Abstract
The subgenomic RNA 2 of tobacco necrosis virus A (TNV sgRNA2) encodes the viral coat protein, is unpolyadenylated and presumably uncapped. Here, we show that TNV sgRNA2 is translated cap independently. This cap-independent translation requires the leader and a 140 nt element of the trailer both in wheat germ extract and in tobacco protoplasts. Similar to barley yellow dwarf virus (BYDV), the TNV 5′ and 3′ elements stimulate translation synergistically. Computer-aided phylogenetic analysis of the secondary structure of the TNV trailer revealed that the 3′ translation element is part of a major conserved stem–loop that contains similarities to structures in the BYDV 3′ translation element. These data suggest that the translation mechanisms of TNV sgRNA2 and BYDV RNA are related. To further characterize this relationship, we tested whether cooperativity exists between TNV sgRNA2 and BYDV 5′ and 3′ elements. We found that the TNV sgRNA2 5′ element stimulates translation synergistically with the BYDV 3′ element in vitro. This finding is the first evidence for conservation of structures that enable a 5′–3′ interaction stimulating cap-independent translation.
INTRODUCTION
Plant viral RNAs employ various, often non-canonical, strategies to synthesize their required proteins. This is most likely also true for the RNAs of tobacco necrosis virus A (TNV-A). TNV-A contains a genomic RNA and two shorter 3′ co-terminal subgenomic RNAs (1). These RNAs are not polyadenylated (2) and the genomic RNA is uncapped (3). The subgenomic RNAs are most likely also uncapped, as primer extension products of these RNAs consist of one single band, and not of two bands, as is often observed for capped RNAs (4).
The TNV-A RNAs contain six different major open-reading frames (1). The most abundant TNV-encoded protein in infected tobacco plants is the coat protein (5). The coat protein coding sequence is present on all three TNV RNAs. It is the fifth open-reading frame of the genomic RNA (1), the third of subgenomic RNA 1 (sgRNA1) and the first of subgenomic RNA 2 (sgRNA2) (4). The coat protein may thus be synthesized via a mechanism of internal initiation of translation from the genomic RNA or sgRNA1, and/or via another, presumably cap-independent mechanism of translation from sgRNA2. Internal initiation of translation of the genomic RNA does occur in vitro. In fact, the coat protein is the most abundant in vitro translation product of the genomic RNA (4,6). Consistently, the region upstream of the coat protein open-reading frame stimulates internal initiation of translation in vitro (4). However, these sequences do not promote a detectable level of internal initiation in vivo in tobacco protoplasts (4). Thus, it is likely that not the genomic RNA or sgRNA1, but sgRNA2 is the major template for coat protein synthesis in vivo. If this is the case, the accumulation levels of sgRNA2 and the coat protein in infected tobacco protoplasts indicate that one coat protein molecule is synthesized per sgRNA2 molecule every 10–12 s (4). This would be comparable with the translation efficiency of efficiently translated mammalian mRNAs, and would imply that TNV sgRNA2 contains RNA elements that enable an efficient cap-independent translation. The mechanism by which TNV sgRNA2 is translated is currently unknown.
In most models for translation initiation, the initial steps of recruitment of translation initiation factors requires binding of these factors to specific RNA elements, a process which is further stimulated through interactions between different RNA elements. The most advanced model for such a mechanism is that of cap- and poly(A)-dependent translation. In the current model, the cap recruits the translational machinery via binding of the cap-binding complex [eukaryotic initiation factor 4F (eIF4F), consisting of eIF4E, eIF4G and eIF4A]. eIF4F unwinds the secondary structure in a manner that is stimulated by eIF4B, allowing binding of the 40S ribosomal subunit. The further enhancement of cap-dependent translation by the poly(A) tail may be mediated by the structural interaction of cap and poly(A) tail via binding of eIF4E, eIF4G and the poly(A) binding protein PABP (7). The resulting circular mRNA may enhance ribosome recruitment by recycling of the 40S ribosomal subunit from the 3′ UTR to the 5′ UTR (8). In addition, interactions between PABP and eIF4G/eIF4B may serve to increase their binding to the mRNA, resulting in a more efficient recruitment of 40S subunits (9,10).
Models for translation of uncapped and unpolyadenylated RNAs share aspects with those of cap-dependent translation, but are generally less advanced. In plants, the best-described cap-independent translation mechanisms are for RNAs of viral origin. Some mechanistic aspects of translation of the uncapped and unpolyadenylated RNAs of barley yellow dwarf virus (BYDV) and satellite tobacco necrosis virus (STNV) have been investigated. Both BYDV and STNV contain translational enhancer sequences in their 5′ and 3′ UTRs (11–15). For STNV, the translational enhancer domain in the trailer is sufficient for cap-independent translation in plant cells, and the 5′ sequences further promote translation synergistically with the 3′ sequences (12). Although sequence complementarity exists between STNV 5′ and 3′ elements, its relevance for translation is unclear (12). The current model for synergistic action by STNV 5′ and 3′ elements is that interaction between these two elements promotes ribosome subunit recycling upon translation termination (12). Similarly, the BYDV 5′ and 3′ RNA elements stimulate translation synergistically (14,15). For the synergistic action, base pairing between these two elements is necessary and sufficient (16). This indicates that, similar to capped and polyadenylated RNA, the BYDV RNA forms a closed loop. In the model for translation stimulation by BYDV elements, the closed loop is necessary to deliver either ribosomes or initiation factors to the 5′ UTR (16). Thus, it appears that also for uncapped plant viral RNAs, interaction between 5′ and 3′ RNA elements plays an important role in facilitation of translation initiation.
There are several indications that the translation mechanism of TNV sgRNA2 shares features with that of the BYDV RNA. First, the TNV trailer shares sequence and structural similarities to the BYDV 3′ translation element (15,16). Secondly, both TNV sgRNA2 and BYDV have base pairing capacity between 5′ and 3′ UTR sequences (16). In this study, we investigate whether this structural conservation also reflects a functional conservation.
We first investigated the sequence requirements for cap-independent translation of TNV sgRNA2. We found that, similar to BYDV, sequences in both 5′ and 3′ UTR are required for cap-independent translation in vitro and in vivo. Phylogenetic analysis revealed that TNV 3′ elements contain structural similarities to BYDV 3′ elements. Moreover, we found that the TNV sgRNA2 leader works synergistically with the BYDV trailer in vitro. These data show a compatibility of unrelated translation-promoting cis elements and suggest that TNV sgRNA2 and BYDV have similar modes of translation stimulation.
