Abstract
We report on a genome-independent microbial fingerprinting method using nucleic acid microarrays for microbial forensics and epidemiology applications and demonstrate that the microarray method provides high resolution differentiation between closely related microorganisms, using Salmonella enterica strains as the test case. In replicate trials we used a simple 192 probe nonamer array to construct a fingerprint library of 25 closely related Salmonella isolates. Controlling false discovery rate for multiple testing at α = 0.05, at least 295 of 300 pairs of S.enterica isolate fingerprints were found to be statistically distinct using a modified Hotelling T2 test. Although most pairs of Salmonella fingerprints are found to be distinct, forensic applications will also require a protocol for library construction and reliable microbial classification against a fingerprint library. We outline additional steps required to produce such a protocol.
INTRODUCTION
The pace of technology and methods development for microbial detection is exceptional and encompasses several embodiments of nucleic acid microarrays, mass spectrometry, microfabricated and/or fully automated PCR instrumentation, capillary electrophoresis devices and a host of other on-chip detection methods. However, current epidemiological and forensic investigations of pathogenic microorganisms continue to use fairly standard, gel-based DNA fingerprinting techniques (1–10).
In most cases, current DNA typing methods access a limited complement of genetic information and the fingerprint is based on DNA fragment sizing technology (i.e. gels). Despite the widespread acceptance of gel-based DNA fingerprinting techniques, however, they frequently fail to answer fundamental epidemiological questions. For example, Hancock et al. identified multiple sources of Eschericia coli O157:H7 in feedlots and dairy farms, but were unable to discriminate between isolates using PFGE (11). Thus, higher resolving power is required to identify the source of disease outbreaks, to determine how pathogens disseminate in the environment and to investigate how genomic structure (or nucleic acid signatures) change with time and cellular propagation. DNA microarrays are one possible technology platform that addresses the need for improved resolving power. More importantly, however, microarray probes are fixed in (physical) space and the hybridization signal contains primary genetic information (rather than size information). We therefore believe that DNA microarrays have the potential to overcome most of the limitations of gel-based, DNA fragment sizing methods in common use for DNA fingerprinting and epidemiological questions.
In order to move beyond microbial identification into microbial forensics, the attendant technology also requires a level of objectivity, quantitation and inferential rigor that can withstand scrutiny in a court of law. Characterizing or classifying a true unknown also implies that the technology should not rely on a priori knowledge of the unknown’s suspected DNA sequence. Beattie et al. (12) were the first to use oligonucleotide microarrays for genomic fingerprinting applications in a technique very similar to the nucleic acid scanning-by-hybridization membranes of Salazar and Caetano-Anollés (13) or the octamer genome scanning gels described by Kim et al. (14). Nevertheless, microarrays have not yet been developed for fingerprinting of closely related microorganisms in the absence of specific DNA signature sequences (i.e. SNPs), nor have the quantitative analysis and statistical tools been developed to use microarrays for forensic analysis of microorganisms. The objective of this study was therefore to develop a generic microbial fingerprinting method with the required statistical foundations for quantitatively comparing fingerprints of closely related microorganisms. The resulting methods are generally applicable to any microorganism, without requiring a priori knowledge of specific nucleic acid signatures.
MATERIALS AND METHODS
Bacterial isolates
A diverse panel of Salmonella enterica strains was assembled from a large bank of isolates maintained by the Field Disease Investigation Unit at Washington State University (Pullman, WA) (Table 1). Isolates were originally collected from outbreak and surveillance work in the Pacific Northwest between 1987 and 2000 and represent no more than one isolate from any single sampling event (15,16). Isolates were propagated as described in detail elsewhere (15,16) and bacterial serotype was determined by the National Veterinary Service Laboratories (Ames, IA). All five isolates having a Typhimurium serotype were phage typed as DT104.
Table 1. Isolates utilized in this study.
| Blocka | Isolate | Serotype |
|---|---|---|
| 1 | 1 | Enteriditis |
| 20 | Arizona | |
| 21 | Typhimurium | |
| 2 | 22 | Dublin |
| 29 | Enteriditis | |
| 34 | Arizona | |
| 3 | 35 | Meleagridis |
| 43 | Typhimurium | |
| 45 | Meleagridis | |
| 4 | 60 | Hadar |
| 78 | Meleagridis | |
| 80 | Arizona | |
| 5 | 92 | Meleagridis |
| 107 | Hadar | |
| 115 | Hadar | |
| 6 | 116 | Typhimurium |
| 117 | Typhimurium | |
| 125 | Enteriditis | |
| 7 | 141 | Typhimurium |
| 163 | Arizona | |
| 165 | Hadar | |
| 8 | 186 | Meleagridis |
| 191 | Arizona | |
| 194 | Hadar | |
| 9 | 198 | Enteriditis |
aBlock refers to the experimental design and statistical methods for creating microbial fingerprints, as described in Materials and Methods.
PCR amplification
Repetitive extragenic palindromic (REP) consensus PCR primers (17) were used to sample microbial genomes and generate amplified fragments for subsequent analysis on the oligonucleotide microarray. Two PCR amplifications were performed for every isolate. Cy3-labeled PCR primers (REP1R-Dt 5′, CY3-IIINCGNCGNCATCNGGC; REP2-D 5′, Cy3-RCGYCTTATCVGGCCTAC, where I = inosine, R = A or G, Y = C or T, V = G, A or C and N = A, C, G or T) were obtained from Biosource International (Camarillo, CA). PCR reagents were from a Qiagen HotStart Taq kit (Valencia, CA), except for the dNTPs (Amersham-Pharmacia Biotech, Piscataway, NJ). PCR amplification was performed in 50 µl total volume, using an MJ Research Tetrad Thermal cycler and 96-well plates (MJ Research, Watertown, MA). Final reaction conditions were 150 ng genomic DNA or 3 µl cell suspension and 1× PCR buffer (Qiagen), 2.5 mM Mg2+, 200 µM each dNTP, 1 U Taq polymerase and 0.6 µM each REP primer. Reagent grade water was used as a negative control and Geobacter chapellei (a Gram-negative, metal-reducing bacterium) served as the out group. Thermal cycling conditions were 95°C for 15 min, followed by 40 cycles of 95°C for 30 s, 40°C for 45 s, 72°C for 3 min and cooling to 4°C. PCR amplification was confirmed by analyzing 20 µl aliquots of the amplification reaction on a 2% agarose gel in 1× TAE running buffer. Aliquots of 20 µl of the remaining labeled amplification products were hybridized directly to microarrays without further manipulation, as described below. For conventional gel-based fingerprinting, primer-labeled Salmonella REP–PCR amplification products were separated at 1–2 V/cm on 1.5% gels composed of a 50:50 mixture of SeaKem GTG:Metaphor agarose (FMC Bioproducts, Rockland, ME) in 1× TAE running buffer, both containing 3 µg/ml ethidium bromide.
