ABSTRACT
Vibrio cholerae is naturally competent when grown on chitin. It is known that expression of the major regulator of competence, TfoX, is controlled by chitin; however, the molecular mechanisms underlying this requirement for chitin have remained unclear. In the present study, we identify and characterize a membrane-bound transcriptional regulator that positively regulates the small RNA (sRNA) TfoR, which posttranscriptionally enhances tfoX translation. We show that this regulation of the tfoR promoter is direct by performing electrophoretic mobility shift assays and by heterologous expression of this system in Escherichia coli. This transcriptional regulator was recently identified independently and was named “TfoS” (S. Yamamoto et al., Mol. Microbiol., in press, doi:10.1111/mmi.12462). Using a constitutively active form of TfoS, we demonstrate that the activity of this regulator is sufficient to promote competence in V. cholerae in the absence of chitin. Also, TfoS contains a large periplasmic domain, which we hypothesized interacts with chitin to regulate TfoS activity. In the heterologous host E. coli, we demonstrate that chitin oligosaccharides are sufficient to activate TfoS activity at the tfoR promoter. Collectively, these data characterize TfoS as a novel chitin-sensing transcriptional regulator that represents the direct link between chitin and natural competence in V. cholerae.
IMPORTANCE
Naturally competent bacteria can take up exogenous DNA from the environment and integrate it into their genome by homologous recombination. This ability to take up exogenous DNA is shared by diverse bacterial species and serves as a mechanism to acquire new genes to enhance the fitness of the organism. Several members of the family Vibrionaceae become naturally competent when grown on chitin; however, a molecular understanding of how chitin activates competence is lacking. Here, we identify a novel membrane-bound transcriptional regulator that is required for natural transformation in the human pathogen Vibrio cholerae. We demonstrate that this regulator senses chitin oligosaccharides to activate the competence cascade, thus, uncovering the molecular link between chitin and natural competence in this Vibrio species.
INTRODUCTION
The ability of a bacterium to take up exogenous DNA is known as natural genetic competence. This ingested DNA can be a potential source of nutrients and can also be integrated into the genome by homologous recombination in the presence of sufficient homology (1). Induction of competence in diverse bacteria is commonly a highly regulated event. Recently, members of the family Vibrionaceae have been demonstrated to be naturally competent and transformable when grown on chitin (2–5).
Chitin is a naturally occurring biopolymer of β1-4-linked GlcNAc residues. It is the primary component of the cell walls in fungi and in the exoskeletons of crustaceans, which include copepods. Vibrio cholerae has previously been demonstrated to colonize the surfaces of copepods and copepod exoskeletons, and this association has been shown to enhance the survival of V. cholerae in the aquatic environment (6, 7). Thus, chitin is likely an important carbon and nitrogen source in this niche
The genes required for degradation and utilization of chitin as a carbon source in V. cholerae are regulated by the histidine kinase ChiS (VC0622) (8, 9). This protein is negatively regulated by the periplasmic chitin binding protein CBP (VC0620) in the absence of chitin, and this repression is relieved when CBP is bound to chitin oligomers (8). When active, ChiS potentially phosphorylates an unknown response regulator to control the expression of genes involved in chitin degradation, uptake into the periplasm, and subsequent uptake into and utilization within the cytoplasm (9).
In addition to its role as a carbon and nitrogen source in the aquatic environment, chitin signals V. cholerae to become naturally competent (4). Elegant studies have demonstrated the regulatory events downstream of chitin sensing that are critical for natural competence (4, 10–12). The major regulators involved in competence activation are TfoX (VC1153) and HapR (VC0583), which regulate the genes required for DNA uptake (11, 12). The expression of HapR is controlled by quorum sensing in V. cholerae, while the production of TfoX is controlled by chitin (11, 13, 14). The molecular mechanisms linking chitin sensing and the activation of TfoX, however, have remained unclear. In the present study, we identify and characterize a novel chitin-sensing regulator that provides the direct link between chitin and natural competence in V. cholerae.
RESULTS
Identification of a membrane-bound transcriptional regulator, TfoS, required for natural transformation in Vibrio cholerae.
