Abstract
We introduced a series of Pro substitutions within and near the α4 helix, a part of the breakage/rejoining region, in human DNA topoisomerase IIα, and analyzed if this region is involved in determination of anti-cancer drug sensitivity in a temperature- sensitive yeast strain (top2-4 allele). Among the 19 mutants generated, H759P and N770P showed resistance to etoposide and doxorubicin at the non-permissive temperature, where cell growth depends on activity of the human enzyme. For these residues, mutants with an Ala substitution were further created, in which H759A also showed resistance to etoposide. H759P, H759A and N770P were expressed, purified and subjected to in vitro measurement of drug sensitivity. They generated lower amounts of the etoposide-induced cleavable complexes, and were also found to have lower decatenation activity than the wild-type. In the crystal structure, the yeast equivalent of His759 is found in the vicinity of the Arg713, a putative anchoring residue of the 3′-side of cleaved DNA strands. These results suggest that His759 and the other α4 helix residues are involved in the enzymatic activity and drug sensitivity of human DNA topoisomerase IIα, via interaction with cleaved DNA.
INTRODUCTION
DNA molecules are compacted in nuclei, forming supercoiled and catenated structures. DNA topoisomerases catalyze a reaction that induces a transient cut and rejoining of DNA, and function in maintaining the chromosomal structure during DNA replication, transcription and other types of DNA transactions. The type I enzyme cuts a single DNA strand, whereas the type II enzyme cuts both strands and makes a gate, allowing another DNA segment to pass through (1). Among species, type II DNA topoisomerases share a highly conserved domain/subunit structure that involves the breakage/rejoining domain (2–4), which is presumably important for the enzyme to interact with DNA (2,4,5).
The breakage/rejoining domain is also the major target for nalidixic acid and quinolones in prokaryotes, and anticancer drugs such as etoposide and doxorubicin in eukaryotes (6–18). These drugs bind, stabilize and leave the reaction intermediate of the cleavable complex (7,19), which may inhibit various DNA transactions and ultimately lead to double-stranded breaks of chromosomal DNA.
In the Escherichia coli type II DNA topoisomerase of gyrase, quinolone resistance has often been mapped to Ser-83, Asp-87 or other single amino acids in the α4 helix of the breakage/rejoining domain (6,9,10). In the corresponding α4 helix in Saccharomyces cerevisiae, Ser740→Trp, Thr744→Pro and Gly747→Glu mutants are associated with resistance/hypersensitivity to CP-115,953, etoposide, mAMSA and doxorubicin (14,17,18). Mutations in this helix are associated with both alteration in the drug sensitivity and/or change in the DNA cleavage pattern (20,21). To our knowledge, despite the obvious implications for anticancer treatment, the functions of the α4 helix in the human enzyme have not been elucidated yet. To address this question, we have scanned the target region by introducing Pro substitutions, and subsequently compared the mutational effect with Ala mutants.
Pro has a ring structure that makes a covalent side-chain bridge between the α carbon and the nitrogen atom, resulting in the lowest free energy among the 20 amino acids (22). In an α helix, usually amino acids form intrahelical hydrogen bonds between the amino and carboxyl groups. These do not form at the Pro residue, resulting in the kink formation when located at or near the center of the helix (23,24). On the other hand, located at ‘turn’ regions, Pro stabilizes the local structure and may increase the heat stability of an enzyme (25).
Interestingly, Pro mutants have often been found with full activity and interesting characteristics after screening from libraries that contain randomized amino acids (12,26,27). Furthermore, previous studies have found that Pro mutants in DNA topoisomerase II alter sensitivity to various types of drugs (12,18,21,28).
In the present study, we identified residues that appeared to confer drug resistance on human DNA topoisomerase IIα (hTOP2α). Mutant proteins were purified and characterized to allow discussion from the viewpoint of structure and function.
