Abstract
Chemical, pharmacokinetic, and pharmacodynamics properties are available in the package inserts of every Food and Drug Administration (FDA) approved prescription drug, including all available chemotherapy drugs. These inserts follow a specific format imposed by the FDA. Whether chemotherapy drugs are administered via the parenteral route or alimentary tract, a significant factor affecting their bioavailability, elimination and consequently the drug’s effectiveness and potency, is its state of aqueous solubility. Water solubility has always lent itself poorly to the different predictive and experimental measures employed in the determination of a useful quantitative assessment. In this project, we first built a chemical structure based searchable database for 85 FDA approved chemotherapy drugs and then used Bio-Rad’s KnowItAll® Informatics suite to focus on the drugs pH-dependent water solubility prediction. We compared the predicted values for water solubility to the available values reported in the drug inserts, testing the practical utility and the predictive ability of our model in reporting such a clinically relevant, underreported pharmacokinetic parameter. A relational cancer drug database (MySQL) was created to further facilitate analysis and/or prediction of a chemotherapy compound’s missing pharmacokinetic properties.
Keywords: Drug database, FDA, pH profiles, drug solubility, ADMET, ADME/Tox, data management, clinical data, undergraduate research
INTRODUCTION
Since 2003, Wesley College (Wesley; www.wesley.edu) undergraduates in the Science, Technology, Engineering, and Mathematics (STEM) related fields of biological chemistry, biology, environmental studies, mathematics, and medical technology, are afforded an opportunity to participate in biomedically related research projects (D’Souza, Dwyer et al., 2011; D’Souza and Wang, 2012) through an Institutional Development Award (IDeA) Networks of Biomedical Research Excellence (INBRE) grant funded by the National Center for Research Resources (NCRR) at the National Institutes of Health (NIH). In 2012, this federally funded national INBRE program contrived to augment and strengthen the IDeA-eligible state’s biomedical research capacity was assigned to the National Institute of General Medical Sciences at NIH (NIGMS).
In Delaware (DE), the University of Delaware (UD) leads and administers the DE-INBRE (www.inbre.udel.edu) program. The other partners in this project include the Christiana Care Health System (Christiana), Delaware State University (DSU), Delaware Technical & Community College (DTCC), the Nemours/A.I. DuPont Hospital for Children (Nemours), and Wesley College.
EVOLUTION OF TEAM DEVELOPMENT OF PHARMACEUTICAL DATABASES AT WESLEY
In the mid-1990s, big pharma got involved with the management, searching, and integration of large molecular data files to discover, design and optimize a diverse library of biology active compounds (Banville, 2006). Such innovative and efficient Computer-Aided Drug Design (CADD) strategies exploit state-of-the-art computational technologies (in silico) to accelerate the drug development and structure optimization process (Mandal et al., 2009). Quantitative structure-activity relationships (QSAR) utilizing in silico tools are now typically employed to understand the relationships between biological activity and chemical drug structure (Ekins et al. 2007).
To introduce core competencies and to increase awareness in biomedical, chemical, and health informatics, students at Wesley get exposed to such cutting-edge computing fields via interdisciplinary undergraduate research projects in chemistry, biology, electronic data-mining, computer programming, statistics, and medicine (D’Souza and Koyoshi, 2008; D’Souza et al., 2009; D’Souza et al., 2009; D’Souza and AlAbed, 2010; D’Souza and Gerges, 2010; D’Souza, AlAbed et al., 2011). Numerous public or commercial computational tools and databases are available to identify mechanistic patterns that can provide structure-activity relationships of biologically active molecules for the early detection of toxicity prior to significant human exposure (Rusyn and Daston, 2010; Reddy et al. 2011). One such commercial solution is the Bio-Rad’s KnowItAll® Informatics System for spectroscopy, cheminformatics, in silico ADME/Tox (absorption, distribution, metabolism, excretion and toxicity, or ADMET) prediction, and lead optimization (D’Souza, 2005; D’Souza, 2007).
Since 1968 the Food and Drug Administration (FDA) approved drug package inserts profile drugs by chemical structure and contain high quality prescribing, safety, efficacy, consumer self-care, and detailed product information about the US marketed pharmaceuticals (Hartgraves, 2002; Donohue, 2006; Shrank and Avorn, 2007; Watson and Barash, 2009; de Leon, 2011). These publically available inserts containing important protocol-required clinical data are created by unrelated pharmaceutical companies, and as a result tend to be very different in the way the required information is reported (Shrank and Avorn, 2007; D’Souza and Koyoshi, 2008; Zarin and Tse, 2008; D’Souza et al., 2009; D’Souza et al., 2009; Watson and Barash, 2009; D’Souza and AlAbed, 2010; D’Souza and Gerges, 2010; D’Souza, AlAbed et al., 2011; de Leon, 2011).
