Significance
Chitin perception by plant receptors triggers various defense responses important for plant immunity. We show the molecular basis of chitin recognition by the rice receptor, CEBiP (chitin-elicitor binding protein), and following receptor dimerization based on the results of biochemical studies, epitope mapping by saturation transfer difference NMR spectroscopy and molecular modeling/docking studies. These results clearly indicated that two CEBiP molecules simultaneously bind to one N-acetylchitoheptaose/octaose from the opposite side, through a binding site in the central lysin motif region, resulting in the dimerization of CEBiP. Based on these observations, we proposed a hypothetical model for the ligand-induced activation of a receptor complex, involving CEBiP and Oryza sativa chitin-elicitor receptor kinase 1 (OsCERK1).
Keywords: plant immunity, MTI/PTI, chitin signaling, receptor–ligand interaction, LysM-receptor
Abstract
Perception of microbe-associated molecular patterns (MAMPs) through pattern recognition receptors (PRRs) triggers various defense responses in plants. This MAMP-triggered immunity plays a major role in the plant resistance against various pathogens. To clarify the molecular basis of the specific recognition of chitin oligosaccharides by the rice PRR, CEBiP (chitin-elicitor binding protein), as well as the formation and activation of the receptor complex, biochemical, NMR spectroscopic, and computational studies were performed. Deletion and domain-swapping experiments showed that the central lysine motif in the ectodomain of CEBiP is essential for the binding of chitin oligosaccharides. Epitope mapping by NMR spectroscopy indicated the preferential binding of longer-chain chitin oligosaccharides, such as heptamer-octamer, to CEBiP, and also the importance of N-acetyl groups for the binding. Molecular modeling/docking studies clarified the molecular interaction between CEBiP and chitin oligosaccharides and indicated the importance of Ile122 in the central lysine motif region for ligand binding, a notion supported by site-directed mutagenesis. Based on these results, it was indicated that two CEBiP molecules simultaneously bind to one chitin oligosaccharide from the opposite side, resulting in the dimerization of CEBiP. The model was further supported by the observations that the addition of (GlcNAc)8 induced dimerization of the ectodomain of CEBiP in vitro, and the dimerization and (GlcNAc)8-induced reactive oxygen generation were also inhibited by a unique oligosaccharide, (GlcNβ1,4GlcNAc)4, which is supposed to have N-acetyl groups only on one side of the molecule. Based on these observations, we proposed a hypothetical model for the ligand-induced activation of a receptor complex, involving both CEBiP and Oryza sativa chitin-elicitor receptor kinase-1.
Plants have the ability to detect potential pathogens through the recognition of microbe-associated molecular patterns (MAMPs; also known as pathogen-associated molecular patterns), such as flagellin, elongation factor Tu (EF-Tu), peptidoglycan, LPS, chitin, and β-glucan, which are typical molecular signatures for whole classes of microbes (1, 2). MAMP-triggered defense is the first barrier to prevent the invasion of pathogens and plays a major role in the basal resistance of plants against various pathogens. It is also well known that this defense system is strikingly similar to the innate immunity of animals (1, 3, 4).
Leucine-rich repeat receptor-like kinases, flagellin-sensetive 2 (FLS2), and EF-Tu receptor, have been shown to recognize bacterial flagellin and EF-Tu, respectively, and serve as receptors for these MAMPs (5). On the other hand, two types of lysin motif (LysM) proteins, CEBiP (chitin-elicitor binding protein) and CERK1 (chitin-elicitor receptor kinase-1), were identified as the cell-surface receptor for chitin, a representative fungal molecular pattern (6–8). Knockout/-down experiments of these genes showed that both of these LysM proteins are required for chitin perception and signaling in rice, whereas CEBiP-type molecules are not involved in chitin signaling in Arabidopsis, indicating the difference between the chitin receptor systems in these model plants (8). Additionally, another LysM receptor-like kinase, LYK4, was also indicated to contribute to chitin signaling in Arabidopsis (9). In the case of rice, it was also shown that CEBiP and Oryza sativa (Os)CERK1 form a heterooligomeric receptor complex ligand dependently (10).
Both CEBiP and OsCERK1 have LysMs, which have been known to bind peptidoglycan and chitin (11), in their ectodomains. In Arabidopsis, CERK1 was shown to bind chitin and trigger immune responses as a kind of “all-in-one” receptor. On the other hand, CEBiP seems to play a major role in the perception of chitin in rice, as the knockdown of CEBiP almost abolished the binding of a radio-labeled chitin oligosaccharide to the plasma membrane, whereas OsCERK1 was shown not to bind chitin (6, 12). Liu et al. recently reported that two other CEBiP homologs, OsLYP4 and -6, also bind chitin and contribute to chitin responses and disease resistance in rice (13), although it is not clear to what extent these proteins contribute as the cell surface receptor for chitin oligosaccharides.
It was also shown that the perception of peptidoglycan in Arabidopsis requires CEBiP-like molecules (14). Arabidopsis homologs of CEBiP, LYM1 and LYM3, play a major role for the binding of peptidoglycan and activation of downstream defense responses through the receptor kinase, CERK1. These results showed that the receptor kinase CERK1 is required for both chitin and peptidoglycan signaling, at least in Arabidopsis. The peptidoglycan receptor system in Arabidopsis seems similar to the rice chitin receptor for the requirement of a binding protein and a receptor kinase, although the receptor complex formation by these two proteins was not confirmed.
Thus, the detailed analysis of ligand recognition by these CEBiP-like molecules and succeeding formation and activation of receptor complex is critically important to understand the molecular mechanisms leading to the activation of downstream defense responses triggered by these MAMPs. Such information would also contribute to the design of novel receptor molecules suitable for future biotechnological application. We show herein the results obtained by biochemical studies on the binding site of CEBiP, epitope mapping of chitin oligosaccharides by saturation transfer difference (STD) NMR spectroscopy, and molecular modeling/docking studies combined with site-directed mutagenesis of the ectodomain of CEBiP. These results clearly indicated that two CEBiP molecules simultaneously bind to one N-acetylchitoheptaose/octaose, (GlcNAc)7/8, through a binding site located in the central LysM region of the ectodomain, resulting in the dimerization of CEBiP. Based on these observations, we proposed a hypothetical model for the ligand-induced activation of a receptor complex, involving both CEBiP and OsCERK1.
Results
Functional Dissection of the Ectodomain of CEBiP.
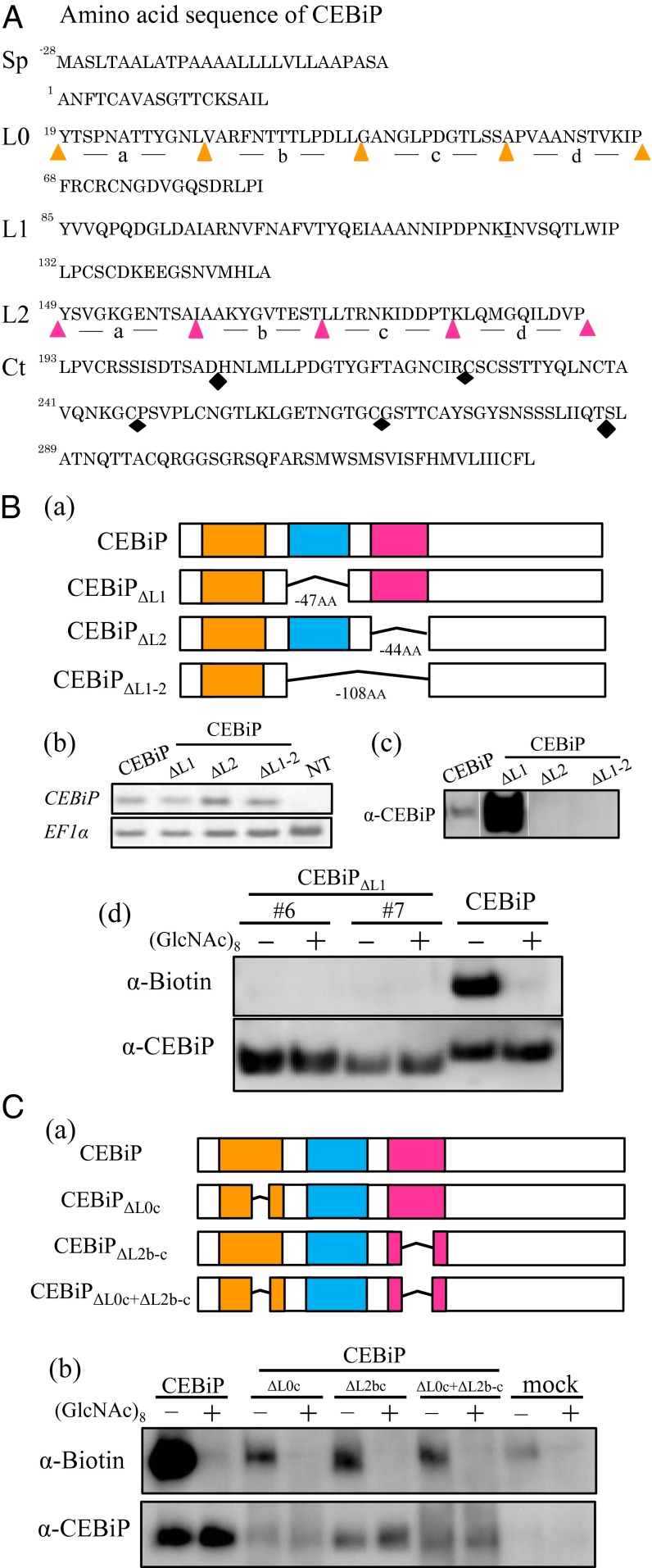
As described previously, CEBiP has two LysMs in the region of Y85-P131 (LysM1) and Y149 to P192 (LysM2) of the ectodomain (Fig. 1A) (6). To evaluate the contribution of these LysMs for chitin binding, deletion mutants for these LysMs were constructed and expressed in tobacco BY-2 cells (Fig. 1B, a). Expression of these mutant genes in the corresponding tobacco BY-2 cell lines was confirmed by RT-PCR (Fig. 1B, b). Binding of these mutant proteins to chitin oligosaccharides were examined by using affinity labeling with biotinylated (GlcNAc)8 (GN8-Bio) (15).
Fig. 1.
