Significance
Cancer progression, as an evolutionary process, should accelerate if higher cellular variability is present. However, the sources of nongenetic variability in metastatic progression are largely unknown. To address this question, we characterized a transcriptional regulatory network for the metastasis suppressor Raf kinase inhibitory protein (RKIP). We previously showed that the transcription factor BACH1 is negatively regulated by RKIP and promotes breast cancer metastasis. Here we reveal a network architecture between the metastasis suppressor RKIP and the metastasis promoter BACH1 that enables single cells to generate a stable subpopulation of prometastatic cells without any genetic changes.
Keywords: HDAC, EZH2, mathematical model, dynamics
Abstract
The sources and consequences of nongenetic variability in metastatic progression are largely unknown. To address these questions, we characterized a transcriptional regulatory network for the metastasis suppressor Raf kinase inhibitory protein (RKIP). We previously showed that the transcription factor BACH1 is negatively regulated by RKIP and promotes breast cancer metastasis. Here we demonstrate that BACH1 acts in a double-negative (overall positive) feedback loop to inhibit RKIP transcription in breast cancer cells. BACH1 also negatively regulates its own transcription. Analysis of the BACH1 network reveals the existence of an inverse relationship between BACH1 and RKIP involving both monostable and bistable transitions that can potentially give rise to nongenetic variability. Single-cell analysis confirmed monostable and bistable-like behavior. Treatment with histone deacetylase inhibitors or depletion of the polycomb repressor enhancer of zeste homolog 2 altered relative RKIP and BACH1 levels in a manner consistent with a prometastatic state. Together, our results suggest that the mutually repressive relationship between metastatic regulators such as RKIP and BACH1 can play a key role in determining metastatic progression in cancer.
Cancer progression is an evolutionary process of variant cells competing to expand first locally and then distally within the human body. Ultimately, tumor evolution generates metastatic lesions that account for over 90% of cancer deaths (1). Whereas the requirement of heritably variant cells for tumor progression is undisputed, how they emerge is much less clear. Traditionally, genetic mutations causing oncogene activation or tumor suppressor loss were considered essential (2). However, accumulating evidence for tumor-promoting epigenetic, microenvironmental, and stochastic forms of heritable variation is challenging the traditional view (3, 4). Genetically identical tumor cells show highly variable responses to apoptosis-inducing ligands (5) and chemotherapeutic drugs (6, 7) attributable to cell–cell differences in gene expression and pathway activity. Tumor cells diversify in vitro (8) and in vivo (9) showing different levels of stem cell marker expression. However, the molecular mechanisms and interactions controlling such nongenetic forms of diversity are not fully known (10, 11).
Multiple studies have implicated positive feedback loops in amplifying and maintaining stochastic fluctuations, creating heritable diversity in genetically homogenous cell populations (12, 13). This heritable nongenetic diversity then generates random subpopulations that can survive during various forms of environmental stress (14, 15), enabling subsequent evolutionary adaptation (16). Understanding how feedback loops and other network structures may affect nongenetic heterogeneity and contribute to metastatic cancer progression will be crucial for combating the disease (3, 10). However, the regulation of metastasis-related genes is incompletely understood, and the role of regulatory network-mediated nongenetic variation in metastatic progression remains unclear.
Tumor metastasis suppressors are critical regulators of metastatic progression. Raf kinase inhibitory protein (RKIP, also called PEBP1), an inhibitor of the MAPK signaling cascade, has well-defined expression in differentiated tissues (17) and has been implicated as a metastasis suppressor (18, 19). Loss of RKIP expression correlates with poor prognosis in a variety of tumor types including prostate and breast cancer (20). Exogenous expression of RKIP in metastatic breast cancer cells is sufficient to prevent invasion, intravasation, and metastasis in xenograft mouse models (18, 19). Thus, elucidating mechanisms that maintain or restore RKIP expression is highly relevant for inhibiting breast cancer metastasis.
In recent studies, we showed that RKIP induces the microRNA let-7 via inhibition of the MAPK pathway, leading to altered expression of key metastatic genes and suppression of breast cancer metastasis. One of the downstream targets inhibited by RKIP through let-7 is BACH1, a basic leucine zipper transcription factor that regulates oxidative stress and stress-induced senescence (21, 22). BACH1 promotes metastasis of triple-negative breast cancers (TNBCs) but does not significantly affect primary tumor growth (23). We recently generated a gene signature based upon RKIP and BACH1 that is prognostic for metastasis-free survival of breast cancer patients (23).
Here we show that BACH1 can also act as an inhibitor of RKIP transcription. The double-negative (overall positive) feedback architecture between RKIP and BACH1 implies that genetically identical cells can stably maintain high or low BACH1 levels when RKIP expression is compromised. In addition, our results demonstrate that BACH1 not only negatively regulates RKIP, but also negatively regulates itself. BACH1 auto-regulation provides a “safety switch” to reestablish the nonmetastatic state if RKIP becomes unstable or BACH1 increases.
We also found that different corepressors, histone deacetylases (HDACs) and the polycomb repressor enhancer of zeste homolog 2 (EZH2), mediate the inhibition of BACH1 and RKIP promoters by BACH1, respectively. Computational models suggest that the RKIP–BACH1 network that we identified may flip in response to microenvironmental effects or nongenetic cellular variability in network regulators (including EZH2 or HDAC), turning a fraction of initially noninvasive cells into a prometastatic state and accelerating tumor progression. We performed single-cell-level measurements that further validated the mathematical model and provided additional insights into metastatic transitions.
Taken together, these results indicate that the BACH1–RKIP regulatory architecture can play an important role in creating nongenetic heterogeneity, accelerating breast cancer progression. However, attempts to shift the balance in favor of RKIP by BACH1 corepressors must be considered carefully with respect to the specifics of BACH1 regulatory connections and their role in a network context.
Results
BACH1 Negatively Regulates RKIP Expression in TNBC Cell Lines.
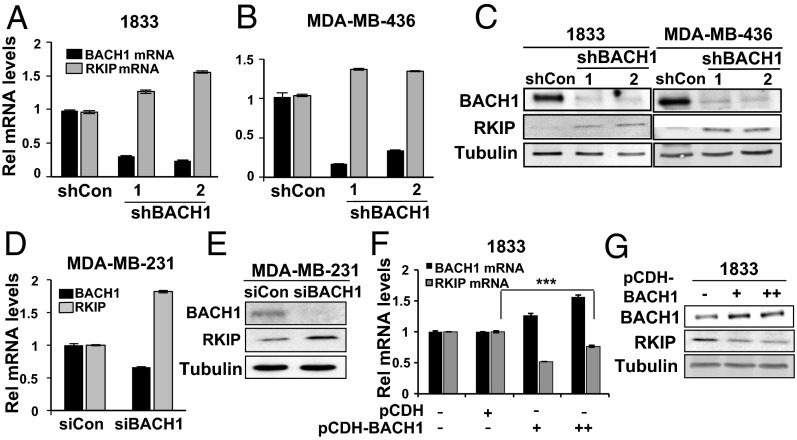
We have previously shown that the metastasis suppressor RKIP negatively regulates expression of BACH1 that, in turn, is a positive regulator of genes that promote breast cancer metastasis to the bone (23). Analysis of gene expression data from human breast tumors revealed that RKIP expression is inversely proportional to that of BACH1 in human TNBC cell lines and primary human breast tumors (SI Appendix, Fig. S1), consistent with a mutually repressive relationship. We therefore hypothesized that BACH1 might also function as a repressor of RKIP expression. We used two independent TNBC cell lines to test this hypothesis: 1833, a bone-tropic line that is derived but distinct from MDA-MB-231, and MDA-MB-436. Stable knockdown of BACH1 via shRNA transduction with two independent shRNAs caused derepression of RKIP expression as measured by mRNA and protein levels in both cell lines (Fig. 1 A–C). We also observed similar regulation in MDA-MB-231 cells; transient depletion of BACH1 via siRNAs caused a twofold increase in RKIP mRNA and protein levels (Fig. 1 D and E). Moreover, transient overexpression of BACH1 in 1833 cells induced a consistent repression of RKIP levels (Fig. 1 F and G). Together, these results indicate that BACH1 represses RKIP expression in TNBC cell lines.
Fig. 1.
BACH1 depletion causes an increase in RKIP expression in TNBC cells. (A and B) The indicated cell lines were stably depleted for BACH1 via shRNAs as described in Materials and Methods. BACH1 and RKIP RNA and protein levels were analyzed by quantitative (q) RT-PCR (A and B) and Western blot (C), respectively. (D and E) MDA-MB-231 cells were transiently depleted for BACH1 via siRNAs as above. BACH1 and RKIP RNA and protein levels were analyzed by qRT-PCR and Western blot, respectively. (F and G) 1833 cells were transiently transfected with a control vector (150 ng, pCDH) or BACH1 expression vector (75 and 150 ng, pCDH-BACH1). RNA and protein levels of RKIP and BACH1 were analyzed as above. Error bars represent SEM from three independent experiments. ***P < 0.001 with Student t test.