Microarray probes
A list of nonamer microarray capture probes was generated by random computer selection based on the sequence of the E.coli K12 genome (GenBank accession no. U00096). The selected nonamer probes (Table 2) occur (on average) 35 times each within the E.coli genome, with nearly equal probability of hybridizing to each strand of the genome. In addition to the nonamer capture probes, the microarray contained Cy3-labled quality control probes (5′-Cy3-TTGTGGTGGTGGTGTGGTGGTGGGGTTGGG TGGTGG-3′) that served as positional reference and spotting quality points and negative control buffer blanks to test for non-specific interactions and residual fluorescence on the microarray surface.
Table 2. Probe list.
| No. | Sequence | No. | Sequence | No. | Sequence | No. | Sequence |
|---|---|---|---|---|---|---|---|
| 1 | GGCGATTAC | 49 | CCGCATATT | 97 | GACGGTTTC | 145 | TAATGTCGC |
| 2 | TATCCGCGT | 50 | GCTTACGCA | 98 | TTGTACCAG | 146 | GTGTTGTAC |
| 3 | CCAGCGATA | 51 | GTTCCACTG | 99 | TGTAGCGTT | 147 | TCTTGGCAT |
| 4 | CTTTGCCTG | 52 | TCTTCCACA | 100 | ATGTGACCA | 148 | GCCAAATGA |
| 5 | TAAACTGCC | 53 | GGTTTCCAC | 101 | GCGGCATAA | 149 | TCACGGTAG |
| 6 | TCGACAGTG | 54 | AGGCAATGA | 102 | ATCGTTGCA | 150 | CGAAGAAGG |
| 7 | TCACCACCT | 55 | GCGATGACA | 103 | CAGAACGAC | 151 | CGTAACCAT |
| 8 | CGGAACGTA | 56 | GCGCTGTAA | 104 | GAATGACCA | 152 | GGTGTACCA |
| 9 | TTATGCCGA | 57 | TCTATCTGC | 105 | GTTCAAGGT | 153 | CCCGCAAAT |
| 10 | AAGATGCCA | 58 | TTGGTCAGC | 106 | CGATGACTG | 154 | TTGGCATCC |
| 11 | AGGCCAGTT | 59 | CGTGGTATG | 107 | CGTCAACTT | 155 | ATAACGGCG |
| 12 | CTTTGCCCT | 60 | GTGGTTTCC | 108 | GCAGCAATT | 156 | CACCGCAAT |
| 13 | GATGTCGGT | 61 | GTGGTTTCC | 109 | ACCATTGTC | 157 | GTCAACTTC |
| 14 | TCGGCTTCT | 62 | ACTGACGCA | 110 | ATCGTGGTC | 158 | TTCTCGACA |
| 15 | GTTTCCTGT | 63 | CGAAGTGTT | 111 | ACTTCCGGT | 159 | CAACGGCTT |
| 16 | GGGCAATAC | 64 | GCAGACAAT | 112 | AGGAAGTGG | 160 | CCTCAGCAA |
| 17 | GCAAACAGC | 65 | CAGTACGTG | 113 | GACGCCATT | 161 | CTGGTCCAT |
| 18 | TGGCAACAC | 66 | ATCCAGACT | 114 | TTACCCACG | 162 | GCTTCGGTA |
| 19 | CACGGGTTA | 67 | GTTTGAGCG | 115 | ACGGTCGAT | 163 | CTGGTCGTT |
| 20 | GACAGAAGA | 68 | GCAGTAAGC | 116 | CGTAGCGTT | 164 | CGTTAGAAC |
| 21 | GCAAAGAGT | 69 | TTCAGCCAA | 117 | ACACGCAGT | 165 | TGCGACCAT |
| 22 | GGTTGCCAT | 70 | CGGGTAAAG | 118 | AGCCCATTA | 166 | GCTATTGCC |
| 23 | TGACTGATG | 71 | ACACAGCAG | 119 | CAACCCAAC | 167 | TAGCGGCTT |
| 24 | TGACGGTAA | 72 | AGAAAGCCT | 120 | CAGACAGAC | 168 | GCTAACTTC |
| 25 | CTGTAATGC | 73 | CATTGACGG | 121 | CCATGCGAA | 169 | CGACTGGTT |
| 26 | AAGCCTTTC | 74 | CACACCACA | 122 | CGAAAGCCA | 170 | GCCGTAAAG |
| 27 | TCCATCGGT | 75 | ACACAGCGA | 123 | GAAAGGCAG | 171 | CGACACGTT |
| 28 | TCACTTTGC | 76 | CTGCAAAGG | 124 | GCTGGTATA | 172 | GGGCCATAA |
| 29 | CCATGCAGT | 77 | TTCGGCAGT | 125 | GGTTTCGTC | 173 | CACGCGTAA |
| 30 | CTGTTGGTG | 78 | GTTGCCGAA | 126 | GTTGAGTTG | 174 | ACCGTTGGT |
| 31 | AATGAGCCA | 79 | ACCACCATG | 127 | TATACAGCC | 175 | ACGAGCATT |
| 32 | GAGGTTGTC | 80 | ATGCTCGTC | 128 | AATTGCACC | 176 | ATGGCACCT |
| 33 | TGGTGTCAC | 81 | AACCGATGT | 129 | CGTACCAAT | 177 | AGTAAGCGA |
| 34 | TGGCAATGC | 82 | AAGAAGAGG | 130 | TATATCGGC | 178 | TGTCGCCAA |
| 35 | ACAATCGCT | 83 | TGCAGAAGC | 131 | CAACCAACG | 179 | TGGTGAAGT |
| 36 | CGAGATGCA | 84 | TTCCAGTCA | 132 | TGCCATTGG | 180 | AGTGACCGA |
| 37 | TGCCGTTAA | 85 | TACGAATGC | 133 | CGAAGAGTG | 181 | TCGTTTCCA |
| 38 | CGTTATGCT | 86 | CTTCAATGG | 134 | AACTGCAAC | 182 | CCGTCTTTC |
| 39 | TCTGGTAAC | 87 | AACGTAACG | 135 | AACGCAGTA | 183 | AGTGGAGTA |
| 40 | TATCGTGGT | 88 | AGCGGCATA | 136 | TTAGCCACA | 184 | TACAGCGGA |
| 41 | TAACCAGGC | 89 | GCGAGAATG | 137 | CGGCTAAAC | 185 | GTCGTCAAT |
| 42 | GTTACAGGG | 90 | CGCTATCTC | 138 | TTACGCGAA | 186 | CAATGACAG |
| 43 | GTTGAAGGC | 91 | TCCGTCAGT | 139 | GCGTAACGA | 187 | CAGCTAATG |
| 44 | AGGGAATGC | 92 | GGCAAATGG | 140 | AATGCGGGT | 188 | CGTGCATAA |
| 45 | ATTTCGCAG | 93 | CTCAAGCCA | 141 | TCCATTTGC | 189 | ACGACTTCA |
| 46 | ATAACGCCT | 94 | GCCGTATCA | 142 | TCGGTTAGC | 190 | GACCACTTC |
| 47 | ACTGTTCCA | 95 | GGTGAAGTG | 143 | GAAGCAGGT | 191 | CGGTAACTC |
| 48 | CAGCCTTTG | 96 | ATGGGTGCT | 144 | TGGTGGCTT | 192 | ACGGAGTTA |
Microarray fabrication