To characterize novel factors involved in competence induction and natural transformation of V. cholerae on chitin flakes, we performed a genome-wide transposon sequencing (Tn-seq) screen (15). Factors known to be involved in competence were depleted when selecting for transformants and served as positive controls for this screen (Table 1). Through this screen, we identified the gene VC2080, recently named tfoS (16), as being required for competence induction and transformation (Table 1). TfoS is a predicted transmembrane protein, containing a large periplasmic domain, a transmembrane alpha-helix, and a cytoplasmic AraC-like helix-turn-helix DNA-binding domain (Fig. 1A). To confirm this hit from our screen, we generated a tfoS deletion mutant and found that it is nontransformable when grown under competence-inducing conditions on chitin flakes (Fig. 1B). Additionally, we showed that the periplasmic domain of TfoS is critical for its function because a strain in which the periplasmic domain is deleted is nontransformable (Fig. 1B). Next, we wanted to determine where in the natural competence regulatory cascade TfoS acts. Competence activation is controlled by two major regulators: TfoX and HapR. TfoX regulates genes important for binding and uptake of exogenous DNA into the periplasm (i.e., pilA, the major pilin of the competence pilus) (11, 17). Also, TfoX works in conjunction with HapR to indirectly regulate genes responsible for DNA stabilization in the periplasm and uptake through the inner membrane (i.e., comEA and comEC) via coactivation of the transcriptional regulator QstR (VC0396) (11, 12). To determine whether TfoS acts upstream of TfoX or QstR, we performed epistasis analysis with these two transcriptional regulators. Overexpression of TfoX rescues a TfoS mutant strain, while overexpression of QstR does not, indicating that TfoS likely acts upstream of TfoX (Fig. 1C). To further define where in the regulatory cascade TfoS acts, we performed quantitative reverse transcription-PCR (qRT-PCR) assays to define the transcriptional responses in a TfoS deletion strain under competence-inducing and noninducing conditions. To measure transcriptional responses under inducing conditions, bacteria were grown in a minimal medium supplemented with the chitin disaccharide (GlcNAc)2, as such chitin oligomers were previously shown to activate competence uniformly in a population of cells (11). The noninducing condition used was growth in minimal medium plus glucose. Under the inducing condition, we find that genes downstream of TfoX, namely pilA (VC2423) and comEA (VC1917), fail to become induced in the TfoS mutant (Fig. 1D). Surprisingly, tfoX is still partially transcriptionally upregulated in the TfoS mutant, although not to wild-type (WT) levels (Fig. 1D). Thus, TfoS could regulate TfoX expression by altering tfoX transcription and/or could alter TfoX expression posttranscriptionally.
TABLE 1 .
Contribution of select genes from our transposon screen to natural transformation
| Gene name | Locus | Avg fold depletiona |
|---|---|---|
| tfoX | VC1153 | 287.8 |
| dprA | VC0048 | 156.7 |
| qstR | VC0396 | 85.7 |
| recA | VC0543 | 85.7 |
| pilA | VC2423 | 74.8 |
| tfoS | VC2080 | 45.8 |
Calculated by averaging the relative representation of each gene in the absence of kanamycin divided by that in the presence of kanamycin (to select transformants) from two independent experiments.
FIG 1 .
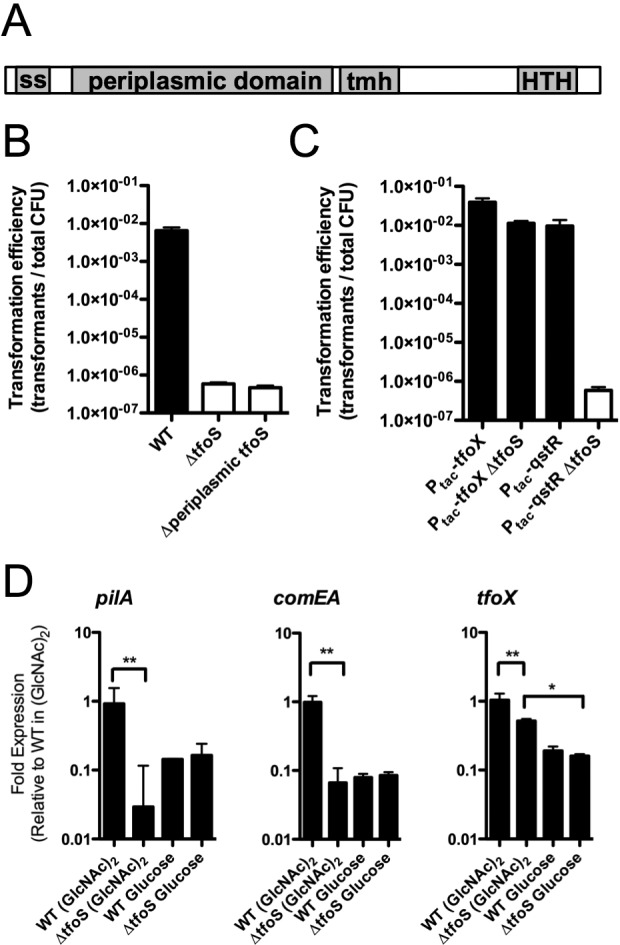
An inner membrane transcriptional regulator in V. cholerae, TfoS, is critical for natural competence and acts upstream of TfoX. (A) Architecture of the tfoS gene product. ss, putative signal sequence; tmh, transmembrane helix; HTH, helix-turn-helix DNA-binding domain. (B) Natural transformation of WT and tfoS deletion strains on chitin flakes with 500 ng of a PCR product that confers resistance to kanamycin. (C) Natural transformation on chitin flakes of WT and TfoS mutant strains, each containing IPTG (isopropyl-β-d-thiogalactopyranoside)-regulated Ptac upstream of the endogenous copy of either tfoX or qstR on the genome in the presence of 1 mM IPTG with a PCR product that confers resistance to spectinomycin. White bars in panels B and C indicate that no transformants were obtained, and those data are presented at the limit of detection. Data in panels B and C are shown as means ± standard deviations (SD). (D) Transcript abundance of the indicated gene is shown for WT and TfoS deletion strains grown under competence-inducing [(GlcNAc)2] and noninducing (glucose) conditions. Data are normalized to the transcript abundance of RpoB and then again to the expression level of the WT under inducing conditions. Data are shown as median ± range. Statistical comparisons were made by nonparametric Mann-Whitney test. All data are from at least three independent biological replicates. *, P < 0.05; **, P < 0.01.
