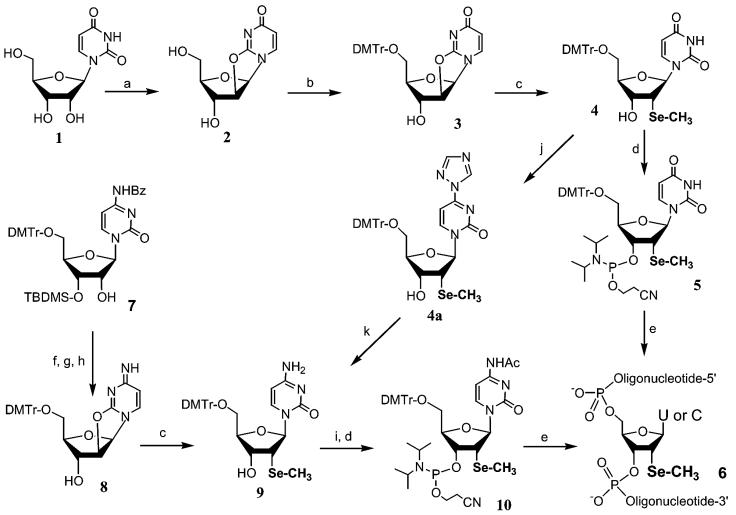
On the basis of the synthesis of 2′-methylseleno uridine phosphoramidite 5 (20), the corresponding 2′-methylseleno cytidine derivative was first attempted from cytidine derivative 7 (Scheme 1). This partially protected derivative was first mesylated at the 2′-position, followed by the displacement of the 2′-mesyl group with the cytosine exo-2-oxygen under basic conditions. To obtain compound 8 for the incorporation of the selenium functionality, the 3′-TBDMS group was removed using tetrabutylammonium fluoride. Unfortunately, a low yield in selenium incorporation was obtained when sodium methyl selenide was used to generate cytidine derivative 9. The poor substitution by sodium methyl selenide is probably because the exo-2-oxygen in cytosine is a much poorer leaving group in comparison to that in uridine. An alternative approach, via conversion of uridine derivative 4 to cytidine 9, was later explored successfully (29).
Scheme 1. (a) (Ph)2CO3, Na2CO3, DMF. (b) DMTr-Cl, Py. (c) NaSeCH3, EtOH-THF. (d) 2-Cyanoethyl N,N-diisopropyl-chlorophosphoramidite and N,N-diisopropylethylamine in CH2Cl2. (e) Synthesis of oligonucleotides on solid phase. (f) Ms-Cl, TEA, THF. (g) Toluene/tetrahexylammonium hydrogen sulfate, Na2CO3 (sat.). (h) (Bu)4N+ F–, THF. (i) TMS-Im, then Ac2O, TEA and DMAP in THF. (j) TMS-Im, then POCl3-triazole-TEA in CH3CN. (k) NH4OH.
We reported previously the synthesis of 2′-methylseleno uridine phosphoramidite 5 (Scheme 1) using a relatively expensive uridine derivative (5′-O-DMTr-3′-O-TBDMS uridine). This synthesis required four steps to generate the key intermediate 4 (20). To improve the synthesis of this intermediate, we started the reaction using uridine, a relatively inexpensive starting material (29). Thus, intermediate 4 was made in three simple steps and the synthesis has been run at a 50 g scale. This uridine intermediate 4 was converted to cytidine derivative 9 via triazolide 4a that was generated in situ. After acetylation of the base, 2′-methylseleno cytidine phosphoramidite 10 was made as in the case of 5. This new advancement in the synthesis of intermediate 4 has facilitated the preparation of the 2′-methylseleno pyrimidine phosphoramidites in large scales.
Design of 2′-Se-derivatized RNAs and DNAs
Collaborative investigation with Egli and co-workers has previously demonstrated that the 2′-deoxyriboses containing the 2′-methylseleno functionality in the furanose ring displayed 3′-endo conformations in the crystal structure of the selenium-derivatized oligonucleotides, which is consistent with the geometry that is adopted by both RNA and A-form DNA (21). These studies indicated that this type of selenium derivatization may not significantly perturb the structure and stability of RNA and A-form DNA duplexes.