In 2007 using the chemical, pharmacokinetic, and pharmacodynamic data obtained from the FDA drug profiles and together with the KnowItAll® Informatics System, we created an FDA Consumer Drug Database© containing 75 orally administered drugs (D’Souza and Koyoshi, 2008). This database is available as one of many training datasets for benchmarking experiments in drug discovery, and for interpreting the success and building of ADME/Tox predictive models (KnowItAll® Informatics System – Experimental ADME/Tox Databases, 2008). We found that amongst the various drug companies there was a significant lack of uniformity in the reporting of clinical data within the FDA-mandated consumer drug information packets (D’Souza and Koyoshi, 2008; D’Souza et al. 2009; D’Souza et al. 2009).
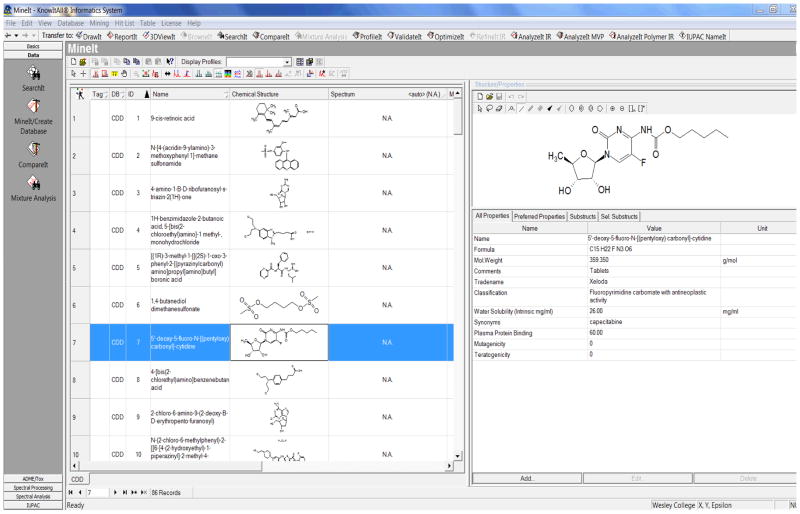
A second project evaluated fourteen chemical and pharmacological properties from 85 FDA cancer drug profiles (D’Souza and AlAbed, 2010; D’Souza, AlAbed et al., 2011). The drugs were selected on the basis of the their published chemical structure and a cancer drug database (shown in Figure 1) containing this information was created utilizing the KnowItAll® Informatics System (D’Souza, AlAbed et al., 2011). We discovered, that here too there were substantial deficiencies in the reporting of some clinical parameters that are essential for the dosage calculations, and for the safety and effectiveness of the FDA approved cancer drugs (D’Souza and AlAbed, 2010; D’Souza and Gerges, 2010; D’Souza, AlAbed et al., 2011). These important findings were recently featured on the Word on Health blog (SRxA’s Word on Health, 2010) of the Strategic Pharmaceutical Advisors (SRxA).
Figure 1.
Screen Print Example From Bio-Rad Knowitall® Informatics System Output
However, public usage of the information from our cancer drug database was heavily limited due to access of the underlying commercial platform. Hence using the information from the KnowItAll® Informatics System as a starting point, a relational cancer drug database (MySQL) was created as a result of a collaborative effort of researchers from Wesley College and Center for Bioinformatics & Computational Biology (CBCB) at the University of Delaware (D’Souza, AlAbed et al., 2011). One purpose of this expanded project was to further facilitate the analysis and/or the prediction of the chemotherapy compound’s missing pharmacokinetic properties.
DESIGN OF A PUBLIC CANCER DRUG DATABASE AND THE PREDICTION OF AQUEOUS DRUG SOLUBILITY
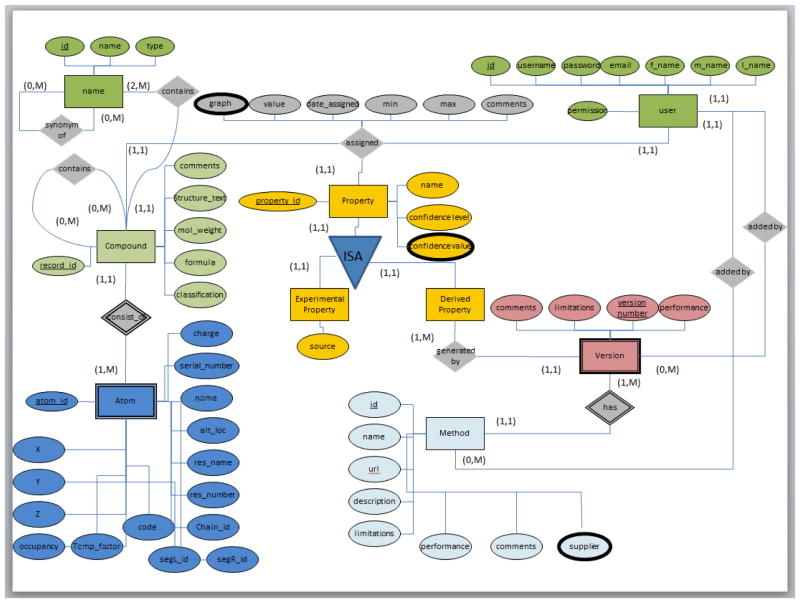
In order to accomplish universal acceptance of a public cancer drug database, our goal was to design the database, load all existing compounds, and implement a web application that will allow users to view and edit the content of the database. The 85 FDA cancer drugs were initially chosen on the basis of the presence of at least one carbonyl containing functional group in the drugs chemical structure and on the availability of documented physicochemical parameters. We first designed a schema of the normalized relational database (D’Souza, AlAbed et al., 2011). The database design was aiming in achieving detailed information about each compound, their properties, as well as the methods that were used to collect those properties.