MATERIALS AND METHODS
Templates for in vitro transcription
pAB02 is a pGEM3Z-derived (Promega) plasmid constructed such that T7-driven run-off transcripts of BsaI-digested pAB02 correspond exactly to the full-length TNV sgRNA2 (4) (André Boorsma, personal communication). pAB01 differs from pAB02 in that nucleotides 1–133 of TNV sgRNA2 are replaced by the 23 nt of the pGEM3Z polylinker sequence immediately downstream of the T7 transcription start site (André Boorsma, personal communication). The template for synthesis of a TNV sgRNA2 derivative lacking the leader was BsaI-digested pAB01. Templates for synthesis of TNV sgRNA2 derivatives lacking the trailer were made by PCR on pAB02 (with leader) or pAB01 (no leader) with upstream primer T7 (5′-CGACGGCCAGTGAATTGTAATACG-3′) and downstream primer PCR-1 (5′-GCCAAGTTACACGT ACAAAGAACTAGAC-3′) [complementary to sequence –3 to +24 relative to the TNV CP stop codon and with a 1 nt insertion (underlined)]. The templates for the 3′ truncated TNV sgRNA2 derivatives were ApaLI(1151)-, BspEI(1104)-, BamHI(1048)-, BsmAI(1038)- and Bsu36I(1014)-digested pAB02 (the position in TNV sgRNA2 of the 3′ terminal nucleotide of the T7 run-off transcripts is indicated between parentheses).
Codon 195 of the CP coding sequence was converted into a TAA stop codon by filling-in and ligating EcoRI-digested pAB02, resulting in plasmid pRD01. Deletion mutants of the 5′ end of the trailer were generated by cleavage of pRD01 with BstBI together with BsaAI, Bsu36I or BamHI, filling in the overhangs with Klenow enzyme, and ligation resulting in plasmids pRD03 (deletion of nucleotides 738–938), pRD04 (deletion of nucleotides 738–1011), pRD05 (deletion of nucleotides 738–1044), respectively. BsaI cleavage of these plasmids yielded templates for the TNV sgRNA2 derivatives with 5′ deletions of the trailer. The template for the RNA with the frameshift mutation and lacking the trailer was a PCR product on pRD01 with primers T7 upstream and PCR-1.
Chimeric cat RNAs with the complete TNV sgRNA2 trailer were synthesized using T7 RNA polymerase from the following BsaI-cleaved plasmids: pFM188H (nucleotides 1–138 of TNV sgRNA2—CGCG—cat coding sequence—112 nt vector sequence—nucleotides 883–1224 of TNV sgRNA2; Fig. 2A), pVE190 (nucleotides 1–138 of TNV sgRNA2—CGCG—cat coding sequence—22 nt vector sequence—nucleotides 939–1224 of TNV sgRNA2; Fig. 2B), pFM188G (nucleotides 1–158 of TNV sgRNA2 leader—12 nt polylinker sequence—cat coding sequence—112 nt vector sequence—nucleotides 883–1224 of TNV sgRNA2; Fig. 2C), and pFM188I (19 nt polylinker sequence—cat coding sequence—112 nt vector sequence—nucleotides 883–1224 of TNV sgRNA2; Fig. 2A, trailer). RNA containing only TNV sgRNA2 leader was synthesized from BsuI-cleaved pVE190. RNA containing the TNV sgRNA2 leader and 3′ translation-stimulating element was synthesized from ApaLI-cleaved pVE196 (nucleotides 1–138 of TNV sgRNA2—CGCG—cat coding sequence—GCTAGC—nucleotides 1012–1151 of TNV sgRNA2).
Figure 2.
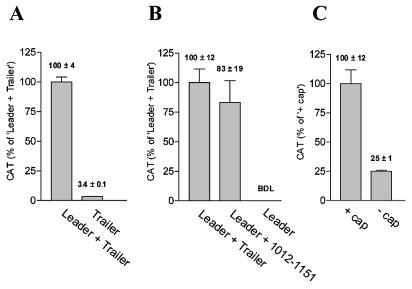
TNV sgRNA2 leader and a 140 nt element in the trailer promote cap-independent translation in tobacco protoplasts. (A) TNV sgRNA2 leader promotes cap-independent translation. Translation of uncapped cat RNAs with TNV sgRNA2 trailer, and with (Leader + Trailer) or without (Trailer) TNV sgRNA2 leader in tobacco protoplasts. CAT protein levels were determined at 5.5 h after RNA introduction. (B) A 140 nt element of the TNV sgRNA2 trailer promotes cap-independent translation. Translation in tobacco protoplasts of uncapped cat RNAs with TNV sgRNA2 leader, and with as trailer the complete TNV sgRNA2 trailer (Leader + Trailer), nucleotides 1012–1151 (Leader + 1012–1151), or vector sequences (Leader). The CAT protein levels were determined at 5 h after RNA introduction. BDL, below detection limit. (C) The cap stimulates translation of RNA with TNV sgRNA2 leader and trailer 4-fold. Translation of capped and uncapped versions of chimeric cat RNA with TNV sgRNA2 leader and trailer in tobacco protoplasts. The CAT protein levels were determined at 5.5 h after RNA introduction.
The templates for the chimeric luciferase RNAs were generated via PCR. The PCR products contained upstream of the luciferase coding region (and downstream of the T7 RNA polymerase promoter) either an 18 nt polylinker sequence (Fig. 4, vector 5′ UTR), nucleotides 1–143 of TNV sgRNA2 followed by CAAAACC (Fig. 4, TNV 5′ UTR), or BYDV nucleotides 2–137 preceded by GA and followed by GCGC (Fig. 4, BYDV 5′ UTR). Downstream of the luciferase coding region, the PCR products contained either a 125 nt vector sequence (Fig. 4, vector 3′ UTR), TTCTAGC followed by nucleotides 969–1151 of TNV sgRNA2 (Fig. 4, TNV 3′ UTR), or nucleotides 4808–5677 of BYDV followed by one G (Fig. 4, BYDV 3′ UTR).
Figure 4.