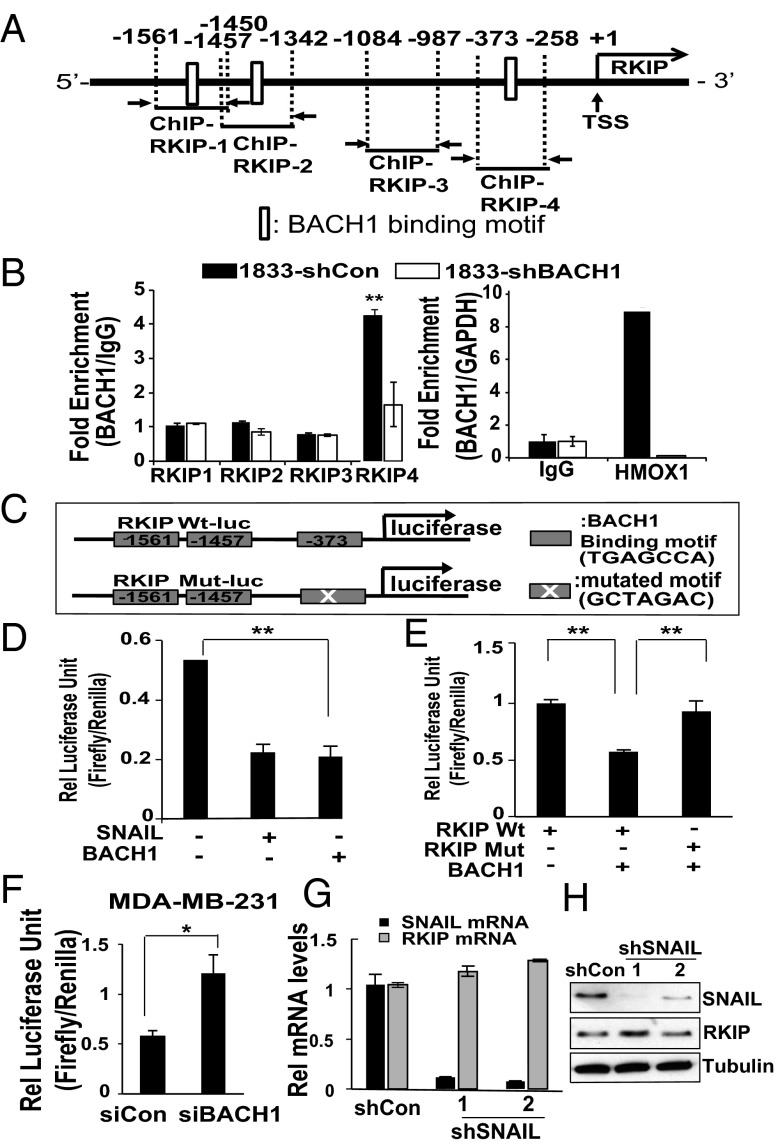
To understand whether BACH1 directly regulates RKIP expression by binding to its promoter region, we analyzed a region −3,000 bp upstream of the RKIP transcription start site (TSS) and identified three BACH1 (AP-1–like) binding motifs (TGAGCCA) (21) (Fig. 2A). Direct recruitment of BACH1 to these sites was tested by ChIP assays using BACH1 antibody and primers designed as shown in Fig. 2A (SI Appendix, Table S1, sequences for qPCR primer). We found that BACH1 was significantly recruited only to the −373-bp upstream regions (RKIP 4) in 1833 cells (Fig. 2B). Cells depleted for BACH1 (1833-shBACH1) or IgG were used as a negative control, whereas heme oxygenase 1 (HMOX1), a well-characterized BACH1 target, served as a positive control. To validate that BACH1 binding to the RKIP promoter is functional, we cloned −2,200 bp of the human Wt RKIP promoter region into a pGL2 vector (RKIP Wt-luc) and performed luciferase assays (Fig. 2C) (24). We also tested SNAIL, which binds to an E-box element in the RKIP promoter region and represses RKIP transcription in prostate cells (24). Luciferase activity was significantly inhibited when 1833 cells were cotransfected with either a BACH1 or SNAIL expression vector (250 ng) (Fig. 2D), suggesting that both BACH1 and SNAIL have the potential to repress RKIP promoter activity in breast cancer cells. Mutation of the BACH1 binding motif at a random position in the sequence (RKIP Mut-luc) restored luciferase activity to control levels even in the presence of exogenous BACH1 (Fig. 2E), indicating that an intact upstream −373-bp region is required for BACH1-mediated RKIP repression in these cells. We also performed the complementary experiment; knockdown of BACH1 by siRNA increased the basal activity of the Wt RKIP reporter more than twofold in MDA-MB-231 cells (Fig. 2F). These results indicate that BACH1 is directly recruited to the RKIP promoter region and represses RKIP transcription.
Fig. 2.
BACH1 is directly recruited to the RKIP promoter region and represses RKIP transcription. (A) Schematic representation of the RKIP promoter region depicting the BACH1 binding motif (open boxes) and the ChIP-qPCR regions. (B) 1833 cells stably depleted for BACH1 and their control cells were used for ChIP assays as described in Materials and Methods using BACH1 antibodies and the RKIP primers illustrated in A or HMOX1 primers as a positive control. ChIP with IgG was used as a negative control. (C) The Wt (RKIP Wt-luc) or mutant (RKIP Mut-luc) RKIP promoter regions (upstream −2,200 bp) were cloned into the pGL2 vector for luciferase assays. A schematic representation is shown. (D) 1833 cells were transfected with control vector (Con), SNAIL-expressing vector (SNAIL), or BACH1-expressing vector (BACH1) and luciferase activity from the RKIP Wt-luc vector was measured as described in Materials and Methods. Relative luciferase unit (RLU) of firefly/Renilla was indicated. (E) 1833 cells were transfected with Wt or Mut RKIP-luc and with the BACH1 expression vector (BACH1) or control vector as indicated. (F) MDA-MB-231 cells were depleted for BACH1 by siRNAs and luciferase activity measured as above. (G and H) 1833 cells were stably depleted for SNAIL via shRNAs as described in Materials and Methods. SNAIL and RKIP RNA and protein levels were analyzed by qRT-PCR (G) and Western blot (H), respectively. All of the experiments were repeated three times at least and error bars represent SEM. *P < 0.05, **P < 0.01 with Student t test.
Because SNAIL regulates the truncated RKIP promoter contained in the reporter construct, we determined whether SNAIL also regulates endogenous RKIP transcription in TNBCs. Using two different shRNAs, we depleted SNAIL from two separate cell lines (1833 and MDA-MB-231). Surprisingly, RKIP mRNA and protein levels were not significantly changed in these cells following SNAIL knockdown (Fig. 2 G and H and SI Appendix, Fig. S2). These results suggest that although both BACH1 and SNAIL can potentially bind and repress RKIP transcription, one or the other (or both) may function as dominant regulators of RKIP dependent upon the specific different cellular context.
BACH1 Binds to Its Own Promoter and Represses BACH1 Transcription in Breast Cancer Cell Lines.
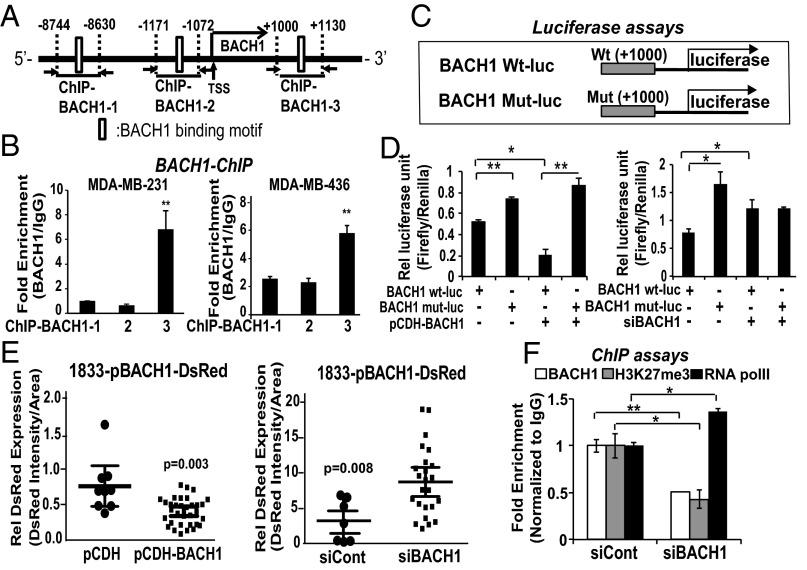
Considering that both SNAIL and BACH1 have regulatory sites around RKIP, we asked whether they may also share other regulatory characteristics, such as SNAIL auto-regulation. To investigate BACH1 auto-regulation at the transcriptional level, we searched for transcription factor binding motifs in its promoter region. We identified two regions upstream (−8,744 bp and −1,171) and one region downstream (+1,000) that contained BACH1 binding motifs, as illustrated in Fig. 3A. We performed ChIP assays using two TNBC cell lines (MDA-MB-231 and MDA-MB-436) and identified the downstream region (+1,000 bp) of the BACH1 promoter as the major binding site for BACH1 (Fig. 3B). Interestingly, ChIP assays using 1833 cells showed that BACH1 was largely recruited to a different site in the upstream region (−8,744 bp) of the BACH1 promoter (SI Appendix, Fig. S3). As expected, no recruitment was observed in BACH1-depleted cells, a negative control. These results indicate that BACH1 is able to bind in vivo to its own promoter at two, and possibly three, different sites. We speculate that the relative affinity of BACH1 for these regions differs dependent upon the nature of the multiprotein transcriptional complexes in the different TNBC cell lines.