Microarrays were manufactured with amine-modified oligonucleotides and 6-well Teflon-masked slides (Erie Scientific, Portsmouth, NH) as previously described (18). Oligo nucleotide capture probes were resuspended in reagent grade water and the concentration of each was measured in triplicate by UV absorption (Bio-Rad Smartspec 3000; Bio-Rad, Hercules, CA). Oligonucleotide capture probes were diluted to 80–100 µM in 0.01% SDS, 50 mM NaOH print buffer. Probes were printed with an Affymetrix 417 Pin and Ring™ arrayer (Affymetrix, Santa Clara, CA), with two complete 192 probe microarrays contained within each well of a Teflon-masked slide. After printing, the slides were baked for 30 min at 130°C and stored at room temperature in the dark.
Experimental design and microarray hybridization
In a prior work (19) we generated a binary fingerprint signature for each array by measuring signal intensities and declaring a probe spot ‘on’ if pixels in the expected spot location were more intense than adjacent pixels, so that the hypothesis of a uniform neighborhood is rejected; otherwise the spot was declared ‘off’ for that replicate. For this study we performed preliminary experiments of microarray fabrication and method level variability before establishing the experimental design outlined below. Twenty-four replicates were required to begin to achieve a statistically reproducible binary array signature for each organism and the set of 192 hybridized nonamer probes utilized herein. For the results presented here, then, the microarray fingerprinting procedure was defined by 24 microarray replications per isolate as {2 PCR amplifications per isolate × 3 slides per amplification reaction × 2 hybridization wells per slide × 2 microarrays per hybridization well}. The 25 Salmonella isolates were organized into nine separate ‘isolate blocks’ (Table 1), where each block (except for the last block) contains three isolates. Isolates from the same block were compared directly on the same slides (the slides thus provide a ‘blocking’ effect for the isolate triples).
Twenty microliters of Cy3-labeled REP–PCR products were diluted to 70 µl in hybridization buffer to achieve a final concentration of 4× SSC (1× SSC = 0.15 M NaCl, 0.015 M trisodium citrate, pH 7.0), 5× Denhardt’s solution (1 g/l Ficoll 400, 1 g/l polyvinylpyrrolidone and 1 g/l ultra-pure bovine serum albumin). Amplification products were heat denatured for 5 min at 95°C, snap cooled on ice and divided evenly between two replicate wells per slide. Independent hybridizations were performed using six slides for each isolate block, split evenly between two independent PCR preparations. The final study design allowed decomposition of experimental variability into three separately estimable components: (i) variability between arrays within a slide; (ii) variability between slides for the same PCR preparation; (iii) variability between different PCR preparations.
Denatured amplicons (in hybridization buffer) were hybridized overnight at 4°C and the slides washed five times in an ice-cold solution of 1× SSC. Slides were dried with compressed air and imaged directly on an ArrayWoRx Microarray Imager (Applied Precision, Issaquah, WA) using 548 nm excitation/595 nm emission filters and a 1.5 s exposure time. ArrayWoRx analysis software was used to identify the location and size of every spot in the array pattern using a fixed grid and to extract an average pixel intensity value for every spot and for the local background around every spot.
Statistics
The initial goal of the statistical development was to determine whether the isolates have distinct microarray fingerprints. A background-corrected intensity value was computed for each spot by taking the (variance stabilizing) log transform of the ratio of the mean spot pixel intensity to the mean background pixel intensity, i.e. log(mean spot pixel intensity) – log(mean background pixel intensity). Following background correction, intensity values for each array were linearly transformed to have mean 0 and standard deviation 1 to correct for variations in brightness between arrays. The linear transformation was not performed across entire slides, but only on individual arrays within slides (note, there are 12 arrays on each slide, representing three isolates). This self- normalization to mean 0 and standard deviation 1 is somewhat conservative and might mask real differences between isolate fingerprints. In fact, analysis of variance comparing array average intensities for different isolates on the same slide revealed a systematic difference in overall signal intensity between isolates, suggesting that some discriminatory information is lost in the self-normalization (not shown).