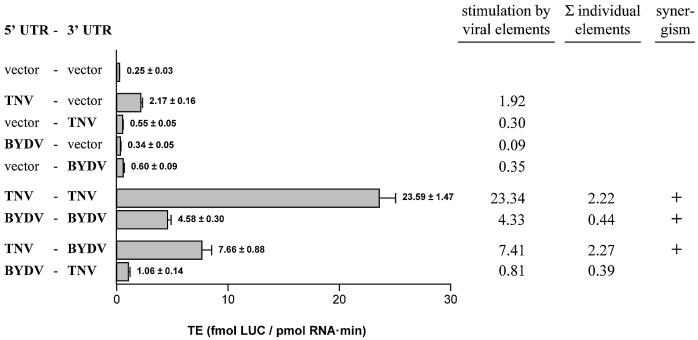
TNV sgRNA2 leader and BYDV trailer functionally interact. TE of uncapped RNAs containing vector-derived UTR sequences (vector), TNV sgRNA2 UTR sequences (TNV) or BYDV UTR sequences (BYDV) in wheat germ extract. Stimulation by viral elements is the increase in TE as compared with the RNA lacking viral sequences (vector–vector). The sum of the individual elements is the sum of (the stimulation of the 5′ viral element with the vector sequences as 3′ element) and (the stimulation of the 3′ viral element with the vector sequences as 5′ element). Synergism between viral 5′ and 3′ UTR sequences is observed when the stimulation of the two elements together is higher than the sum of the stimulation by the individual elements (30).
In vitro transcription and translation
In vitro synthesis of capped and uncapped RNAs was performed essentially as described by Meulewaeter et al. (12). In vitro translation of RNAs was done essentially as described by Meulewaeter et al. (17). For the translation of RNAs encoding luciferase, some modifications were made in the protocol. Protein synthesis occurred in the absence of [35S]methionine and with 0.5 mM of unlabeled methionine. The amount of luciferase protein was quantified using the Luciferase assay system (Promega). Briefly, 4 µl of in vitro translation mixture was diluted in 40 µl of 1× lysis buffer (Promega). Twenty microliters was added to 100 µl of Luciferase Assay Reagent I (Promega) and measured immediately for 10 s in a Turner designs TD-20/20 luminometer. The values were standardized to a dilution series of recombinant Luciferase protein.
Protein accumulation was determined at eight time points between 16 and 80 min (TNV CP, CAT), or at 10 time points between 28 and 150 min (LUC) of incubation. Protein accumulation (P) in function of time (t) was analyzed using the mathematical description as described by Danthinne et al. (11):
P(t) = (aR0 / b)(1 – e–b(t–T)) 1
in which T corresponds to the time point at which the first translation product is completed, a is the translational efficiency (TE) of the mRNA (= protein molecules synthesized per mRNA molecule per time unit at t = T), R0 is the initial RNA input (at t = 0), and b is a constant that is inversely proportional to the functional half-life of the mRNA (= t1/2) according to the relation t1/2 = ln 2 / b (11). The functional half-life is the time in which the protein accumulation rate halves and thus measures the stability of the mRNA that is actively translated, as opposed to the chemical stability that measures the physical integrity of the transcript. As in these experiments the input of translatable mRNA is equal under all conditions, the product aR0 = A (= protein synthesis rate at t = T) also reflects the TE of the mRNA. Equation 1 can also be written as:
P(t) = (A·t1/2 / ln 2)(1 – e–(ln 2 / t1/2)(t–T)) 2
From equation 2, it can be deduced that P(∞) = (A·t1/2) / ln 2, showing that the protein peak level is proportional to both the TE and the functional half-life of the mRNA. By non-linear regression using equation 2 and the GraphPad Prism 3.0 software, a best-fitting curve to the experimental data points was calculated and values for A, t1/2 and T were obtained. The TE as shown in the Figures corresponds to A.
Determination of translation and RNA stability in tobacco protoplasts
RNA introduction was essentially as described by Meulewaeter et al. (12). Typically, 10–15 pmol of synthetic cat transcript was introduced in 106 mesophyl protoplasts of Nicotiana tabacum cv. Petit Havanna SR1. Total RNA was isolated from the protoplasts 130–140 min after electroporation. Total protein was isolated from the protoplasts 5–6 h after electroporation in duplo. RNA and protein analysis was done according to Meulewaeter et al. (12).
RNA secondary structure predictions
RNA secondary structures were predicted by folding simulations using a genetic algorithm (18), implemented in the package STAR (http://wwwbio.leidenuniv.nl/~batenburg/STAR.html). The thermodynamic parameters used were from the version 2.3 with Mfold web server of M. Zucker (http://bioinfo.rpi.edu/applications/mfold). Simulations were performed at various lowered (10–25°C) temperatures in order to mimic the conditions of the natural environment for the plant virus RNAs.
RESULTS
Cap-independent translation of TNV sgRNA2 requires the leader and a 140 nt sequence of the trailer
The sgRNA2 of TNV-A (TNV sgRNA2) is most likely uncapped and encodes the coat protein (4) (see Fig. 1A for a schematic representation). Whether TNV sgRNA2 is indeed translated cap independently, and what cis elements are involved, is currently unknown. As a first step in unraveling the translation mechanism of TNV sgRNA2, we verified whether TNV sgRNA2 carries cis elements promoting cap-independent translation. Therefore, we translated uncapped TNV sgRNA2 in a wheat germ extract that does not allow efficient translation of uncapped RNAs lacking cis elements for cap-independent translation (17). We found that the uncapped version of TNV sgRNA2 had the same translational effiicency (TE) (Fig. 1B) and functional half-life (data not shown) as its capped counterpart in a wheat germ extract, showing that TNV sgRNA2 carries RNA sequences that are sufficient for cap-independent translation in vitro.
Figure 1.