Functional analysis of LysM regions in the ectodomain of CEBiP by deletion experiments. (A) Amino acid sequence of CEBiP. Sp, signal peptide; L0, LysM0; L1, LysM1; L2, LysM2; Ct, C-terminal region. LysM0 and LysM2 were divided into four parts, from L0a to L0d and L2a to L2d, for deletion experiments. The C-terminal region was also divided into four parts for similar experiments (black diamonds indicate the border of each segment). (B) Effect of the deletion of LysM regions in CEBiP on GN8-Bio binding. (a) Constructs for the deletion experiments; (b and c) Expression of the deletion mutants in the tobacco BY-2 cells was analyzed by RT-PCR or Western blotting with an anti-CEBiP antibody (α-CEBiP); (d) Affinity cross-linking of GN8-Bio to the microsomal fractions (MF) from the tobacco BY-2 cells expressing deletion mutants. GN8-Bio (0.4 µM) was mixed with the MF in the presence (+) or absence (−) of excess (40 µM) unlabeled (GlcNAc)8 as a competitor. Cross-linking with EGS and the detection with anti-biotin antibody (α-Biotin) were performed as described in Materials and Methods. (C) Effect of the partial deletion of LysM0 and LysM2 on GN8-Bio binding. (a) Constructs for the deletion experiments; (b) affinity cross-linking of GN8-Bio to the MF from the transgenic N. benthamiana leaves expressing the deletion mutants.
Although the mutant protein with LysM1 deleted (CEBiPΔL1) was detected in the microsome from the BY-2 cells with an anti-CEBiP antibody, the other two mutant proteins, deletion of the LysM2 (CEBiPΔL2) or deletion of both LysM1 and LysM2 (CEBiPΔL1-2), could not be detected, indicating the deletion of LysM2 affected the expression of mutant proteins, probably because of the instability of the expressed proteins (Fig. 1B, c). Affinity-labeling experiments with the microsomal membranes isolated from two independent BY-2 cell lines expressing CEBiPΔL1 showed that the CEBiPΔL1 protein did not bind GN8-Bio, although the mutant protein itself was highly expressed in these cell lines (Fig.1B, d). The result suggested the importance of LysM1 for the chitin binding activity of CEBiP.
To further characterize the contribution of these LysMs for chitin binding, we examined their interchangeability in the ectodomain of CEBiP. We designed the chimeric constructs such as CEBiPL2→L1 (LysM2 was replaced with LysM1) and CEBiPL1→L2 (LysM1 was replaced with LysM2), and expressed with a transient expression system of Nicotiana benthamiana (SI Appendix, Fig. S1A, a). The expression of these chimeric genes in the transfected leaves was confirmed by RT-PCR (SI Appendix, Fig. S1A, b). Although the presence of CEBiPL1→L2 protein was detected by the anti-CEBiP antibody, CEBiPL2→L1 protein could not be detected, again indicating the importance of LysM2 region for the expression of these proteins (SI Appendix, Fig. S1A, c). Replacement of LysM1 with LysM2 in CEBiPL1→L2 abolished the detection by affinity labeling with GN8-Bio, further supporting the importance of LysM1 for chitin binding (SI Appendix, Fig. S1A, d).
In addition to LysM1 and LysM2, the analysis of the hidden Markov model (HMM) in this study evidenced the presence of third LysM domain at the N-terminal side of the LysM1, although not identified as LysM by domain prediction algorithms (PFAM, www.sanger.ac.uk) (16). Actually, this region was described as a further LysM in CEBiP in a recent report by Fliegmann et al. (17). Thus, we designated this region as LysM0 and examined its contribution to chitin binding. Unfortunately, the deletion mutant that lacked whole LysM0 could not be expressed in N. benthamiana. We then divided LysM0 into four parts (designated as L0a to L0d) (Fig. 1A and SI Appendix, Fig. S1B, a) and deleted each of them from CEBiP. The construct lacking L0d (CEBiPΔL0d) resulted in the failure of the expression of mutant protein (SI Appendix, Fig. S1B). On the other hand, mutant proteins with the deletion of L0a, L0b, and L0c (CEBiPΔL0a, CEBiPΔL0b, and CEBiPΔL0c) were successfully expressed and shown to bind GN8-Bio (SI Appendix, Fig. S1B). CEBiPΔL0c especially showed the highest binding activity (Fig. 1C, b and SI Appendix, Fig. S1B, d). Similarly, a deletion mutant lacking a portion of LysM2 (CEBiPΔL2b-c) was successfully expressed and shown to bind GN8-Bio (Fig. 1C, b). Finally, a deletion mutant in which both of these regions were deleted (CEBiPΔL0c+ΔL2b-c) showed the binding to GN8-Bio, again indicating the central role of LysM1 in the recognition of chitin oligosaccharides (Fig. 1C, b).
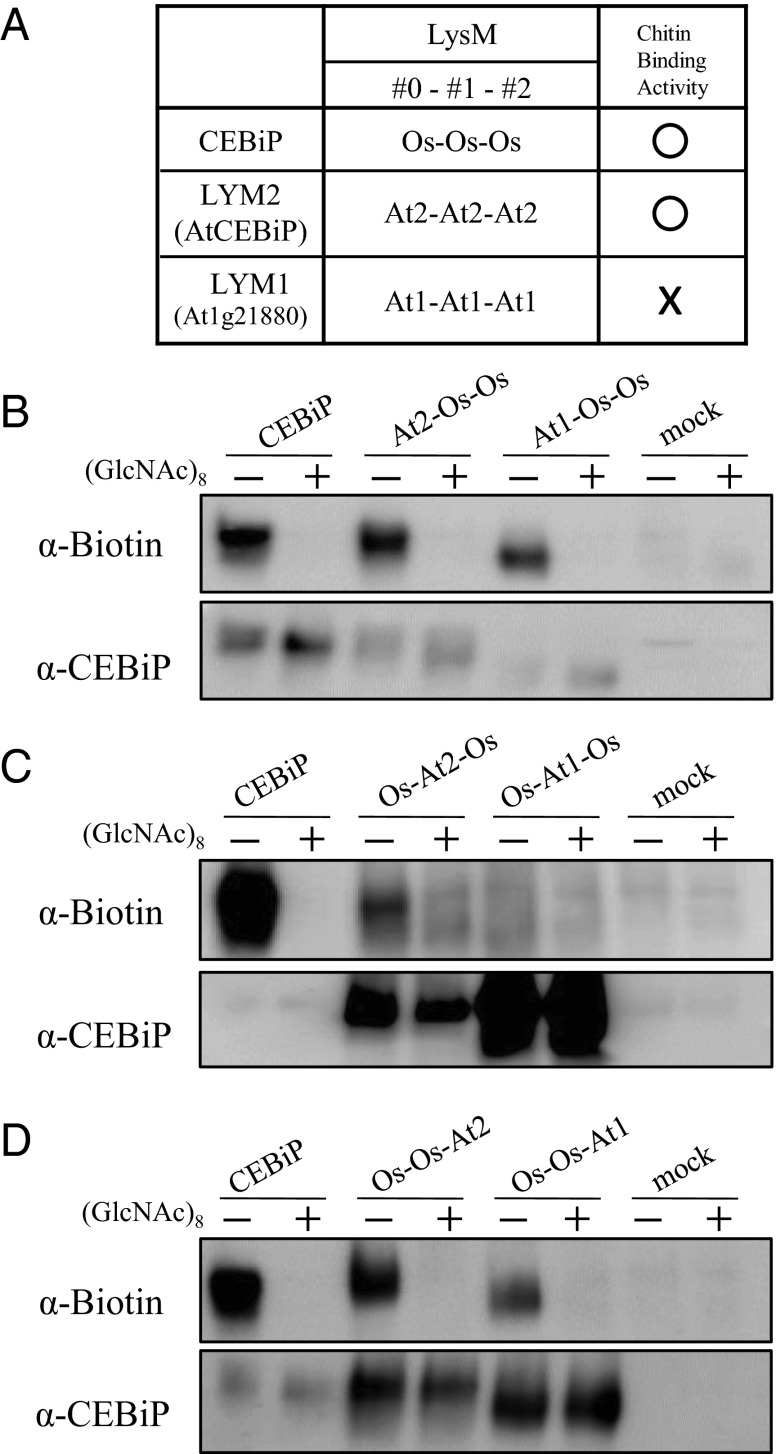
We also analyzed the function of each LysM in CEBiP by replacing them with the corresponding regions of an Arabidopsis CEBiP homolog, which does not bind chitin. Among the three Arabidopsis CEBiP homologs, it was shown previously that LYM2 (AtCEBiP) binds chitin oligosaccharides, whereas LYM1 and LYM3 bind peptidoglycan but do not bind chitin (Fig. 2A) (12, 14).
Fig. 2.
Functional analysis of LysM regions of CEBiP by replacement experiments with the corresponding regions of Arabidopsis CEBiP homologs. (A) Designation of LysMs in CEBiP and the Arabidopsis homologs (LYM2/AtCEBiP and LYM1/At1g21880). Chitin oligosaccharide binding ability of each protein was also shown. (B–D) Chitin oligosaccharide binding ability of chimeric proteins analyzed by affinity labeling with GN8-Bio.
Chimeric constructs, in which a LysM motif of CEBiP was replaced by the corresponding region of LYM1 or LYM2, were generated and expressed in N. benthamiana. Chimeric proteins with the replacement of LysM0 or LysM2 with the corresponding region of LYM1 bound GN8-Bio as similar to the wild-type CEBiP, whereas the chimeric CEBiP protein replaced for the LysM1 with the part of LYM1 completely lost the binding activity (Fig. 2 B–D). On the other hand, all of the constructs in which each LysM of CEBiP was replaced with the chitin-binding homolog of Arabidopsis, LYM2 (AtCEBiP), retained their ability to bind GN8-Bio. These results clearly demonstrated that LysM1 plays a critical role for the chitin recognition, supporting the results obtained by the deletion experiments.
In addition to the analysis of LysM domains, we also evaluated the possible contribution of an extended C-terminal region in the CEBiP ectodomain, which possesses 136 amino acid residues with high cysteine content, for chitin binding. We divided the region between H207 to T286 into four parts [deleted region; G267 to T286 (ΔC-20), P247 to T286 (ΔC-40), C227 to T286, (ΔC-60), H207 to T286 (ΔC-80)] (Fig. 1A and SI Appendix, Fig. S1C, a) and expressed the corresponding deletion mutants in N. benthamiana. All deletion mutants completely retained the chitin binding activity (SI Appendix, Fig. S1C, b), indicating that this C-terminal region of CEBiP ectodomain does not affect chitin binding activity significantly.
STD NMR Epitope Mapping of Chitin Oligosaccharides in the Presence of the Ectodomain of CEBiP.