MATERIALS AND METHODS
Yeast strains, oligonucleotides and plasmid construction
Yeast strains SD1-4 (MATa, ade1, ade2, ura3-52, top2-1) and JN394t2-4 (MATa, ura3-52, leu2, trp1, his7, ade1-2 ISE2, rad52::LEU2 top2-4) were as described previously (12). DNA oligomers were synthesized and purified by Amersham Pharmacia Biotech or NK Products (Osaka, Japan). Wild-type plasmid constructions (p’YES-PGAL1-hTOP2 and YCp-PADH1-hTOP2) and non-functional dummy derivatives (Dummy Vector and YCp-PADH1-dum) were described previously (29). DNA topoisomerase IIα constructs that complemented the temperature sensitivity of JN394t2-4 also complemented SD1-4. Etoposide and doxorubicin were purchased from Sigma (St. Louis, MO).
Pro scanning
PCR mutagenesis was carried out using 19 primers, 5′-CA GTG GCT GAA ATG TCT TCT TAT CAT CAT GGT GAG ATG TCA CTA ATG ATG ACC ATT ATC AAT TTG GCT CAG AAT TTT GTG GGT AGC A, in which each of the underlined codons was replaced with a Pro codon of CCT. The reverse primer, 5′-C ATG TAG CCT GGT ACC AAA C, is complementary to the KpnI site of human DNA topoisomerase IIα. Using LLpYES-PGAL1-hTOP2 as a template, 30 cycles of PCR were carried out (94°C for 30 s, 54°C for 30 s and 72°C for 30 s) using the proofreading-competent Pyrobest DNA polymerase (TaKaRa, Kyoto, Japan). A 138-bp PCR product was isolated by 1.2% agarose gel electrophoresis and purified with a QIAEXII gel extraction kit (Qiagen, Valencia, CA). In an independent tube, a 143-bp fragment was synthesized using another set of primers, 5′-CT GAT AAC GAG AGA TCT ATC C, which contains a BglII site, and 5′-AGA AGA CAT TTC AGC CAC TG. The two PCR fragments were mixed and further amplified using the above-mentioned primers containing the restriction sites. The 260mer fragment was digested by BglII and KpnI, and ligated with the large BglII–KpnI fragment of the Dummy Vector. In some experiments, His759 or Asn770 were replaced with Ala using the same procedure.
Genetic selection of functional mutants
Complementation assays were performed as described previously (12). Plates were cultured at 35°C for 10 days and functional mutants were detected in three independent experiments.
Screening of drug-resistant mutants
Wild-type and functional mutants in multicopy pYES vector (p’YES-PGAL1-hTOP2 and its mutants) were used for transformation of JN394t2-4 cells. Screening for drug-resistant mutants was carried out as described previously (28). Briefly, transformed JN394t2-4 cells were streaked on a SCgal-URA plate (6.7 g/l yeast nitrogen base without amino acids, 5 g/l casamino acids, 20 g/l galactose, 20 mg/l adenine sulfate and 20 mg/l tryptophan) containing 100 µg/ml etoposide or 2 µg/ml doxorubicin and assessed for colony formation.
Complementation efficiency
The smaller NotI–KpnI fragment of YCp-PADH1-dum was replaced with the corresponding fragment from each p’YES-PGAL-hTOP2 mutant. JN394t2-4 with wild-type or mutant DNA topoisomerase IIα was grown in 10 ml of YPD (10 g/l yeast extracts, 20 g/l pepton, 20 g/l glucose) liquid medium at 27°C for 2 days. Aliquots were plated onto SCglu-URA plates (same as SCgal-URA except that 20 g/l glucose is used for sugar source) and incubated at 27 or 35°C (permissive or non-permissive temperature) for 10 days. Complementation efficiency at 35°C was determined as the colony number relative to that at 27°C taken as 100%.
Drug sensitivity
Etoposide or doxorubicin sensitivity was determined using the JN394t2-4 carrying either wild-type or mutant YCp-PADH1-hTOP2. Cells were plated on SCglu-URA with 0, 4, 8, 16, 32, 64, 128 or 256 µg/ml etoposide, or 0, 1, 2, 4, 8 or 16 µg/ml of doxorubicin or solvent. Visible colonies were counted after they were cultured at 35°C for 10 days. Drug sensitivity was determined as colony number relative to that of the control plate taken as 100%.