Consequently, we have derivatized a number of biologically and structurally important RNAs using this derivatization strategy for structural analysis. Though the 3D structure of an RNA molecule is difficult to foretell, its secondary structure is relatively easy to predict. In order to avoid structural perturbation, we decided to incorporate this 2′-Se derivatization into a stem region, which is a major element in the secondary structure of RNA.
To illustrate this principle, several RNA oligonucleotides were chosen and derivatized at specific positions through the solid phase synthesis. A 12mer RNA fragment of the HIV-1 Rev binding element (30) was selected and derivatized with both 2′-Se-U and 2′-Se-C at three different positions (Fig. 1A). To better understand G-U wobble base-pairing of RNA (31), (GGCGUGCC)2 was selected for selenium derivatization at both U and C sites (Fig. 1B). Derivatization of this duplex RNA 8mer has been attempted previously using conventional approaches without success in the structure determination. Likewise, other RNAs were selected for the selenium derivatization on their stems (Fig. 1C and D).
Figure 1.
Secondary structures of selected RNAs containing the selenium derivatization. The underlined C and U are derivatized with the 2′-selenide functionality.
There are many DNA short repeats in human genomic sequences that are implicated in some genetic diseases (32,33). Studies of the structures and functions of these DNA repeats can provide invaluable insights into the disease mechanisms. As an exploration, two DNA purine repeats (TGGAGGAGGAT and TAGGAGGAGGAT) with unknown structures have been derivatized using this novel Se-derivatization strategy. The selenium modification was introduced at their 5′-termini to minimize possible perturbation. Similarly, a purine-rich DNA (GGAAGGTTTGGGAT) was derivatized internally (bold T) for the structural analysis. In order to further study DNA structure and to understand possible perturbation caused by the 2′-Se derivatization, other functional DNAs (see Table 1), including Z-DNA (34), A-form DNA (35), and transcription promoter DNA 32mer (36), were also derivatized analogously using the 2′-Se-C and 2′-Se-U. As a demonstration, seven selenium labels were successfully incorporated into the promoter DNA, which was confirmed by MS analysis (see Table 1).
Table 1. Electrospray MS analysis of RNA and DNA oligonucleotides derivatized with the 2′-selenium functionality.
| RNA and DNA oligonucleotides | Molecular formula | LC-MS measured (calc.) m/z |
|---|---|---|
| 5′-GCUSeGGACGCAGG-3′ (RNA 12mer, motif of Rev binding element of HIV-1) | C117H146N51O80P11Se (isotopic mass: 3965.5) | M3–: 1320.8 (1320.8); M4–: 990.4 (990.4); M5–: 792.1 (792.1); M6–: 660.0 (659.9). |
| 5′-GCSeUGGACGCAGG-3′ (RNA 12mer, motif of Rev binding element of HIV-1) | C117H146N51O80P11Se (isotopic mass: 3965.5) | M3–: 1321.0 (1320.8); M4–: 990.6 (990.4) |
| 5′-CCSeUGACGAUACAGC-3′ (RNA 14mer, motif of Rev binding element of HIV-1) | C134H169N54O93P13Se (isotopic mass: 4504.6) | M3–: 1500.8 (1500.5); M4–: 1125.3 (1125.2) |
| 5′-GGCGUSeGCC-3′ (RNA 8mer containing the G-U wobble base pairs) | C77H98N31O54P7Se (isotopic mass: 2617.3) | M2–: 1307.5 (1307.7); M3–: 871.4 (871.4); M4–: 653.4 (653.3) |