Fig. 3.
BACH1 binds to its own promoter region and represses BACH1 transcription in breast cancer cell lines. (A) Schematic representation of the BACH1 promoter region depicting the BACH1 binding motif (open boxes) and the ChIP-qPCR regions. (B) The indicated cells were used for ChIP assays as described in Materials and Methods using BACH1 antibody and the three BACH1 primer sets shown in A. (C) Schematic representation of luciferase constructs. Wt (BACH1 Wt-luc) or mutant (BACH1 Mut-luc) BACH1 promoter regions (downstream +1,000 bp) were cloned into the pGL2 vector for luciferase assays. BACH1 binding motif (TGAGTCA) was mutated to ACGTCAG. (D) 1833 cells were transfected with BACH1 Wt- or Mut-luciferase clones along with BACH1 expression vector (pCDH-BACH1) or siBACH1 (100 nM) as indicated. Relative luciferase units (RLU) of firefly/Renilla measured are shown. (E) Wt BACH1 promoter regions were cloned into DsRed Expression 2 vector for cytosolic DsRed expression analysis. Intensity of DsRed and size of cells in each single cell in random fields (>10) was quantified and shown (DsRed intensity per cell area). (F) BACH1-depleted and control MDA-MB-231 cells were used for ChIP assays using antibodies against BACH1, Histone 3 Lysine 27 trimethylation (H3K27me3) as a transcriptional repression maker, and RNA polymerase II as a transcriptional activation marker. Relative fold enrichment that is normalized by IgG recruitment is shown. Error bars represent SEM from three independent experiments. *P < 0.05, **P < 0.01 with Student t test.
To understand whether BACH1 regulates BACH1 expression by binding to its own promoter region, we performed reporter assays using the BACH1 binding region. The Wt BACH1 binding site (TGAGTCA at +1,000 bp) was mutated at a random position in the sequence to ACGTCAG (Mut), and both Wt and Mut promoters were cloned into pGL2 for luciferase assays (Fig. 3C). The luciferase activity of the BACH1 reporter (pGL2-Wt) was significantly reduced upon exogenous BACH1 expression in MDA-MB-231 cells (Fig. 3D). In contrast, the luciferase activity of the mutant BACH1 reporter (pGL2-Mut) was not changed following exogenous BACH1 expression. Similarly, BACH1 depletion increased luciferase activity of the Wt BACH reporter but had no significant effect on the Mut BACH1 reporter (Fig. 3D). In addition, the BACH1 promoter region was cloned into a DsRed-Expression 2 vector (25). Stable 1833 cells expressing DsRed linked to the BACH1 promoter were generated as described in Materials and Methods. Control vector with pCMV promoter was used as a positive control to monitor the efficiency of viral infection, which approached 96% of the cells. The relative expression of cytosolic DsRed in single cells expressing the DsRed–BACH1 reporter was significantly increased (P = 0.008) with siBACH1 treatment and reduced (P = 0.003) with exogeneous BACH1 expression (Fig. 3E). These data indicate that BACH1 represses its own expression by binding to its promoter region. Consistent with these results, ChIP assays showed reduced recruitment of the repression marker H3K27me3 and enhanced recruitment of the transcriptional activator marker RNA Pol II (Fig. 3F). Together these results indicate that BACH1 is a direct transcriptional repressor of BACH1.
HDACs Mediate BACH1 Binding and Repression of BACH1 but Not RKIP Transcription.
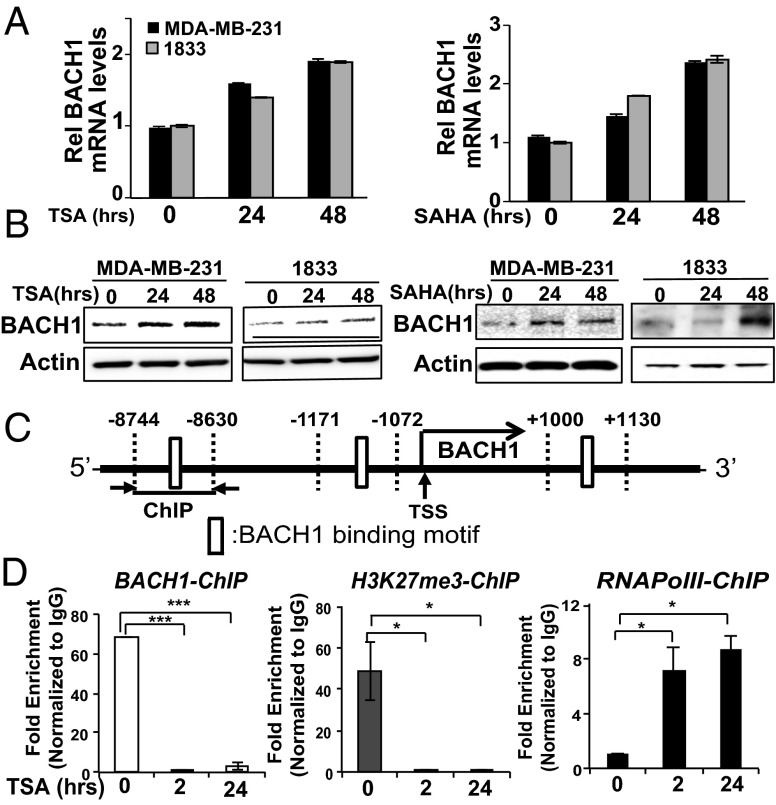
BACH1 was previously shown to form a repressive complex containing p53, HDAC1, and the nuclear cofactor NcoR that regulates cellular senescence in mouse embryonic fibroblasts (21). To investigate the involvement of HDACs in BACH1 regulation of itself and RKIP, we tested two histone deacetylase inhibitors (HDACi) that inhibit class I and II deacetylases: trichostatin A (TSA), a natural compound with high cell toxicity, and vorinostat (SAHA), a synthetic derivative of hydroxamic acid that is the first HDACi approved by the Food and Drug Administration for clinical use in cancer patients (26). Because inhibition of HDACs causes acetylation of lysine residues on histones, and thus transcriptional activation of regulated genes, we anticipated a derepression of BACH1 transcription following HDACi treatment. As expected, exposing MDA-MB-231 and 1833 cells to either of these HDAC inhibitors for 24 and 48 h caused a time-dependent induction of BACH1 mRNA levels (Fig. 4A) and a corresponding increase in BACH1 protein levels (Fig. 4B). This regulation by class I or II HDACs is likely to be indirect because >6 h of treatment is required (SI Appendix, Fig. S4). Treatment of MDA-MB-436 cells with TSA caused a similar induction of BACH1 mRNA and protein levels; however, SAHA did not show any effect in this cell line (SI Appendix, Fig. S5 A and B). Thus, an additional element may be necessary in some cell lines to derepress BACH1 transcription in response to SAHA. To confirm that HDACs participate in the formation of a repressive complex on the BACH1 promoter, we performed BACH1 ChIP assays in 1833 cells treated with TSA for different times using primers shown in Fig. 4C. BACH1 recruitment to the upstream region (−8,744 bp) of its own promoter was dramatically inhibited when 1833 cells were treated with TSA for 2 or 24 h (Fig. 4D). Concomitantly, the repressive histone mark H3K27me3 was reduced, whereas the activation mark RNA pol II was increased. The data suggest that BACH1 is recruited to its own promoter region to repress its own expression, and this repressive complex can be inhibited indirectly by TSA treatment in breast cancer cells.
Fig. 4.
HDAC inhibitors increase BACH1 expression and prevent BACH1 from binding to its own promoter. (A and B) MDA-MB-231 and 1833 cells were treated for the indicated times with TSA or SAHA as described in Materials and Methods. RNA and protein levels were analyzed by qRT-PCR (A) and Western blot (B), respectively. (C) Schematic diagram depicting ChIP-qPCR regions with indicated primers. (D) 1833 cells were treated with TSA as before for the indicated times and ChIP assays were performed using antibodies against BACH1, RNA Pol II, or H3K27me3 and the primers shown in C. Relative fold enrichment that is normalized by IgG recruitment is shown. Error bars represent SEM from three independent experiments. *P < 0.05, ***P < 0.001 with Student t test.