TfoS indirectly controls TfoX translation by regulating the sRNA TfoR.
It has previously been shown that translation initiation of tfoX mRNA is regulated posttranscriptionally via a small RNA (sRNA), TfoR (18). TfoR is Hfq dependent and base pairs with a stem-loop structure in the tfoX transcript to help expose its Shine-Dalgarno sequence to positively regulate tfoX translation. To determine if TfoS regulates this sRNA, we generated PtfoR-lacZ transcriptional fusions in both WT and TfoS mutant backgrounds and tested their activity when grown on chitin flakes (competence-inducing condition). Under this condition, we find that there is significantly more LacZ activity in the WT than in the TfoS mutant, indicating that TfoS is a positive regulator of the TfoR sRNA (Fig. 2A). Since it has previously been shown that TfoR acts posttranscriptionally to enhance tfox translation, we next wanted to assess whether TfoS alters tfoX translation. To that end, we generated a translational fusion of tfoX to lacZ in both WT and TfoS mutant backgrounds and tested the activity of these strains under competence-inducing conditions. Indeed, when these strains are grown on chitin, we find that the activity of this reporter is significantly higher in the WT than in the TfoS mutant, indicating that TfoS positively regulates TfoX translation (Fig. 2A).
FIG 2 .
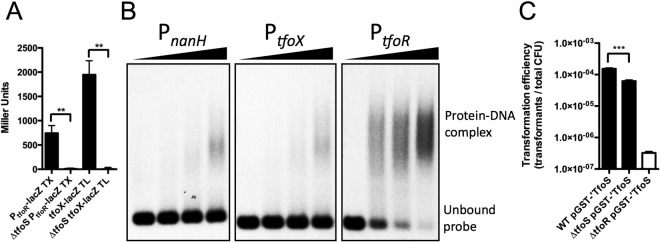
TfoS directly regulates the sRNA TfoR to indirectly regulate tfoX translation. (A) Miller assay for V. cholerae WT and TfoS mutant strains containing either a PtfoR-transcriptional (TX) fusion or a tfoX-translational (TL) fusion to the endogenous copy of lacZ in the chromosome. (B) EMSAs using 0.5 nM the indicated probe and purified GST-tagged cytoplasmic TfoS protein. The amount of protein added increases from left to right, as indicated by the black triangles above the panels, and corresponds to final concentrations of 0, 80, 160, and 320 nM protein. Data in panel B are representative of at least three independent EMSAs. (C) Transformation of WT, TfoS mutant, and TfoR mutant strains, each containing pGST-′TfoS and grown in M9 plus GlcNAc in the presence of 1 µM IPTG with 500 ng of a PCR product conferring kanamycin resistance. White bars in panel C indicate no transformants were obtained, and those data are presented at the limit of detection. Statistical comparisons were made by Student’s t test. Data from panels A and C are shown as means ± SD and are from at least three independent biological replicates. **, P < 0.01; ***, P < 0.001.
Since we found that TfoS upregulates TfoR on chitin, we next sought to define whether TfoS was a direct regulator of the TfoR sRNA. It is known that helix-turn-helix domains require dimerization to efficiently bind to DNA, so we generated a construct to express a glutathione S-transferase (GST)-tagged cytoplasmic portion of the TfoS protein (pGST-′TfoS), which includes the putative DNA-binding domain. GST naturally forms a dimer and thus is predicted to bring the DNA-binding domains of TfoS into close proximity, potentially mimicking the conformation of this protein in its active DNA-binding state (19). To determine if pGST-′TfoS could directly bind to the tfoR promoter, we expressed and purified this protein from Escherichia coli to perform electrophoretic mobility shift assays (EMSAs). We tested the ability of this purified protein to bind to the nanH, tfoX, and tfoR promoters. The nanH promoter was used as a negative control, since this gene is not differentially regulated by chitin (9). GST-′TfoS did not bind appreciably to the negative-control nanH promoter or the tfoX promoter, but it did bind to and shift the mobility of the tfoR promoter fragment (Fig. 2B). This indicates that TfoS likely directly regulates transcription of the TfoR sRNA by binding to its promoter and recruiting RNA polymerase. Artificial overexpression of TfoR has previously been shown to promote competence in V. cholerae grown on GlcNAc, a condition that does not induce competence (18). To test if TfoS-mediated upregulation of TfoR is sufficient for competence under noninducing conditions, we performed transformation assays with the WT, TfoS mutant, and TfoR mutant strains containing the constitutively active pGST-′TfoS construct when strains were grown in GlcNAc. Under these conditions, WT and TfoS mutant strains were transformable, while the TfoR deletion mutant was not (Fig. 2C). This result suggests that TfoS-mediated upregulation of TfoR is sufficient to render strains competent independent of the presence of chitin and also suggests that TfoS acts upstream of TfoR since the constitutively active pGST-′TfoS construct did not rescue a TfoR mutant for competence and transformability. We also found that the WT strain expressing GST-′TfoS is significantly more transformable than the TfoS mutant strain. This may be due to the fact that the WT strain still expresses the endogenous full-length copy of TfoS, which may interact with the GST-tagged ′TfoS to further increase TfoS activity in this strain.
Chitin is sufficient to activate TfoS in the heterologous host E. coli.