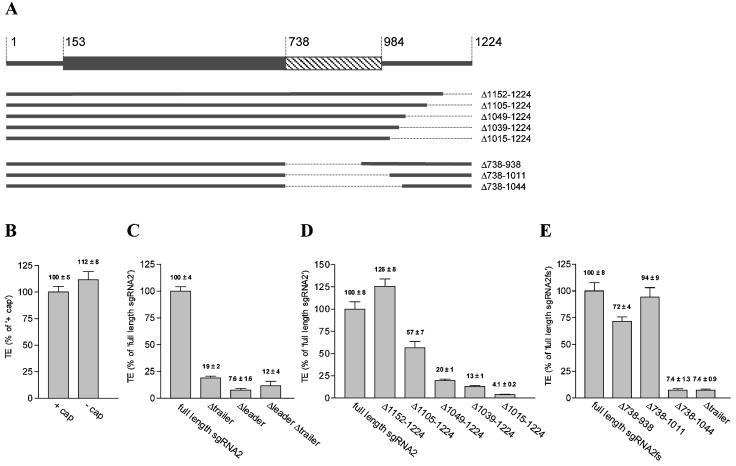
Cap-independent translation of TNV sgRNA2 in wheat germ extract requires the leader and a 140 nt sequence in the trailer. (A) Schematic representation of TNV sgRNA2 and the deletion mutants. The box (nucleotides 153–983) indicates the coat protein open-reading frame. The gray and the hatched parts of the box (nucleotides153–737 and 738–983, respectively) indicate those parts of the coat protein coding region that are within or downstream of, respectively, the open-reading frame in the frameshift mutant sgRNA2fs. (B) TNV sgRNA2 is translated cap independently in wheat germ extract. Capped and uncapped versions of TNV sgRNA2 were translated in wheat germ extract and the TE was calculated. (C) Both the leader and the trailer of TNV sgRNA2 contain sequence information required for cap-independent translation. TE of TNV sgRNA2 derivatives with both leader and trailer (full-length sgRNA2), lacking the trailer, lacking the leader, or lacking both leader and trailer was determined in wheat germ extract. (D) Nucleotides 1152–1224 of TNV sgRNA2 are not required for cap-independent translation. 3′ Deletion derivatives of TNV sgRNA2 were translated in wheat germ extract and the TE was determined. (E) Nucleotides 738–1011 are not required for cap-independent translation. TE of TNV sgRNA2 frameshift mutants without (full-length sgRNA2fs) or with different 5′ deletions of the trailer in wheat germ extract. The indicated numbers refer to the deleted TNV sgRNA2 nucleotide. All nucleotides are numbered according to their position in the full-length TNV sgRNA2 (4).
We next investigated whether leader and trailer sequences contribute to cap-independent translation of TNV sgRNA2 in vitro. Figure 1C shows that TNV sgRNA2 derivatives lacking either the leader or trailer are translated with at least a 5-fold lower efficiency than full-length TNV sgRNA2. Thus, both the leader and the trailer contain sequence information required for translation of uncapped TNV sgRNA2. Moreover, the trailer only stimulates translation in the presence of the leader, whereas the stimulatory effect of the leader is significantly higher in the presence than in the absence of the trailer. This observation implies that the TNV sgRNA2 leader and trailer act synergistically in translation.
To analyze in more detail what sequences within the 241 nt trailer contain information for cap-independent translation, the translation of a 3′ and 5′ deletion series of the trailer was compared in wheat germ extract. The 3′ deletion analysis showed that removal of the 3′ 73 nt (Δ1152–1224) did not reduce translation, and that further removal of 3′ nucleotides reduced the TE step-wise (Fig. 1D). To allow mapping of the 5′ border without changing the coding region in every deletion mutant, we generated a TNV sgRNA2 frameshift mutant with a premature translation stop codon. This frameshift mutation did not affect the efficiency of translation in vitro (data not shown). The progressive 5′ deletion analysis of the trailer revealed that deletions from nucleotide 738 (in the former coat protein coding region) up to nucleotide 1012 did not affect the TE. A further deletion up to nucleotide 1045 reduced translation to levels similar as for RNAs lacking the trailer (Fig. 1E). These data show that in the TNV sgRNA2 trailer, sequences upstream of nucleotide 1012 and sequences downstream of nucleotide 1151 are dispensable for cap-independent translation. Sequences between nucleotides 1012 and 1151 thus contain information that is required for cap-independent translation in vitro.
We next validated the sequence elements involved in cap-independent translation of TNV sgRNA2 in vivo. To allow for an accurate detection of the encoded protein in vivo, we replaced the coat protein coding sequence of TNV sgRNA2 by the cat coding sequence. In the resulting chimeric cat RNAs, the stimulatory effect of TNV sgRNA2 leader and trailer on cap-independent translation in vitro was similar as in wild-type TNV sgRNA2 (data not shown).
To determine the impact of TNV sgRNA2 leader and trailer elements on cap-independent translation in vivo, we translated uncapped cat RNAs with and without TNV sgRNA2 leader and trailer in tobacco protoplasts. Figure 2A and B show that for uncapped RNAs lacking either the leader or the trailer, the CAT protein accumulation was at least ∼30-fold lower than for RNA containing both leader and trailer. Addition of a cap to the RNA with both leader and trailer increased the CAT protein levels ∼4-fold (Fig. 2C), which is considerably less than the >20-fold stimulation of the cap for RNAs lacking cis elements required for cap-independent translation (12). As the cat RNA levels in the tobacco protoplasts did not vary significantly between the RNAs, these data show that both the TNV sgRNA2 leader and trailer contain information for stimulation of translation of uncapped RNA in vivo, and the cap stimulates translation further. When only the 3′ information required for cap-independent translation in vitro (nucleotides 1012–1151; see above) was used as trailer, the protein accumulation in tobacco protoplasts was similar to that of RNAs with the complete trailer (Fig. 2B). These data show that the TNV sgRNA2 leader and a 140 nt element (nucleotides 1012–1151) of the trailer are sufficient for stimulation of translation of uncapped RNAs in vivo in tobacco protoplasts. Moreover, the presence of both leader and trailer directed CAT protein levels that were at least 30-fold higher than the sum of the contribution of the individual elements. This finding is in line with our observations in vitro, and implies that both in vitro and in vivo, TNV sgRNA2 leader and trailer stimulate cap-independent translation synergistically.
The 3′ translation-stimulatory sequences can fold into a phylogenetically conserved stem–loop structure
The involvement of leader and trailer sequences in cap-independent translation and their synergistic function is not unique for the TNV sgRNA2. Also, the RNA of the plant virus BYDV requires a synergistic interaction between 5′ and 3′ elements for cap-independent translation (14,19). These similarities raise the question whether BYDV and TNV sgRNA2 use similar translation mechanisms.