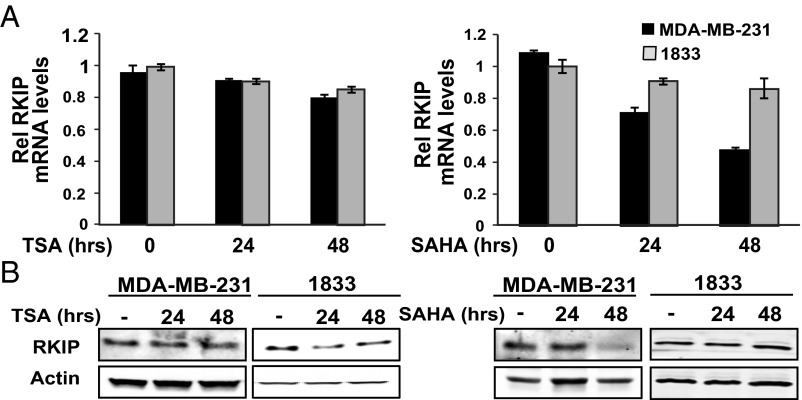
We next determined whether HDACs are also involved in RKIP transcriptional regulation by BACH1. For this purpose, we analyzed RKIP mRNA and protein levels following TSA and SAHA treatment in multiple TNBC cell lines. In contrast to the induction we observed for BACH1, we saw no significant increase in RKIP expression in TSA- or SAHA-treated MDA-MB-231, 1833 (Fig. 5 A and B), or MDA-MB-436 cells (SI Appendix, Fig. S5 C and D). Instead, we observed a small but significant reduction in RKIP expression in MDA-MB-231 and 1833 cells treated respectively with TSA and SAHA (Fig. 5 A and B). This result could be explained by the fact that the HDACi treatments induce BACH1 protein (Fig. 4 A and B), which in turn can repress RKIP. This effect is not evident in all cell lines and may depend on the level of induction of BACH1 as well as the differential sensitivity of RKIP to BACH1 repression.
Fig. 5.
HDACs are not involved in direct repression of RKIP. (A and B) MDA-MB-231 and 1833 cells were treated for the indicated times with TSA or SAHA as described in Materials and Methods. RNA and protein levels were analyzed by qRT-PCR (A) and Western blot (B), respectively. Error bars represent SEM from three independent experiments.
Together the results presented here suggest that HDACs are directly involved in BACH1 repression of its own transcription. However, even when we tried a class III HDAC inhibitor, nicotinamide, we did not observe up-regulation of RKIP mRNA levels (SI Appendix, Fig. S6). Thus, BACH1 repression of RKIP does not seem to depend upon recruitment of HDACs, suggesting that different BACH1-based repressive complexes assemble on BACH1 versus RKIP promoter regions to regulate gene expression.
A Polycomb Repressive Component, EZH2, Represses RKIP at the BACH1 Binding Site.
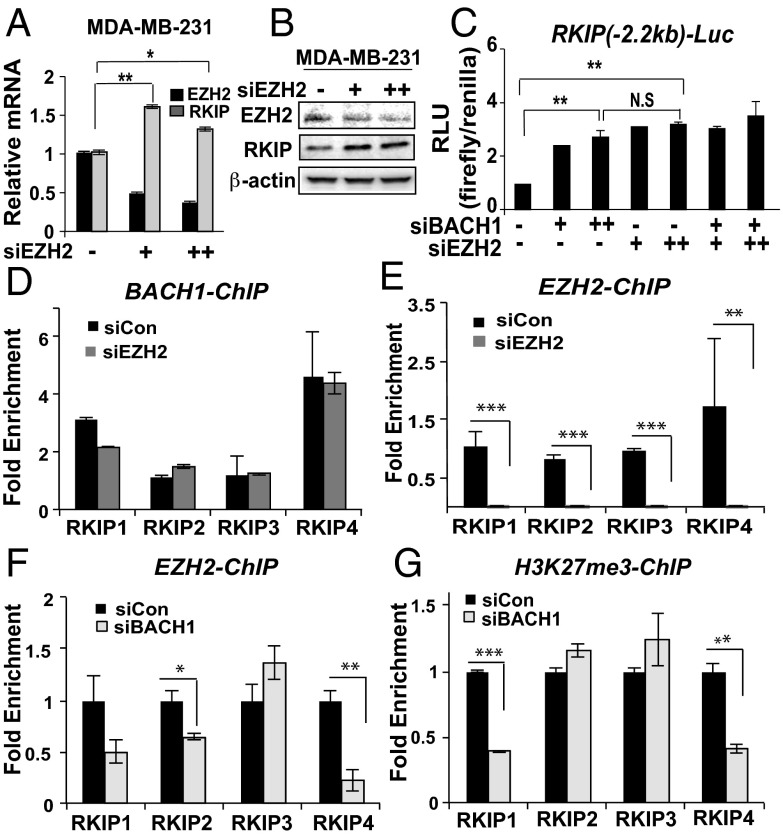
A polycomb repressive component, EZH2, negatively regulates RKIP expression in prostate tumor and MDA-MB-231 cells (27). This study, which was primarily based on prostate tumor cells, demonstrates that SNAIL can recruit EZH2 to the RKIP promoter. EZH2, which requires a sequence-specific DNA-binding partner to complex with DNA, is the histone methyltransferase component of the polycomb repressive complex 2. Because SNAIL does not seem to play a major role in repressing RKIP expression in breast cancer systems, we hypothesized that BACH1 could be the primary mediator of EZH2 binding to the RKIP promoter in TNBC cells. EZH2 knock-down by siRNAs significantly increased levels of RKIP transcripts and protein in MDA-MB-231, as observed previously (Fig. 6 A and B) and also in MDA-MB-436 cells (SI Appendix, Fig. S7A). By contrast, EZH2 depletion did not significantly affect BACH1 levels in these cell lines (SI Appendix, Fig. S7 B and C). These results suggest that EZH2 can contribute to repression of RKIP transcription in TNBC cell lines; however, there is no evidence that EZH2 similarly regulates BACH1.
Fig. 6.
EZH2 represses RKIP at the BACH1 binding site. (A and B) Cells were transiently depleted for EZH2 by siRNA in MDA-MB-231 cells as described in Materials and Methods. RNA and proteins levels of RKIP and EZH2 were analyzed by qRT-PCR (A) and Western blot (B), respectively. (C) Luciferase activity using the RKIP Wt-luc clone was measured in 1833 cells depleted for BACH1 and/or EZH2 as indicated. Relative luciferase unit (RLU) of firefly/Renilla measured was shown in C. N.S, nonsignificant results. (D and E) ChIP assays using antibodies against BACH1 and EZH2 and the RKIP primers shown in Fig. 2A were performed in control or EZH2-depleted MDA-MB-231 cells. (F and G) ChIP assays using antibodies against EZH2 and H3K27me3 were performed in BACH1-depleted and control MDA-MB-231 cells. Error bars represent SEM from three independent ChIP experiments. *P < 0.05, **P < 0.01, ***P < 0.001 with Student t test.
To determine whether BACH1 and EZH2 cooperate in repressing RKIP transcription, we performed luciferase assays using the RKIP-luciferase vector (Wt) that we used above in BACH1- or/and EZH2-depleted cells. Knockdown of BACH1 or EZH2 alone caused a two- to threefold increase in luciferase activity driven by the RKIP promoter (Fig. 6C). Thus, both EZH2 and BACH1 bind to and repress the RKIP promoter within the reporter construct when expressed in 1833 cells. However, simultaneous depletion of the repressors did not further increase induction of luciferase activity. These data indicate that the effect of their concomitant depletion is not additive, consistent with a common mechanism of action. In addition, BACH1 ChIP assays in MDA-MB-231 showed that BACH1 binding to the RKIP promoter is not affected by EZH2 depletion (siEZH2), indicating that EZH2 is not required for BACH1 binding to the RKIP promoter (Fig. 6D). By contrast, EZH2 recruitment was dramatically reduced in MDA-MB-231-treated with siEZH2 as a control (Fig. 6E). EZH2 ChIP assays in BACH1-depleted MDA-MB-231 cells demonstrated that BACH1 binding to the RKIP promoter is required for EZH2 to associate (Fig. 6F). Finally, ChIP analysis of the repressive histone mark for transcription (H3K27me3) following treatment of cells with siBACH1 shows a similar dependency of binding on BACH1 (Fig. 6G). These results confirm that EZH2 repression occurs via association with BACH1.
BACH1 and RKIP Participate in a Bistable Circuit.
The experimental results we obtained above indicate that RKIP and BACH1 participate in a double-negative (overall positive) feedback loop by mutually repressing each other’s expression (Fig. 7A). A synthetic toggle switch gene circuit with similar architecture could flip between two stable gene expression states following transient perturbations (28). Double-negative feedback is also common in natural decision-making networks (12, 29, 30), suggesting that in certain conditions the RKIP–BACH1 network may support the coexistence of two stable states (bistability) within a cancer cell population. Consequently, cells in the noninvasive, “high RKIP–low BACH1” state (which we will call here antimetastatic) could respond to transient perturbations by switching to a prometastatic “low RKIP–high BACH1” state, which could be stable for prolonged periods of time. Thus, single-cell switching could generate a stable subpopulation of invasive cells that accelerate tumor progression toward metastasis without any genetic changes (3). Likewise, prometastatic cells could switch back and remain antimetastatic after receiving a transient stimulus.
Fig. 7.