The information collected for each drug was first tabulated within a database created using the KnowItAll® ADME/Tox Edition (Figure 1). This program gave an output of the drug that included the data about the different properties, such as the pH profile, and the source of the data itself. From that output, we collected our data and then began the process of editing the preexisting MySQL-database (D’Souza, AlAbed et al., 2011). We took the pH profiles for each drug and saved it into a separate file in order to add it to the database. When adding new data to this database, the schema for the database itself had to be modified (Figure 2, changes are in bold).
Figure 2.
E/R Diagram For The Cancer Drug Database
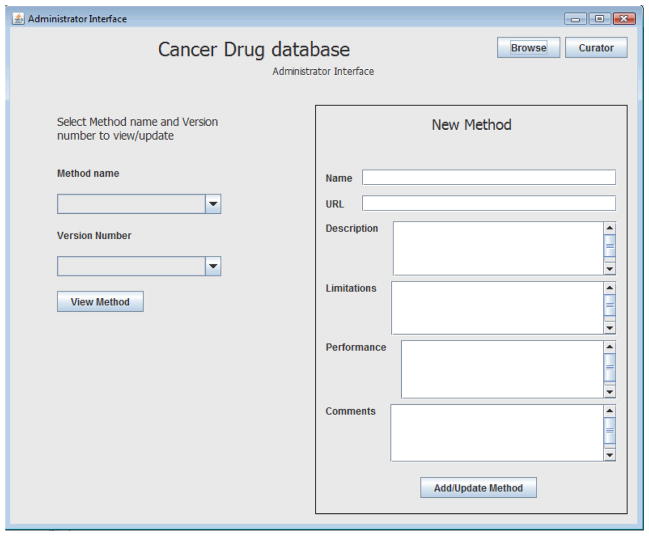
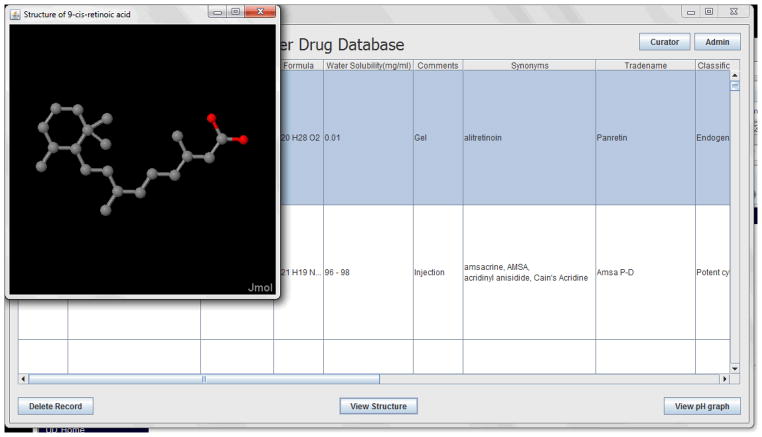
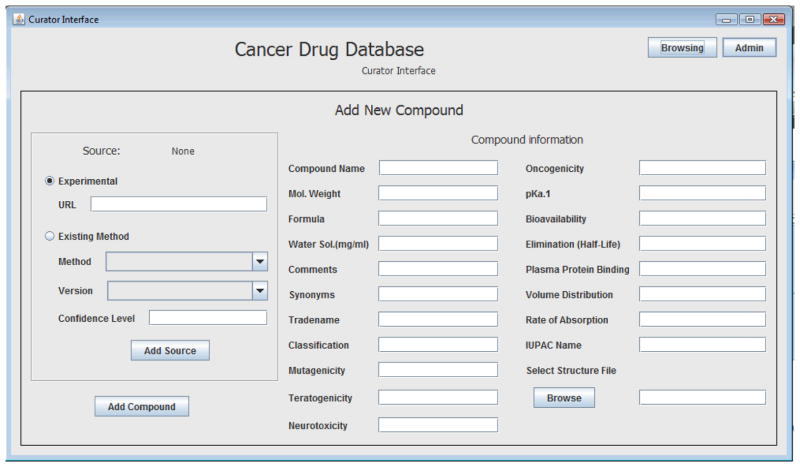
As a result of this work, a relational database has now been created (Figures 3–5). The database interface allows the user to add new possible measurements of the compound properties coupled with the detailed description of the each method (Figure 3), to browse among stored compounds and to visualize the structure (Figures 4, 5), and to import new compounds and their corresponding structures (Figure 4).