Enzyme purification
Expression and purification of wild-type and mutant hTOP2α were performed as described previously (29). Briefly, histidine-tagged wild-type hTOP2α and H759P, N770P and H759A mutants were expressed using the BAC-TO-BAC HT Baculovirus Expression System (Life Technologies, MD). Proteins of post-infected cells were loaded onto an In-resin column (His-Bind Resin, Novagen, Madison, WI), and bound protein was eluted with 1 M imidazole and 500 µl fractions were collected. Peak fractions were dialyzed and the protein concentration was determined with Bradford reagent (Bio-Rad, Richmond, CA). The enzyme was stored at –80°C for at least 3 months without loss of activity.
Decatenation activity assay
Decatenation activity was measured using kinetoplast DNA (kDNA, TopoGEN, Columbus, OH) as described previously (29); the 20 µl decatenation reaction mixture contained 100 ng kDNA in 50 mM Tris–HCl (pH 8.0), 120 mM KCl, 10 mM MgCl2, 0.5 mM DTT, 30 µg/ml BSA and 1 mM ATP. Enzyme was added and followed by incubation for 15 min at 37°C. The reaction was terminated with 2 µl loading dye (1% SDS, 50% glycerol, 0.05% Bromophenol Blue), and products were separated by electrophoresis in a 1% agarose gel in the presence of 1.0 µg/ml ethidium bromide. Gels were destained in water for 45 min and the reaction products were visualized under UV light. One unit of decatenation activity was defined as the amount of enzyme that decatenates 100 ng kDNA under these conditions.
DNA cleavage assay
Amounts of cleaved DNA were measured using plasmid DNA pRYG (TopoGEN). The 20 µl cleavage assay reaction mixture contained 480 ng of pRYG in 50 mM Tris–HCl (pH 8.0), 120 mM KCl, 10 mM MgCl2, 0.5 mM DTT, 30 µg/ml BSA and 1 mM ATP. Reactions were carried out in the presence of 0, 64, 128 or 256 µg/ml etoposide. A 200 ng aliquot of wild-type or mutant hTOP2α was added. After incubation at 37°C for 15 min, reaction was terminated by 0.5% SDS, followed by further incubation with 80 µg/ml proteinase K for 15 min, and mixing vigorously with 25 µl of phenol/chloroform/isoamyl alcohol (25:24:1, Nacalai Tesque, Kyoto, Japan). The aqueous phase was recovered by centrifugation and used for electrophoresis as described above. Amounts of linear DNA product were quantified using a Densitograph and Lane analyzer (ATTO, Tokyo, Japan).
RESULTS
Screening for the drug-resistant mutants in human DNA topoisomerase IIα
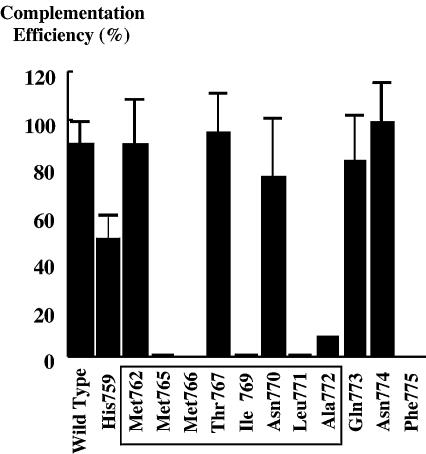
In human DNA topoisomerase IIα, amino acids between 760 and 772 correspond to the α4 helix in S.cerevisiae DNA topoisomerase II (3). We introduced Pro substitutions into the putative α helix and nearby residues (AA757–775). Human DNA topoisomerase IIα mutants were cloned into a multicopy vector pYES (29) and used for genetic complementation of a temperature-sensitive strain SD1-4 (12,30,31). In this screening, 12 mutants functionally complemented the strain at the non-permissive temperature. These mutants were further used for transformation of the JN394t2-4 drug permeable yeast strain. Each transformant was subsequently streaked on plates containing 100 µg/ml etoposide or 2 µg/ml doxorubicin (12). Under conditions where the wild-type did not form colonies, nine mutants (H795P, M762P, M765P, M766P, T767P, N770P, L771P, A772P and F775P) survived and grew. However, in the multicopy pYES vector, these mutants showed very poor complementation efficiency and did not yield reliable results in the subsequent experiments. Therefore, the 12 mutant cDNA sequences were transferred into the YCp-PADH1 vector (29) (see Materials and Methods). This vector contains the centromere sequence, and is equally distributed to daughter cells. If the poor complementation efficiency was caused by unequal plasmid distribution and high frequency of plasmid loss, the new construct may increase the complementation efficiency. In accordance with our expectation, six mutants complemented the JN394t2-4 strain with reasonably high efficiency (Fig. 1), and were able to be studied further.