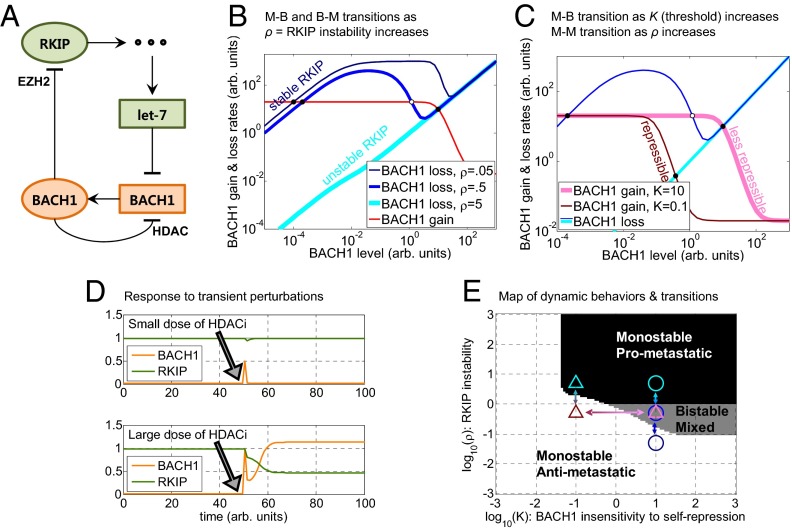
Modeling RKIP-BACH1 network dynamics in breast cancer. (A) Schematic illustration of the molecular interactions included in the mathematical model. Pointed arrows and blunted arrows indicate activation and repression, respectively. Green and orange nodes suppress and promote metastasis, respectively. Circles represent proteins, and rectangles represent RNAs. The three dots represent a cascade of interactions that mediate let-7 regulation by RKIP. (B) Rate-balance plots showing BACH1 gain (shades of red) and loss (shades of blue) rates as functions of BACH1. When BACH1 gain is above BACH1 loss, BACH1 levels grow. BACH1 levels drop when the opposite is true. Steady states (equilibria) where the red curve intersects each blue curve correspond to equal loss and gain rates and can be stable (filled circles) or unstable (open circles). Increasing RKIP degradation rate (ρ) causes a downward shift and alters the shape of the BACH1 loss curves (lighter shades of blue), whereas the BACH1 gain curve is unaffected. Stable RKIP (dark blue) causes fast BACH1 loss through let-7, allowing only a single low BACH1 steady state. Destabilizing RKIP slows BACH1 loss, enabling bistability through an M–B transition. Finally, highly unstable RKIP (light blue) causes a B–M transition to a single, high BACH1 steady state. (C) An example of the switch associated with a change of two parameters (K and ρ). Decreasing BACH1 dissociation constant (K) causes a leftward shift of the BACH1 gain curve (pink to red). Weak BACH1 autoregulation (pink, K = 10) maintains bistability, whereas strong BACH1 autoregulation (red, K = 0.1) forces the system into a monostable low-BACH1 state (B–M transition). From here, the system can transition directly to a monostable prometastatic state (M–M transition) if RKIP becomes unstable (dark blue line corresponding to ρ = 0.5 shifts to the light blue line corresponding to ρ = 5). (D) An example of a switch between two equilibrium states following a sufficiently large perturbation under conditions of bistability. The upper and lower panels illustrate the effects of small and large HDACi pulses (modeled as a spike of BACH1 concentration), respectively, applied at time t = 50. Other parameters were s = 0.1, S = 1.5, c = 100, m = 1, r = 5, ρ = 1, and a = 5. (E) Map of dynamic behaviors and possible transitions in the (ρ, K) parameter space. The circles correspond to BACH1 loss curves, plotted by using the same colors as in B. The triangles correspond to BACH1 gain and loss curves using the same colors as in C. The arrows represent examples of M–B, B–M, and M–M transitions.
To explore the existence of bistability and the effect of perturbations on the mutual interaction between BACH1 and RKIP, we developed a mathematical model of the RKIP–BACH1 network (Fig. 7A) using typical parameter values extracted from the literature (SI Appendix, Figs. S8–S10). As explained in detail in SI Appendix, the model describes how BACH1 (B), RKIP (R), and let-7 (L) evolve in time and reach equilibrium in the presence of multiple feedback interactions (Fig. 7A). For the original parameter values, the unique equilibrium state was an antimetastatic, low BACH1 state. We investigated how this equilibrium state changes as we alter various parameters (e.g., rates of RKIP protein degradation, BACH1 protein synthesis, etc.; SI Appendix, Figs. S10–S14) using rate-balance plots (31). The mathematical analysis revealed either one or two stable equilibrium steady states, corresponding to monostable or bistable behavior, respectively, depending on the parameter values. Overall, we found that moderate parameter changes had minimal effect on system behavior, indicating that the antimetastatic state is robust to perturbations (SI Appendix, Fig. S10). However, large changes of certain parameters (such as RKIP stability, BACH1 synthesis, or BACH1 auto-regulation strength) caused the emergence of a stable, prometastatic high BACH1 (and low RKIP) steady state, and an unstable state separating the two stable states. Further parameter changes caused the antimetastatic steady state to vanish, giving way to a single prometastatic steady state (SI Appendix, Figs. S13 and S14).
For example, to illustrate how decreasing RKIP protein stability affects system behavior, we studied the rates of BACH1 gain (red) and loss (different shades of blue) at various BACH1 levels (Fig. 7B) on rate-balance plots (31). Intersections of the red and blue curves correspond to equilibria (steady states) where cells can reside, having BACH1 gain and loss rates in balance. Normally, RKIP is a very stable protein (32), permitting only a single, low BACH1, antimetastatic steady state (single intersection at low BACH1 level). However, lowering RKIP levels causes the emergence of two additional steady states, indicating a transition to bistability (three intersections at low, middle, and high BACH1 levels), and finally to prometastatic monostability (a single intersection at high BACH1 level).
One additional feature of the RKIP–BACH1 network is the self-repression of BACH1. Whereas bistability was expected from the double-negative feedback architecture, it is less clear how negative BACH1 auto-regulation may affect network behavior. Negative feedback causes the downslope of the red “gain” curve at high BACH1 levels on all rate-balance plots (Fig. 7 B and C). To understand the role of negative BACH1 auto-regulation, we increased its strength by lowering the threshold level of BACH1 (the effective dissociation constant K) needed for self-repression. Considering a system with destabilized RKIP, weak self-repression (pink line, Fig. 7C) caused bistability. In contrast, strong self-repression (dark red line, Fig. 7C) caused BACH1 to shut down its own expression at very low levels, enforcing a single, antimetastatic, low BACH1 steady state. Therefore, negative auto-regulation can be essential for cells to avoid or leave the prometastatic high BACH1 state, providing a safety switch that can reestablish the antimetastatic state even after other perturbations have moved the system into a bistable or prometastatic regime.
An interesting consequence of bistability is the possibility of a switch between two equilibrium states under a sufficiently large perturbation (e.g., transient RKIP instability, BACH1 increase, EZH2 increase, or HDACi decrease). An example using HDACi as a perturbation is illustrated in Fig. 7D, where a large perturbation causes a cell with low B to transition to a state with higher B (Lower), whereas a small perturbation has vanishing effect (Upper). A high BACH1 subpopulation could then emerge after transient perturbations and act as a stable reservoir of highly invasive cells that expand into surrounding tissues or migrate to distal sites, fueling metastatic progression.
Overall, the RKIP–BACH1 network could support three different phenotypes defined by the level of BACH1 and the number of stable steady states: monostable antimetastatic, monostable prometastatic, and bistable (mixed). To illustrate how multiple parameters induce phenotype changes, we mapped these behaviors for various parameter pairs, such as RKIP degradation rate (ρ) and BACH1 dissociation constant (threshold) K (Fig. 7E and SI Appendix, Figs. S11 and S12). In these maps the white, black, and gray shadings mark the monostable (M) antimetastatic, monostable prometastatic, and bistable (B) behaviors, respectively. We define “prometastatic” and “antimetastatic” states based on BACH1 being higher or lower relative to a threshold (SI Appendix). Interestingly, these maps indicate that cells can transition between the monostable prometastatic and antimetastatic domains either directly (by M–M transitions) or indirectly, by first crossing into a mixed, bistable region (an M–B transition followed by a B–M transition). The M–M transitions are biologically defined, based on steady-state threshold crossing, and do not correspond to usual dynamical bifurcations. Nevertheless, the multiplicity of phenotype transitions derives from the complexity of the RKIP–BACH1 network structure relative to the classical toggle switch, which must cross the bistable domain to transition between the monostable states if repression is ultrasensitive (β, γ >3) (28). An example of different transitions associated with sequential changes of two parameters (K and ρ) is shown in Fig. 7 C and E. Stronger BACH1 feedback (lower K) causes a B–M transition (intersection of dark red and dark blue lines) followed by a prometastatic M–M transition through a subsequent drop in RKIP stability (intersection of dark red and light blue lines).
The direct adjacency of monostable states suggests an alternate source of nongenetic heterogeneity (besides steady-state switching in the bistable regime). Cells near prometastatic borders can cross the boundary and become prometastatic as they move in the parameter space under the influence of treatment or microenvironmental and random intracellular fluctuations.
Single Cell-Level Imaging Reveals Diversity of BACH1 and RKIP Expression.