The variation between normalized spot intensities can be described using a linear mixed-effects model. Let Yiplcr denote the background-corrected, normalized spot intensity for the ith isolate on the pth probe for the rth replicate array on the cth slide for the lth PCR (i = 1…25, p = 1…192, r = 1…4, c= 1…6 and l = 1,2 within an isolate block). The model is
Yiplcr = µip + θipl + αiplc + εiplcr,
where µip is the average intensity for the ith isolate on the pth probe, θipl ∼ (0,δip2) is a random between-PCR effect, αiplc ∼ (0,τip2) is a random between-slide effect, εiplcr ∼ (0,σip2) is a residual term describing the variability between replicate arrays on the same slide and θipl, αiplc and εiplcr are independent. Variance components δip2, τip2 and σip2 were estimated separately over 24 replicates for each isolate/probe combination using the restricted maximum likelihood method (REML).
The sample mean over 24 replications for the ith isolate, pth probe (denoted Ȳip…) has mean µip and variance Vip = δip2/2 + τip2/6 + σip2/24. If we make the simplifying assumption that the relative proportions of variance components are the same for all i, p, then Vip = ρSip2 for some constant ρ and where Sip2 is the sample variance computed over the 24 replicates. In this case, isolate averages Ȳip…. might be statistically compared using sample standard deviations, thus simplifying computations. Importantly, we do not assume that the 24 replications are independent (clearly, by design, they are not, so that the effective sample size is <24). Instead, we use distribution-free approaches to compare isolate fingerprints.
To identify probes with differential signal intensity between pairs of isolates, we employed an empirical Bayes method following the approach described in Efron et al. (20), which provides a solution to the so-called simultaneous inference problem, i.e. we are making inferences about 192 probes for each of 300 pairs of isolates, or 57 600 total inferences. Failure to account for chance effects due to the large number of comparisons can result in overly optimistic conclusions. Using the linear model (lm) function in the R computing environment (version 1.7.1; R Foundation for Statistical Computing, Auckland, NZ), we computed the difference statistics between pairs of isolates for each probe, pooling variance across all 25 isolates. As an alternative, and if we chose not to make the simplifying assumption about variance component proportions, we could employ the full linear mixed effects model, computing difference statistics using the lme function in R. In the empirical Bayes approach of Efron et al. (20), the probability density of the difference statistic is expressed as a mixture
f(z) = p0f0(z) + (1 – p0)f1(z),
where p0 is the prior probability that there is no difference between two isolates at a probe, f0 is the density of difference statistics when there is no difference (the null density) and f1 is the density when there is a difference. The statistical problem is to estimate the (a posterior) probability that a probe differentiates two isolates,
p1(z) = 1 – p0f0(z)/f(z).
We estimated f by smoothed Poisson regression (B-splines, 6 df) fit to the bin counts of the histogram (with 250 bins) computed from the difference statistics. (Separate analyses were performed for comparisons between isolates from the same isolate block and for comparisons between isolates from different isolate blocks. Test statistics for these two groups, by design, will have different distributions.) The null density f0 can be similarly estimated from empirically derived null difference statistics, computed for example using permutation or re-sampling methods. When p0 is assumed to be near 1 (a conservative assumption), Efron (Technical Report 2003-28B/225, Department of Statistics, Stanford University) showed how to obtain a reasonably accurate empirical null distribution by fitting the central peak of f to a normal density.
Next, we performed a multivariate test, comparing the entire 192 probe profiles for each pair of isolates. Specifically, for isolates i and i′ we test H0: µi = µi′ versus HA: µi ≠ µi′. The usual Hotelling T2 test statistic for multivariate two sample comparisons is proportional to the Mahalanobis distance, dMah2(i,i′) = (Ȳi – Ȳi′)′S–1(Ȳi – Ȳi′), where S is the (pooled) within-isolate sample covariance matrix. Because of the large number of probes relative to the number of samples, this difference statistic can be very unstable and, depending on how the co-variance matrix is defined, might not even be computable (due to a singular covariance matrix). Thus, we computed modified Hotelling T2 difference statistics using a data reduction approach proposed by Langsrud (21). The difference statistic is obtained via singular value decomposition of the combined data matrix for two isolates and is given by:
T2 = [(SS1 + λ + SSk)/k] ÷ [(SSk + 1 + d + λ + SSn)/(n – k – d)],
where SSi is the sum of squares contribution of the ith component, k is the number of retained components and d is the number of buffer components, which were not included in the test statistic [to prevent ‘contamination’ of the numerator, increasing the power of the test; see Langsrud (21)]. Of the 48 components, we (somewhat arbitrarily) retained the first five and used the next 16 as buffers.
To test the significance of the 300 computed T2 values, we constructed an empirical null distribution for T2 (the distribution if there is no difference between isolate pairs) by re-sampling (with replacement) error residuals Ripcr = Yipcr – medianr(Yipcr) and slide residuals Dipc = medianr(Yipcr) –µip, similar to the approach in Amaratunga and Cabrera (22). The corresponding vectors containing residuals for all 192 probes, Dic and Ricr, were sampled and summed to mimic the experimental design and T2 was computed for each simulated experiment. We performed 5000 such experimental simulations for both within-isolate block comparisons and between-isolate block comparisons and computed P values by comparing the 300 computed T2 values with the appropriate empirical null distribution. We adjusted P values to control the false discovery rate for the large number of tests using Benjamini and Hochberg’s sequential algorithm (23): let p(1) ≤ p(2) ≤ … ≤ p(300) be the ordered P values. Find r = max[i:p(i) ≤ (iα/300)] and reject H(1)0 … H(r)0. This algorithm implicitly takes p0, the prior probability that isolate profiles are not different, to be 1, which is conservative.