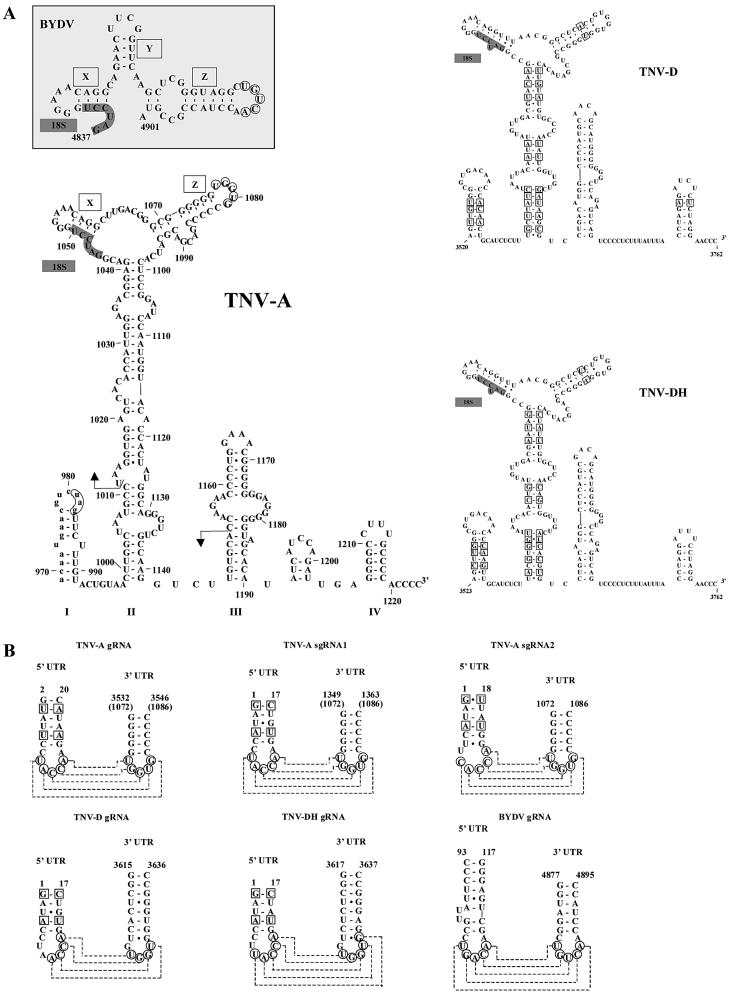
A first insight can be achieved by comparing the primary and secondary structures of the translation-stimulatory elements of these RNAs. Similarities between BYDV and TNV at the sequence level that have been described are a conserved stretch of 18 nt that includes complementarity to the 3′ end of the 18S rRNA (15), and the base pairing capacity between 5′ and 3′ sequences (16). Also at the level of secondary structure, similarities between the predicted folding of part of the TNV trailer and the folding of the BYDV 3′ cap-independent translation element have been reported (16). Here, we further characterize the putative folding of the TNV trailer. We chose for a computer-based phylogenetic analysis in which folding of the complete TNV sgRNA2 trailer was compared with the trailers of the related plant RNA viruses TNV-D (20) and a Hungarian TNV-D isolate [TNV-DH (21)]. We found that the TNV sgRNA2 trailer together with 3′ nucleotides of the coding region is able to form four stem–loop (SL) structures (Fig. 3A, I–IV). SL-I, -II and -IV are conserved among TNV-A, TNV-D and TNV-DH. Moreover, computer-aided folding of the 3′ UTRs of leek white stripe virus (22) and olive latent virus (23) revealed secondary structures similar to those of TNV-A, -D and -DH (A. P. Gultyaev, F. Meulewaeter and C. W. A. Pleij, manuscript in preparation). Importantly, similar secondary structures can be folded despite a significant divergence at the nucleotide level between the analyzed viral RNAs, and the proposed secondary structures are supported by many covariations between the more closely related TNV-D and TNV-DH (squares in Fig. 3A). These data strongly support folding of the TNV-A trailer as depicted in Figure 3A.
Figure 3.
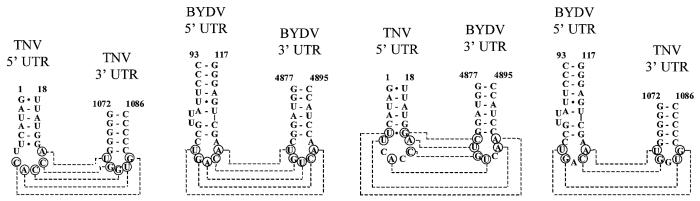
TNV sgRNA2 contains phylogenetically conserved structural elements with similarities to those in the RNA of BYDV. (A) TNV sgRNA2 trailer can fold into a phylogenetically conserved structure that contains similarities to the structure present in the BYDV 3′ translation element. The predicted foldings of TNV-A, TNV-D and TNV-DH, and the folding of the translation-stimulatory nucleotides 4837–4901 of the BYDV RNA (19) are shown. I–IV are the major stem–loops formed in the TNV sgRNA2 trailer. X, Y and Z are the small stem–loop structures in BYDV and their homologous stem–loops in the TNV sgRNA2 trailer (X and Z), comprising the small regions of complementarity to the leader (circled bases) as described earlier by Guo et al. (16). 18S indicates complementary region to the 18S ribosomal RNA. The arrows indicate the TNV sgRNA2 trailer region containing the translation-stimulatory sequences. (B) The proposed leader–trailer interactions are phylogenetically conserved in TNV RNAs. Comparison of the predicted stem–loop structures in the leaders and putative interactions with the trailers for the TNV-A genomic and subgenomic RNAs, BYDV RNA [as described by Guo et al. (16)], and the genomic RNAs of TNV-D, TNV-DH. Circles indicate the complementary nucleotides, and the dashed lines indicate the putative interactions. Covariations between TNV-A, TNV-D and TNV-DH are indicated with squares. Nucleotide numbering for the TNV RNAs is according to the position in sgRNA2 for the full trailer sequence, and indicates the position of the nucleotides within the RNA for which the putative leader–trailer interaction is shown (1,4). Numbering between parentheses indicates the position within TNV sgRNA2. Nucleotide positions in the genomic RNA are indicated for BYDV (29), TNV-D (20) and TNV-DH (21).
The TNV sgRNA2 sequences involved in translation stimulation are located mainly in the proposed SL-II. The upper part of this SL-II contains two minor stem–loop structures (X and Z; Fig. 3A) that are also conserved in the BYDV 3′ cap-independent translation element (16) and that are functional in BYDV translation (19). Importantly, stem–loop X is identical in BYDV and TNVsgRNA2, and contains part of the 18 nt conserved sequence, as well as the complementarity to the 18S rRNA. Moreover, the loop in stem–loop Z contains the sequence complementarity to the leader (circled bases), as described by Guo et al. (16), that in BYDV stimulates translation through base pairing with the leader (16).
The putative interactions between stem–loop Z and the leader would be further supported if all structures involved in such an interaction were conserved. Therefore, we performed a computer-aided phylogenetic analysis in which we investigated whether (i) the stem–loop formation in the leader, and (ii) the base pairing capacity between leader and trailer in TNV-A genomic and subgenomic RNAs that are described by Guo et al. (16), are also conserved in TNV-D and TNV-DH. The predicted stem–loop structures in the 5′ UTRs appeared conserved and supported by covariations (squares in Fig. 3B). Importantly, base pairing capacity between 5′ and 3′ UTR sequences is conserved between RNAs of TNV-A, TNV-D and TNV-DH (Fig. 3B) also. This finding further supports the existence of a 5′–3′ loop–loop ‘kissing’ interaction in TNV sgRNA2.