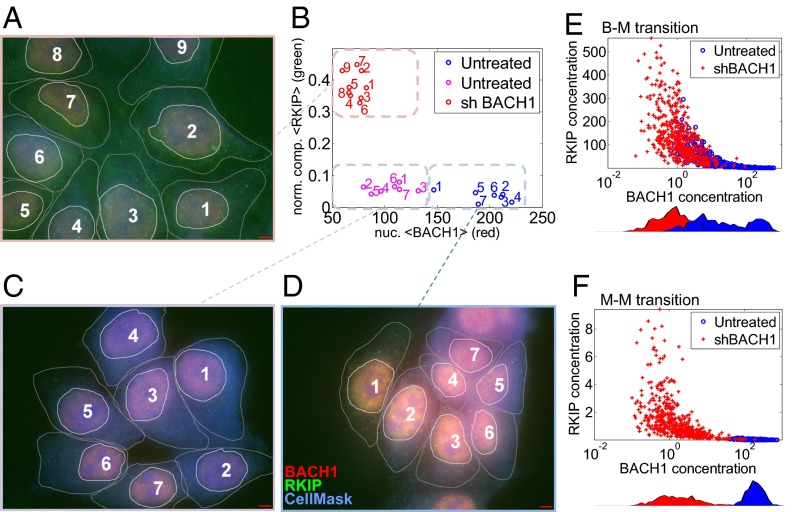
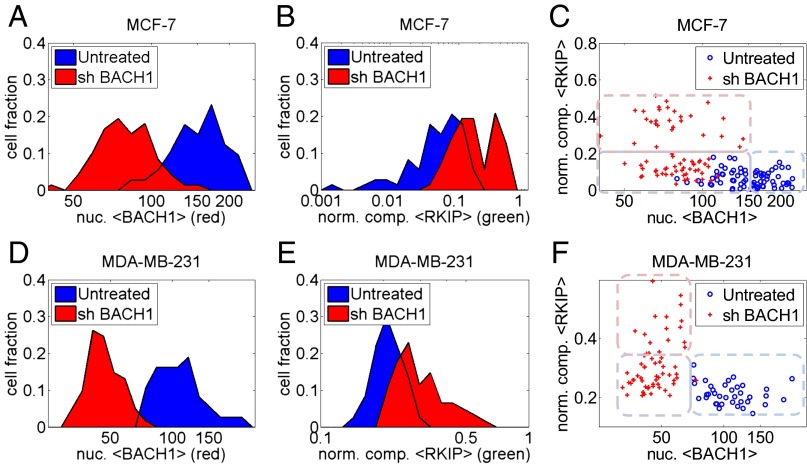
The mathematical model can generate predictions that are testable only by single cell-level measurements. For example, it can infer when and how the inverse RKIP–BACH1 correlation observed in patient samples manifests at the single-cell level. Moreover, it can predict how RKIP and BACH1 protein levels change if BACH1 transcripts are depleted by RNA interference. Such single cell-level predictions and measurements provide mechanistic information without which the connection between molecular interactions, heterogeneity, and tumor progression is unattainable. As a first step toward establishing such a connection, we applied sequential immunofluorescence followed by microscopy and computational image processing to quantitatively estimate RKIP and BACH1 protein levels in single MCF-7 breast cancer cells (Fig. 8 A–D). BACH1 (red) was predominantly nuclear, whereas RKIP (green) was generally distributed in the cell, with a characteristic punctate appearance. To investigate the relationship between BACH1 and RKIP, we developed computational tools to delineate cells and nuclei and estimate BACH1 expression by averaging red pixel intensities within the nuclei. We also inferred RKIP concentration by averaging compensated green pixel intensities normalized by blue pixel intensities (using the CellMask Blue cytoplasmic stain) over the area of each cell. Imaging unperturbed MCF-7 cells revealed highly variable BACH1 expression, whereas RKIP expression was relatively low in all cells (Fig. 8 B–D). However, shBACH1 treatment lowered BACH1 and raised RKIP (Fig. 8A). We used this approach below to characterize single breast cancer cells through BACH1 and RKIP distributions before and after shBACH1 treatment.
Fig. 8.
Single cell-level measurements and simulations. (A–D) Sample immunofluorescence images (100×; red, BACH1; green, RKIP; blue, CellMask Blue cytoplasmic stain) of MCF-7 cells before and 48 h after shBACH1 treatment. (Scale bar, 100 µm.) Nuclear BACH1 and cellular RKIP levels obtained from image processing are shown in the RKIP-BACH1 space (B). (E and F) Simulations of shBACH1 treatment of bistable and monostable prometastatic cell populations. An initially bistable BACH1 population (two blue peaks) transitions to a monostable population (single red peak) following shBACH1 treatment (E). In contrast, a monostable-to-monostable transition in response to BACH1 depletion causes a leftward shift of one peak (blue) along the horizontal axis (red) (F). The full ODE system corresponding to the RKIP-BACH1 network was solved numerically after five model parameters (K, S, ρ, a, and the shRNA synthesis rate α) were randomly drawn from lognormal distributions centered at the following parameter values: K = 100, S = 700, a = 1000, ρ = 0.005 (bistable), or ρ = 0.5 (prometastatic). Blue, untreated cells (α = 0); red, cells after shBACH1 treatment (average α = 5,000). Correlations were r = ‒0.37, P = 4e‒33 (B–M transition), and r = ‒0.45, P = 7.3e‒51 (M–M transition).
To predict how shBACH1 treatment would affect RKIP and BACH1 expression when cell–cell variability is present, we simulated populations of cells in various locations of the parameter space (SI Appendix, Figs. S17–S19). For each simulated cell we drew randomly five model parameters most likely affected by cellular and environmental fluctuations from lognormal distributions and allowed the system to reach steady state (Fig. 8 E and F). These simulations revealed that an initially bistable BACH1 population (two blue peaks) transitions to a monostable population (single red peak) following shBACH1 treatment (Fig. 8E). In contrast, a monostable-to-monostable transition in response to BACH1 depletion involved a leftward shift of one peak (blue) to lower values (red) (Fig. 8F). Overall, cells tended to align along different axes before and after treatment, and no cells had high BACH1 and high RKIP expression simultaneously in these simulations. In addition, there was an intriguing difference between the effect of shBACH1 treatment on bistable and monostable prometastatic cells. Whereas initially bistable cells appeared beneath the vertical, posttreatment population, initially monostable cells did not. There was always a strong negative correlation between RKIP and BACH1 when considering both treated and untreated cell populations together (Fig. 8 E and F).
To determine whether these computationally predicted features of shBACH1 treatment could be observed experimentally, we collected and analyzed multiple fields for two different cell lines (highly aggressive MDA-MB-231 and less aggressive MCF-7 breast cancer cells) by fluorescence microscopy. The initial distribution of BACH1 in MCF-7 cells was broader, more consistent with a noisy bistable scenario. Shifts in single cell-level RKIP and BACH1 distributions (Fig. 9 A, B, D, and E) confirmed RKIP induction following BACH1 knockdown, in agreement with population-average measurements. Cells aligned remarkably well along the axes in the RKIP–BACH1 space, and practically no cells had high BACH1 and high RKIP expression simultaneously, in agreement with the computational predictions. Finally, MCF-7, but not MDA-MB-231, cells showed an increased tendency of the population before shBACH1 depletion (blue, Fig. 9 C and F), to overlap with the population after shBACH1 depletion (red, Fig. 9 C and F), as observed in Fig. 8E. Although there can be multiple causes for such an overlap in the low BACH1 range, it is consistent with the existence of some bistable cells in MCF-7 cell populations, but less in MDA-MB-231 cell populations. Overall, the similarities between the predicted and observed effects of the shRNA treatment indicate that the model has successfully captured some essential features of the experimental network.
Fig. 9.
Single cell-level measurements of RKIP and BACH1 in MCF-7 and MDA-MB-231 cells. (A and D) Treatment with shBACH1 causes a leftward shift in the BACH1 distribution. (B and E) Treatment with shBACH1 causes a rightward shift in the RKIP distribution. (C and F) Scatter plots of pre- and posttreatment cell populations in the RKIP–BACH1 space. BACH1 and RKIP were negatively correlated at the single-cell level for all cells (untreated and shBACH1-treated; MCF-7: r = ‒0.51, P = 4.6e‒11; MDA-MB-231: r = ‒0.42, P = 1.6e‒5). Blue, untreated cells; red, cells after shBACH1 treatment. Mock treatment with luciferase instead of shBACH1 did not cause any change in RKIP or BACH1.
Discussion
Cancer progression, as an evolutionary process, should accelerate if higher heritable cellular variability is present. Accumulating evidence suggests that, in addition to genetic differences, heritable cellular variability can be the consequence of environmental or stochastic fluctuations. However, it is currently unknown how specific regulatory networks may generate or modulate cellular variability. Here we reveal a network architecture between the metastasis suppressor RKIP and the metastasis promoter BACH1 that could enable single cells to expand into prometastatic subpopulations without any genetic changes. In light of recent studies suggesting a positive role of biological noise in cancer progression (3, 4), it is important to identify regulatory networks where nongenetic diversification can take place and to identify actual genes and systems where these ideas can be tested. The RKIP–BACH1 regulatory structure and similar topologies can fulfill such a role.
The results shown here indicate that BACH1 is a key regulator of the metastasis suppressor RKIP in breast cancer cells. Previous studies from our laboratory have shown that RKIP suppresses BACH1 expression indirectly through signaling, transcriptional, and RNA interference (let-7) pathways. We demonstrated that BACH1 promotes invasion and metastasis of breast cancer cells, its expression inversely correlates with that of RKIP, and it is part of a gene signature prognostic for metastasis-free survival in breast cancer (23). Here we show that BACH1 in turn functions to negatively regulate RKIP transcription as well as its own transcription. Thus, BACH1 is part of a double-negative (overall positive) regulatory feedback loop for repression of RKIP transcription, generally known as toggle switch topology. Considering that the original synthetic toggle switch (28) could flip between different states following transient stimuli, these results suggest that the environment- and network-dependent, single cell-level balance of RKIP and BACH1 could trigger prometastatic transitions that drive progression to breast cancer metastasis.