Since we have identified a direct target of TfoS in V. cholerae, we decided to test whether TfoS activity could be studied in a heterologous host. To that end, we generated a transcriptional fusion of the tfoR promoter to lacZ in E. coli. To determine if this fusion was functional, we introduced the constitutively active pGST-′TfoS construct into this strain and showed that expression from PtfoR is ~30-fold higher than that in an empty vector control (Fig. 3A). This result in E. coli further confirms that TfoS directly regulates the tfoR promoter.
FIG 3 .
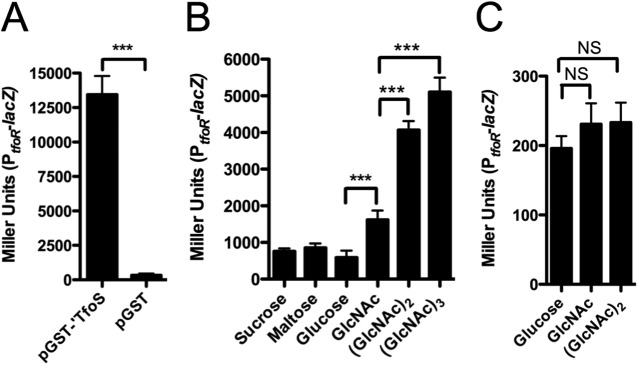
Chitin is sufficient to induce TfoS activity at the tfoR promoter in the heterologous host Escherichia coli. (A) Miller assays for E. coli PtfoR-lacZ fusion strains containing either pGST-′TfoS or pGST (empty vector) grown in M9 plus glucose. (B) Miller assays for the E. coli PtfoR-lacZ fusion strain containing pTfoS grown in M9 plus the indicated carbon source (at 3.33 mM for trisaccharides, 5 mM for disaccharides, and 10 mM for monosaccharides) in the presence of 10 µM IPTG. (C) Miller assays for the E. coli PtfoR-lacZ fusion strain grown under the same conditions outlined in panel B. All data are from at least three biological replicates and are shown as means ± SD. Statistical comparisons were made by Student’s t test. ***, P < 0.001; NS, not significant.
As mentioned previously, TfoS is an inner membrane protein with a large periplasmic domain. Since we found that TfoS activity in V. cholerae is promoted by chitin oligomers, we next sought to determine if chitin oligomers were sufficient for activation of TfoS in the heterologus host E. coli. It has previously been shown that E. coli can grow on short chitin oligomers, and it is expected that the chitin disaccharide can enter the periplasm of V. cholerae and E. coli through nonspecific porins (20, 21). Thus, to test whether TfoS is activated by chitin, the full-length tfoS gene was cloned (pTfoS) and expressed in the E. coli PtfoR-lacZ fusion strain and subsequently grown on minimal medium containing distinct carbon sources. Using this assay, we found that the chitin disaccharide [(GlcNAc)2] and trisaccharide [(GlcNAc)3] are potent activators of TfoS activity, while monomeric GlcNAc is a weak activator compared to the other carbohydrates tested (glucose, maltose, and sucrose) (Fig. 3B). Importantly, this difference in induction was not an inherent property of the PtfoR promoter since the PtfoR-lacZ fusion strain did not display these differences in activity when grown on these carbon sources in the absence of the pTfoS construct (Fig. 3C). Thus, these results suggest that TfoS is directly activated by chitin oligomers to positively regulate the tfoR promoter.
DISCUSSION
The presented data are consistent with a model in which, in the absence of chitin, TfoS is unable to dimerize and bind DNA, while in the presence of chitin, the periplasmic domain of TfoS interacts with chitin oligosaccharides and dimerizes (Fig. 4). This brings the cytoplasmic DNA-binding domains of these monomers into close proximity and allows this regulator to bind DNA at the tfoR promoter to positively regulate transcription of this sRNA by recruiting RNA polymerase (RNAP). Alternatively, it is possible that TfoS in its native form is a dimer in the inner membrane and that binding of chitin oligomers to the periplasmic domain induces a conformational change in the cytoplasmic portion of the protein, which switches it from an inactive to an active DNA-binding state and promotes transcription of tfoR. The Hfq-dependent sRNA TfoR then interacts with the 5′ untranslated region (UTR) of tfoX mRNA to expose a Shine-Dalgarno sequence and allow for efficient translation of this transcript. TfoX then goes on to activate the genes required for binding to and uptake of exogenous DNA.
FIG 4 .