Figure 1.
Complementation efficiency of human TOP2α expressed from YCp-PADH1. JN394t2-4 cells were transformed with wild-type or 12 Pro mutants in the centromeric YCp-PADH1. Solid bars show the relative colony numbers at 35/27°C (permissve/non-permissive temperature), with mutated residues indicated. Complementation efficiency of M766P varied in experiments (10–60%). For mutants with high complementation efficiency, the data represent the averages of 3–7 experiments and are associated with S.D. ranges. Amino acids in the putative α4 helix are boxed.
Drug resistance against etoposide and doxorubicin
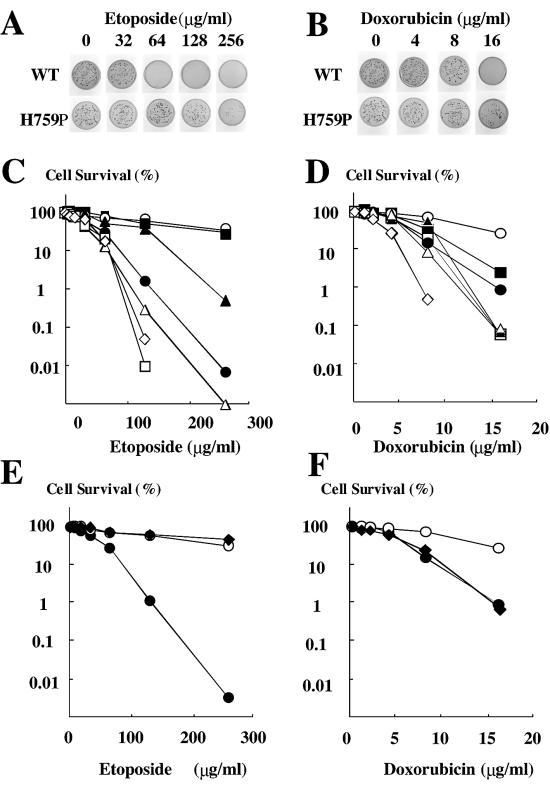
Using these six active mutants, drug resistance was evaluated under the various concentrations of etoposide or doxorubicin (Fig. 2). With etoposide, H759P and N770P survived, with 10-fold more colonies than the wild-type (Fig. 2A and C). The 50% inhibitory concentrations (IC50) were determined to be 40, 180 and 150 µg/ml for wild-type, H759P and N770P, respectively (Table 1). On plates containing doxorubicin, H759P mutant formed 10-fold more colonies than the wild-type, and N770P conferred marginal resistance (Fig. 2B and D). The IC50 values for wild-type, H759P and N770P were 5, 12 and 7 µg/ml, respectively. In this assay, N774P seemed to be hypersensitive to doxorubicin (Fig. 2D).
Figure 2.
Measurement of sensitivity to etoposide. JN394t2-4 cells with either wild-type or mutant TOP2 were spread on plates containing stepwise concentrations of etoposide (A, C and E) or doxorubicin (B, D and F) and incubated at 35°C. (A and B) Examples of colony growth (wild-type and H759P). (C and D) Colony formation of the Pro mutants was plotted as a function of drug concentration. Filled circles, open circles, filled triangles, open triangles, filled squares, open squares and open diamonds are for wild-type, H759P, M762P, T767P, N770P, Q773P and N774P, respectively. (E and F) Colony formation with wild-type (filled circles), H759A (filled diamonds) and H759P (open circles), plotted as a function of drug concentration.