| (5′-GGCSeGUGCC-3′) (RNA 8mer containing the G-U wobble base pairs) | C77H98N31O54P7Se (isotopic mass: 2617.3) | M2–: 1307.8 (1307.7); M3–: 871.5 (871.4) |
| 5′-CGGGGAAGACCGCUCUAUG CCSeCC-3′ | C219H276N88O157P22Se (isotopic mass: 7411.0) | [M+NH3]5–: 1484.9 (1484.6); M6–: 1234.2 (1234.2) |
| 5′-UUGCSeGUCGCUCCGGAAA AGUCGC-3′ | C219H274N86O159P22Se (isotopic mass: 7412.9) | M3–: 2470.2 (2470.0) |
| 5′-USeGGAGGAGGAT-3′ (GGA repeat DNA) | C110H135N49O63P10Se (isotopic mass: 3539.5) | M2–: 1768.7 (1768.8); M3–: 1179.0 (1178.8); M4–: 883.8 (883.9); M5–: 707.0 (706.9); M6–: 588.9 (588.9). |
| 5′-USeAGGAGGAGGAT-3′ (AGG repeat DNA) | C120H147N54O68P11Se (isotopic mass: 3852.6) | M2–: 1925.3 (1925.3); M3–: 1283.3 (1283.2); M4–: 962.2 (962.2); M5–: 769.5 (769.5); M6–: 641.0 (641.1). |
| 5′-GGAAGGTUSeTGGGAT-3′ (purine-rich DNA) | C140H173N58O83P13Se (isotopic mass: 4476.7) | M3–: 1491.4 (1491.2); M4–: 1118.2 (1118.2); M5–: 894.5 (894.3); M6–: 745.2 (745.1). |
| 5′-GUSeGTACAC-3′ (A-form DNA) | C78H99N30O46P7Se (isotopic mass: 2488.4) | M2–: 1243.2 (1243.2); M3–: 828.5 (828.5); M4–: 621.3 (621.1). |
| 5′-USeGCGCA-3′ (Z-form DNA) | C58H74N23O34P5Se (isotopic mass: 1871.3) | M–: 1870.3 (1870.3); M2–: 934.6 (934.7) |
| 5′-ATTCAGCSeG-3′ (DNA 8mer) | C79H101N30O46P7Se (isotopic mass: 2502.4) | M2–: 1250.2 (1250.2); M3–: 833.2 (833.1); M4–: 624.7 (624.6); M5–: 499.5 (499.5) |
| 5′-DMTr-ATCSeAGTAATCAT-3′ (DMTr-on DNA 12mer) | C140H169N44O71P11Se (isotopic mass: 4022.7) | M2–: 2010.6 (2010.4); M3–: 1340.2 (1339.9); M4–: 1004.9 (1004.7); M5–: 803.7 (803.5); M6–: 669.6 (669.5) |
| 5′-ATCSeAGTAATCAT-3′ (DNA 12mer) | C119H151N44O69P11Se (isotopic mass: 3720.6) | M2–: 1859.3 (1859.3); M3–: 1239.4 (1239.2); M4–: 928.6 (929.2); M5–: 743.2 (743.1); M6–: 619.1 (619.1) |
| 5′AAGUSeGUSeCAAGUSeACUSeT USeTUSeTCCUSeAA-AATGTGAT-3′ (transcription promoter, DNA 32mer containing seven selenium labels) | C316H398N113O193P31Se7 (isotopic mass: 10382.1) | [M+NH3]5–: 2078.7 (2078.8); [M+NH3]6–: 1731.5 (1732.2); [M+NH3]7–: 1484.1 (1484.6) |
Synthesis of the 2′-Se-derivatized RNAs and DNAs
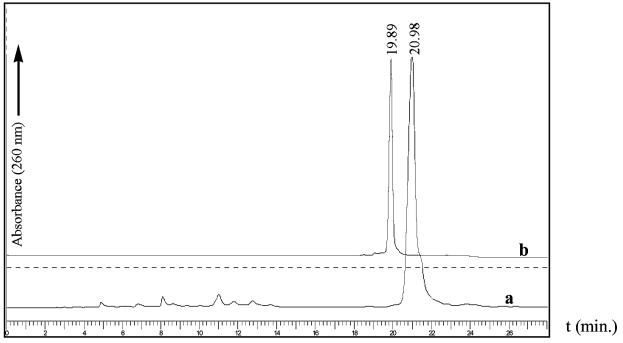
We first attempted the synthesis of selenium-functionalized RNA oligonucleotides using the 2′-O-TBDMS protecting group in combination with 1H-tetrazole as the coupling reagent. Unfortunately, this procedure failed to produce a satisfactory coupling yield under the experimental conditions. Although no further experiments were performed to investigate the specific causes of the low yield, this result prompted us to explore 5-BMT as the coupling reagent in combination with the nucleoside phosphoramidites containing the 2′-O-TOM protection (27). We found that the 2′-methylseleno pyrimidine phosphoramidites were as reactive as the ordinary deoxynucleoside phosphoramidites. Coupling of the Se-phosphoramidites was performed in 25 s, and the coupling yields were over 99% when 5-BMT was used. The yields were confirmed by RP-HPLC analysis. As examples, the HPLC profiles of the crude 2′-Se-U RNA 8mer with the DMTr group and the purified 2′-Se-U RNA 8mer without the DMTr group are shown in Figure 2.