Figure 3.
Screen-Shot Of The Cancer Drug Database GUI View (New Method Of Measurement Addition)
Figure 5.
Screen-Shot Of The Cancer Drug Database GUI View (Visualization Of The Existing Compounds In The Database)
Figure 4.
Screen-Shot Of The Cancer Drug Database GUI View (New Compound Addition)
The visualization of the compounds is accomplished by using Jmol, an interactive web browser applet. Jmol is a free, open source molecule viewer for students, educators, and researchers in chemistry and biochemistry. It is also a cross-platform, running on Windows, Mac OS X, and Linux/Unix systems.
Hence to aid in the use of determining missing clinical data, we have created a viable option that utilizes in-silico tools to predict missing pharmacokinetic parameters. This gives us the ability to enhance better drug delivery in the human body. In order to address pH-sensitive drug pharmacokinetics we show (below) the predictive ability of our model using five examples of well-known cancer drugs that contain at least one carbonyl group in their chemical drug structure.
CLINICAL/PHARMACOKINETICS CORRELATION AS APPLIED TO OUR PREDICTIVE MODEL
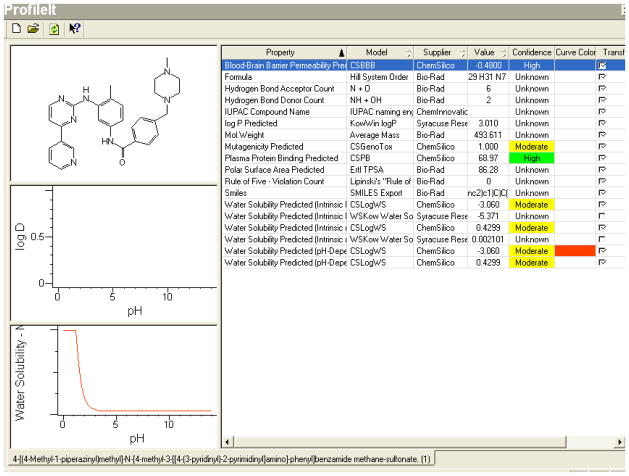
Taxol®
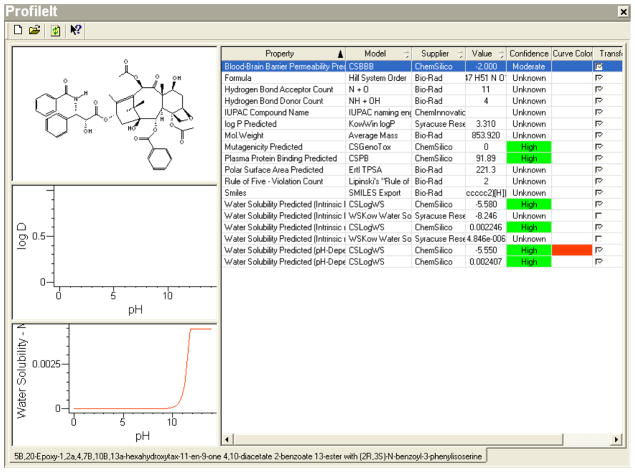
As an indicated first line or adjuvant chemotherapy for solid tumors like ovarian carcinoma, breast carcinoma or non-small cell lung carcinoma, Taxol®(paclitaxel), intravenously administered, targets these tumors by its inherent very high bioavailability. Due to its low water solubility and high protein binding capacity, it directly redistributes from the intravascular compartment into peripheral compartment leading to high-targeted therapy on the intended solid tumor cancer cells. The solubility reported in experimental and in vitro models is mirrored in our in-silico predictions (illustrated in Figure 6) thus indicating the accuracy by which pharmacokinetic properties of such drugs can be predicted in anticipation of its utility. At the normal blood pH (7.4) Taxol® is essentially not water soluble, a function that is predicted by our in-silico model using the functional groups’ chemical characteristics of its parent compound. Extrapolating our model into its predictive utility, the selection of similar solid tumor effector chemotherapy agents can be more accurately achieved via narrowing on the specific pharmacokinetic parameters, in this case water solubility, thus cutting considerably on the cost and time needed to bring similar drugs into clinical utility.
Figure 6.