Taken together, these data indicate that functional elements for TNV sgRNA2 cap-independent translation are located in the upper part SL-II (based on homology to BYDV) and in the middle or lower part of SL-II (our mutational analysis) of the trailer, and that one of these elements interacts with a conserved stem–loop structure in the leader. These data further support the notion by Guo et al. (16) that there may be parallels in the folding of TNV sgRNA2 and BYDV functional elements and, therefore, support the hypothesis that these two viral RNAs may have related translation strategies that are promoted by related structures.
TNV sgRNA2 5′ elements stimulate translation synergistically with BYDV 3′ elements
To further characterize a putative relationship in the mechanisms of translation of TNV sgRNA2 and BYDV, we made use of the observations that both in TNV sgRNA2 (this study) and in BYDV (14), the 5′ and 3′ elements stimulate translation synergistically. If these elements use similar translation mechanisms, the TNV sgRNA2 leader may stimulate translation synergistically with the BYDV trailer, and vice versa. To investigate whether this is the case, we generated chimeric luciferase RNAs with different combinations of TNV sgRNA2, BYDV and vector-derived 5′ and 3′ elements (Fig. 4). These RNAs were translated in wheat germ extract, and the TEs were determined.
Figure 4 shows that TNV sgRNA2 5′ and 3′ elements (TNV-TNV) together stimulate the TE ∼10-fold more than the sum of the individual contributions (TNV-vector plus vector-TNV), confirming that, also for the luciferase open-reading frame, the TNV sgRNA2 5′ and 3′ elements stimulate translation synergistically. The BYDV 5′ and 3′ elements (BYDV-BYDV) also promote the TE ∼10 times more than the sum of the stimulation by the individual elements (BYDV-vector plus vector-BYDV), confirming a synergistic function in our test system, as expected from the observations by Wang and Miller (14).
When the BYDV 5′ and the TNV sgRNA2 3′ elements were placed on the same RNA (BYDV-TNV), the TE was stimulated ∼2-fold more than the sum of the stimulation by the individual elements (BYDV-vector plus vector-TNV), which appeared not significant in repetitions of this experiment (data not shown). When the TNV sgRNA2 5′ and BYDV 3′ elements were present on the same RNA (TNV-BYDV), the situation was different. Namely, the TE was reproducibly stimulated ∼3-fold more than the contribution of the individual elements (TNV-vector plus vector-BYDV). This shows that the TNV sgRNA2 5′ sequences and the BYDV 3′ sequences stimulate translation synergistically, implying that 5′ elements of TNV sgRNA2 interact functionally with 3′ elements of BYDV.
Taken together, the homology at the primary and secondary structure level, as well as the synergism between TNV sgRNA2 5′ elements and BYDV 3′ elements, strongly suggest that TNV sgRNA2 and BYDV use related mechanisms of translation. This would imply that two viral RNAs, that have evolved separately, are not only conserved in the use for translation of two elements that are separated in space, but also in the way in which these two elements functionally interact.
DISCUSSION
Previous studies (4) indicated that the sgRNA2 of TNV-A is a highly efficient messenger for coat protein synthesis. Here, we further investigated the mechanism of TNV sgRNA2 translation. We found that TNV sgRNA2 sequences mediate cap-independent translation in vitro and in vivo, supporting that in infected plant cells TNV sgRNA2 is indeed a messenger for coat protein synthesis, and that the apparent efficient translation of TNV sgRNA2 occurs via a cap-independent mechanism. The requirements for TNV sgRNA2 translation resemble those of the uncapped RNA of the plant virus BYDV at several points. First, both the leader and an element in the trailer stimulate translation synergistically. Secondly, the translation-stimulatory sequences in the TNV sgRNA2 trailer comprise a phylogenetically conserved stem–loop structure that has similarities to the 3′ translation element of BYDV. Thirdly, there is a possible requirement for a 5 nt loop–loop interaction between leader and trailer (16). Remarkably, this loop–loop ‘kissing’ interaction is sufficient for translation stimulation in the BYDV RNA (16). Importantly, we observed that TNV sgRNA2 leader stimulates in vitro translation synergistically with the BYDV trailer, pointing towards a related translation mechanism of TNV sgRNA2 and BYDV that is conserved in evolution.
The data described here not only give insight in the translation mechanism of TNV sgRNA2, but also have implications for how translation-stimulatory sequences of plant viral RNAs evolve and interact. Characteristic for TNV sgRNA2 and BYDV is that they have two separated structural elements (within 5′ and 3′ UTR) that interact to form one functional unit (i.e. translation-stimulating unit). In cases where one functional unit is separated in two structural elements, one may expect two driving forces (or constraints) for evolution. First, the two structural elements evolve for an optimal functional interaction. Here, one element can specifically adapt to the other (in the most simple scenario, nucleotides involved in pairing covary so that they maintain complementarity). If this was the sole driving force for evolution, structural elements of one functional unit would not be able to cooperate with structural elements of another functional unit that evolved separately. Secondly, each of the two structural elements may evolve as a separate functional unit that retains its own characteristics required for an efficient functional interaction. Such a constraint would predict that structural elements of a functional unit are able to cooperate with structural elements of another functional unit that evolved independently.
For evolution of the translation-stimulatory sequences of TNV sgRNA2 and BYDV, both driving forces are likely to be true. Obviously, the two functional elements in each viral RNA (BYDV or TNV sgRNA2) have evolved together to maintain the maximal synergy, and the highest translation stimulation is thus obtained when two elements from the same virus are present in the RNA (∼10-fold more than the contributions of the individual elements). On the other hand, structural elements from the two different viruses can also communicate. The observation that TNV sgRNA2 leader does stimulate translation synergistically with BYDV trailer (∼3-fold more than the contributions of the individual elements) indicates that the evolution of the two structural elements is somehow restricted, suggesting a driving force to keep some of the specifics of the functional interaction unchanged.