Figure 2.
HPLC analysis of Se-U-RNA 8mer. (a) Spectrum of the crude 2′-O-TOM-off product with DMTr-on. (b) Spectrum of the purified product with DMTr-off. The HPLC conditions are listed in Materials and Methods.
Partial detritylation of the purified DMTr-on oligonucleotides was observed during lyophilization. Analysis with HPLC or TLC indicated that ∼50% of the oligonucleotide lost the DMTr group (data not shown). We traced the cause of the detritylation to an acidic environment (pH 4.5) generated during the evaporation of the TEAAc buffer. Degradation of both Se-DNA and Se-RNA oligonucleotides was also observed when these oligonucleotides were treated with the conventional approach to remove the 5′-DMTr groups. We found that treatment using 2% aqueous trichloroacetic acid for only 1.5 min was sufficient to remove the DMTr groups without causing degradation of these selenium-derivatized oligonucleotides.
In order to investigate the stability of the selenium functionality in the iodine oxidation during the solid phase synthesis, we conducted the oxidation using 20 mM I2 for 20 s. Interestingly, no measurable oxidation of the selenide functionality was observed in most of the cases. However, when the selenide moiety was close to the 3′-terminus, 2–5% of the selenoxide oxidized from the selenide product was occasionally observed by LC-MS. This selenoxide displayed an extra 16 Da of mass over the corresponding selenide on MS spectrum (data not shown). We find that the 2′-selenium derivatized oligonucleotides are quite stable, and there is no detectable oxidation or degradation under air for months.
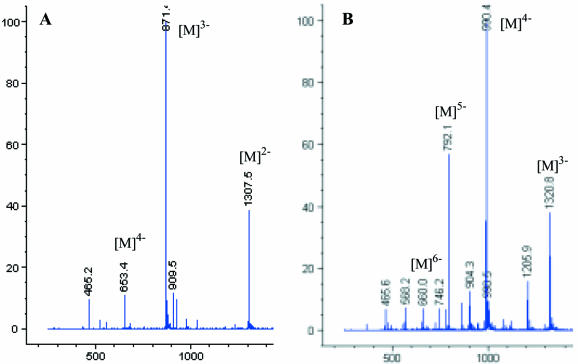
After HPLC purification, the derivatized oligonucleotides were confirmed by electrospray mass spectrometry (negative ion mode). Two typical MS spectra of the derivatized RNA oligonucleotides are shown in Figure 3. The molecular peaks with several different charges are observed. The MS analytical data of all Se-DNA and Se-RNA oligonucleotides synthesized and presented in this report are shown in Table 1. These data confirm the introduction of selenium labels using both 2′-Se-C and 2′-Se-U phosphoramidites, including incorporation of seven selenium labels per nucleic acid molecule in a DNA promoter.
Figure 3.
Electrospray MS spectra of the RNAs derivatized with the 2′-selenium functionality. (A) The RNA 8mer (GGCGUSeGCC), C77H98N31O54P7Se, isotopic mass: 2617.3, measured (calc.) m/z: [M]2–: 1307.5 (1307.7); [M]3–: 871.4 (871.4); [M]4–: 653.4 (653.3). (B) The RNA 12mer (GCUSeGGACGCAGG): C117H146N51O80P11Se, isotopic mass: 3965.5, measured (calc.) m/z: [M]3–: 1320.8 (1320.8); [M]4–: 990.4 (990.4); [M]5–: 792.1 (792.1); [M]6–: 660.0 (659.9).