STD NMR spectroscopy (18–21) was used to map interacting epitopes of chitin oligosaccharides and to reveal the molecular mechanism of interaction with the LysM domains of CEBiP. In detail, we acquired a series of STD NMR spectra using, as ligands, chitin fragments of different length, starting from tri- to octasaccharides, with the aim to determine the best oligosaccharide length in the interaction with the LysM domains. Because the previous experiments indicated that LysM1 is important for chitin binding and LysM2 contributes to stable expression, we decided to use the LysM1-LysM2 region of CEBiP for STD NMR studies. We recombinantly produced LysM1-LysM2 domains (LysM1-2, residues 85–192) as fused to a carrier protein, thioredoxin (Trx-tag), because STD NMR is highly successful for high molecular weight systems. Before NMR experiments, we analyzed the conformational state of LysM1-2 once Trx had been cleaved using far-UV CD spectra (SI Appendix, Fig. S2). Spectra are typical of proteins with a high content of β-sheets, with a minimum at 204 nm. Consistent with the typical LysM fold, CD signal at 222 nm also evidences the existence of an α-helix (SI Appendix, Fig. S2B). Because thioredoxin has been used as a stabilizing partner of LysM1-2, we optimized the STD conditions to perform saturation transfer double-difference (STDD) NMR experiments useful to overcome strong background signals and to subtract possible artifacts because of contingent unspecific interactions between ligands and the stabilizing partner (22–24). The STDD NMR requires the acquisition of an additional STD spectrum of the receptor in absence of ligands, which is then subtracted from the STD spectrum acquired in the presence of ligands. In our case, STDD spectra were obtained by the subtraction between the regular STD NMR spectrum and those performed on the ligand in the presence of thioredoxin alone.
Although a qualitative analysis of the STDD spectra using N-acetylchitotri-, tetra-, and hexaose, (GlcNAc)3,4, or 6, as ligands demonstrated their binding with the LysM domains (SI Appendix, Fig. S3), a deeper look at the spectra showed a clear dependency of binding ability on ligand size. In detail, low STD effects were observed on the tri- and tetrasaccharides (SI Appendix, Fig. S3 B and C), and higher signals were obtained using the hexasaccharide (SI Appendix, Fig. S3D). Although the analysis was only qualitative, it could be inferred that STD enhancements belonged to all of the protons of the sugar units.
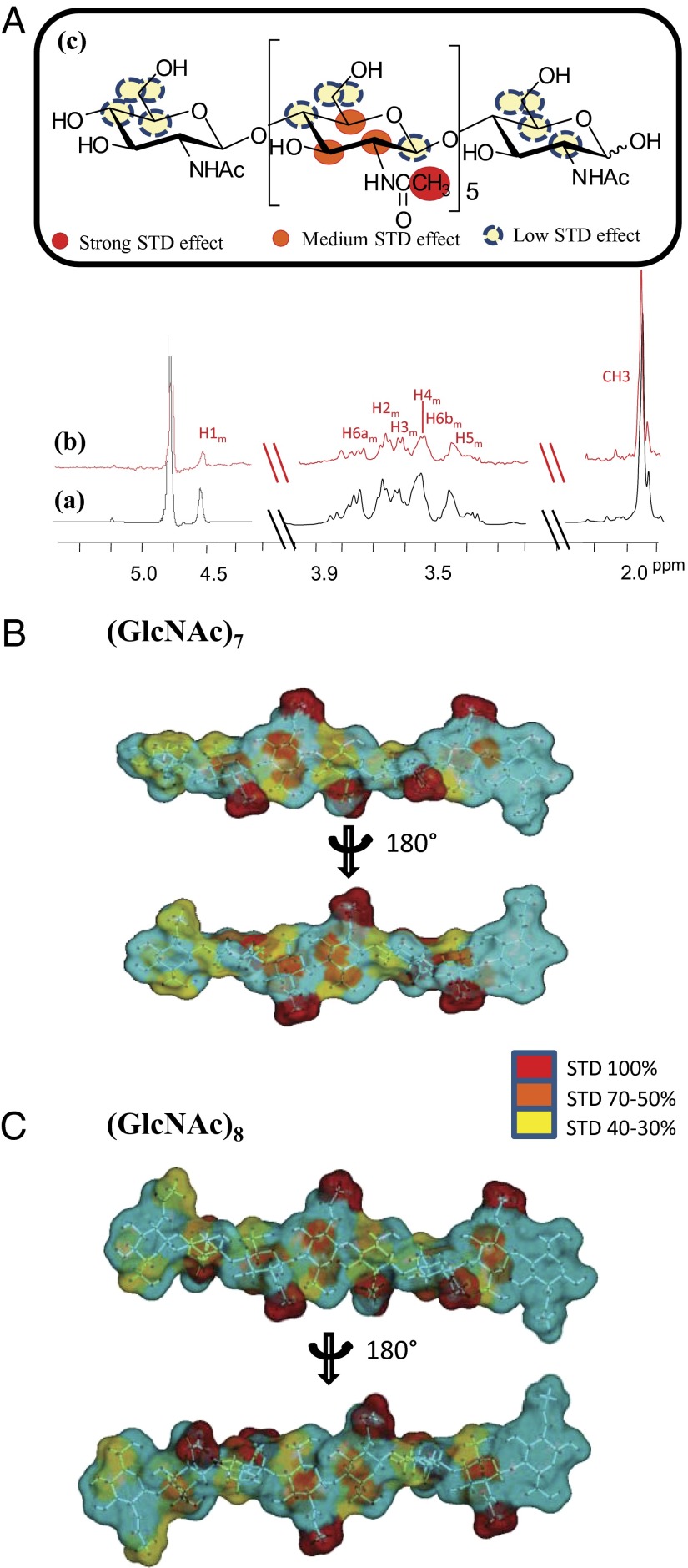
Based on the above findings, we focused our attention on studying chitin oligosaccharides with higher molecular weight: that is hepta- and octasaccharides. The STDD NMR spectrum of the heptasaccharide bound to the protein is reported in Fig. 3A [see SI Appendix for NMR assignment (SI Appendix, Table S1)]. To precisely map ligand epitopes in close contact with the protein, we acquired STD build up curves by collecting spectra at different saturation times (25–28) and fitting experimental data with the monoexponential equation: STD = STDmax[1 – exp (−ksat t)] (see Materials and Methods, NMR Spectroscopic Analysis for complete details). This method avoids artifacts in the epitope definition, which are consequences of differences in ability to accumulate saturation in the free state, allowing to obtain the real STD effect of each proton, independently of T1 bias, and to prevent any misinterpretation of STD enhancements.
Fig. 3.
STD NMR-derived epitope mapping on (GlcNAc)7 and (GlcNAc)8 bound to Trx-LysM1-2. (A) Reference 1H NMR spectrum (a) and STDD NMR spectrum (b) of mixture Trx-LysM1-2: (GlcNAc)7 (1:100). (c) Chemical structure and epitope for binding of (GlcNAc)7 to Trx-LysM1-2; relative STD intensities are color coded according to the scale and refers to the relative STD effects as shown in SI Appendix, Table S2. (B and C) STD quantitative analysis-derived epitope mapping on the molecular envelope of (GlcNAc)7 and (GlcNAc)8 with color coding from the highest (red) to lowest (yellow) observed STD effect.
Thus, the binding epitope of the heptasaccharide is shown in Fig. 3B (SI Appendix, Table S2). The most prominent STD effect belonged to the acetyl group thus contributing primarily to the binding process; additionally, different STD enhancements were observed for all of the protons of internal residues of GlcNAc.
From these results, it was inferred that the acetyl group was in close contact and pivotal in the interaction with the protein and, according to the qualitative analysis, protons H2m, H3m, and H5m of middle residues were strongly involved in the binding with LysM domain as well.
These STD studies were then complemented with the analysis of trNOESY experiments both free in solution and in the presence of the LysM1-2 domain to assess oligosaccharide conformational changes upon binding (27, 29–31). Thus, trNOESY experiments were carried out at a 7:1 molar ratio with different NMR mixing times; negative NOEs were observed for both the free and bound states, and detailed analysis of interresidue cross-peaks in the trNOESY spectrum allowed the deduction of the bioactive conformation of the oligosaccharide product when bound to the protein. Remarkably, key NOE distances obtained in both the free and bound states showed only minor changes, thus suggesting that the bioactive conformation of chitin heptasaccharide resembled the most populated conformational state of the free ligand (SI Appendix, Table S3). The NOE data were used to obtain a conformational model of the oligosaccharide by molecular mechanics and molecular dynamics simulations. A molecular snapshot of a representative conformer is depicted in Fig. 3B, with the STD-derived epitope mapping on the molecular envelope of heptasaccharide with color coding from the highest (Fig. 3B, red) to lowest (Fig. 3B, yellow) observed STD effect.
By using the same strategy, the interaction of CEBiP with (GlcNAc)8 was evaluated and the corresponding epitope map was obtained using a qualitative analysis because of scarce solubility of the substrate, (Fig. 3C and SI Appendix, Fig. S4). The corresponding results were in good agreement with collected data on the heptasaccharide; the acetyl group gave rise to the highest STD effect, indicating that this moiety was fundamental in the binding process with LysM1-2 of CEBiP, and once again protons H2m, H3m, and H5m also significantly contributed to the interaction. Furthermore, both oligosaccharide ends showed in all cases the lowest interaction with the LysM1-2 domains.
Therefore, the binding epitope as derived by NMR analysis provided clear support that CEBiP, with its LysM domains, recognized and bound chitin oligosaccharides and showed a strong preference for longer chain substrates, especially hepta- and octasaccharides.
Docking Model of Chitin Oligosaccharide with LysM1 Domain of CEBiP.