The question then rises what are the structural features that determine the interaction between 5′ and 3′ elements and are conserved between BYDV and TNV sgRNA2. One of the main prerequisites for efficient interaction may be complementarity between loop sequences of the two structural elements. Evolution of sequences involved in loop–loop interactions was analyzed in great detail in the case of antisense primers involved in plasmid copy number control (24). In such antisense RNA-regulated systems an antisense RNA is encoded by the same DNA region that determines its target. For translation of the BYDV RNA, 5 nt of complementarity between the loop of stem–loop Z and the leader are necessary and sufficient for synergism, whereas 4 nt of complementarity are not sufficient for synergism (16). When we looked at the extent of complementarity between BYDV 5′ sequences and TNV sgRNA2 3′ sequences, that do not stimulate translation synergistically, we found that there are 4 nt of complementarity, interrupted by a mismatch, in the region that contains the essential complementarity in BYDV (Fig. 5). In contrast, TNV sgRNA2 5′ sequences and BYDV 3′ sequences, that do stimulate translation synergistically, have 6 nt of complementarity, interrupted by a bulge and a mismatch (Fig. 5). This indicates that, in line with what was observed for BYDV 5′ and 3′ elements (16), the extent of complementarity may play a role in the synergistic function between TNV sgRNA2 and BYDV 5′ and 3′ elements. Importantly, the divergence in sequences between BYDV and TNV shows evolution of this base pairing capacity. However, the retained base pairing capacity between TNV sgRNA2 leader and BYDV trailer suggests that this evolution is somehow restricted, bearing in mind that TNV and BYDV are not closely related viruses.
Figure 5.
Complementarity between loop sequences from stem–loop Z and loop sequences of BYDV and TNV sgRNA2 leader. Circles indicate the complementary nucleotides, and the dashed lines indicate the putative interactions. Numbers indicate the nucleotide positions in the genomic RNA for BYDV (29) and in sgRNA2 for TNV-A (4).
One of the most likely reasons for such a constraint is a requirement for some specific conformations in loops that establish efficient loop–loop interactions (reviewed in 13,25,26). Such conformations expose nucleotides involved in the initial binding steps and usually conform to specific sequence motifs. For instance, in the majority of the naturally occurring loop–loop contacts determining antisense RNA binding, one of the loops contains a so-called U-turn motif (YUNR, where Y = pyrimidine, R = purine, N = any nucleotide), which was shown to facilitate fast RNA/RNA interaction (27). Since many of these systems are unrelated, the conservation of U-turn sequences is the result of convergent evolution due to the structural constraint (25). Examples of other loop–loop interactions also show a restricted number of sequences defining specific motifs. For instance, in RNA dimerization loops of various HIV-1 isolates only two out of 64 potential self-complementary hexanucleotides are preferred (26).
In the case of TNV sgRNA2 and BYDV RNA, the sequences UUCA and CUGA, located at the 5′ sides of the 5′ UTR loops, are probable candidates for the U-turn conformation (Figs 3B and 5). Some other nucleotides could also be important for the loop conformation, for instance CA dinucleotides at the 3′ sides of the leader loops in both RNAs. Apparently, such a similarity determines a pairing of heterogeneous loops that explains the observed synergy of TNV and BYDV structural elements. Indirect evidence for the importance of specific loop conformations in the cooperation between 5′- and 3′-terminal translational enhancers in necroviruses can be derived from mutagenesis experiments on STNV RNA (12). In STNV constructs, some compensatory mutations did not restore the cooperative interaction, probably because of disrupting functional loop structures. This is also consistent with the observation that sequence complementarity is not sufficient for maximal BYDV translation (16). Similar functional deficiencies in compensatory mutants lacking specific motifs are observed in antisense RNA systems and HIV dimerization (26,27).
The apparent restricted evolution of base pairing capacity between leader and trailer may also be determined by a requirement for interaction with host factors. A candidate host factor for mediating a conserved cap-independent translation mechanism is the translation initiation factor eIF4F, as the translation enhancer sequences of both BYDV and STNV appear to require a direct or indirect interaction with eIF4F for their functionality. Namely, the amount of eIF4F required for translation is reduced by the 3′ translation element of both BYDV (15) and STNV (13). Secondly, eIF4F reverses the in trans inhibition by the BYDV 3′ translation element (15). Thirdly, functionality of the STNV translational enhancer domain correlates with affinity for two proteins that arguably correspond to eIF4E and eIFiso4E, which are subunits of eIF4F and eIFiso4F, respectively (28). This apparent conservation in host factor requirements between divergent plant viral RNAs may be a cause for limited evolution of the involved RNA elements.
Based on the above observations and interpretations, we would like to summarize our view of how these viral RNAs adopt a translation promoting configuration as follows. The underlying principle is that the functional, translation promoting ‘unit’ is composed of two structural elements, one in the 5′ UTR and one in the 3′ UTR. Functionality of these structural elements requires not only folding into matching and conserved secondary and tertiary structure, but, in addition, kissing of nucleotides present in the loops of the two elements as well as binding to host factors, possibly eIF4F. The requirements of specific structures necessary for the efficient RNA/RNA interaction and/or binding to the host factor leaves little room for evolution. As a consequence, two structural elements derived from different RNAs can functionally interact, consistent with what was observed in this study for the TNV sgRNA2 leader and the BYDV trailer.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank André Boorsma, Ronnie Duinkerken and Véronique Gosselé for their contributions to the experiments. Allen Miller is thanked for the gift of the BYDV cDNA construct. Part of this work was sponsored with a post-doctoral fellowship to F.M. by the Institute for the Promotion of Innovation by Science and Technology in Flanders (IWT-Flanders).
NOTE ADDED IN PROOF
During the reviewing process of this manuscript, a paper by R. Shen and W. A. Miller was accepted for publication in the Journal of Virology, which provides data that further support conservation of structures of plant viral RNAs that promote synergistic stimulation of cap-independent translation. They show that for tobacco necrosis virus strain D (TNV-D), the 3′ translation-stimulatory sequences comprise the conserved stem–loop (including stem–loops X and Z). The TNV-D 3′ sequences stimulate translation together with the BYDV 5′ sequences, which, similar to our observations, correlates with the ability of base pairing between the loop of stem–loop Z and 5′ loop sequences.