Despite its superficial resemblance to a toggle switch, the BACH1–RKIP network has more regulatory complexity, including microRNA regulation and negative autoregulation. A mathematical model corresponding to this network revealed domains of monostable and bistable dynamics. Monostable dynamics enforces a single cellular phenotype (only low or high BACH1), while bistable cells can assume either a low BACH1 or a high BACH1 state. In contrast to the original toggle switch, in which only monostable-bistable transitions were possible at ultrasensitive repression (β, γ >3) (28), the RKIP-BACH1 network is readily capable of direct transitions between monostable high BACH1 and monostable low BACH1 states. These transitions should provide an additional mechanism for nongenetic diversification within the cell population as environmental or stochastic fluctuations move cells across borders between these monostable regimes. The mathematical model predicts different features of monostable–bistable and monostable–monostable transitions observable at the single-cell level. To quantitatively analyze BACH1 and RKIP expression at the single-cell level, we collected and analyzed microscope images in normal conditions and after shRNA depletion of BACH1. The results were consistent with direct transitions not only between bistable and monostable regimes but also between two monostable regimes. Interestingly, cells could populate states with low RKIP and low BACH1 but avoided states where both protein levels were high both in experiments and simulations. This behavior could reflect the permissiveness of states where both proteins are low: Cells could fluctuate down transiently if they can increase their expression before the other protein turns on. Mechanisms causing such fluctuations could be delayed negative feedback (BACH1 autoregulation) or biological noise. Overall, this analysis has broadened our understanding of metastatic transitions and phenotypic heterogeneity in cell populations beyond bistability. The multiplicity of transition scenarios distinguishes the RKIP–BACH1 network, which has more complexity, from the simple two-gene toggle switch (28), where the set of possible transitions is much more restricted.
Recent studies from our laboratories demonstrated that SNAIL, a transcription factor that promotes the epithelial–mesenchymal transition leading to invasion (33), participates in a network architecture similar to RKIP–BACH1. SNAIL represses RKIP transcription directly, binding proximal to BACH1 in the RKIP promoter. Like BACH1, SNAIL is suppressed downstream of RKIP via let-7 inhibition (19). Together, these regulatory loops comprise a double-negative or overall RKIP–SNAIL positive feedback system. Similar to BACH1, SNAIL negatively regulates its own promoter (34). Thus, as cancer regulatory network mapping advances (35, 36), it will be important to identify network structures with similar dynamics. In particular, the BACH1 (or SNAIL) negative feedback loop could be used as a “safety switch” to force cells out of their metastatic state.
Our data suggest that selective use of HDAC or EZH2 inhibitors could favor BACH1 or RKIP, respectively. Whereas BACH1 has been shown previously to promote senescence in part through recruitment of histone deacetylases (21), it is clear that the interplay of different HDACs is complex and dependent upon the specific promoter as well as the intracellular environment in breast tumor cells. However, TSA seemed to be an effective inducer of BACH1 in all three TNBC lines. Surprisingly, although several HDAC inhibitors regulated BACH1, we were unable to find any among the standard HDAC classes that induced RKIP expression. In fact, TSA and SAHA actually inhibited RKIP expression further in some cells, consistent with the increase in BACH1 transcription upon HDAC inhibition. By contrast, depletion of EZH2 did lead to up-regulation of RKIP transcription in at least two breast cancer cell lines, as previously reported. These observations also suggest that BACH1 regulation of its own promoter occurs through a repressive complex whose components are distinct from those in the BACH1 complex on the RKIP promoter. Taken together, these studies indicate that treatments with drugs that affect such broad-based transcriptional repressors must be considered with care, on a case-by-case basis. For example, applying inappropriate treatment (such as TSA) will promote metastasis by flipping the network into the low RKIP/high BACH1 state. However, treatments selectively increasing RKIP or let-7 levels will have the potential to change the balance from prometastatic to antimetastatic factors.
The studies here have focused on the role of BACH1 in breast cancer cells. However, it is likely that BACH1 is similarly regulated by RKIP in other tumor types as well. Future studies should elucidate the relationship between RKIP and BACH1 as well as the relative roles of BACH1 and SNAIL in regulating the transition to metastatic disease for other cancers.
Materials and Methods
Cell Culture and Generation of Cell Lines.
Human breast cancer cell lines (MDA-MB-231 1833, MDA-MB-436, and MDA-MB-231) with stable knockdown of shBACH1 or shSNAIL were generated using a lenti-viral transduction and maintained with puromycin (0.2 μg/mL) as described previously (19). Puromycin was used for selection of stale cells but removed before all of the cellular analysis.
Reporter and DsRed Expression Assays.
A luciferase plasmid containing human RKIP promoter regions (−2,200 bp upstream from TSS) into pGL2 (Promega) was generated as previously described (24). The BACH1 binding sites at the upstream −373 bp from TSS in the RKIP promoter or at the downstream +1,000 bp from TSS in the BACH1 promoter were mutated from TGAGCCA to GCTAGAC or from TGAGTCA to ACGTCAG, respectively. Then Wt and mutated BACH1 binding sites were subcloned into pGL2 vectors or DsRed Expression 2 vectors for reporter assays. Transient transfection of plasmids (250 ng for pCDH-SNAIL and pCDH-BACH1) or siRNA (100 nM for siBACH1) along with a luciferase plasmid was performed with lipofectamine 2000 (Invitrogen Corp.). Dual-glow luciferase activities (firefly luciferase and Renilla luciferase) were measured by Glowmax according to the manufacturer’s protocol (Promega). DsRed and cell surface marker expression using an Alexa Flour 488 anti-human HLA-a,b,c antibody (BioLegend) were detected using the microscope (Live DSU confocal microscope, Core facility, The University of Chicago). Intensity of DsRed protein expression and the size of each single cell on the random field (>10) was measured using ImageJ and shown (DsRed intensity per cell area).
Immunoblotting.
Whole-cell lysates were prepared and Western blotting was conducted using antibodies against BACH1 (sc-14700; Santa Cruz), EZH2 (4905; Cell Signaling), and SNAIL (sc-28199; Santa Cruz) as previously described (19). Band density was quantified using Licor (Odyssey Fc, dual-mode imaging system).
RNA Analysis and Quantitative RT-PCR.
Total RNA was isolated from cells using TRIzol (Invitrogen Corp.) according to the manufacturer’s instructions. Two micrograms of total RNA were used for reverse transcription using the high-capacity cDNA Reverse Transcription kit (Applied Biosystems), followed by quantitative PCR (Applied Biosystems) with primers for human BACH1, RKIP, SNAIL, and GAPDH (Origene).
Chemicals.
Breast cancer cells were treated with TSA (400 nM; Selleckchem), SAHA (3 μM; Selleckchem), or nicotinamide (20 mM; Sigma–Aldrich) for 24 and 48 h.
ChIP Analysis.
Cells were treated with TSA (400 nM) for 2 or 24 h before ChIP assays. For the transient knockdown of EZH2 or BACH1, cells were transfected with 100 nM of siRNA for EZH2 (SR301494; OriGene Technologies, Inc.) or BACH1 (siGENOME human BACH1 SMARTpool; Thermo Scientific) using Lipofectamin 2000 and incubated for 48 h. Cells were collected and immunoprecipitated with 2 μg of antibodies against BACH1 (sc-14700; Santa Cruz), EZH2 (4905; Cell Signaling), RNA polymerase II (05–952; Upstate Biotech), Histone 3 lysine 27 trimethylation (07–449; Upstate Biotech), and IgG (sc-2028; Santa Cruz). Detailed procedures are described previously (23). Primer sequences for ChIP-qPCR are indicated in SI Appendix, Table S1.
Single-Cell Imaging.
Human breast cancer cell lines were cultured at 37 °C and 5% (vol/vol) CO2 in DMEM or RPMI (Cellgro) with phenol red, supplemented with 5–10% (vol/vol) FBS, 2 mM l-glutamine, and 100 units/mL penicillin–streptomycin. For shRNA transfection, cells were grown in a six-well plate for 24 h in DMEM or RPMI supplemented with 5–10% (vol/vol) FBS without antibiotics. 1 µg of lentiviral-shRNA of human BACH1 was then transfected using the Lipofectamine 2000 transfection reagent (Life Technologies) in Opti-MEM (Invitrogen) for 48 h. The luciferase target sequence was used as nonspecific control siRNA.
In preparation for sequential immunofluorescence, cells were grown on poly-d-lysine coated coverslips and fixed with 4% (vol/vol) paraformaldehyde (ultrapure; Electron Microscopy Sciences) for 30 min on ice. Cells were then permeabilized with 0.5% Triton X-100 for 1 h and blocked with 4% (vol/vol) BSA for 30 min and incubated overnight at 4 °C in 1% BSA with mouse monoclonal primary anti-PEBP1 (anti-RKIP) antibody (clone 2D4 from OriGene, 1:250). Subsequently, cells were washed and incubated with goat anti-mouse Alexa 488 (Invitrogen) secondary antibody (1:1,000) for 1 h at room temperature. After washing three times, cells were incubated overnight with primary rabbit polyclonal anti-BACH1 antibody (HPA003175, 1:250; Sigma–Aldrich) at 4 °C in 1% BSA. Cells were then washed and incubated with donkey anti-rabbit Alexa 546 (Invitrogen) secondary antibody (1:1,000) for 1 h at room temperature. Finally, cells were stained with HCS CellMask Blue (H32720, 1 µg/mL; Life Technologies) for cytoplasm and nucleus.