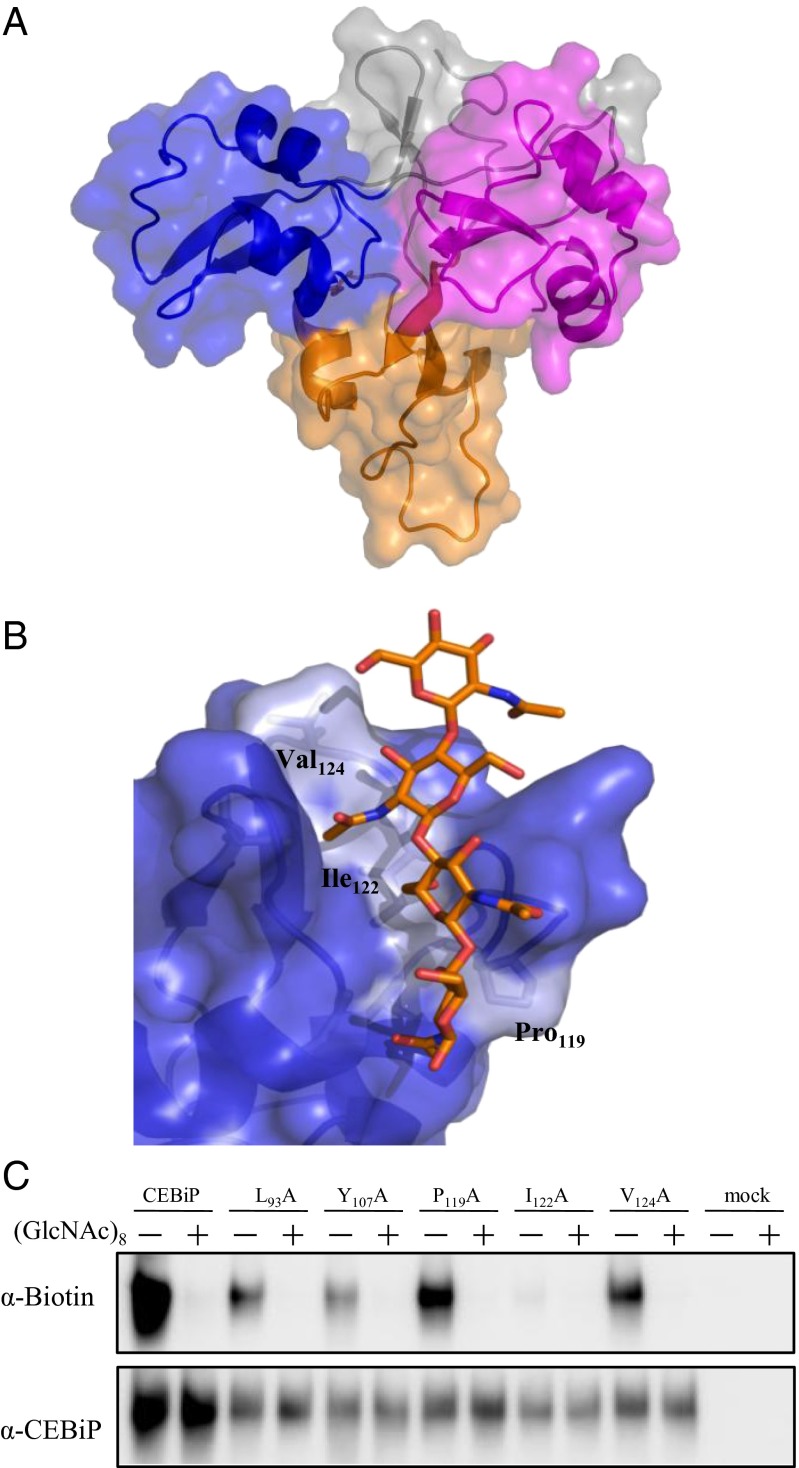
The data so obtained enabled the rational modeling of oligosaccharides into the CEBiP binding site on the LysM1 region. To this aim, we determined the homology model structure of CEBiP, after consensus-based sequence alignment. The entire extracellular region of CEBiP was built using the program MODELER and the structure of CERK1 from Arabidopsis thaliana (32) as a template (Fig. 4A).
Fig. 4.
Structural basis of chitin oligosaccharides binding. (A) Ribbon and surface representation of the homology model of CEBiP ectodomain. LysM0, LysM1, and LysM2 are drawn in orange, blue, and magenta, respectively. Other regions are colored gray. (B) Modeling of binding of N-acetylchitooligosaccharides to the LysM1domain of CEBiP. (C) Binding of GN8-Bio to mutant CEBiP proteins. CEBiP proteins containing the mutation for the indicated amino acid residues were expressed in N. benthamiana, from which MF for affinity labeling was prepared.
The combination of STD NMR epitope mapping data with knowledge of the bound conformation of ligands, obtained by trNOESY experiments, is a powerful method to build up models of protein–ligand interaction. Using the bioactive conformation observed by NMR, we modeled oligosaccharide binding to the LysM1 domain of CEBiP. As shown in Fig. 4B, four GlcNAc moieties can be accommodated in the binding cleft of LysM1 domain. Because STD NMR data show that external sugar moieties of oligosaccharides barely contribute to binding, it can be suggested that the smallest oligosaccharide, which saturates the four binding subsites on LysM1, is (GlcNAc)6. In this interaction model of the hexasaccharide, main interactions involve the three sugars from GlcNAc-2 to GlcNAc-4 (numbered from the nonreducing end) and weaker interactions characterize GlcNAc-5, whereas GlcNAc-1 and GlcNAc-6 do not interact with LysM1. Consistent with STD NMR results, the acetyl moieties of (GlcNAc)6 provide a crucial contribution to binding. Both GlcNAc-2 and GlcNAc-4 sugar moieties are involved in hydrophobic interactions (with Leu93 and Val87, respectively) and in hydrogen bonds with backbone nitrogen atoms (of Leu93 and Gln90, respectively) (SI Appendix, Fig. S5). Apart from hydrogen bonds mediated by the acetyl moieties, (GlcNAc)6 binding is mainly stabilized by hydrophobic interactions involving sugar ring carbons, a finding which well agrees with our STD NMR results (Fig. 3). Our interaction model also explains the low STD effects observed upon binding of tri- and tetrasaccharides (SI Appendix, Fig. S3 B and C), compared with hexasaccharide or larger substrates (SI Appendix, Fig. S3D).
Assessing Key Residues Involved in Chitin Binding.
Modeling of binding of chitin oligosaccharides to the LysM1 domain of CEBiP identified possible residues involved in binding (Fig. 4B). The predicted binding site is composed of hydrophobic residues like Pro119, Ile122, and Val124. Among these residues, Ile122 is located in the central part of the putative binding cleft, a finding which suggested us that this residue may be critical for oligosaccharide binding (Fig. 4B).
To corroborate this hypothesis, Ile122 was mutated to Ala. The mutated protein, LysM1-2 I122A, was first checked by CD spectroscopy to exclude that the mutation could have altered its structural integrity (SI Appendix, Fig. S2) and then analyzed for its interaction with chitin oligosaccharides by STDD NMR experiments under the same conditions used for wild type LysM1-2.
NMR results with the heptasaccharide unequivocally showed that the mutant protein LysM1-2 I122A was not able to bind the heptasaccharide at all. Indeed, no STD signals were observed (SI Appendix, Fig. S6). The inability of the mutant to interact with heptasaccharide definitively identifies Ile122 as a key amino acid in the recognition process of chitin oligosaccharides by CEBiP LysM1.
Mutant CEBiP proteins containing mutation for each of Leu93, Tyr107, Pro119, Ile122, and Val124, which were suggested to have close contact with the heptasaccharide in modeling studies, were also expressed in N. benthamiana and tested for their ability to bind chitin oligosaccharides by using affinity labeling. The microsomal membrane prepared from N. benthamiana leaves expressing the I122A mutant showed the presence of mutant protein but did not show the binding to GN8-Bio, whereas the membrane preparation for the other four mutants showed the binding to GN8-Bio, supporting the conclusion on the critical importance of Ile122 for chitin binding (Fig. 4C). In line with this notion, alignment of amino acid sequences of CEBiP homologs indicated that the chitin binding homologs such as LYM2 (AtCEBiP) and MtLYM2 carried Ile at the position corresponding to Ile122 of CEBiP. On the other hand, peptidoglycan binding homologs, AtLYM1 and AtLYM3, carried Leu instead of Ile. OsLYP4 and -6, which were reported to bind both chitin and peptidoglycan, carried Val and Leu, respectively (SI Appendix, Table S4).
To further analyze the contribution of aliphatic side chain of Ile122 in chitin binding, we substituted Ile122 with Val or Leu and analyzed the binding of GN8-Bio to the mutant protein by affinity labeling. As shown in SI Appendix, Fig. S7, substitution of Ile122 with Leu retained similar or even higher affinity to GN8-Bio but the substitution with Val significantly decreased the affinity. These results indicated that the length of the aliphatic side chains of these amino acid residues is a critical factor for the interaction with chitin oligosaccharides. These results well agree with our modeling studies, which show interactions of the bound oligosaccharide with the side chain of the hydrophobic residue at position 122. These results and the presence of Leu/Val in the corresponding positions of peptidoglycan binding homologs may suggest that these residues are also involved in the interaction with the glycan portion of peptidoglycan, although the epitopes involved in the interaction with these proteins are not yet clear (13, 14).
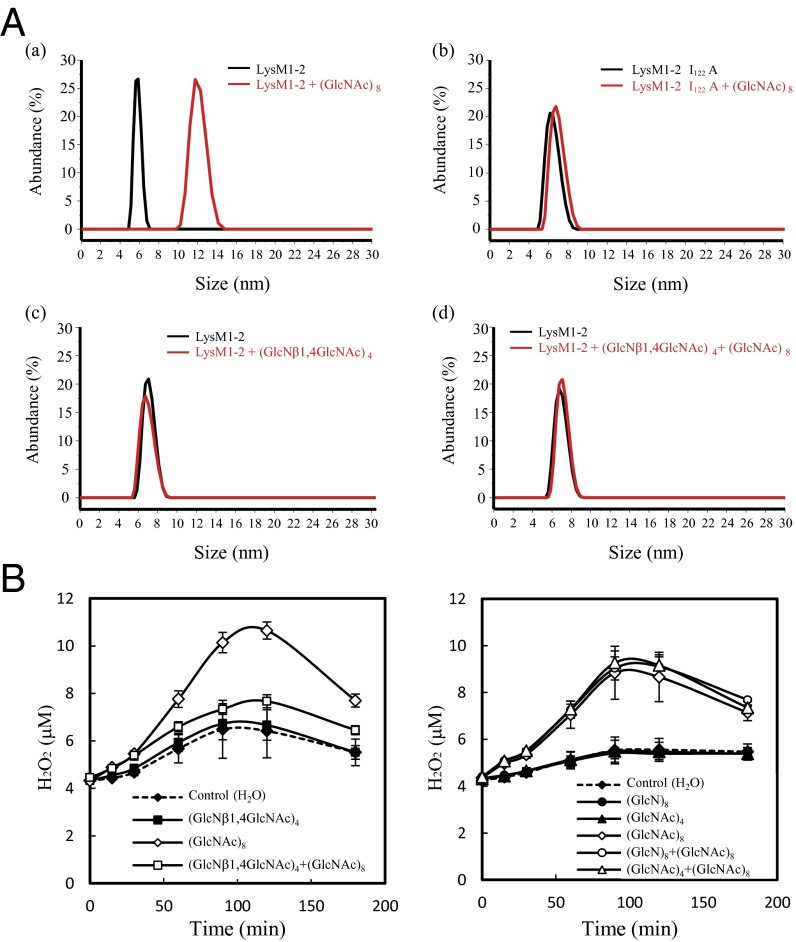
Binding of (GlcNAc)8 Dimerizes the Ectodomain of CEBiP.
Size-measurement studies of LysM1-2 were carried out to investigate the effect of (GlcNAc)8 on its oligomerization state. LysM1-2 samples before and after incubation with (GlcNAc)8 were measured using light-scattering experiments. As shown in Fig. 5A, a, (GlcNAc)8 induces a significant increase of LysM1-2 size distribution, with an average molecular diameter raising from 6.12 ± 0.55 nm before incubation to 12.45 ± 1.61 nm. Consistently, weight average molar mass values measured for LysM1-2 before and after (GlcNAc)8 treatment are 11,680 ± 116.8 Da and 22,610 ± 226.1 Da, corresponding to a monomeric and dimeric organization of the molecule, respectively (Fig. 5A, a). These results unambiguously show that LysM1-2 dimerizes in the presence of (GlcNAc)8. Parallel studies were carried out on LysM1-2 I122A mutant. Consistent with the observed inability of this mutant to interact with the sugar, these studies showed that (GlcNAc)8 is unable to induce dimerization (Fig. 5A, b).