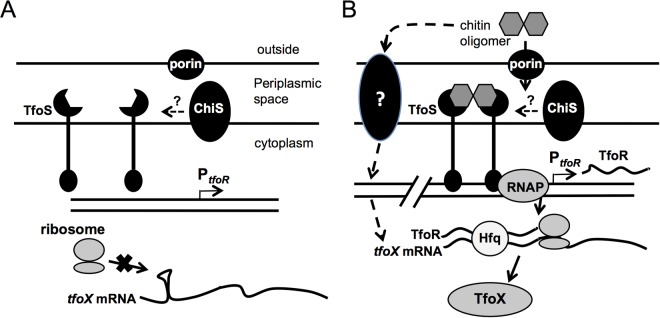
Proposed model for the role of TfoS in activating competence in V. cholerae. (A) In the absence of chitin oligomers, TfoS cannot dimerize and activate TfoR transcription. Alternatively, in the absence of chitin oligimers, dimerized TfoS is in an inactive state and the DNA-binding domains of the protein are unable to upregulate TfoR. In the absence of TfoR, TfoX translation is inhibited by a stem-loop structure in the tfoX mRNA 5′ UTR that occludes the Shine-Dalgarno sequence required for translation initiation (18). (B) In the presence of chitin, tfoX mRNA is upregulated by an unknown mechanism. Also, short chitin oligomers enter the periplasm by nonspecific porins or via the V. cholerae chitporin (VC0972) (9). These chitin oligomers interact with and dimerize TfoS protein (and/or induce a conformational change in TfoS dimers) in the inner membrane, which allows the cytoplasmic DNA-binding domains to bind to the tfoR promoter, recruit RNAP, and positively regulate TfoR transcription. ChiS may also act to enhance TfoS activity by an unknown mechanism (16). TfoR then interacts with Hfq and the 5′ UTR of tfoX mRNA to expose the Shine-Dalgarno sequence of the tfoX mRNA to positively regulate tfoX translation. TfoX then goes on to activate the genes necessary for competence.
Natural competence is observed in several species of the family Vibrionaceae, including but not limited to, Vibrio parahaemolyticus, Vibrio vulnificus, and Vibrio fischeri (2, 3, 5). In these other species, activation of competence is also linked to growth on chitin (2, 3, 5). It has previously been shown that the sRNA TfoR is conserved in other naturally competent Vibrio species (18). Using computational searches, we have identified homologs of TfoS in other Vibrio species, and it is tempting to speculate that these homologs also function to sense chitin and activate competence in these bacteria. These homologs include VP0854 in V. parahaemolyticus RIMD2210633, VV1039 in V. vulnificus YJ016 and VF_0832 in V. fischeri ES114 (22–24).
It has previously been shown that V. cholerae encodes ChiS (VC0622), which is a membrane-bound histidine kinase that senses and responds to chitin (8, 9). ChiS presumably phosphorylates a cognate response regulator to activate the genes required for chitin degradation and utilization; however, the cognate response regulator for ChiS has not been identified. In this study, we provide evidence that suggests that TfoS is an independent chitin sensor in V. cholerae. It is clear that ChiS and TfoS are distinct sensors of chitin, because phenotypically a ChiS mutant does not grow on chitin oligomers, while a TfoS mutant can (8). This further demonstrates that TfoS is not the response regulator for ChiS activity since these mutants are phenotypically distinct. Additionally, we show that in the heterologous host E. coli, chitin oligomers are sufficient to promote TfoS activity at the tfoR promoter. Thus, these data are consistent with a model in which TfoS is a sensor of chitin degradation products that specifically activates competence in V. cholerae. Regulatory separation of chitin utilization and competence genes may provide kinetic control of these pathways (i.e., activation of chitin utilization genes prior to activation of competence) or may indicate that there is a benefit to activation of only one activity (chitin utilization or competence) under specific conditions.
While our manuscript was under review, another research group independently identified this membrane-bound transcriptional regulator as being critical for regulating the small RNA TfoR (16). Our data are in agreement with relation to the activity of TfoS at the tfoR promoter; however, the mechanism by which TfoS is activated differs between these two studies. In the study by Yamamoto et al. (16), TfoS activity at the tfoR promoter in E. coli required coexpression of TfoS and ChiS and was independent of the presence of chitin oligosaccharides. In this article, however, we show in E. coli that TfoS activity at the tfoR promoter can be promoted by expression of TfoS and the presence of chitin oligosaccharides. This result does not, however, eliminate a potential role for ChiS in this system. It is possible that ChiS stabilizes the interaction of TfoS with chitin oligosaccharides or, alternatively, promotes dimerization of TfoS to allow for optimal activity at the tfoR promoter (Fig. 4). Regardless, these two studies independently identified TfoS via distinct approaches and characterized its role in regulating the sRNA TfoR. Our work complements that of Yamamoto et al. (16) by demonstrating that this regulator is directly responsive to chitin and further demonstrates how chitin closely regulates the expression of competence in V. cholerae. It will be of interest to define the chitin oligomer-binding site of TfoS in future work and determine how this mechanistically results in activation of this protein.
It is clear from this and other studies that TfoX is regulated at multiple levels (18, 25). We found that TfoS upregulates TfoR to increase translation of TfoX mRNA. We also observed, however, that TfoX transcript levels in the TfoS mutant were significantly higher when grown under inducing conditions than under noninducing conditions. This indicates that TfoX is under transcriptional control by an additional as yet unidentified regulator. The partial transcriptional upregulation of TfoX in the TfoS mutant is not, however, sufficient to promote competence. Conversely, we show that TfoS activation is sufficient to promote competence under noninducing conditions since the constitutively active GST-tagged DNA binding domain of TfoS was able to render V. cholerae transformable when grown in the absence of chitin. This suggests that translational activation of TfoX is sufficient to induce competence even under noninducing conditions.