TE2000 or TiE (Eclipse; Nikon) inverted microscopes were used with a 100× N.A. 1.3 oil objective for obtaining high-resolution images. The Image Processing Toolbox from Matlab (MathWorks, Inc.) was used for image analysis. Scripts allowed manual cell and nucleus delineation, from where BACH1 intensities were obtained by averaging over nuclei in the red channel. RKIP intensities were compensated using control images with BACH1 staining only and then normalized by the CellMask Blue intensity for every pixel and finally averaged over individual cell areas.
Mathematical Models.
The details of mathematical modeling can be found in SI Appendix. Briefly, to analyze RKIP–BACH1 network dynamics, we established a system of three nonlinear ordinary differential equations (ODEs) using basic biochemical considerations. These equations described the time dependences of RKIP protein (R), let-7 microRNA (L), and BACH1 mRNA (B) concentrations based on the synthesis and degradation rates of these molecules. The original system of three ODEs was first rescaled then converted into a system of two ODEs, and finally to a single ODE by sequential elimination of variables. Steady states of these systems were obtained by setting time derivatives to 0 and seeking solutions of the resulting algebraic equations. We generated rate-balance plots (31) in Matlab (MathWorks, Inc.) to graphically analyze the gain and loss of BACH1 as a function of BACH1 level and various parameters. Steady states were calculated numerically using the fzero function in Matlab. We also used Matlab to generate nullclines and analyzed the dynamics of the two ODE system in the RKIP–BACH1 phase space. Steady-state stability was determined based on the maximum eigenvalue of the two ODE system’s Jacobian at each steady state.
Statistical Analysis.
Samples were analyzed by the two-sample Student t test. P values were calculated for samples from three independent experiments unless otherwise indicated.
Supplementary Material
Acknowledgments
We thank Jack Skinner for helpful discussion and assistance. This work was supported by National Institutes of Health (NIH) Grant GM 87630 (to M.R.R.) and NIH Grants DP2 OD006481 04 (NIH Director’s New Innovator Award Program), NIGMS 1R01GM106027-01, and NCI U54 CA112970 08 (to G.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304840111/-/DCSupplemental.
References
- 1.Brabletz T, Lyden D, Steeg PS, Werb Z. Roadblocks to translational advances on metastasis research. Nat Med. 2013;19(9):1104–1109. doi: 10.1038/nm.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194(4260):23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 3.Brock A, Chang H, Huang S. Non-genetic heterogeneity—A mutation-independent driving force for the somatic evolution of tumours. Nat Rev Genet. 2009;10(5):336–342. doi: 10.1038/nrg2556. [DOI] [PubMed] [Google Scholar]
- 4.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: A looking glass for cancer? Nat Rev Cancer. 2012;12(5):323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 5.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459(7245):428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141(1):69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen AA, et al. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322(5907):1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- 8. Gupta PB, et al. (2011) Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell 146(4):633–644, and erratum (2011) 147(5):1197. [DOI] [PubMed]
- 9.Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: An evolving concept. Nat Rev Cancer. 2012;12(2):133–143. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- 10.Frank SA, Rosner MR. Nonheritable cellular variability accelerates the evolutionary processes of cancer. PLoS Biol. 2012;10(4):e1001296. doi: 10.1371/journal.pbio.1001296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snijder B, Pelkmans L. Origins of regulated cell-to-cell variability. Nat Rev Mol Cell Biol. 2011;12(2):119–125. doi: 10.1038/nrm3044. [DOI] [PubMed] [Google Scholar]
- 12.Veening JW, Smits WK, Kuipers OP. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol. 2008;62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. [DOI] [PubMed] [Google Scholar]
- 13.Nevozhay D, Adams RM, Van Itallie E, Bennett MR, Balázsi G. Mapping the environmental fitness landscape of a synthetic gene circuit. PLOS Comput Biol. 2012;8(4):e1002480. doi: 10.1371/journal.pcbi.1002480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blake WJ, et al. Phenotypic consequences of promoter-mediated transcriptional noise. Mol Cell. 2006;24(6):853–865. doi: 10.1016/j.molcel.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 15.Smith MC, Sumner ER, Avery SV. Glutathione and Gts1p drive beneficial variability in the cadmium resistances of individual yeast cells. Mol Microbiol. 2007;66(3):699–712. doi: 10.1111/j.1365-2958.2007.05951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charlebois DA, Abdennur N, Kaern M. Gene expression noise facilitates adaptation and drug resistance independently of mutation. Phys Rev Lett. 2011;107(21):218101. doi: 10.1103/PhysRevLett.107.218101. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L, et al. Raf kinase inhibitory protein inhibits beta-cell proliferation. Surgery. 2004;136(3):708–715. doi: 10.1016/j.surg.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 18.Fu Z, et al. Effects of raf kinase inhibitor protein expression on suppression of prostate cancer metastasis. J Natl Cancer Inst. 2003;95(12):878–889. doi: 10.1093/jnci/95.12.878. [DOI] [PubMed] [Google Scholar]
- 19.Dangi-Garimella S, et al. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 2009;28(4):347–358. doi: 10.1038/emboj.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng L, Imamoto A, Rosner MR. Raf kinase inhibitory protein (RKIP): A physiological regulator and future therapeutic target. Expert Opin Ther Targets. 2008;12(10):1275–1287. doi: 10.1517/14728222.12.10.1275. [DOI] [PubMed] [Google Scholar]
- 21.Dohi Y, et al. Bach1 inhibits oxidative stress-induced cellular senescence by impeding p53 function on chromatin. Nat Struct Mol Biol. 2008;15(12):1246–1254. doi: 10.1038/nsmb.1516. [DOI] [PubMed] [Google Scholar]
- 22.Warnatz HJ, et al. The BTB and CNC homology 1 (BACH1) target genes are involved in the oxidative stress response and in control of the cell cycle. J Biol Chem. 2011;286(26):23521–23532. doi: 10.1074/jbc.M111.220178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yun J, et al. Signalling pathway for RKIP and Let-7 regulates and predicts metastatic breast cancer. EMBO J. 2011;30(21):4500–4514. doi: 10.1038/emboj.2011.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beach S, et al. SNAIL is a repressor of RKIP transcription in metastatic prostate cancer cells. Oncogene. 2008;27(15):2243–2248. doi: 10.1038/sj.onc.1210860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strack RL, et al. A noncytotoxic DsRed variant for whole-cell labeling. Nat Methods. 2008;5(11):955–957. doi: 10.1038/nmeth.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-Iglesias O, et al. Histone deacetylase inhibitors: Mechanism of action and therapeutic use in cancer. Clin Transl Oncol. 2008;10(7):395–398. doi: 10.1007/s12094-008-0221-x. [DOI] [PubMed] [Google Scholar]
- 27.Ren G, et al. Polycomb protein EZH2 regulates tumor invasion via the transcriptional repression of the metastasis suppressor RKIP in breast and prostate cancer. Cancer Res. 2012;72(12):3091–3104. doi: 10.1158/0008-5472.CAN-11-3546. [DOI] [PubMed] [Google Scholar]
- 28.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403(6767):339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 29.Ferrell JE., Jr Self-perpetuating states in signal transduction: Positive feedback, double-negative feedback and bistability. Curr Opin Cell Biol. 2002;14(2):140–148. doi: 10.1016/s0955-0674(02)00314-9. [DOI] [PubMed] [Google Scholar]
- 30.Balázsi G, van Oudenaarden A, Collins JJ. Cellular decision making and biological noise: From microbes to mammals. Cell. 2011;144(6):910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferrell JE, Xiong W. Bistability in cell signaling: How to make continuous processes discontinuous, and reversible processes irreversible. Chaos. 2001;11(1):227–236. doi: 10.1063/1.1349894. [DOI] [PubMed] [Google Scholar]
- 32.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 33.Baritaki S, et al. Mechanisms of nitric oxide-mediated inhibition of EMT in cancer: Inhibition of the metastasis-inducer SNAIL and induction of the metastasis-suppressor RKIP. Cell Cycle. 2010;9(24):4931–4940. doi: 10.4161/cc.9.24.14229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peiró S, et al. SNAIL1 transcriptional repressor binds to its own promoter and controls its expression. Nucleic Acids Res. 2006;34(7):2077–2084. doi: 10.1093/nar/gkl141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saadatpour A, et al. Dynamical and structural analysis of a T cell survival network identifies novel candidate therapeutic targets for large granular lymphocyte leukemia. PLOS Comput Biol. 2011;7(11):e1002267. doi: 10.1371/journal.pcbi.1002267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu M, Jolly MK, Levine H, Onuchic JN, Ben-Jacob E. MicroRNA-based regulation of epithelial–hybrid–mesenchymal fate determination. Proc Natl Acad Sci USA. 2013;110(45):18144–18149. doi: 10.1073/pnas.1318192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.