Fig. 5.
CEBiP dimerization and activation of chitin receptor complex by chitin oligosaccharides. (A) Molecular size-shift measurements of the effect of (GlcNAc)8 on the oligomerization state of LysM1-2 (a) and LysM1-2 I122A mutant (b), respectively. (c) Effect of (GlcNβ1,4GlcNAc)4 on molecular size. (d) Effect of (GlcNAc)8 after pretreatment with an equimolar amount of (GlcNβ1,4GlcNAc)4. All measurements were conducted by light scattering using a 100-μM protein concentration and a protein:ligand ratio of 1:2. (B) Specific inhibition of (GlcNAc)8-induced ROS generation by (GlcNβ1,4GlcNAc)4 in rice cells. Concentration of (GlcNAc)8 was 0.1 nM and the other oligosaccharides were 100nM. Competing oligosaccharides, (GlcNβ1,4GlcNAc)4, (GlcNβ1,4GlcN)4 or (GlcNA)4 were pretreated 10 min before the addition of (GlcNAc)8.
A Unique Oligosaccharide, (GlcNβ1,4GlcNAc)4 Acts as an Antagonist for the Binding of GN8-Bio, (GlcNAc)8-Induced Dimerization and Reactive Oxygen Species Generation.
Chitin, like cellulose, has every sugar unit flipped by 180° with respect to its neighbors to give a chitobiose repeat segment. Because molecular modeling suggested a key role of acetyl moieties in LysM1 binding, we further investigated the binding/dimerization mechanism of CEBiP using a unique oligosaccharide, (GlcNβ1,4GlcNAc)4, in which sugar residues are alternately N-acetylated, therefore carrying N-acetyl moieties only on one side. Using affinity labeling, we observed that (GlcNβ1,4GlcNAc)4 inhibits binding of GN8-Bio to CEBiP, indicating the competition of this oligosaccharide with the labeled ligand for the chitin binding site (SI Appendix, Fig. S8). Therefore, we performed light-scattering experiments upon incubation with (GlcNβ1,4GlcNAc)4. Results showed that, unlike (GlcNAc)8, (GlcNβ1,4GlcNAc)4 does not induce changes in the average molecular diameter of LysM1-2, proving that this sugar is unable to induce receptor dimerization (Fig. 5A, c). We also observed that pretreatment of LysM1-2 with (GlcNβ1,4GlcNAc)4 completely inhibited the ability of (GlcNAc)8 to induce receptor dimerization (Fig. 5A, d). In addition, we evaluated the biological activity of (GlcNβ1,4GlcNAc)4 using ROS generation in suspension-cultured rice cells. As shown in Fig. 5B, the oligosaccharide did not induce ROS generation by itself but inhibited the ROS generation induced by (GlcNAc)8, in other words, it acted as an antagonist. On the other hand, related oligosaccharides, (GlcNβ1,4GlcN)4 or (GlcNAc)4, did not show such inhibition of (GlcNAc)8-induced ROS generation (Fig. 5B). Taken together, these results show that the presence of N-acetyl groups in the alternating sugar residues of (GlcNβ1,4GlcNAc)4 enables the sugar to interact with the binding site of CEBiP but fails to trigger both dimerization and activation processes.
Discussion
CEBiP Binds Chitin Oligosaccharides Using the Binding Site Located in the Central LysM Domain.
As described in Results and also recently reported by Fliegmann et al. (17), CEBiP carries three LysM domains, namely LysM0 to LysM2. Biochemical studies with various deletion mutants, as well as chimeric proteins, clearly showed that the central LysM domain (LysM1) in the ectodomain of CEBiP plays a major role for the perception of chitin oligosaccharides. Of special note, the fact that the replacement of only LysM1 with the corresponding region of an Arabidopsis CEBiP homolog, which does not bind chitin (LYM1), resulted in the complete loss of chitin binding, whereas similar substitution with the region from a chitin-binding homolog (LYM2) maintained the ability to bind chitin, strongly supports the conclusion. Molecular modeling and site-directed mutagenesis studies confirmed the presence of the chitin binding site in LysM1. This conclusion matches the observation on the chitin binding site in the ectodomain of Arabidopsis CERK1 receptor kinase based on the crystal structure of CERK1 ectodomain with (GlcNAc)4, where the chitin oligosaccharide also bound to the central LysM domain of the ectodomain (32).
Molecular Basis of the Ligand Recognition by CEBiP.
Prompted by the above biological data, a combinatorial approach based on advanced STD and NOE-based NMR spectroscopic techniques and molecular dynamics allowed us to get fresh insight into the binding mechanism between the LysM domain in the ectodomain of CEBiP and chitin oligosaccharides. We have provided a molecular description of binding of the central LysM domain (LysM1) in the ectodomain of CEBiP to chitin oligosaccharides by revealing the conformations and epitope patterns of a plethora of chitin oligosaccharides bound to LysM1-2 domains.
STD NMR experimental data, gathered on chitin fragments, clearly showed that short-length oligosaccharides only weakly bound to LysM1, whereas a strong binding was detectable for (GlcNAc)7 and (GlcNAc)8. Both qualitative and quantitative analysis, based on the construction of STD build-up curves, allowed to precisely map ligand epitopes in contact with the protein. These studies demonstrated that N-acetyl groups of chitin oligosaccharides are crucial moieties for binding. Likely, N-acetyl groups are key molecular signatures that allow LysM1 to distinguish chitin from glucose-containing polymers, such as cellulose. Furthermore, the inner units of the chitin oligosaccharides exhibited stronger interaction with LysM motifs, whereas both the outer residues seemed to be less involved in the interaction, likely because more exposed to the solvent.
Using all of this information, we used modeling studies to produce a 3D model of LysM1 interaction with chitin oligosaccharides. These studies demonstrate that LysM1 domain of CEBiP accommodates chitin oligosaccharides in a hydrophobic cleft that hosts four GlcNAc moieties. Of these, two GlcNAc moieties mainly interact through hydrogen bonding and hydrophobic interactions mediated by their acetyl groups. Because STD NMR data show that outer ends of oligosaccharides barely contribute to binding, (GlcNAc)6 is the shortest oligosaccharide to saturate the four sugar binding subsites in LysM1 cleft. This finding well explains the significantly stronger binding of hexasaccharide or larger substrates and also their biological activities (15, 33). Our modeling study further indicated that several amino acid residues are in close contact with the bound chitin oligosaccharides. The analysis of chitin binding site in comparison with that observed in the crystal structure of CERK1 (32) shows that only two residues involved in sugar binding, Val87 and Ile122, are conserved (SI Appendix, Fig. S9). Of these residues, Ile122 is located in the central region of LysM1 binding cleft and provides a large hydrophobic interaction surface with the sugar. Consistently, site-directed mutagenesis of Ile122 to alanine completely hampered (GlcNAc)8 binding (Fig. 4C), indicating that Ile122 plays an important role in ligand recognition. The importance of Ile122 in chitin binding was further supported by the conservation of this residue at the corresponding positions of chitin binding homologs, LYM2 (AtCEBiP) and MtLYM2 (SI Appendix, Table S4).
Binding of (GlcNAc)7 or (GlcNAc)8 to LysM1 Domains Dimerizes CEBiP.
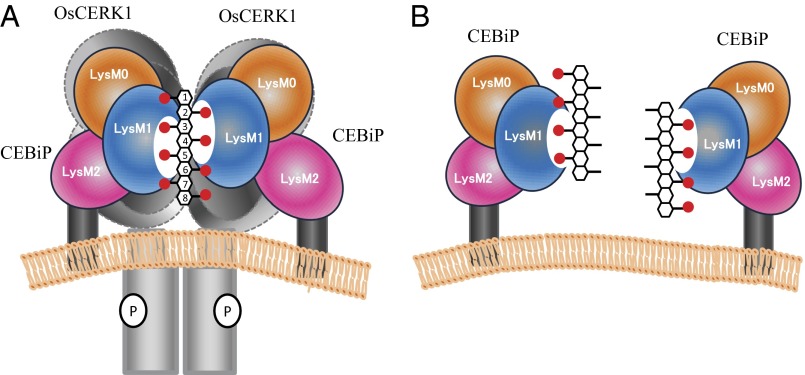
Light-scattering studies clearly evidence that (GlcNAc)8 is able to induce dimerization of LysM1-2 (Fig. 5A, a), whereas it fails to induce dimerization of the LysM1-2 I122A mutant (Fig. 5A, b), which is unable to bind (GlcNAc)8 (Fig. 4C). These results, together with STD NMR experiments and modeling studies, suggested a “sandwich-like” model in which two CEBiP molecules bind a single (GlcNAc)8 chain. In this model, LysM1 domains of two CEBiP molecules anchor N-acetyl moieties from opposite sides to produce a one-sugar staggered dimer (Fig. 6). Therefore, dimerization requires at least five internal GlcNAc moieties, four per monomer, with three shared between the two CEBiP molecules (3, 4, and 5 in Fig. 6A). Given this model, optimal dimerization can be induced by sugars larger than (GlcNAc)7, which contain an optimal number of internal GlcNAc moieties for cross-linking. However, elicitor activities were previously observed for both hexa- and heptamers (34, 35). Likely, the sandwich-like dimeric structure in Fig. 6 may assemble also in the presence of (GlcNAc)6, albeit leading to a less-stable complex. Consistently, the stronger binding we observe for (GlcNAc)7 and (GlcNAc)8, compared with (GlcNAc)6, may be ascribed to the formation of more stable CEBiP sandwich-like dimers.
Fig. 6.
Hypothetical model of the activation of CEBiP-OsCERK1 complex by (GlcNAc)8. (A) Sandwich-like model of activation of CEBiP-OsCERK1 receptor complex by (GlcNAc)8. (B) Model of dimerization/activation inhibition by (GlcNβ1,4GlcNAc)4.
To corroborate this “sandwich-like” dimerization, we measured the effect on receptor dimerization and activation of a unique oligosaccharide, (GlcNβ1,4GlcNAc)4, in which the four alternated N-acetyl groups are predicted to point only to one side of the molecule. (GlcNβ1,4GlcNAc)4 was able to bind the receptor but unable to induce its dimerization. In addition, a pretreatment of LysM1-2 with (GlcNβ1,4GlcNAc)4 hampered receptor dimerization in response to (GlcNAc)8 (Fig. 5A, d). Furthermore, (GlcNβ1,4GlcNAc)4 did not induce ROS generation in rice cells but inhibited the ROS generation by (GlcNAc)8. These results are fully consistent with our model, because (GlcNβ1,4GlcNAc)4 locks CEBiP through its N-acetyl groups but lacks the set of N-acetyl groups necessary to engage a further CEBiP molecule to form a CEBiP dimer. Because of this unique property, (GlcNβ1,4GlcNAc)4 acts as an antagonist by hampering receptor dimerization (Fig. 5A, d) and inhibiting ROS generation (Fig. 5B).