RESULTS
Statistical analyses
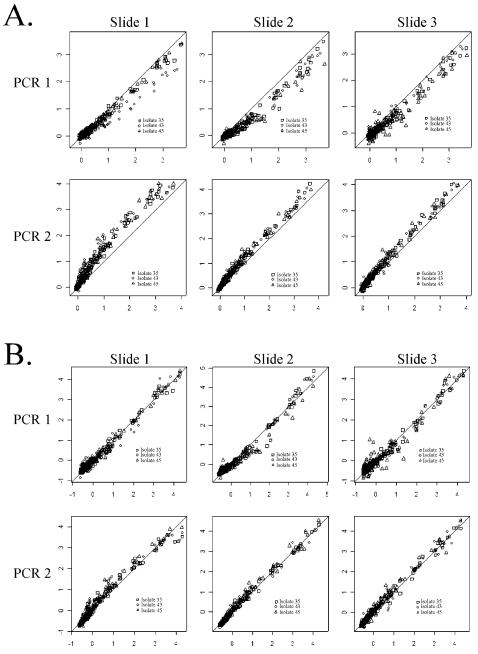
Figure 1 shows the need for normalization across replicate arrays. Scatter plots of median intensity values (by isolate) across arrays on a slide (y-axis) versus median intensity over all slides (x-axis) for the ‘isolate block’ {35,43,45} are given before (Fig. 1A) and after (Fig. 1B) self-normalization. The un-normalized plots reveal a systematic difference in overall brightness between two independent PCR preparations. Self-normalization removes these differences. Similar results were observed for the other isolate blocks (not shown).
Figure 1.
Plots of median chip intensity (y-axis) versus median intensity over all chips (x-axis) for the ‘isolate block’ {36,43,45}before (A) and after (B) self-normalization. Plotting characters represent isolates. Before normalization, there appears to be a significant lot effect (A). The normalized plots (B) suggest that between slide repeatability is high and that a linear normalization is appropriate. The lots represent independent PCR preparations. The three slides (independent hybridizations) within a lot were prepared on three separate days.
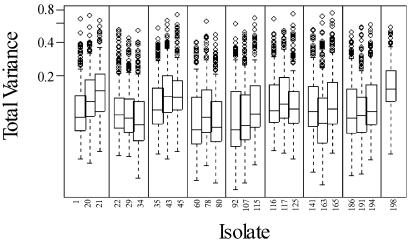
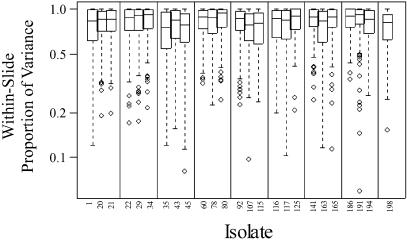
The distribution of total variance for 192 probes, computed separately for each isolate, is shown in Figure 2. Approximately 20 probes have consistently larger than average variance. There are additional differences between individual isolates. On average, 80% of variability is attributed to within-slide variation, 11% to variation between slides for the same PCR preparation and 9% to variation between independent PCRs (Fig. 3). Self-normalization successfully removed most of the differences between slides and PCR preparations.
Figure 2.
Box plots of total variance for 192 probes, computed separately for each isolate. The median value is represented by a line within the rectangular box, which captures half of the 192 observations (the lower and upper edges of the rectangle represent the first and third quartiles, respectively). The ‘whiskers’ in each box plot extend to the extremes of the data, and very extreme points (individual probes with extremely high variance) are represented as individual data points (circles). Vertical lines delineate ‘isolate blocks’.
Figure 3.
Proportion of total variance attributed to variation between arrays within slides, computed separately for each isolate using REML. Each box plot summarizes the distribution over 192 probes. On average, 80% of the variance is attributed to within-slide variation, 11% to variation between slides for the same PCR preparation and 9% to variation between independent PCR amplifications. Vertical lines delineate ‘isolate blocks’.
Pairwise comparisons
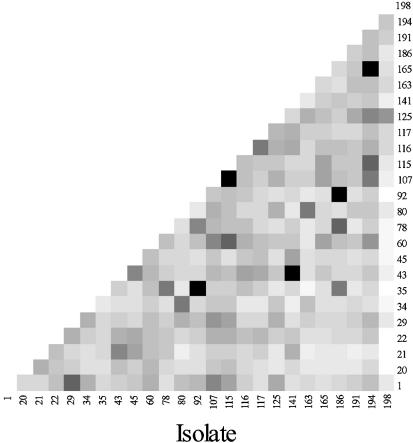
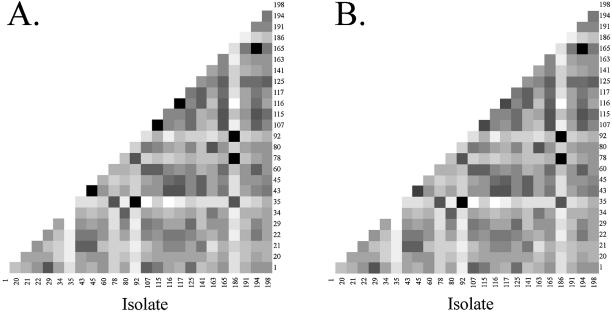
Using the empirical Bayes method to identify probes with differential signal intensity between pairs of isolates, and making the conservative assumption that very few of the differences are significant, we found that all but five of the 300 isolate pairs have at least one discriminating probe, i.e. for 295 of the 300 isolate pairs we conclude that Pr(two fingerprints differ at one or more probes) > 0.95. Figure 4 displays the number of discriminating probes for each pair of isolates calculated in this manner.
Figure 4.
Matrix of discriminating probes for each pair of isolates from no discriminating probes (black) to many discriminating probes (white).