Thermodenaturization and stability of oligonucleotide duplexes containing the 2′-methylseleno derivatization
To further study the stability of the oligonucleotide duplexes containing the 2′-selenium derivatization, one set of A-form DNA oligonucleotides (GTGTACAC)2 containing different modifications was synthesized (Table 2). Though the melting temperature of the 2′-MeSe octamer was less than that of the 2′-MeO octamer, it was slightly higher than that of the native octamer. This result suggests that the two derivatized USeMe residues have no significant effect on duplex stability of the A-form DNA. This result is consistent with the previous UV melting study of the A-form DNA duplex [(GCGTAdUSeMeACGC)2] (21).
Table 2. UV melting temperatures of the A-form DNAs.
| A-form octamer DNAs | Melting temperature (°C) |
|---|---|
| Native octamer (5′-GTGTACAC-3′) | 21.2 |
| 2′-MeO-octamer (5′-GUOMeGTACAC-3′) | 24.8 |
| 2′-MeSe-octamer (5′-GUSeMeGTACAC-3′) | 21.5 |
Crystallization of selenium-derivatized oligonucleotides
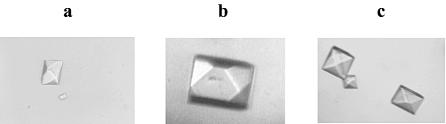
An A-form DNA with a self-complementary sequence (5′-GTGTACAC-3′) (35) was chosen for selenium derivatization and crystallization studies. It was found that these oligonucleotides (Table 2) were able to crystallize in many identical conditions. As examples, photos of crystals of the octamer (native, 5′-GUOMeGTACAC-3′), the selenium-derivatized octamer (Se-Oct, 5′-GUSeMeGTACAC-3′), and the selenium-bromide derivatized octamer (Se/Br-Oct, 5′-GUSeMeGdUBrACAC-3′) are shown in Figure 4. These native and derivatized crystals were crystallized in the same conditions, and they appear to have the same morphology.
Figure 4.
Photos of crystals of the native and Se-derivatized octamers. (a) Native-Oct. (b) Se-Oct. (c) Se/Br-Oct. Sizes of the crystals range from 0.1 × 0.1 to 0.4 × 0.4 mm.
CONCLUSIONS
Derivatization of DNA and RNA with selenium represents a new strategy to facilitate structural determination by X-ray crystallography via MAD phasing. We have recently achieved the covalent incorporation of selenium into nucleic acids for MAD phasing through collaboration with Egli and coworkers (19–23). This strategy involves the replacement of specific oxygen atoms in the nucleotide building blocks with selenium, followed by chemical or enzymatic incorporation of the modified building blocks into DNAs or RNAs. As an important part of our ongoing research on derivatizing DNA and RNA molecules for crystallographic phasing, we have reported here the chemical synthesis of selenium-labeled oligonucleotides with important structural or biological properties. To achieve the Se-derivatization in more desired sites, the novel 2′-methylseleno cytidine phosphoramidite has been synthesized, and the accessibility of the 2′-methylseleno uridine phosphoramidite has been advanced. DNA and RNA oligonucleotides derivatized with selenium at the specific cytidine and uridine sites have been prepared in large scales using the solid phase approach. In this novel strategy, specific incorporation of the 2′-selenium functionality to the stems of RNA molecules can avoid structural perturbation. In addition, the crystallization result indicates that the oligonucleotides derivatized with the 2′-selenium functionality are crystallizable. Therefore, this derivatization strategy should significantly facilitate nucleic acid X-ray crystallography.
ACKNOWLEDEGEMENTS
We thank Drs Helen Berman, Martin Egli, Stephen R. Holbrook, Catherine L. Lawson, Benoit Masquida, Dinshaw Patel and Eric Westhof for their discussions, enthusiasm and screenings on these RNA and DNA oligonucleotides derivatized with selenium. We also thank Dr Clifford E. Soll at Hunter College for assisting in MS data collection. This work was supported by PSC-CUNY Awards (64203-00-33 and 65356-00-34), CUNY Project Programs (80210-0302 and 80209-0408) and National Institute of Health grant (GM069703).