Ligand-induced dimerization of receptors is a critical event in various receptor systems (36–38), especially as a trigger of autophosphorylation of receptor kinase as classically reported for receptor tyrosine kinases. Recently, Liu et al. reported that (GlcNAc)8-induced dimerization of CERK1 receptor kinase is mainly based on immunoprecipitation analysis, but the detailed molecular mechanism leading to the dimerization was not clear (32). We showed herein the molecular mechanism of ligand-induced dimerization of CEBiP by combining a large set of chemical and biological techniques.
We recently reported that the chitin receptor systems in rice and Arabidopsis are significantly different (12). In the case of Arabidopsis, CERK1 receptor kinase seems to function as a ligand-binding protein and as a signaling molecule with the kinase activity. CEBiP homolog in Arabidopsis, LYM2 (AtCEBiP), seems not to contribute to chitin signaling, although the protein binds chitin oligosaccharides with a high affinity (9, 12). Interestingly, it was recently reported that LYM2 contributes to disease resistance of Arabidopsis independent of chitin signaling by CERK1 (39, 40). Faulkner indicated that the plasmodesmata-localized LYM2 is involved in chitin-induced plasmodesmata closure, which seems to be important for chitin-triggerd immunity (39). Thus, the ligand-induced dimerization of CERK1 directly links to the downstream signaling in Arabidopsis. On the other hand, both CEBiP and OsCERK1 are required for chitin signaling in rice. It was also shown that CEBiP binds chitin oligosaccharides with high affinity, whereas OsCERK1 does not. Moreover, it was shown that CEBiP and OsCERK1 form a heterooligomeric receptor complex in the presence of biologically active chitin oligosaccharide, (GlcNAc)8 (10). These results indicated that in the case of rice and other plant species equipped with a similar complex receptor system, ligand binding by CEBiP must lead to the activation of the receptor kinase, OsCERK1.
The simplest hypothesis to link the dimerization of CEBiP to the activation of OsCERK1 is that the dimerization of CEBiP leads to the dimerization of OsCERK1, which is expected to be closely associated with CEBiP. Actually, we showed that the ectodomain of CEBiP could interact with that of OsCERK1 by using a yeast two-hybrid assay (10). In addition, it was observed that a part of OsCERK1 was coimmunoprecipitated with CEBiP, even in the absence of (GlcNAc)8. Although the amount of coimmunoprecipitated OsCERK1 increased with the addition of (GlcNAc)8, these results indicated that at least a part of CEBiP and OsCERK1 interact with each other on the plasma membrane even in the absence of chitin oligosaccharides, probably through the interaction of their ectodomains. Thus, we surmise that the dimerization of CEBiP molecules brings their partnering OsCERK1 in close contact, resulting in the autophosphorylation of the receptor kinase (Fig. 6). Although the model is still hypothetical, it seems to be a promising scenario and we hope to examine the validity of this model further in future studies.
Materials and Methods
Elicitors and Plant Material.
Chitoheptaose and octaose were kindly supplied by the Yaizu Suisankagaku Industry and reacetylated before use. N-Acetylchitotetraose was purchased from SEIKAGAKU Co. (GlcNβ1,4GlcNAc)4 was chemo-enzymatically synthesized through three steps (41). Briefly, a di-N-acetylchitobiose derivative having an oxazoline skeleton was chemically synthesized according to a method of Kobayashi et al. (42), and its N-acetyl group was enzymatically removed by chitin deacetylase from Colletotrichum lindemuthianum ATCC 56676. The product, 2-methyl-{4-O-(2-amino-2-deoxy-β-glucopyranosyl)-1,2-dideoxy-α-glucopyrano}(2,1-D)-2-oxazoline was used as a monomer for the synthesis of (GlcNβ1,4GlcNAc)4 via a ring-opening polyaddition reaction with the aid of chitinase from Bacillus sp. (SEIKAGAKU). (GlcNβ1,4GlcNAc)4 was purified using an HPLC column, Asahipak NH2-P50 (Shimadzu) with the eluate, water/acetonitrile 25/75 (flow rate: 1 mL/min). Elution was monitored by a UV-detector at 210 nm. The fractions corresponding to the product were collected and lyophilized, and the resultant white powders were stored at room temperature.
Suspension-cultured rice cells (Oryza sativa L. cv. Nipponbare) were maintained using modified N-6 medium, as described previously (43). Rice cells were incubated on a rotary shaker at 25 °C and 120 rpm in the dark and were transferred to fresh medium every week. After every other transfer to new medium, the cell clusters were filtered through a 20-mesh screen to generate fine aggregates, and then were used for the next culture. Cells harvested 4 or 5 d after transfer to the new medium were used in the experiments.
Construction of Various Types of CEBiP Mutant Genes.
The expression vectors were constructed using the Gateway pENTR/d-TOPO cloning kit (Life Technologies). pENTR/d-TOPO-CEBiP, pENTR/d-TOPO-AtCEBiP and pENTR/d-TOPO-LYM1 were subcloned as previously described (6, 12). Binary vectors, pMDC32 destination vector (44) and pEAQ-HT-DEST1 destination vector, were kindly provided by Mark Curtis (University of Zurich, Zurich, Switzerland) and George Lomonossoff (John Innes Center, Norwich, United Kingdom), respectively.
Deletion and site-directed mutagenesis of CEBiP were carried out by inverse PCR with the corresponding primer sets (SI Appendix, Table S5) using pENTR/d-TOPO-CEBiP as template. The PCR products were treated with DpnI (TOYOBO) for cutting template, and then self-ligated using T4 DNA ligase/T4 polynucleotide kinase (Takara Bio).
For the construction chimeric CEBiP plasmid, amplified LysM DNA fragments were introduced into corresponding LysM deleted pENTR/d-TOPO-CEBiP by using the In-Fusion cloning kit (Clontech). The DNA fragments encoding each LysM were amplified from pENTR/d-TOPO-CEBiP, pENTR/d-TOPO-AtCEBiP and pENTR/d-TOPO-LYM1 using the primer sets (SI Appendix, Table S5). For the LysM-swapping experiments, LysM deleted pENTR/d-TOPO-CEBiP was amplified by inverse PCR using the primer sets (SI Appendix, Table S3) and pENTR/d-TOPO-CEBiP as a template. The amplified LysM DNA flagments were introduced into the LysM-deleted pENTR/d-TOPO-CEBiP by using the In-Fusion cloning kit. Both PCR products were treated with the cloning enhancer (Clontech) before in-fusion cloning reaction.
Plasmids of CEBiP and CEBiP mutants were introduced into pMDC32 or pEAQ-HT-DEST1 by using the LR reaction (Life Technologies). pMDC32 and pEAQ-HT-DEST1 were used for the tobacco BY-2 cells and N. benthamiana expression system, respectively. Before the LR reaction with pEAQ-HT-DEST1, entry vectors of CEBiP mutants were first digested with EcoRV or NruI to disable the kanamycin-resistance gene in the vector. The expression vectors thus obtained were transformed into Agrobacterium tumefaciens C58C1 by electroporation.
Expression of CEBiP and CEBiP Mutants in N. benthamiana and Tobacco BY-2 Cells.
N. benthamiana plants were grown for 4–6 wk in a 14-/10-h light/dark cycle at 20 °C (light) or 22 °C (dark). To express the CEBiP and various types of CEBiP mutants, transformed A. tumefaciens was pressure-infiltrated into N. benthamiana leaves as reported by Sainsbury et al. (45). Leaves were harvested at 6 d after infiltration and used for the preparation of microsomal fractions.
Suspension-cultured tobacco BY-2 cells were maintained in LSD medium. To generate BY-2 cells expressing LysM-deleted CEBiP, tobacco BY-2 cells were cocultivated with the transformed A. tumefaciens. After 2 d of cocultivation, transformed BY-2 cell lines were selected on LSD agar medium containing 50 mg/L hygromycin and 12.5 mg/L Meropenem for 2–3 wk. Selected cell lines were transferred into LSD liquid medium and used for the preparation of microsomal fractions.
Expression levels of CEBiP and CEBiP mutants were analyzed by Western blotting. Microsomal membrane fractions were prepared from the BY-2 cells harvested 6–7 d after transfer to the new medium or N. benthamiana leaves harvested 6 d after infiltration (46). After SDS/PAGE, Western blotting was performed using an Immun-Blot PVDF Membrane (Bio-Rad). Detection of CEBiP and CEBiP mutants were performed by using rabbit antisera against CEBiP as a primary antibody (6) and horseradish peroxidase-conjugated goat anti-rabbit IgG (MP Biomedicals) as a secondary antibody. Proteins were detected by the chemiluminescence with Immobilon Western Detection reagents (Millipore) and recorded by LAS4000 camera system (GE Healthcare).
The expression level of each gene was measured by RT-PCR. Total RNA was isolated from the BY-2 cells or N. benthamiana leaves using an RNeasy Plant Mini kit (Qiagen) and first-strand cDNA synthesis was performed using a QuantiTect Reverse Transcription Kit (Qiagen). RT-PCR was performed with Takara Ex Taq (Takara Bio) using gene-specific primers described in SI Appendix, Table S5.
Affinity Labeling with Biotinylated-GN8.
Affinity labeling of GN8-Bio, the conjugate of biocytin hydrazide and (GlcNAc)8, was performed as described previously (15). Microsomal membrane preparation was mixed with 0.04 or 0.4 µM of GN8-Bio with or without competing sugars and adjusted to 30 µL with binding buffer. After incubation for 1 h on ice, 3 µL of 3% EGS (ethylene glycol bis[succinimidylsuccinate], wt/vol) solution was added to the mixture and left to stand for 30 min. The reaction was stopped by the addition of 1 M Tris, mixed with SDS/PAGE sample buffer, boiled for 5 min, and used for SDS/PAGE. After the blotting onto PVDF membrane, detection of biotinylated proteins was performed by using a rabbit antibody against biotin (Rockland or Bethyl) as a primary antibody and horseradish peroxidase-conjugated goat anti-rabbit IgG (MP Biomedicals) as a secondary antibody.
Analysis of Chitin-Induced ROS Generation in Rice Cells.