Utilizing the modified Hotelling T2 difference statistic, 296 of 300 pairwise comparisons were found to have distinctly different fingerprints at α = 0.05. Figure 5A shows the pairwise distances between isolates, from black (not significantly different) to white (very different); Figure 5B shows the results if we exploit the blocking advantage enjoyed by isolate pairs found in the same ‘isolate block’. This analysis subsumed between-PCR variability in the slide residuals, largely to generate a large pool of between-slide residuals from which to sample. We therefore performed another analysis in which both between-slide (within PCR) and between-PCR residuals were sampled. The effect on number of significantly different pairs was minimal: a few more samples were found not different, but after adjusting p0 from 1 to its estimated upper bound according to p0 ≤ min(f/f0) (20), we found over 295 pairs to be statistically different.
Figure 5.
(A) Matrix of pair-wise comparisons using a modified Hotelling T2 test to correct for multiple testing (α = 0.05) and using the conservative p0 = 1. (B) The effect of exploiting blocking structure for comparisons within an ‘isolate block’. The gray scale corresponds to the distance between isolate profiles, from white (large) to black (small). All but four of the 300 pairs are statistically distinct after correction for false discovery. The four non-significant pairs are colored black.
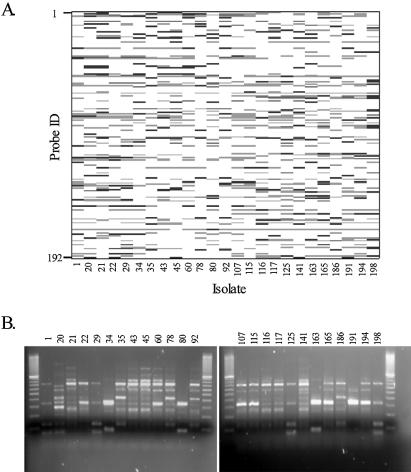
Isolate profile plots
We constructed a synthetic gel image or isolate profile plot to visually compare microarray data to conventional gel images (Fig. 6). For a given isolate, probe spots shaded black differentiate the isolate from the average of all other isolates (Fig. 6A). These are probe spots for which the contrast between the isolate and the average of all other isolates (computed using the lm function in R) is significantly larger than 0 (α = 0.01). An ANOVA-based clustering of isolates (again for each probe) was obtained by rank ordering the average intensity levels for each isolate and segmenting into two groups to maximize their contrast (using t values from group contrasts in the ANOVA model). Probes falling into the high intensity group are shaded gray in Figure 6A (if they have not already been shaded black). The profiles are relative, because they depend on which other isolates are present in the study. Alternative views can be obtained, for example, by comparing each isolate to a standard reference isolate.
Figure 6.
< (A) Relative profiles for 25 Salmonella isolates. Black bands (or probes) differentiate the isolate from the average of all other isolates. These are probes for which the difference between the isolate and the average of all other isolates is significantly larger than 0 (α = 0.01). Probes falling into the high intensity group from the ANOVA-based clustering are shaded gray (if they have not already been shaded black). This signature is useful for making relative comparisons, but it is not the signature used for classification. (B) REP–PCR agarose gel fingerprints from the same 25 isolates.
The number of discriminating probes in the (relatively simple) microarray fingerprints (Fig. 6A) far exceeds the number of discriminating bands in the corresponding standard REP–PCR gel (Fig. 6B). The pattern of discriminating probes shows (qualitatively) that a relatively simple fingerprinting chip and protocol can detect and project differences between S.enterica strains. In contrast, the REP–PCR gel fingerprints did not even qualitatively distinguish between several of the strains (Fig. 6B, e.g. isolates 43, 45, 60 and 92 and 115 and 117).
Towards a classification protocol
Although we have evidence that most pairs of Salmonella fingerprints are distinct, it does not necessarily follow that unknown samples can be reliably classified. Dudoit et al. (24), for example, described some of the challenges of microarray classification and performed a comparison of well-known classification algorithms. In forensic applications, the number of classes (isolates) is potentially very large, which increases the likelihood of misclassification. In addition, some unknowns do not belong to any of the pre-defined classes, requiring an approach to identify new classes. In forensic applications of microarray technology we have the luxury of replication (to a degree), i.e. given an unknown sample, we can perform independent PCR amplifications and obtain multiple hybridizations across multiple slides. We can more accurately classify the average over replicate hybridizations than a single hybridization.
We performed simple classification experiments separately for each isolate block. Using ridge discriminant analysis and self-normalized arrays, we used leave-one-out-cross- validation to assess the performance of classifying: (i) individual arrays; (ii) the average of four arrays across a slide. Classifying each ‘unknown’ to one of three groups, we correctly classified: (i) 84% of individual arrays; (ii) 90% of array averages (132/144). However, nine of the 12 misclassifications resulted from confusion between the isolate pairs (43,45) and (107,115), which were barely distinct in the multivariate test (Fig. 5). In fact, they are not distinct in the multivariate test that ignores blocking; likewise, the discrimination ignores blocking. The other three misclassifications are from the isolate block {116,117,125}, where isolate pair (116,117) is not significant in the multivariate test that ignores blocking. Thus, for five of the eight blocks tested we achieved 100% classification. If we remove the 24 ‘unknowns’ corresponding to isolates (43,45) and (107,115), we might claim a success rate of 97% (117/120).
It is important to note that there might be a small upward bias in the statistical classification estimates used to identify an unknown relative to a reference library. In the work presented here, for example, there are six slides for each isolate block, three slides from each of two independent PCR preparations. In the cross-validation assessment, five slides were used to construct a ‘library’ at each step and one slide was treated as the ‘unknown’ to be classified (actually three unknowns, as there are three isolates per slide). Thus, two samples used to construct the library came from the same PCR as the unknown sample, suggesting a possible upward bias in the estimated success rate. Therefore, classification performance might be improved by averaging across slides and PCR amplifications (we did not have a large enough sample to fairly test this hypothesis). If we try to classify individual arrays to one of 25 isolates, for example, we correctly classify 48% (which is much higher than the 4% expected under random chance, but still inadequate for forensic applications). Using the average of four arrays improves performance to 63% correctly classified isolates.