Screen-Shot Of The Knowitall® Predictor Profileittm For Taxol® Is Represented Below, And The Required Water Solubility Data Can Be Extracted From These Predictions
Matulane®
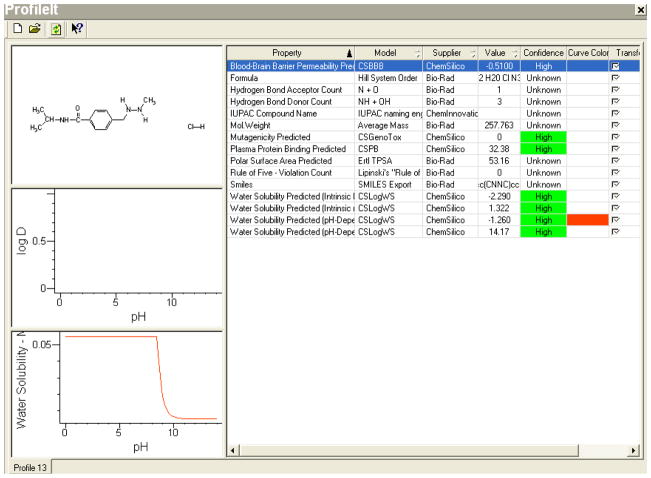
Hematopoietic malignancies, in their advanced stages, tend to involve other solid organs outside the lymph nodes and marrow spaces. As such they need to be targeted in the intravascular and extravascular compartment including across the blood brain barrier. Chemotherapeutic agents that are used in these settings, such as Matulane® for advanced stage Hodgkin’s Lymphoma, are chosen for their specific pharmacokinetic parameters notably their aqueous solubility. Due to its good solubility at gastric pH as well as at plasma pH, Matulane® is the perfect candidate. It is can be readily absorbed when given orally, thus there is an ease of administration. Completely soluble in blood, cross the blood brain barrier, and thus has the ability to be readily bioavailable in all potential compartments. The kidneys, decreasing the amount of potential toxicities, also can readily eliminate it. All these characteristics are mainly due to its water solubility. The accuracy by which our computer model was able to predict its solubility is documented in Figure 7. Mirroring the utility and applicability of our findings on formulating similar drugs, agents with similar solubility can be predicted with measurable accuracy using our model. Thus in filtering through potential chemotherapeutic candidates with proven in-vitro clinical efficacy, our model can help in narrowing down the search.
Figure 7.
Screen-Shot Of The Knowitall® Predictor Profileittm For Matulane® Is Represented Below, And The Required Water Solubility Data Can Be Extracted From These Predictions
Nexavar®
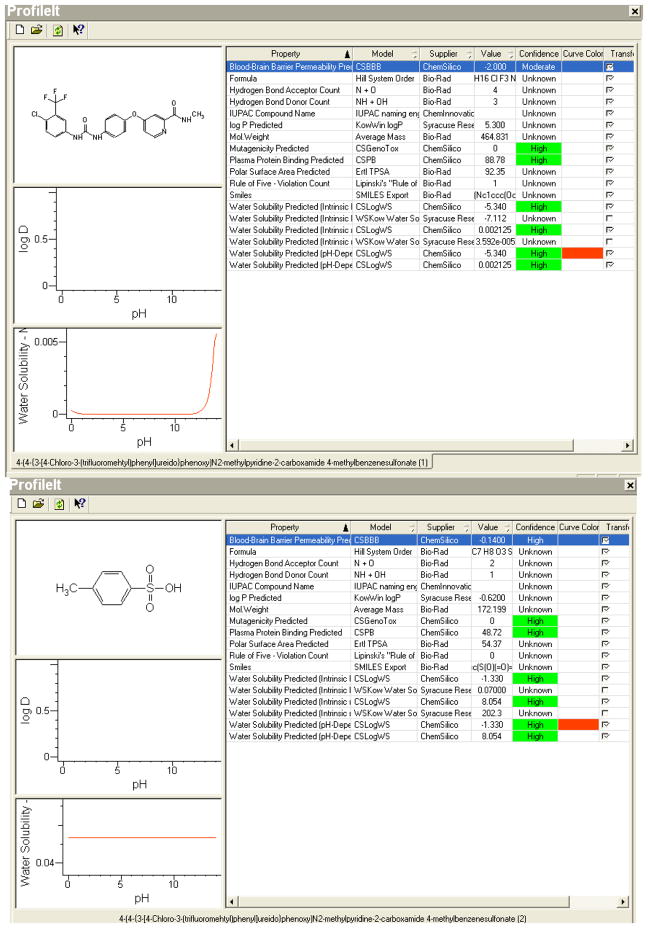
In formulating drugs that are intended for treatment of solid tumor with the primary metabolite or molecule being insoluble in aqueous solutions, a parenteral rout of administration is not very practical. The drug has to be administered in a continuous mode to achieve comparable result to its in vitro action, and a functional group needs to be added to make the whole compound water soluble. An example is Nexavar®, a chemotherapeutic agent used primarily to treat renal cell carcinoma and unresectable hepatocellular carcinoma. The active ingredient, sorafenib tosylate is a kinase that achieves its steady state blood level after 7 days of continuous oral administration of 400 mg twice daily. This needs to be maintained for a total of 28 days to produce its optimum inhibitory effect on tumor growth. Thus the best way to achieve this is through the oral route, a potential problem with an insoluble molecule like sorafenib; a parameter that we were able to predict with high comparable accuracy using our computer model. We were also able to predict the solubility at gastric pH and plasma pH after pairing the active ingredient with the functional group (Figure 8). It also follows that the applicability of our model is of practical utility in formulating similar drugs with specific clinical/pharmacokinetic parameters.