REFERENCES
- 1.Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 2.McKay D.B. and Wedekind,J.E. (1999) Small Ribozymes. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 265–286. [Google Scholar]
- 3.Bartel D.P. (1999) Recreating an RNA Replicase. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 143–162. [Google Scholar]
- 4.Storz G. (2002) An expanding universe of noncoding RNAs. Science, 296, 1260–1263. [DOI] [PubMed] [Google Scholar]
- 5.Lee R.C. and Ambros,V. (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science, 294, 862–864. [DOI] [PubMed] [Google Scholar]
- 6.Holbrook S.R. and Kim,S.H. (1997) RNA crystallography. Biopolymers, 44, 3–21. [DOI] [PubMed] [Google Scholar]
- 7.Hendrickson W.A. (1999) Maturation of MAD phasing for the determination of macromolecular structures. J. Synchrotron Radiat., 6, 845–851. [Google Scholar]
- 8.Hendrickson W.A. (2000) Synchrotron crystallography. Trends Biochem. Sci., 25, 637–643. [DOI] [PubMed] [Google Scholar]
- 9.Ealick S.E. (2000) Advances in multiple wavelength anomalous diffraction crystallography. Curr. Opin. Chem. Biol., 4, 495–499. [DOI] [PubMed] [Google Scholar]
- 10.Hendrickson W.A., Horton,J.R. and LeMaster,D.M. (1990) Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J., 9, 1665–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deacon A. and Ealick,S.E. (1999) Selenium-based MAD phasing: setting the sites on larger structures. Structure, 7, R161–R166. [DOI] [PubMed] [Google Scholar]
- 12.Ferre-D’Amare A.R., Zhou,K. and Doudna,J.A. (1998) Crystal structure of a hepatitis delta virus ribozyme. Nature, 395, 567–574. [DOI] [PubMed] [Google Scholar]
- 13.Rupert P.B. and Ferre-D’Amare,A.R. (2001) Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature, 410, 780–786. [DOI] [PubMed] [Google Scholar]
- 14.Batey R.T., Rambo,R.P., Lucast,L., Rha,B. and Doudna,J.A. (2000) Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science, 287, 1232–1239. [DOI] [PubMed] [Google Scholar]
- 15.Nikulin A., Serganov,A., Ennifar,E., Tishchenko,S., Nevskaya,N., Shepard,W., Portier,C., Garber,M., Ehresmann,B., Ehresmann et al. (2000) Crystal structure of the S15-rRNA complex. Nature Struct. Biol., 7, 273–277. [DOI] [PubMed] [Google Scholar]
- 16.Shah K., Wu,H. and Rana,T.M. (1994) Synthesis of uridine phosphoramidite analogs: reagents for site-specific incorporation of photoreactive sites into RNA sequences. Bioconjug. Chem., 5, 508–512. [DOI] [PubMed] [Google Scholar]
- 17.Gott J.M., Willis,M.C., Koch,T.H. and Uhlenbeck,O.C. (1991) A specific, UV-induced RNA-protein cross-link using 5-bromouridine-substituted RNA. Biochemistry, 30, 6290–6295. [DOI] [PubMed] [Google Scholar]
- 18.Ennifar E., Carpentier,P., Ferrer,J.L., Walter,P. and Dumas,P. (2002) X-ray-induced debromination of nucleic acids at the Br K absorption edge and implications for MAD phasing. Acta Crystallogr. D Biol. Crystallogr., 58, 1262–1268. [DOI] [PubMed] [Google Scholar]
- 19.Carrasco N., Ginsburg,D., Du,Q. and Huang,Z. (2001) Synthesis of selenium-derivatized nucleosides and oligonucleotides for X-ray crystallography. Nucleosides Nucleotides Nucleic Acids, 20, 1723–1734. [DOI] [PubMed] [Google Scholar]
- 20.Du Q., Carrasco,N., Teplova,M., Wilds,C.J., Egli,M. and Huang,Z. (2002) Internal derivatization of oligonucleotides with selenium for X-ray crystallography using MAD. J. Am Chem. Soc., 124, 24–25. [DOI] [PubMed] [Google Scholar]