In the present study, we demonstrate that TfoS positively regulates TfoR to induce competence in V. cholerae. Furthermore, we demonstrate that this regulation of TfoR is required for natural competence since the constitutively active GST-tagged DNA-binding domain of TfoS could not rescue a TfoR mutant strain. It remains possible, however, that TfoS also regulates additional genes that play a role in competence or other aspects of V. cholerae biology. Defining the TfoS consensus binding site and identifying additional targets of TfoS may uncover additional roles for this chitin-sensing transcriptional regulator and will be the focus of future work.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
All V. cholerae strains were derivatives of E7946, and all E. coli strains were derivatives of MG1655 and are described in detail in Table S1 in the supplemental material. The primers and recombinant DNA techniques used to generate these strains and plasmids are detailed in Table S2 in the supplemental material. Bacteria were commonly grown at 37°C in lysogeny broth (LB) and M9 minimal medium (M9 salts [Sigma-Aldrich], 2 mM MgSO4, and 100 µM CaCl2) supplemented with various carbon sources. Medium was supplemented with tetracycline (2 µg/ml), ampicillin (100 µg/ml), kanamycin (50 µg/ml), and/or spectinomycin (100 µg/ml) when appropriate.
Generation of Tn-seq libraries and performing the genetic screen.
Libraries were generated by in vivo transposon mutagenesis using a Tn10 vector (pDL1098), which conferred spectinomycin resistance. The map of this vector, the methods for generating and sequencing these libraries are described in a separate manuscript (E. K. McDonough, D. W. Lazinski, and A. Camilli, submitted for publication). The libraries for this study were prepared in a complex medium we have termed “spectacular broth” (SB): 1× M9 salts (Sigma-Aldrich), 1.2% peptone (Bacto), 2.4% yeast extract (Fisher Scientific), 5 mM MgSO4, 200 µM CaCl2, and 0.4% glucose. Genomic DNA from this library was isolated and served as the “input.” Then, this library was either diluted back into SB to allow for 13 generations of growth or placed onto chitin flakes for transformation assays. Outgrowth of the library on SB for 13 generations served as an “outgrowth control” to identify genes with a growth defect in SB. As per our normal transformation assays, we placed ~108 CFU of this library washed in defined artificial seawater (DASW) onto 80 mg of chitin flakes in a 1-ml reaction volume and performed transformation assays as previously described. To prevent a “bottleneck” due to the low number of bacteria in a single transformation reaction, we performed 36 independent 1-ml transformations on chitin. The PCR product used in transformation assays replaced the coding sequence of lacZ with an FLP recombination target (FRT)-Kan-FRT cassette to confer resistance to kanamycin. After incubation of cells with this PCR product for 24 h, all 36 reaction mixtures were vortexed vigorously, and the bacterium-containing supernatants were removed and pooled. Cells in this supernatant were then either grown for 13 generations in plain SB (“chitin”) or grown for 21 generations in SB containing kanamycin (“chitin transformants”) to select for transformants. DNA was subsequently isolated from both of these cultures to prepare sequencing libraries. All sequencing libraries were then run on an Illumina HiSeq2000. We analyzed data essentially as previously described (26). Comparisons were primarily made treating “chitin transformants” as the output and “chitin” as the input to identify genes that were involved in natural transformation. Comparisons were also made where “chitin” was the output and “input” was the input, as well as where “outgrown control” was the output and “input” was the input to characterize and exclude genes that were defective for survival on chitin and growth in SB, respectively.
Generation of mutant strains.
Deletion mutants were generated by splicing overlap extension (SOE) PCR and natural competence exactly as previously described (27). Where indicated, the resistance cassette was removed by FLP recombination (resulting in an in-frame FRT scar) exactly as previously described (28). Overexpression constructs for Ptac-tfoX and Ptac-qstR were generated by SOE PCR. The middle fragments for these constructs were derived from a kanamycin-resistant derivative (pDL1093) of the Tn10 plasmid used to generate Tn-seq libraries described earlier (pDL1098). Specifically, using the primers ABD624 and ABD625, we amplified a fragment of the transposon containing the following from 5′ to 3′: a kanamycin resistance marker in the reverse orientation and lacI in the reverse orientation, the Ptac promoter in the forward orientation, and the rrnB antiterminator sequence in the forward orientation (see Fig. S1 in the supplemental material). The E. coli PtfoR-lacZ fusion strain was generated by recombineering exactly as previously described (29). The primers used for all mutant constructs are given in Table S2 in the supplemental material. All mutant strains were confirmed by PCR and/or sequencing.
Generation of recombinant plasmids.
The pGST-′TfoS construct was generated by amplifying the cytoplasmic portion of the tfoS gene using ABD721 and ABD722 and cloned into the NdeI and BamHI sites of the pGex-tev expression vector(30) The pTfoS construct was generated in the host vector pDL993 (see Fig. S2 in the supplemental material). This vector has a pACYC origin of replication, lacI, a tetracycline resistance marker, and the Plac promoter followed by a multicloning site. To generate pTfoS, we amplified the full-length tfoS gene using ABD787 and ABD788 and then cloned it into the EcoRI and XhoI sites of pDL993.
Transformation assays.