Figure 8.
Screen-Shots Of The Knowitall® Predictor Profileittm For Nexavar® Is Represented Below, And The Required Water Solubility Data Can Be Extracted From These Predictions
Gleevec®
While the above mentioned kinase is insoluble at acidic as well as basic pH and can only be clinically useful if a functional group is added, other kinase inhibitors such as Gleevec® (Imatinib) lends itself perfectly due to its excellent solubility at low pH and very low solubility at neutral pH. Gleevec® is very effective in treating Philadelphia chromosome positive hematologic malignancies such as CML and ALL. It can also be used in other sarcomas such as GIST. Being very soluble at low pH it can be given orally with excellent absorption, yet with its insolubility at blood pH it can attain therapeutic levels fast, get slowly eliminated and have a high volume of distribution. As illustrated below (Figure 9), our model accurately predicts this biphasic solubility as a function of the pH. This also lends itself easily to pre-experimental phases in selecting drugs that are intended to be similarly administered with a comparable eventual volume of distribution.
Figure 9.
Screen-Shots Of The Knowitall® Predictor Profileittm For Gleevec® Is Represented Below, And The Required Water Solubility Data Can Be Extracted From These Predictions
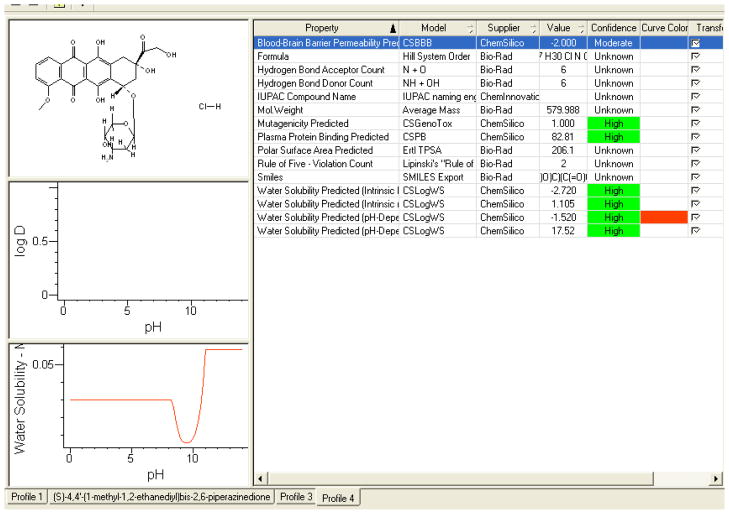
Doxil®
Perhaps the best practical application for our model is in formulating targeted therapy whereby effector agents are known for their in-vitro action yet their solubility prohibits effective delivery to the target organs/tumors. Having the ability to accurately predict the behavior of the major molecule/metabolite at different pH levels, make it possible to manipulate the drugs in its pre-experimental form, thus giving us an edge in predicting the final product behavior at the given pH. This concept is best-illustrated by applying our computer model to a drug like Doxil®; a parenteraly administered chemotherapeutic agent used to treat ovarian cancers and AIDS related Kaposi’s Sarcoma. The active ingredient is Doxorubicin, which is encapsulated, in Doxil®, in a liposome lipid bilayer. Studies have shown that, in contrast to Doxorubicin, Doxil®, and due to the liposome delivery system, have a very low volume of distribution confined to the vascular fluid volume. This gives this agent it targeted cytotoxic effect within the neovascularization inherently pronounced in ovarian carcinoma and Kaposi’s sarcoma. If we apply our prediction algorithm to Doxil® it shows its stable low solubility at plasma pH and is even less soluble at higher pH up to extremely high pH where the lipid bilayer is broken exposing the soluble Doxorubicin. This is exactly mirrored clinically and correlates accurately to the predicted values (Figure 10). This parameter insures the stability of the delivery system until targeted areas are reached releasing the active ingredient. Our model’s applicability in formulating such drugs thus transcends its in-silico applicability into experimental utility.
Figure 10.
Screen-Shots Of The Knowitall® Predictor Profileittm For Doxil® Is Represented Below, And The Required Water Solubility Data Can Be Extracted From These Predictions
CONCLUSIONS
Even though the FDA periodically updates its guidance relevant to the disclosure of risk information in prescription drug product labels and inserts, no consumer-information standards have been implemented for drugs (Shrank and Avorn, 2007). A lack of pH-sensitive pharmacokinetic information hinders the ability of providing quality clinical care. To overcome such challenges we have definitively shown that in silico tools could be employed to predict the potential pH-sensitive biological and biochemical interactions between the molecular chemical drug entities and targets in the patients. The implementation of the web application for clinicians, researchers, and healthcare providers to add real-world data is available at http://annotation.dbi.udel.edu/CancerDB/.