- 21.Teplova M., Wilds,C.J., Wawrzak,Z., Tereshko,V., Du,Q., Carrasco,N., Huang,Z. and Egli,M. (2002) Covalent incorporation of selenium into oligonucleotides for X-ray crystal structure determination via MAD: proof of principle. Biochimie, 84, 849–858. [DOI] [PubMed] [Google Scholar]
- 22.Wilds C.J., Pattanayek,R., Pan,C., Wawrzak,Z. and Egli,M. (2002) Selenium-assisted nucleic acid crystallography: use of phosphoroselenoates for MAD phasing of a DNA structure. J. Am. Chem. Soc., 124, 14910–14916. [DOI] [PubMed] [Google Scholar]
- 23.Carrasco N. and Huang,Z. (2004) Enzymatic synthesis of phosphoroselenoate DNA using thymidine 5′-(α-P-seleno)triphosphate and DNA polymerase for X-ray crystallography via MAD. J. Am. Chem. Soc., 126, 448–449. [DOI] [PubMed] [Google Scholar]
- 24.Ogilvie K.K., Sadana,K.L., Thompson,E.A., Quilliam,M.A. and Westmore,J.B. (1974) The use of silyl groups in protecting the hydroxyl functions of ribonucleosides. Tetrahedron Lett., 15, 2861–2863. [Google Scholar]
- 25.Scaringe S.A., Francklyn,C. and Usman N. (1990) Chemical synthesis of biologically active oligoribonucleotides using beta-cyanoethyl protected ribonucleoside phosphoramidites. Nucleic Acids Res., 18, 5433–5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt C., Welz,R. and Muller,S. (2000) RNA double cleavage by a hairpin-derived twin ribozyme. Nucleic Acids Res., 28, 886–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welz R. and Mueller,S. (2002) 5-(Benzylmercapto)-1H-tetrazole as activator for 2′-O-TBDMS phosphoramidite building blocks in RNA synthesis. Tetrahedron Lett., 43, 793–797. [Google Scholar]
- 28.Puglisi J.D. and Tinoco,I.,Jr (1989) Absorbance melting curves of RNA. Methods Enzymol., 180, 304–25. [DOI] [PubMed] [Google Scholar]
- 29.McGee D.P.C., Vanghn-Settle,A., Vargeese,C. and Zhai,Y. (1996) 2′-Amino-2′-deoxyuridine via an intramolecular cyclization of a trichloroacetimidate. J. Org. Chem., 61, 781–785. [DOI] [PubMed] [Google Scholar]
- 30.Hung L.W., Holbrook,E.L. and Holbrook,S.R. (2000) The crystal structure of the Rev binding element of HIV-1 reveals novel base pairing and conformational variability. Proc. Natl Acad. Sci. USA, 97, 5107–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X., McDowell,J.A., Kierzek,R., Krugh,T.R., Turner,D.H. (2000) Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G·U pairs: one hydrogen bond for each G·U pair in r(GGCGUGCC)2 and two for each G·U pair in r(GAG UG CUC)2. Biochemistry, 39, 8970–8982. [PubMed] [Google Scholar]
- 32.Astolfi P., Bellizzi,D. and Sgaramella,V. (2003) Frequency and coverage of trinucleotide repeats in eukaryotes. Gene, 317, 117–125. [DOI] [PubMed] [Google Scholar]
- 33.Delatycki M.B., Williamson,R. and Forrest,S.M. (2000) Friedreich ataxia: an overview. J. Med. Genet., 37, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harper A., Brannigan,J.A., Buck,M., Hewitt,L., Lewis,R.J., Moore,M.H. and Schneider,B. (1998) Structure of d(TGCGCA)2 and a comparison to other DNA hexamers. Acta Crystallogr. D Biol. Crystallogr., 54, 1273–1284. [DOI] [PubMed] [Google Scholar]
- 35.Jain S.C., Zon,G. and Sundaralingam,M. (1989) Base only binding of spermine in the deep groove of the A-DNA octamer d(GTGTACAC). Biochemistry, 28, 2360–2364. [DOI] [PubMed] [Google Scholar]
- 36.Benoff B., Yang,H., Lawson,C., Parkinson,G., Liu,J., Blatter,E., Ebright,Y.W., Berman,H.M. and Ebright,R.H. (2002) Structural basis of transcription activation: The CAP-alphaCTD-DNA complex. Science, 297, 1562–1566. [DOI] [PubMed] [Google Scholar]