Table 1. 50% inhibitory concentrations (IC50) for wild type and mutant derivatives.
| Etoposide (µg/ml) | Doxorubicin (µg/ml) | |
|---|---|---|
| WT | 40 | 5 |
| H759P | 180 | 12 |
| M762P | 100 | 8 |
| T767P | 32 | 5 |
| N770P | 150 | 7 |
| Q773P | 28 | 5 |
| N774P | 38 | 3 |
| H759A | 230 | 5 |
H759A resistance to etoposide
Pro substitution may affect enzymes by various mechanisms. To understand what caused the mutational effect, we created Ala mutants at His-759 and Asn-770, and quantified complementation efficiency and drug sensitivity (Fig. 2E and F and Table 1). In this assay, N770A did not complement the temperature-sensitive strain, although H759A was found to have 90% complementation efficiency and also to be resistant to etoposide (IC50; 230 µg/ml). Interestingly, H759A showed wild-type level sensitivity to doxorubicin (IC50 5 µg/ml) in contrast to that found with Pro substitution (Table 1).
In vitro drug sensitivity
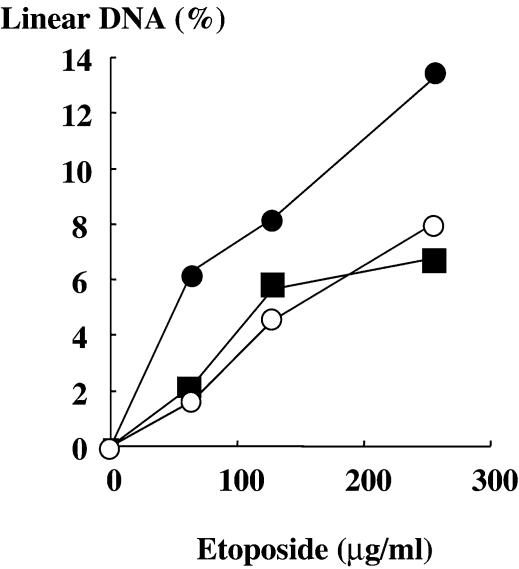
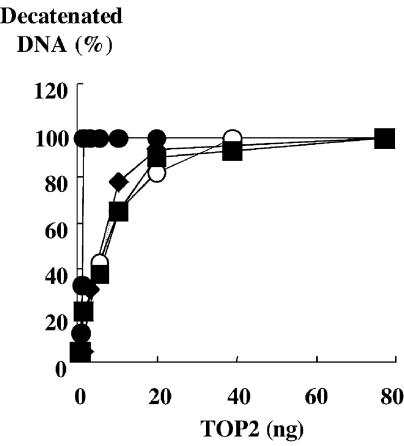
We expressed and purified histidine-tagged enzymes, and quantified the in vitro etoposide sensitivity using a DNA cleavage assay. Compared to the wild-type, H759P, H759A and N770P produced only half or one-third of the amount of cleaved DNA (Fig. 3), consistent with their drug resistance phenotype in yeast. We also quantified the activity of each mutant using a decatenation activity assay (Fig. 4). The results showed that H759P and N770P were responsible for the reduction in enzymatic activity.
Figure 3.
In vitro drug sensitivity of purified enzymes. Cleavable complex formation was quantitated and plotted relative to the total DNA. Filled circles, open circles and filled squares are for wild-type, H759P and N770P, respectively. In this assay, cleavable complex formation by H759A was under the detection limit, and was not plotted.
Figure 4.
In vitro activity of purified TOP2α. Activities of wild-type and mutants were measured at 37°C for 15 min. Decatenated product was quantified and plotted as a relative value to the total DNA. Filled circles, open circles, filled squares and filled diamonds are for wild-type, H759P, N770P and H759A, respectively.
DISCUSSION
In type II DNA topoisomerase, the amino acid sequence in the α4 helix of the breakage/rejoining region is one of the determinants of various drug resistance and hypersensitivity (Fig. 5). If one knows the specific relationship between amino acid identity and drug sensitivity, chemotherapeutic effects might be predicted prior to medication. From the pharmaceutical point of view, such information would also be useful to design new drug derivatives. To identify mutant alleles, one might isolate and analyze clinical samples from patients who show drug resistance. This strategy could directly indicate a relationship between drug effect and identity of specific amino acids. However, it is not as easy as the study of bacterial counterparts of gyrase, because in cancer chemotherapy, multiple drugs are usually used for single patients. In these cases, the effect of single drugs is hard to evaluate. Furthermore, clinical patients are often not appropriate for initial trials because they have various and complex factors that affect drug application and sensitivity.
Figure 5.