The accumulation of ROS in the medium of suspension-cultured rice cells by elicitor treatment was measured by the luminol assay (47) using the microtiter plate-based method (48). Briefly, 40 mg of cultured cells were transferred to 1 mL of fresh medium in a 2-mL centrifuge tube and preincubated for 30 min at 25 °C on a thermomixer shaken at 750 rpm. For the competition assay with various oligosaccharides, each 100 nM of (GlcNβ1,4GlcNAc)4, (GlcNβ1,4GlcN)4, or (GlcNAc)4 was added to the reaction mixtures 10 min before the addition of 0.1 nM (GlcNAc)8. Ten-microliter aliquots of the medium were then transferred to 96-well microtiter plates at various time points and immediately supplemented with 50 μL of 1.1 mM luminol and 100 μL of 14 mM potassium hexacyanoferrate solution using a programmable injector attached to the luminometer. Chemiluminescence was measured by a microplate luminometer model TR717 (Berthold Technologies). The amount of ROS was estimated by using a standard curve for hydrogen peroxide.
Homology Modeling of LysM Domains and Mutant Design.
Based on the PFAM database (16), CEBiP comprises a signal peptide region, two LysM domains and a C-terminal transmembrane region (SI Appendix, Fig. S2). Because we were interested in the understanding of molecular mechanisms that regulate chitin oligosaccharides elicitation, we focused on LysM regions. The best template was identified using profile HMMs and the program HMMer. Once the best template was identified, the model of CEBiP extracellular region was generated by MODELER 9V9. Stereo chemical quality of predicted models was improved by energy minimization using GROMACS. Modeling of chitin binding was performed based on the recent structure of CERK1 from A. thaliana, which shares a sequence identity of 24.9% with the extracellular region of CEBiP (residues 9–192). The model of this complex was energy minimized using GROMACS (49). Based on this model, important residues for binding were identified.
NMR Spectroscopic Analysis.
All NMR experiments were performed on a Bruker 600-MHz DRX equipped with a cryo probe at 298 k. All of the samples were dissolved in deuterated phosphate buffer with 0.005% CHAPS and 10 mM DTT (pH 7.4) and spectra were calibrated with internal [D4](trimethylsilyl)propionic acid sodium salt (TSP, 10 μM).
All free ligand proton resonances were assigned by using COSY, TOCSY, NOESY, ROESY, and heteronuclear single quantum coherence (HSQC) experiments. 1H-NMR spectra were recorded with 32 k and 64 k datapoints. Double quantum-filtered phase-sensitive COSY spectra were performed by using data sets of 4,096 × 512 (t1 × t2) points; TOCSY spectra were performed with a spin lock time of 100 ms, by using datasets of 4,096 × 256 points. NOESY and ROESY spectra were measured by using datasets of 4,096 × 256 points; mixing times between 100 and 400 ms were used. In all homonuclear spectra the data matrix was zero-filled in the F1 dimension to give a matrix of 4,096 × 2,048 points and resolution was enhanced in both dimensions by a cosine-bell function before Fourier transformation. HSQC experiments were measured in the 1H-detected mode via single quantum coherence with proton decoupling in the 13C domain, by using data sets of 2,048 × 256 points. Experiments were carried out in the phase-sensitive mode according to the method of States et al. (50).
All STD NMR experiments were recorded with 16- or 32-k datapoints; the original FID was zero-filled to 32 or 64 k and Fourier transformation was applied. To increase the S/N ratio, the FIDs were multiplied with an exponential function (lb = 1–2 Hz). For the STD spectra acquired on the bound ligands the pseudo 2D pulse program stddiff.3 was used and the unwanted broad resonance signals of the protein were avoided by using a spin lock pulse of 50 ms. For protein saturation, a 40-Gauss pulse with a length of 50 ms and an attenuation of 50 db were used. The on resonance pulse was at −1 or 6.8 ppm, and 40 ppm was set as off resonance pulse frequency. Reference experiments were carried out to assure the absence of direct irradiation of the ligand. On the mixtures, STD NMR experiments were performed using protein-ligand molar ratio varied from 1:50–1:100 and saturation times between 0.5 and 5 s were used.
To overcome false-positives in STD NMR spectra, STDD NMR experiments were performed. The STDD was obtained acquiring STD NMR spectra, under the same conditions, on ligands in presence of LysM1-2 fused to a carrier protein (Trx) and in presence of thioredoxin alone. The first difference was taken internally through phase cycling and the second difference was given by manual subtraction between the normal STD NMR spectrum and those performed on the ligand in the presence of thioredoxin alone.
The STD effect was calculated by (I0 − Isat)/I0, where (I0 − Isat) is the intensity of the signal in the STD NMR spectrum and I0 is the peak intensity of an unsaturated reference spectrum (off-resonance). The STD signal with the highest intensity was set to 100% and the others were normalized to this peak. Data acquisition and processing were performed with TOPSPIN software.
STD build up curves were constructed by data processing. In detail, STDD NMR spectra at different saturation times have been acquired and, for each proton that gave rise to STD effects, the corresponding STD build up curve has been constructed by fitting experimental data with the monoexponential equation: STD = STDmax[1 – exp (−ksat t)] where STD stands for the STD signal intensity of a given proton at a saturation time t, STDmax is the asymptotic maximum of the curve that corresponds to the maximal STD intensity obtainable when long saturation time are used, and ksat represents the observed saturation rate constant. Once constructed STD build up curves it is possible to process resulting values of STDmax and Ksat to determine the slope of each curve at zero saturation time (SI Appendix, Table S2). The values of resulting STD fit are very informative because they are affected only by the vicinity of ligand protons to the binding site of the protein and reflect the real STD effect of each proton. To obtain the epitope map and compare the results, all of the values were normalized to the highest STD fit giving the final percentages of STD, namely STD epitope fit.
Tr-NOESY experiments on the oligosaccharide in both free and bound state (in the presence of LysM1-2), were measured by using data sets of 4,096 × 256 points; mixing times were between 150 and 400 ms. In all homonuclear spectra the data matrix was zero-filled in the F1 dimension to give a matrix of 4,096 × 2,048 points and resolution was enhanced in both dimensions by a cosine-bell function before Fourier transformation. A molar ratio of 7:1 (substrate-to-protein) was chosen for the experiments in the bound state.
Conformational Studies.
Molecular mechanics calculations were performed using the MM3* force field as included in MacroModel 8.0. A dielectric constant of 80 was used. For the disaccharide structure, both Φ and Ψ were varied incrementally using a grid step of 18°, each (Φ, Ψ) point of the map was optimized using 2,000 P.R. conjugate gradients. The molecular dynamic simulations were run by using the MM3* force field; bulk water solvation was simulated by using MacroModel generalized Born GB/SA continuum solvent model. All simulations were performed at 300 K, structures were initially subjected to an equilibration time of 300 ps, then a 5,000-ps molecular dynamic simulation was performed with a dynamic time-step of 1.5 fs, a bath constant t of 0.2 ps, and the SHAKE protocol to the hydrogen bonds. Trajectory coordinates were sampled every picosecond, and a total of 5,000 structures were collected for every simulation (51, 52). Ensemble average-interproton distances were calculated using the NOEPROM program applying the isolated spin pair approximation as previously described (51, 53). Coordinate extractions were performed with the program SuperMap, supplied with the NOEPROM package, and data visualized with ORIGIN software. Solvent-accessible surfaces were calculated with the Surface utility of MacroModel and with Molecular Surface displays of the Chem3D package (Cambridge Software).
Light-Scattering Experiments.
Size measurements were performed on a Zetasizer, Malvern Nano-ZS spectrometer. LysM1-2 (100 μM) was dissolved with PBS, pH 7.4, containing 0.01% CHAPS and 2 mM DTT. The wavelength of the laser was 632.8 nm, and the scattering angles 90° and 175°. The molecular diameter is calculated from the autocorrelation function of the intensity of light scattered from the particles assuming a spherical form of particles. For each sample, the mean value of particles diameters was calculated from five replicate determinations. In parallel experiments, LysM1-2 (100 μM) was treated with: (i) a 2-M excess of (GlcNAc)8, (ii) a 2-M excess of (GlcNβ1,4GlcNAc)4, (iii) a pretreatment with a 2-M excess of (GlcNβ1,4GlcNAc)4 followed by a treatment with a 2-M excess of (GlcNAc)8. Measurements were carried out at 22 °C in the same conditions used for the protein alone. Size measurement experiments to evaluate the effect of (GlcNAc)8 on the oligomerization state of LysM1-2 I122A mutant were performed in the same conditions used for the unmutated protein.
For weight average molar masses measurements, size-exclusion chromatography coupled to a DAWN MALS instrument (Wyatt Technology) and an OptilabTM rEX (Wyatt Technology). Three hundred micrograms of sample, either alone or after incubation with a 2-M excess of (GlcNAc)8, was loaded on a WTC 015S5a column (Wyatt Technology), equilibrated in PBS, pH 7.4, 0.01% CHAPS, 2 mM DTT. A constant flow rate of 0.5 mL/min was applied. Weight average molar masses were computed using the Astra 5.3.4.14 (Wyatt Technologies) software.
Supplementary Material
Acknowledgments
We thank Drs. Tomonori Shinya, Shigeru Kuwata, and Mr. Kota Kamiya for the help in the transformation of Nicotiana benthamiana and tobacco BY-2 cells; Dr. Mark Curtis of University of Zurich and Dr. George Lomonossoff of John Innes Center for the supply of pMDC32 and pEAQ-HT-DEST1 vectors, respectively; and Ms. Yuki Kitagawa for the help in the oligosaccharide synthesis. This work was supported in part by Ministry of Education, Culture, Sports, Science and Technology, Japan Grants 22248041 and 25114516 (to N.S.) and 22570052 (to H.K.); Mizutani Foundation for glycoscience Grant 120012 (to R.B.); and by the Program for Promotion of Basic Research Activities for Innovative Bioscience (N.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1312099111/-/DCSupplemental.