REFERENCES
- 1.Meulewaeter F., Seurinck,J. and Van Emmelo,J. (1990) Genome structure of tobacco necrosis virus strain A. Virology, 177, 699–709. [DOI] [PubMed] [Google Scholar]
- 2.Condit C. and Fraenkel-Conrat,H. (1979) Isolation of replicative forms of 3′ terminal subgenomic RNAs of tobacco necrosis virus. Virology, 97, 122–130. [DOI] [PubMed] [Google Scholar]
- 3.Lesnaw J.A. and Reichmann,M.E. (1970) Identity of the 5′-terminal RNA nucleotide sequence of the satellite tobacco necrosis virus and its helper virus: possible role of the 5′-terminus in the recognition by virus-specific RNA replicase. Proc. Natl Acad. Sci. USA, 66, 140–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meulewaeter F., Cornelissen,M. and Van Emmelo,J. (1992) Subgenomic RNAs mediate expression of cistrons located internally on the genomic RNA of tobacco necrosis virus strain A. J. Virol., 66, 6419–6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones I.M. and Reichmann,M.E. (1973) The proteins synthesized in tobacco leaves infected with tobacco necrosis virus and satellite tobacco necrosis virus. Virology, 52, 49–56. [DOI] [PubMed] [Google Scholar]
- 6.Salvato M.S. and Fraenkel-Conrat,H. (1977) Translation of tobacco necrosis virus and its satellite in a cell-free wheat germ system. Proc. Natl Acad. Sci. USA, 74, 2288–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wells S.E., Hillner,P.E., Vale,R.D. and Sachs,A.B. (1998) Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell, 2, 135–140. [DOI] [PubMed] [Google Scholar]
- 8.Mazumder B., Seshadri,V. and Fox,P.L. (2003) Translational control by the 3′-UTR: the ends specify the means. Trends Biochem. Sci., 28, 91–98. [DOI] [PubMed] [Google Scholar]
- 9.Gallie D.R. (2002) Protein–protein interactions required during translation. Plant Mol. Biol., 50, 949–970. [DOI] [PubMed] [Google Scholar]
- 10.Wilkie G.S., Dickson,K.S. and Gray,N.K. (2003) Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci., 28, 182–188. [DOI] [PubMed] [Google Scholar]
- 11.Danthinne X., Seurinck,J., Meulewaeter,F., Van Montagu,M. and Cornelissen,M. (1993) The 3′ untranslated region of satellite tobacco necrosis virus RNA stimulates translation in vitro. Mol. Cell. Biol., 13, 3340–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meulewaeter F., Danthinne,X., Van Montagu,M. and Cornelissen,M. (1998) 5′- and 3′-sequences of satellite tobacco necrosis virus RNA promoting translation in tobacco [published erratum appears in Plant J. (1998), 15, 153–154]. Plant J., 14, 169–176. [DOI] [PubMed] [Google Scholar]
- 13.Timmer R.T., Benkowski,L.A., Schodin,D., Lax,S., Metz,A.M., Ravel,J.M. and Browning,K.S. (1993) The 5′ and 3′ untranslated regions of satellite tobacco necrosis virus RNA affect translational efficiency and dependence on a 5′ cap structure. J. Biol. Chem., 268, 9504–9510. [PubMed] [Google Scholar]
- 14.Wang S. and Miller,W.A. (1995) A sequence located 4.5 to 5 kilobases from the 5′ end of the barley yellow dwarf virus (PAV) genome strongly stimulates translation of uncapped mRNA. J. Biol. Chem., 270, 13446–13452. [DOI] [PubMed] [Google Scholar]
- 15.Wang S., Browning,K.S. and Miller,W.A. (1997) A viral sequence in the 3′-untranslated region mimics a 5′ cap in facilitating translation of uncapped mRNA. EMBO J., 16, 4107–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo L., Allen,E. and Miller,W.A. (2001) Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol. Cell, 7, 1103–1109. [DOI] [PubMed] [Google Scholar]
- 17.Meulewaeter F., Van Montagu,M. and Cornelissen,M. (1998) Features of the autonomous function of the translational enhancer domain of satellite tobacco necrosis virus. RNA, 4, 1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gultyaev A.P., van Batenburg,F.H.D. and Pleij,C.W.A. (1995) The computer simulation of RNA folding pathways using a genetic algorithm. J. Mol. Biol., 250, 37–51. [DOI] [PubMed] [Google Scholar]
- 19.Guo L., Allen,E. and Miller,W.A. (2000) Structure and function of a cap-independent translation element that functions in either the 3′ or the 5′ untranslated region. RNA, 6, 1808–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coutts R.H., Rigden,J.E., Slabas,A.R., Lomonossoff,G.P. and Wise,P.J. (1991) The complete nucleotide sequence of tobacco necrosis virus strain D. J. Gen. Virol., 72, 1521–1529. [DOI] [PubMed] [Google Scholar]
- 21.Molnar A., Havelda,Z., Dalmay,T., Szutorisz,H. and Burgyan,J. (1997) Complete nucleotide sequence of tobacco necrosis virus strain DH and genes required for RNA replication and virus movement. J. Gen. Virol., 78, 1235–1239. [DOI] [PubMed] [Google Scholar]
- 22.Lot H., Rubino,L., Delecolle,B., Jacquemond,M., Turturo,C. and Russo,M. (1996) Characterization, nucleotide sequence and genome organization of leek white stripe virus, a putative new species of the genus Necrovirus. Arch. Virol., 141, 2375–2386. [DOI] [PubMed] [Google Scholar]
- 23.Grieco F., Savino,V. and Martelli,G.P. (1996) Nucleotide sequence of the genome of a citrus isolate of olive latent virus 1. Arch. Virol., 141, 825–838. [DOI] [PubMed] [Google Scholar]
- 24.Tomizawa J.-I. (1993) Evolution of functional structures of RNA. In Gesteland,R.F. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 419–445. [Google Scholar]
- 25.Franch T. and Gerdes,K. (2000) U-turns and regulatory RNAs. Curr. Opin. Microbiol., 3, 159–164. [DOI] [PubMed] [Google Scholar]
- 26.Brunel C., Marquet,R., Romby,P. and Ehresmann,C. (2002) RNA loop–loop interactions as dynamic functional motifs. Biochimie, 84, 925–944. [DOI] [PubMed] [Google Scholar]
- 27.Franch T., Petersen,M., Wagner,E.G.H., Jacobsen,J.P. and Gerdes,K. (1999) Antisense RNA regulation in prokaryotes: rapid RNA/RNA interaction facilitated by a general U-turn loop structure. J. Mol. Biol., 294, 1115–1125. [DOI] [PubMed] [Google Scholar]
- 28.van Lipzig R., Van Montagu,M., Cornelissen,M. and Meulewaeter,F. (2001) Functionality of the STNV translational enhancer domain correlates with affinity for two wheat germ factors. Nucleic Acids Res., 29, 1080–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller W.A., Waterhouse,P.M. and Gerlach,W.L. (1988) Sequence and organization of barley yellow dwarf virus genomic RNA. Nucleic Acids Res., 16, 6097–6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herschlag D. and Johnson,F.B. (1993) Synergism in transcriptional activation: a kinetic view. Genes Dev., 7, 173–179. [DOI] [PubMed] [Google Scholar]