DISCUSSION
Objective fingerprinting
A central tenet of forensics is that genetic data withstand the scrutiny of a trial in a court of law. From our perspective, this tenet is manifest in more conventional microbiology applications as the need to objectify and quantify the DNA fingerprinting analytical process, data extraction and profile analysis procedures. Traditional PCR or multi-locus fingerprinting techniques are relatively simple to objectify and quantify, because the target signatures are discreet, known and limited in number, and the basal detection limit for the analytical method (e.g. single-locus PCR) is easily discerned. The detection and identification of true ‘unknowns’, however, more often requires genome scanning techniques such as amplified fragment length polymorphism, mini- and microsatellite fingerprinting and similar data-rich analysis methods (3,25). The inability to quantify multiplex (genome scanning) PCR detection limits for all amplified fragments is but one complication of DNA fingerprinting that is not inherent in conventional nucleic acid signature analysis. Thus, traditional gel-based fingerprinting methods are presently descriptive, not quantitative, which can limit their utility for some microbial forensics, epidemiology and source attribution applications. Our overarching objective is therefore to convert from a descriptive to quantitative microbial fingerprinting method that is reproducible through time and space and across laboratories and users. It is only through method level replication and objective data analysis that this objective will be realized.
From gels to microarrays
Objective identification or definition of a gel band continues to be problematical (25) (Fig. 6B), especially with smeared backgrounds (e.g. isolate 141) or low and high intensity bands (e.g. isolates 34 and 191). Criteria for including or excluding bands above or below a given size are arbitrary and a single gel cannot simultaneously resolve low and high molecular weight bands. Gels are also susceptible to warps, bubbles, distortions and other anomalies that are difficult to objectively correct within or between gels, even with internal standards and advanced computer software (3). For these reasons, gel electrophoresis (fragment sizing) frequently cannot even descriptively resolve near neighbors, as illustrated for S.enterica isolates 43, 45, 60 and 92 in Figure 6B. More importantly, the migrational variability of nucleic acids in sieving media make gels ill-suited for automated, objective band scoring across gels for forensic applications. The fundamental problem of positional variability in gel-based fragment sizing techniques therefore led us to develop microarrays for genomic fingerprinting.
Although Figure 6A is conceptually similar to a standard fingerprinting gel (bands and estimates of median or average band intensity), it is important to re-emphasize that the microarray fingerprint profiles were generated from 24 replicates arrays (recognizing that the effective number of replicates is less than 24) and only reflect those probes (or bands in the on–off plot) that are objectively determined to be discriminatory. Replication allows the opportunity to quantitatively assess the significance of observed differences between isolate fingerprint profiles, in contrast to simply visualizing differences between gel-based fingerprints via dendrograms, principal component analysis or cluster analysis.
The conceptual similarity between the biochemistry of microarray and gel fingerprinting also translates into similar sources of measurement variability, including variable backgrounds, identifying and defining a ‘band’ (or spot) amidst a variable background, a low signal-to-noise ratio and variable performance across gels, microarrays or users. Thus, while the linkage between microarrays and gel fingerprinting is obvious and a natural extension of prior work (12), the statistical foundation for image analysis, assay replication, defining a microarray DNA fingerprint and quantitatively comparing fingerprint profiles is not.
Quantitative fingerprint comparisons
Because we cannot know or quantify a priori the presence, amplification efficiency or hybridization efficiency of every REP-based amplicon in every genome, any detectable microarray signal above background is, in principle, a significant datum in a microbial fingerprint. However, it is well known that variability in microarray manufacture, data and image analysis is significant (for reviews see 26,27). The challenge for quantitative microarray-based microbial fingerprinting therefore becomes one of scoring reproducible hybridization events, such that true biological variability exceeds the inherent noise of the analytical process. Only then can fingerprints generated on one day be reliably classified to a fingerprint reference library. Process improvements for reducing method level variability may include non-contact microarray printing, alternative microarray substrates, increased image acquisition times and/or amplicon fragmentation and labeling prior to hybridization. We are confident that continued process level improvements in microarray manufacturing and use will ultimately result in a very practical microarray fingerprinting protocol (much fewer than 24 replicate arrays) that can be easily and readily applied to the analysis of unknown isolates in a high throughput manner.
The ability to quantitatively compare fingerprints in this manner is a significant advance over gel-based dendrograms and comparative analyses and provides the basis and direction for the development of quantitative microbial forensics tools. Developing a protocol for comparing unknown samples with a reference library, however, will require careful consideration of replication requirements, both at the library construction stage and at the classification stage. Establishing useful (practical) replication requirements for library construction and library comparisons will also require continual monitoring of process controls: as the process improves, replication requirements might be relaxed. A common hybridization control, for example, may more faithfully preserve true fingerprint differences (in particular, differences due to elevation and scatter effects). Additional improvement might be obtained by averaging data at multiple levels (e.g. slides), increasing the number of probes (e.g. several of the isolates had very few discriminating probes) or increasing the number of samples in training library (thus increasing the precision of isolate library ‘profiles’). Future efforts will focus on improving microarray fabrication and process controls, increasing the number of probes on the array, expanding the fingerprint library and developing the statistical algorithms to quantitatively compare new fingerprint profiles against a reference library.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from the US Department of Homeland Security [DHS-Chemical and Biological National Security Program (CBNP)] and the National Institute of Health (NIH). The Argonne National Laboratory is operated for the US DOE by the University of Chicago under contract W-31-109-ENG-38. The Pacific Northwest National Laboratory is operated for the US DOE by Battelle Memorial Institute under contract DE-AC06-76RLO 1830.