Acknowledgments
The Wesley College Directed Research Program is supported by - grants from the National Institute of General Medical Sciences - NIGMS (8 P20 GM103446-13) at the National Institutes of Health (NIH); a National Science Foundation (NSF) Delaware EPSCoR grant EPS-0814251; and an NSF ARI-R2 grant 0960503. The authors also sincerely acknowledge the efforts of the numerous faculty, staff, and administrators at Wesley, UD and the other partner institutions who are strongly committed to making these inter-institutional commitments succeed in Delaware.
Biographies
Malcolm J. D’Souza is professor of chemistry at Wesley College, in Dover, Delaware. He is the principal investigator on the Wesley College DE-INBRE program and serves as Director of Sponsored Research. In 2009, he was nominated and identified by a selection committee for the 2009 Northern Illinois University (NIU) Golden Anniversary Alumni Award. In 2012, he was awarded the American Chemical Society’s (ACS) E. Emmett Reid Award, which recognizes high-quality teaching in chemistry at small colleges in the ACS Mid-Atlantic region. In addition to his research in physical organic chemistry, he also has projects, presentations, and publications in the area of chemometrics, developing commercial databases that assist in the development of new pharmaceutical and agricultural products. E-mail: malcolm.dsouza@wesley.edu
Ghada J. AlAbed extracted the necessary drug profile data and completed this database project as an undergraduate research assistant in the Wesley College Directed Research Program in Chemistry. In April 2010, a component of this project was one of just 60 from across the USA that was chosen to be showcased as a poster at the 2010 Council of Undergraduate Research (CUR) Posters on the Hill event. Ghada is employed as an administrative manager at ABC Pediatrics in Dover Delaware.
Melissa Earley completed the design of relational database as an INBRE-supported summer intern at the Center for Bioinformatics & Computational Biology (CBCB) at the University of Delaware. On graduation in May 2013, she secured employment as a software developer at Highmark Blue Cross Blue Shield, in Wilmington, Delaware.
Natalia Roberts is an assistant professor of biochemistry at Georgetown University, Washington DC, and a senior bioinformatics scientist at the Protein Information Resource (PIR). She supervised the design and implementation of the database. Her professional interests include research in the intersection of AI, machine learning, bioinformatics, and biochemistry. E-mail: np6@georgetown.edu
Fady J. Gerges graduated from the American University Beirut class of 1995. This was followed by residency and chief residency at Boston University graduating in 2003. Dr. Gerges has specialized in cytopathology and hematopathology with a special interest in gastrointestinal pathology. In 2005, he joined Doctors Pathology Services, PA, in Delaware, and is currently serving as the Director of Surgical Pathology. E-mail: fgerges@dpspa.com
Contributor Information
Malcolm J. D’Souza, Wesley College, USA
Ghada J. AlAbed, Wesley College, USA
Melissa Earley, Wesley College, USA.
Natalia Roberts, Georgetown University, USA.
Fady J. Gerges, Doctors Pathology, USA
References
- 1.Banville DL. Mining Chemical Structural Information from the Drug Literature. Drug Discovery Today. 2006;11 (1–2):35–42. doi: 10.1016/S1359-6446(05)03682-2. [DOI] [PubMed] [Google Scholar]
- 2.de Leon J. Highlights of Drug Package Inserts and the Website DailyMed: The Need for Further Improvement in Package Inserts to Help Busy Prescribers. Journal of Clinical Psychopharmacology. 2011;31(3):263. doi: 10.1097/JCP.0b013e318218f3e4. [DOI] [PubMed] [Google Scholar]
- 3.Donohue J. A History of Drug Advertising: The Evolving Roles of Consumers and Consumer Protection. Milbank Quarterly. 2006;84 (4):659–699. doi: 10.1111/j.1468-0009.2006.00464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D’Souza MJ. KnowItAll® - Software Reviews, Chemistry World. 2005;2(9):70–71. [Google Scholar]
- 5.D’Souza MJ. KnowItAll® U System - Software Reviews, Chemistry World. 2007;4(11):70–72. [Google Scholar]
- 6.D’Souza MJ, Koyoshi F. Extracting Relevant Information from DFA Drug Files to Create a Structurally Diverse Drug Database Using KnowItAll®. Pharmaceutical Reviews. 2008;7(3) Retrieved from: http://www.pharmainfo.net/reviews/extracting-relevant-information-fda-drug-files-create-structurally-diverse-drug-database-usi. [PMC free article] [PubMed] [Google Scholar]
- 7.D’Souza MJ, Koyoshi F, Everett LM. Bioinformatics, Computational Biology, Genomics, and Chemoinformatics. 2009. Structure Activity Relationship (SAR) Patterns Observed Within a Series of Unrelated Common Consumer Drugs; pp. 1–6. [Google Scholar]
- 8.D’Souza MJ, Koyoshi F, Everett LM. Structure Activity Relationships (SARs) Using a Structurally Diverse Drug Database: Validating Success of Predictor Tools. Pharmaceutical Reviews. 2009;7(5) Retrieved from: http://www.pharmainfo.net/reviews/structure-activity-relationships-sars-using-structurally-diverse-drug-database-validating-su. [PMC free article] [PubMed] [Google Scholar]
- 9.D’Souza MJ, AlAbed GJ. Deficiencies in the Reporting of VD and t1/2 in the FDA approved chemotherapy drug inserts. Pharmaceutical Reviews. 2010;8(1) Retrieved from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3106297/ [PMC free article] [PubMed] [Google Scholar]
- 10.D’Souza MJ, Gerges FJ. Raising a Red Flag: Deficiencies found in the Reporting of Important Parameters in FDA Approved Drug Profiles. Pharma IQ. 2010 July issue, Retrieved from: http://www.pharma-iq.com/article.cfm?externalID=2850.