References
- 1.Boller T, He SY. Innate immunity in plants: An arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science. 2009;324(5928):742–744. doi: 10.1126/science.1171647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zipfel C. Pattern-recognition receptors in plant innate immunity. Curr Opin Immunol. 2008;20(1):10–16. doi: 10.1016/j.coi.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Kwon C, Panstruga R, Schulze-Lefert P. Les liaisons dangereuses: Immunological synapse formation in animals and plants. Trends Immunol. 2008;29(4):159–166. doi: 10.1016/j.it.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Nürnberger T, Brunner F, Kemmerling B, Piater L. Innate immunity in plants and animals: Striking similarities and obvious differences. Immunol Rev. 2004;198:249–266. doi: 10.1111/j.0105-2896.2004.0119.x. [DOI] [PubMed] [Google Scholar]
- 5.Boller T, Felix G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 6.Kaku H, et al. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci USA. 2006;103(29):11086–11091. doi: 10.1073/pnas.0508882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miya A, et al. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA. 2007;104(49):19613–19618. doi: 10.1073/pnas.0705147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wan J, et al. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell. 2008;20(2):471–481. doi: 10.1105/tpc.107.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan JR, et al. LYK4, a lysin motif receptor-like kinase, is important for chitin signaling and plant innate immunity in Arabidopsis. Plant Physiol. 2012;160(1):396–406. doi: 10.1104/pp.112.201699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimizu T, et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010;64(2):204–214. doi: 10.1111/j.1365-313X.2010.04324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buist G, Steen A, Kok J, Kuipers OP. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol. 2008;68(4):838–847. doi: 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- 12.Shinya T, et al. Functional characterization of CEBiP and CERK1 homologs in arabidopsis and rice reveals the presence of different chitin receptor systems in plants. Plant Cell Physiol. 2012;53(10):1696–1706. doi: 10.1093/pcp/pcs113. [DOI] [PubMed] [Google Scholar]
- 13.Liu B, et al. Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. Plant Cell. 2012;24(8):3406–3419. doi: 10.1105/tpc.112.102475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willmann R, et al. Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc Natl Acad Sci USA. 2011;108(49):19824–19829. doi: 10.1073/pnas.1112862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shinya T, et al. Characterization of receptor proteins using affinity cross-linking with biotinylated ligands. Plant Cell Physiol. 2010;51(2):262–270. doi: 10.1093/pcp/pcp185. [DOI] [PubMed] [Google Scholar]
- 16.Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40(Database issue):D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fliegmann J, et al. Biochemical and phylogenetic analysis of CEBiP-like LysM domain-containing extracellular proteins in higher plants. Plant Physiol Biochem. 2011;49(7):709–720. doi: 10.1016/j.plaphy.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Meyer B, Peters T. NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors. Angew Chem Int Ed Engl. 2003;42(8):864–890. doi: 10.1002/anie.200390233. [DOI] [PubMed] [Google Scholar]
- 19.Squeglia F, et al. Chemical basis of peptidoglycan discrimination by PrkC, a key kinase involved in bacterial resuscitation from dormancy. J Am Chem Soc. 2011;133(51):20676–20679. doi: 10.1021/ja208080r. [DOI] [PubMed] [Google Scholar]
- 20.Marchetti R, et al. Burkholderia cenocepacia lectin A binding to heptoses from the bacterial lipopolysaccharide. Glycobiology. 2012;22(10):1387–1398. doi: 10.1093/glycob/cws105. [DOI] [PubMed] [Google Scholar]
- 21.Silipo A, et al. NMR spectroscopic analysis reveals extensive binding interactions of complex xyloglucan oligosaccharides with the Cellvibrio japonicus glycoside hydrolase family 31 α-xylosidase. Chemistry. 2012;18(42):13395–13404. doi: 10.1002/chem.201200488. [DOI] [PubMed] [Google Scholar]
- 22.Claasen B, Axmann M, Meinecke R, Meyer B. Direct observation of ligand binding to membrane proteins in living cells by a saturation transfer double difference (STDD) NMR spectroscopy method shows a significantly higher affinity of integrin alpha(IIb)beta3 in native platelets than in liposomes. J Am Chem Soc. 2005;127(3):916–919. doi: 10.1021/ja044434w. [DOI] [PubMed] [Google Scholar]
- 23.Haselhorst T, et al. Direct detection of ligand binding to Sepharose-immobilised protein using saturation transfer double difference (STDD) NMR spectroscopy. Biochem Biophys Res Commun. 2007;359(4):866–870. doi: 10.1016/j.bbrc.2007.05.204. [DOI] [PubMed] [Google Scholar]
- 24.Assadi-Porter FM, et al. Direct NMR detection of the binding of functional ligands to the human sweet receptor, a heterodimeric family 3 GPCR. J Am Chem Soc. 2008;130(23):7212–7213. doi: 10.1021/ja8016939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szczepina MG, Bleile DW, Pinto BM. Investigation of the binding of a carbohydrate-mimetic peptide to its complementary anticarbohydrate antibody by STD-NMR spectroscopy and molecular-dynamics simulations. Chemistry. 2011;17(41):11446–11455. doi: 10.1002/chem.201100222. [DOI] [PubMed] [Google Scholar]
- 26.Angulo J, et al. Saturation transfer difference (STD) NMR spectroscopy characterization of dual binding mode of a mannose disaccharide to DC-SIGN. ChemBioChem. 2008;9(14):2225–2227. doi: 10.1002/cbic.200800361. [DOI] [PubMed] [Google Scholar]
- 27.Enríquez-Navas PM, Marradi M, Padro D, Angulo J, Penadés S. A solution NMR study of the interactions of oligomannosides and the anti-HIV-1 2G12 antibody reveals distinct binding modes for branched ligands. Chemistry. 2011;17(5):1547–1560. doi: 10.1002/chem.201002519. [DOI] [PubMed] [Google Scholar]
- 28.Mayer M, James TL. NMR-based characterization of phenothiazines as a RNA binding scaffold. J Am Chem Soc. 2004;126(13):4453–4460. doi: 10.1021/ja0398870. [DOI] [PubMed] [Google Scholar]
- 29.Johnson MA, Pinto BM. NMR spectroscopic and molecular modeling studies of protein-carbohydrate and protein-peptide interactions. Carbohydr Res. 2004;339(5):907–928. doi: 10.1016/j.carres.2003.12.028. [DOI] [PubMed] [Google Scholar]
- 30. Jimenez-Barbero J (2002) in NMR Spectroscopy of Glycoconjugates, ed Peters T (Wiley, Weinheim)
- 31. Neuhaus D (2000) in The Nuclear Overhauser Effect in Structural and Conformational Analysis, ed Williamson MP (Wiley, New York)
- 32.Liu TT, et al. Chitin-induced dimerization activates a plant immune receptor. Science. 2012;336(6085):1160–1164. doi: 10.1126/science.1218867. [DOI] [PubMed] [Google Scholar]
- 33.Shibuya N, Minami E. Oligosaccharide signalling for defense responses in plant. Physiol Mol Plant Pathol. 2001;59(5):223–233. [Google Scholar]
- 34.Yamada A, Shibuya N, Kodama O, Akatsuka T. Induction of phytoalexin formation in suspension-cultured rice cells by N-acetyl-chitooligosaccharides. Biosci Biotechnol Biochem. 1993;57(3):405–409. [Google Scholar]
- 35.Nishizawa Y, et al. Regulation of the chitinase gene expression in suspension-cultured rice cells by N-acetylchitooligosaccharides: Differences in the signal transduction pathways leading to the activation of elicitor-responsive genes. Plant Mol Biol. 1999;39(5):907–914. doi: 10.1023/a:1006161802334. [DOI] [PubMed] [Google Scholar]
- 36.Hubbard SR, Till JH. Protein tyrosine kinase structure and function. Annu Rev Biochem. 2000;69:373–398. doi: 10.1146/annurev.biochem.69.1.373. [DOI] [PubMed] [Google Scholar]
- 37.Heldin CH. Dimerization of cell surface receptors in signal transduction. Cell. 1995;80(2):213–223. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- 38.Botos I, Segal DM, Davies DR. The structural biology of Toll-like receptors. Structure. 2011;19(4):447–459. doi: 10.1016/j.str.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faulkner C, et al. LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proc Natl Acad Sci USA. 2013;110(22):9166–9170. doi: 10.1073/pnas.1203458110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Narusaka Y, et al. Presence of LYM2 dependent but CERK1 independent disease resistance in Arabidopsis. Plant Signal Behav. 2013;8(9):e25345. doi: 10.4161/psb.25345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tokuyasu K, Mori Y, Kitagawa Y, Hayashi K (1999) 2-methyl-(4-O-(2-amino-2-deoxy-.beta.- glucopyranosyl)-1,2-dideoxy-.alpha.-glucopyrano)(2,1-D)-2-oxazoline and its salt, 50% deacetylated chitin or its oligosaccharide and salt thereof. US Patent 6,437,107.
- 42.Kobayashi S, Kiyosada T, Shoda S. Synthesis of artificial chitin: Irreversible catalytic behavior of a glycosyl hydrolase through a transition state analogue substrate. J Am Chem Soc. 1996;118(51):13113–13114. [Google Scholar]
- 43.Tsukada K, et al. Rice receptor for chitin oligosaccharide elicitor does not couple to heterotrimeric G-protein: Elicitor responses of suspension cultured rice cells from Daikoku dwarf (d1) mutants lacking a functional G-protein α-subunit. Physiol Plant. 2002;116(3):373–382. [Google Scholar]
- 44.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133(2):462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sainsbury F, Thuenemann EC, Lomonossoff GP. pEAQ: Versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol J. 2009;7(7):682–693. doi: 10.1111/j.1467-7652.2009.00434.x. [DOI] [PubMed] [Google Scholar]
- 46.Shibuya N, Kaku H, Kuchitsu K, Maliarik MJ. Identification of a novel high-affinity binding site for N-acetylchitooligosaccharide elicitor in the membrane fraction from suspension-cultured rice cells. FEBS Lett. 1993;329(1-2):75–78. doi: 10.1016/0014-5793(93)80197-3. [DOI] [PubMed] [Google Scholar]
- 47.Schwacke R, Hager A. Fungal elicitors induce a transient release of active oxygen species from cultured spruce cells that is dependent on Ca(2+) and protein-kinase activity. Planta. 1992;187(1):136–141. doi: 10.1007/BF00201635. [DOI] [PubMed] [Google Scholar]
- 48.Desaki Y, et al. Bacterial lipopolysaccharides induce defense responses associated with programmed cell death in rice cells. Plant Cell Physiol. 2006;47(11):1530–1540. doi: 10.1093/pcp/pcl019. [DOI] [PubMed] [Google Scholar]
- 49.Van Der Spoel D, et al. GROMACS: Fast, flexible, and free. J Comput Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 50.States DJ, Haberkorn RA, Ruben DJ. A two-dimensional nuclear overhauser experiment with pure absorption phase in 4 quadrants. J Magn Reson. 1982;48(2):286–292. [Google Scholar]
- 51.Asensio JL, Jimenez-Barbero J. The use of the AMBER force field in conformational analysis of carbohydrate molecules: Determination of the solution conformation of methyl α-lactoside by NMR spectroscopy, assisted by molecular mechanics and dynamics calculations. Biopolymers. 1995;35(1):55–73. doi: 10.1002/bip.360350107. [DOI] [PubMed] [Google Scholar]
- 52.Bernardi A, et al. Second-generation mimics of ganglioside GM1 oligosaccharide: A three-dimensional view of their interactions with bacterial enterotoxins by NMR and computational methods. Chemistry. 2002;8(20):4598–4612. [PubMed] [Google Scholar]
- 53.Corzana F, et al. The pattern of distribution of amino groups modulates the structure and dynamics of natural aminoglycosides: Implications for RNA recognition. J Am Chem Soc. 2007;129(10):2849–2865. doi: 10.1021/ja066348x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.