V. cholerae strains were tested for natural transformation on chitin flakes from shrimp shells essentially as previously described (28). Briefly, 80 mg of autoclaved chitin (from shrimp shells; Sigma-Aldrich) was inoculated with 1 ml of V. cholerae at an optical density at 600 nm (OD600) of 0.1 (~108 CFU) in defined artificial seawater (DASW: 7 g/liter Instant Ocean [Aquarium Systems]) and incubated statically at 30° C for 16 to 24 h. Then, the supernatant was exchanged for fresh DASW, and 500 ng of a PCR product was added. This PCR product inserts a kanamycin or spectinomycin resistance cassette into the coding sequence of lacZ (VC2338) (see Table S2 in the supplemental material). Reaction mixtures were incubated at 30°C for an additional 16 to 24 h in the presence of DNA and then plated onto selective and nonselective media to assess transformation efficiency.
Where indicated, V. cholerae cells were tested for transformability when grown in M9 plus GlcNAc at 37°C. Cells were grown in this medium to an OD600 of 0.2, and then 500 ng of a PCR product was added (described above). Cells were incubated for 16 h statically and subsequently plated on selective and nonselective media to assess transformation efficiency. No transformants were ever obtained for WT V. cholerae under these conditions.
qRT-PCR assays.
V. cholerae strains were grown in either M9 plus (GlcNAc)2 or M9 plus glucose to an OD600 of 0.2 to 0.5. Cells were then harvested, and RNA was isolated using a Qiagen RNEasy minikit according to the manufacturer’s instructions. Samples were then prepared exactly as previously described (27), and quantitative PCR was performed on a Stratagene Mv3005P via the dye incorporation method (SYBR) and analyzed with MxPro qPCR software (Stratagene).
Miller assays.
Cells were harvested either from overnight cultures or from cells in mid-logarithmic growth, and β-galactosidase activity assays were performed exactly as previously described (31).
Purification of GST-tagged cytoplasmic TfoS and EMSAs.
The GST-tagged cytoplasmic TfoS (pGST-′TfoS) was expressed and purified from E. coli BL21(DE3) essentially as previously described (30). For EMSAs, probes were generated by PCR (for primers, see Table S2 in the supplemental material) using Cy5-labeled dCTP as previously described (27). Binding reactions were performed in 20 µl in the presence of 0.5 nM probe, 10 mM Tris-HCl (pH 8.0), 100 mM KCl, 10% glycerol, 1 mM dithiothreitol (DTT), 100 µg/ml bovine serum albumin (BSA), 50 µg/ml sheared salmon sperm DNA, and the indicated concentration of protein. Reaction mixtures were incubated at room temperature for 20 min and then electrophoretically separated on a 2% agarose gel in Tris-acetate-EDTA buffer.
SUPPLEMENTAL MATERIAL
mTn10 transposon fragment used for overexpression. Download
Vector map of pDL993. Download
Strains used in this study.
Primers used in this study.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant AI055058 to A.C. A.C. is a Howard Hughes Medical Institute Investigator.
Footnotes
Citation Dalia AB, Lazinski DW, Camilli A. 2014. Identification of a membrane-bound transcriptional regulator that links chitin and natural competence in Vibrio cholerae. mBio 5(1):e01028-13. doi:10.1128/mBio.01028-13.
REFERENCES
- 1. Redfield RJ. 1993. Genes for breakfast: the have-your-cake-and-eat-it-too of bacterial transformation. J. Hered. 84:400–404 [DOI] [PubMed] [Google Scholar]
- 2. Chen Y, Dai J, Morris JG, Jr, Johnson JA. 2010. Genetic analysis of the capsule polysaccharide (K antigen) and exopolysaccharide genes in pandemic Vibrio parahaemolyticus O3:K6. BMC Microbiol. 10:274. 10.1186/1471-2180-10-274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gulig PA, Tucker MS, Thiaville PC, Joseph JL, Brown RN. 2009. User friendly cloning coupled with chitin-based natural transformation enables rapid mutagenesis of Vibrio vulnificus. Appl. Environ. Microbiol. 75:4936–4949. 10.1128/AEM.02564-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824–1827. 10.1126/science.1120096 [DOI] [PubMed] [Google Scholar]
- 5. Pollack-Berti A, Wollenberg MS, Ruby EG. 2010. Natural transformation of Vibrio fischeri requires tfoX and tfoY. Environ. Microbiol. 12:2302–2311. 10.1111/j.1462-2920.2010.02250.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huq A, Small EB, West PA, Huq MI, Rahman R, Colwell RR. 1983. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl. Environ. Microbiol. 45:275–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nahar S, Sultana M, Naser MN, Nair GB, Watanabe H, Ohnishi M, Yamamoto S, Endtz H, Cravioto A, Sack RB, Hasan NA, Sadique A, Huq A, Colwell RR, Alam M. 2011. Role of shrimp chitin in the ecology of toxigenic Vibrio cholerae and cholera transmission. Front. Microbiol. 2:260. 10.3389/fmicb.2011.00260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li X, Roseman S. 2004. The chitinolytic cascade in vibrios is regulated by chitin oligosaccharides and a two-component chitin catabolic sensor/kinase. Proc. Natl. Acad. Sci. U. S. A. 101:627–631. 10.1073/pnas.0307645100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meibom KL, Li XB, Nielsen AT, Wu CY, Roseman S, Schoolnik GK. 2004. The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. U. S. A. 101:2524–2529. 10.1073/pnas.0308707101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Antonova ES, Bernardy EE, Hammer BK. 2012. Natural competence in Vibrio cholerae is controlled by a nucleoside scavenging response that requires CytR-dependent anti-activation. Mol. Microbiol. 86:1215–1231. 10.1111/mmi.12054 [DOI] [PubMed] [Google Scholar]