- 11.D’Souza MJ, AlAbed GJ, Wheatley JM, Roberts N, Veturi Y, Bi X, Continisio CH. A Database Developed with Information Extracted from Chemotherapy Drug Package Inserts to Enhance Future Prescriptions. The 2011 International Conference on Computational Intelligence and Software Engineering (CiSE 2011); 2011. IEEE Conference Record #17768; IEEE Catalog Number: CFP1160F-PRT. [PMC free article] [PubMed] [Google Scholar]
- 12.D’Souza MJ, Dwyer P, Allison BE, Miller JM, Drohan J. Wesley College Ignites Potential with Undergraduate Student Research Program. Council of Undergraduate Research Quarterly. 2011;32 (2):41–45. [PMC free article] [PubMed] [Google Scholar]
- 13.D’Souza MJ, Wang Q. Inter-Institutional Partnerships Propel A Successful Collaborative Undergraduate Degree Program In Chemistry. Journal of College Teaching and Learning. 2012;9 (4):245–252. doi: 10.19030/tlc.v9i4.7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ekins S, Mestres J, Testa B. In silico Pharmacology for Drug Discovery: Methods for Virtual Ligand Screening and Profiling. British Journal of Pharmacology. 2007;152 (1):9–20. doi: 10.1038/sj.bjp.0707305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartgraves T. DTC Prescription Drug Advertising: The History and Impact of FDA Regulation. 2002. [Google Scholar]
- 16.KnowItAll® Informatics System Desktop Solutions. Retrieved from: http://www.knowitall.com/
- 17.KnowItAll® Informatics System – Experimental ADME/Tox Databases. 2008 Retrieved from: http://www.knowitall.com/literature/docs/96199-Bio-Rad_KnowItAll_ADME_Databases_Datasheet.pdf.
- 18.KnowItAll® U System. Available: http://www.knowitallu.com/
- 19.Mandal S, Moudgil M, Mandal SK. Rational Drug Design. European Journal of Pharmacology. 2009;625:90–100. doi: 10.1016/j.ejphar.2009.06.065. [DOI] [PubMed] [Google Scholar]
- 20.Reddy S, Reddy TS, Krushna GS. Advantages of Bio and Cheminformatics tools in Drug Design. Current Research in Drug Targeting. 2011;1 (2):6–17. [Google Scholar]
- 21.Rusyn I, Datson GP. Computational Toxicology: Realizing the Promise of the Toxicity Testing in the 21st Century. Environmental Health Perspectives. 2010;118 (8):1047–1050. doi: 10.1289/ehp.1001925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shrank WH, Avorn J. Educating Patients About Their Medications: The Potential And Limitations Of Written Drug Information. Health Affairs. 2007;26(3):731–740. doi: 10.1377/hlthaff.26.3.731. [DOI] [PubMed] [Google Scholar]
- 23.SRxA’s Word on Health. 2010 Retrieved from: http://srxawordonhealth.com/2010/07/26/fda-criticized-by-the-government-accountability-office/
- 24.The Bio-Rad KnowItAll®. website: http://www.bio-rad.com/evportal/en/US/INF/Category/203353/Academic.
- 25.The Delaware-INBRE. website: www.inbre.udel.edu.
- 26.The FDA. website: www.fda.gov.
- 27.The National Institutes of General Medicine. website: http://www.nigms.nih.gov/Training/IDeA/
- 28.The University of Delaware. website: www.udel.edu.
- 29.The Wesley College. website: www.wesley.edu.
- 30.The Wesley College INBRE. website: http://www.wesley.edu/academics/grants/inbre.html.
- 31.Watson KT, Barash PG. The New Food and Drug Administration Drug Package Insert: Implications for Patient Safety and Clinical Care. Anesthesia & Analgesia. 2009;108(1):211–218. doi: 10.1213/ane.0b013e31818c1b27. [DOI] [PubMed] [Google Scholar]
- 32.Zarin DA, Tse T. Moving Towards Transparency of Clinical Trials. Science (New York, NY) 2008;319(5868):1340. doi: 10.1126/science.1153632. [DOI] [PMC free article] [PubMed] [Google Scholar]