REFERENCES
- 1.Baggesen D.L., Sandvang,D. and Aarestrup,F.M. (2000) Characterization of Salmonella enterica serovar typhimurium DT104 isolated from Denmark and comparison with isolates from Europe and the United States. J. Clin. Microbiol., 38, 1581–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waterhouse R.N. and Glover,L.A. (1993) Identification of procaryotic repetitive DNA suitable for use as fingerprinting probes. Appl. Environ. Microbiol., 59, 1391–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ticknor L.O., Kolstø,A.-B., Hill,K.K., Keim,P., Laker,M.T., Tonks,M. and Jackson,P.J. (2001) Fluorescent amplified fragment length polymorphism analysis of Norwegian Bacillus cereus and Bacillus thuringiensis soil isolates. Appl. Environ. Microbiol., 67, 4863–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Louws F.J., Fulbright,D.W., Stephens,C.T. and de Bruijn,F.J. (1994) Specific genomic fingerprints of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl. Environ. Microbiol., 60, 2286–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson P.J., Walthers,E.A., Kalif,A.S., Richmond,K.L., Adair,D.M., Hill,K.K., Kuske,C.R., Andersen,G.L., Wilson,K.H., Hugh-Jones,M.E. and Keim,P. (1997) Characterization of the variable-number tandem repeats in vrrA from different Bacillus anthracis isolates. Appl. Environ. Microbiol., 63, 1400–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johansson M.L., Molin,G., Pettersson,B., Uhlen,M. and Ahrne,S. (1995) Characterization and species recognition of Lactobacillus plantarum strains by restriction fragment length polymorphism (RFLP) of the 16S rRNA gene. J. Appl. Bacteriol., 79, 536–541. [Google Scholar]
- 7.Navarro E., Simonet,P., Normand,P. and Bardin,R. (1992) Characterization of natural populations of Nitrobacter spp. using PCR/RFLP analysis of the ribosomal intergenic spacer. Arch. Microbiol., 157, 107–115. [DOI] [PubMed] [Google Scholar]
- 8.McClelland M., Arensdorf,H., Cheng,R. and Welsh,J. (1994) Arbitrarily primed PCR fingerprints resolved on SSCP gels. Nucleic Acids Res., 22, 1770–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muyzer G., De Waal,E.C. and Uitterlinden,A.G. (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol., 59, 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Wurff A.W.G., Chan,Y.L., van Straalen,N.M. and Schouten,J. (2000) TE-AFLP: combining rapidity and robustness in DNA fingerprinting. Nucleic Acids Res., 28, e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hancock D.D., Besser,T.E., Rice,D.H., Ebel,E.D., Herriott,D.E. and Carpenter,L.V. (1998) Multiple sources of Escherichia coli O157 in feedlots and dairy farms in the northwestern USA. Prev. Vet. Med., 35, 11–19. [DOI] [PubMed] [Google Scholar]
- 12.Beattie K.L. (1997) Genomic fingerprinting using oligonucleotide arrays. In Caetano-Anolles,G. and Gresshoff,P.M. (eds) DNA Markers: Protocols, Applications and Overviews. John Wiley & Sons, New York, NY, pp. 213–224. [Google Scholar]
- 13.Salazar N.M. and Caetano-Anollés,G. (1996) Nucleic acid scanning-by-hybridization of enterohemorrhagic Escherichia coli isolates using oligodeoxynucleotide arrays. Nucleic Acids Res., 24, 5056–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J., Nietfeldt,J. and Benson,A.K. (1999) Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc. Natl Acad. Sci. USA, 96, 13288–13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Besser T.E., Goldoft,M., Pritchett,L.C., Khakhria,R., Hancock,D.D., Rice,D.H., Gay,J.M., Johnson,W. and Gay,C.C. (2000) Multiresistant Salmonella typhimurium DT104 infections of humans and domestic animals in the Pacific Northwest of the United States. Epidemiol. Infect., 124, 193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis M.A., Hancock,D.D., Besser,T.E., Rice,D.H., Gay,J.M., Gay,L., Gearhart,L. and DiGiacomo,R. (1999) Changes in antimicrobial resistance among Salmonella enterica serovar typhimurium isolates from humans and cattle in the Northwestern United States, 1982–1997. Emerg. Infect. Dis., 5, 802–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Versalovic J., Koeuth,T. and Lupski,J.R. (1991) Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res., 19, 6823–6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Call D.R., Chandler,D.P. and Brockman,F.J. (2001) Fabrication of DNA microarrays using unmodified oligomer probes. Biotechniques, 30, 368–379. [DOI] [PubMed] [Google Scholar]
- 19.Kingsley M.T., Straub,T.M., Call,D.R., Daly,D.S., Wunschel,S.C. and Chandler,D.P. (2002) Fingerprinting closely related Xanthomonas pathovars with random nonamer oligonucleotide microarrays. Appl. Environ. Microbiol., 68, 6361–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Efron B., Tibshirani,R., Storey,J.D. and Tusher,V. (2001) Empirical Bayes analysis of a microarray experiment. J. Am. Statist. Assoc., 96, 1151–1160. [Google Scholar]
- 21.Langsrud O. (2002) 50–50 multivariate analysis of variance for collinear responses. Statistician, 51, 305–317. [Google Scholar]
- 22.Amaratunga D. and Cabrera,J. (2001) Analysis of data from viral DNA microchips. J. Am. Statist. Assoc., 96, 1161–1170. [Google Scholar]
- 23.Benjamini Y. and Hochberg,Y. (1995) Controling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B, 57, 289–300. [Google Scholar]
- 24.Dudoit S., Fridlyand,J. and Speed,T. (2002) Comparison of discrimination methods for the classification of tumors using gene expression data. J. Am. Statist. Assoc., 97, 77–87. [Google Scholar]
- 25.Brumlik M.J., Szymajda,U., Zakowska,D., Liang,X., Redkar,R.J., Patra,G. and Del Vecchio,V.G. (2001) Use of long-range repetitive polymorphism-PCR to differentiate Bacillus anthracis strains. Appl. Environ. Microbiol., 67, 3021–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schuschhardt J., Beule,D., Malik,A., Wolski,E., Eickhoff,H., Lehrach,H. and Herzel,H. (2000) Normalization strategies for cDNA microarrays. Nucleic Acids Res., 28, e47i–e47v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herzel H., Beule,D., Kielbasa,S., Korbel,J., Sers,C., Malik,A., Eickhoff,H., Lehrach,H. and Schuchhardt,J. (2001) Extracting information from cDNA arrays. CHAOS, 11, 98–107. [DOI] [PubMed] [Google Scholar]