- 11. Lo Scrudato M, Blokesch M. 2012. The regulatory network of natural competence and transformation of Vibrio cholerae. PLoS Genet. 8:e1002778. 10.1371/journal.pgen.1002778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lo Scrudato M, Blokesch M. 2013. A transcriptional regulator linking quorum sensing and chitin induction to render Vibrio cholerae naturally transformable. Nucleic Acids Res. 41:3644–3658. 10.1093/nar/gkt041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69–82. 10.1016/j.cell.2004.06.009 [DOI] [PubMed] [Google Scholar]
- 14. Zhu J, Miller MB, Vance RE, Dziejman M, Bassler BL, Mekalanos JJ. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 99:3129–3134. 10.1073/pnas.052694299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods 6:767–772. 10.1038/nmeth.1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yamamoto S, Mitobe J, Ishikawa T, Wai SN, Ohnishi M, Watanabe H, Izumiya H. 2013. Regulation of natural competence by the orphan two-component system sensor kinase ChiS involves a non-canonical transmembrane regulator in Vibrio cholerae. Mol. Microbiol., in press. 10.1111/mmi.12462 [DOI] [PubMed] [Google Scholar]
- 17. Seitz P, Blokesch M. 2013. DNA-uptake machinery of naturally competent Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 110:17987–17992. 10.1073/pnas.1315647110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yamamoto S, Izumiya H, Mitobe J, Morita M, Arakawa E, Ohnishi M, Watanabe H. 2011. Identification of a chitin-induced small RNA that regulates translation of the tfoX gene, encoding a positive regulator of natural competence in Vibrio cholerae. J. Bacteriol. 193:1953–1965. 10.1128/JB.01340-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maru Y, Afar DE, Witte ON, Shibuya M. 1996. The dimerization property of glutathione S-transferase partially reactivates bcr-abl lacking the oligomerization domain. J. Biol. Chem. 271:15353–15357. 10.1074/jbc.271.26.15353 [DOI] [PubMed] [Google Scholar]
- 20. Keyhani NO, Roseman S. 1997. Wild-type Escherichia coli grows on the chitin disaccharide, N,N′-diacetylchitobiose, by expressing the cel operon. Proc. Natl. Acad. Sci. U. S. A. 94:14367–14371. 10.1073/pnas.94.26.14367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hunt DE, Gevers D, Vahora NM, Polz MF. 2008. Conservation of the chitin utilization pathway in the Vibrionaceae. Appl. Environ. Microbiol. 74:44–51. 10.1128/AEM.01412-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen CY, Wu KM, Chang YC, Chang CH, Tsai HC, Liao TL, Liu YM, Chen HJ, Shen AB, Li JC, Su TL, Shao CP, Lee CT, Hor LI, Tsai SF. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577–2587. 10.1101/gr.1295503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, Tagomori K, Iijima Y, Najima M, Nakano M, Yamashita A, Kubota Y, Kimura S, Yasunaga T, Honda T, Shinagawa H, Hattori M, Iida T. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet 361:743–749. 10.1016/S0140-6736(03)12659-1 [DOI] [PubMed] [Google Scholar]
- 24. Ruby EG, Urbanowski M, Campbell J, Dunn A, Faini M, Gunsalus R, Lostroh P, Lupp C, McCann J, Millikan D, Schaefer A, Stabb E, Stevens A, Visick K, Whistler C, Greenberg EP. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl. Acad. Sci. U. S. A. 102:3004–3009. 10.1073/pnas.0409900102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yamamoto S, Morita M, Izumiya H, Watanabe H. 2010. Chitin disaccharide (GlcNAc)2 induces natural competence in Vibrio cholerae through transcriptional and translational activation of a positive regulatory gene tfoXVC. Gene 457:42–49. 10.1016/j.gene.2010.03.003 [DOI] [PubMed] [Google Scholar]
- 26. Klein BA, Tenorio EL, Lazinski DW, Camilli A, Duncan MJ, Hu LT. 2012. Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis. BMC Genomics 13:578. 10.1186/1471-2164-13-578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dalia AB, Lazinski DW, Camilli A. 2013. Characterization of undermethylated sites in Vibrio cholerae. J. Bacteriol. 195:2389–2399. 10.1128/JB.02112-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. De Souza Silva O, Blokesch M. 2010. Genetic manipulation of Vibrio cholerae by combining natural transformation with FLP recombination. Plasmid 64:186–195. 10.1016/j.plasmid.2010.08.001 [DOI] [PubMed] [Google Scholar]
- 29. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pratt JT, Ismail AM, Camilli A. 2010. PhoB regulates both environmental and virulence gene expression in Vibrio cholerae. Mol. Microbiol. 77:1595–1605. 10.1111/j.1365-2958.2010.07310.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nickels BE. 2009. Genetic assays to define and characterize protein-protein interactions involved in gene regulation. Methods 47:53–62. 10.1016/j.ymeth.2008.10.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
mTn10 transposon fragment used for overexpression. Download
Vector map of pDL993. Download
Strains used in this study.
Primers used in this study.