Relationship between mutations and drug sensitivity in the α4 helix. Protein sequences around the putative α4 helices of human, S.cerevisiae and E.coli type II topoisomerases (HsTop2α, ScTop2 and EcGyrA, respectively) are aligned (3). Amino acid numbers are also indicated. The α helices are shaded in boxes. Residues that confer drug sensitivity are in italics: (a) S740W, hypersensitive to etoposide and resistant to CP-115,953 (14); (b) T744P, hypersensitive to CP-115,953 (18); (c) G747E, resistant to doxorubicin (17); (d–h) G81C, G81D (9,10,34), D82A (10,34), S83W, S83L, S83A, A84P (9), D87Y, D87N, D87V, D87T, D87G and D87H (9,34); resistant to quinolones.
We have therefore adopted an alternative approach, in which a series of amino acid substitutions was introduced into the putative α4 helix and nearby regions of human DNA topoisomerase IIα. We chose Pro to substitute for the target residues, because mutations with interesting characteristics have often been reported for Pro substitutions (12,18,21,26–28). Using these mutants and a drug-permeable yeast strain, we identified new residues that confer etoposide and doxorubicin resistance.
Possible mechanisms of drug resistance
In DNA topoisomerase II, drug resistance is often associated with a reduction in enzymatic activity. With the His759 mutation, enzyme activity was found to be reduced (Fig. 4). However, yeast cells with H759P were resistant to doxorubicin whereas those with H759A were sensitive (Fig. 2). This result suggests that drug resistance is not a simple outcome of reduced activity, but that His759 could be directly involved in the formation of ternary complexes with drugs and substrate DNA.
Structural implications
In the crystal structure of S.cerevisiae DNA topoisomerase II, His736 (corresponding to His759 in human DNA topoisomerase II), is located at –1 upstream of the N-terminal residues of the α4 helix, and Gly747 (Asn770) is found within the helix. In this model, His736 is in the vicinity of Arg690 (Arg713, Fig. 6), which is the putative anchoring residue of the 3′-side of cleaved DNA strands (32,33). In our experiments, both Ala and Pro substitutions similarly affected the functions of human DNA topoisomerase IIα. Therefore, the mutation may be associated with loss of His function. It is possible that direct interaction of His759 and the 3′-terminus of the cleaved DNA plays a role in enzyme activity. Alternatively, His759 could be involved in the proper positioning of the essential Arg713. In either case, waiving the His759 charge might have decreased the DNA anchoring ability of Arg713.
Figure 6.
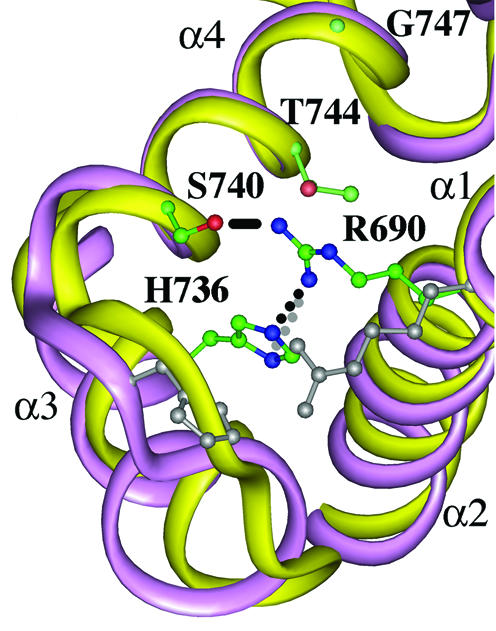
Structural model of S.cerevisiae DNA topoisomerase II. Yellow and purple ribbons represent a part of the breakage/rejoining domain of S.cerevisiae DNA topoisomerase II (α1–α4 helices, 2) and the corresponding domain of E.coli gyrase (4), respectively. S.cerevisiae Arg690 (corresponding to human Arg713), His736 (His759), Ser740, Thr744 and Gly747 (Asn770) are visualized with balls and sticks. Carbon, nitrogen and oxygen atoms are in green, blue and red, respectively. The distances between the guanidinium nitrogen of Arg690 and the δ1 nitrogen of His736, the guanidinium nitrogen of Arg690 and the ε2 nitrogen of His736, and the guanidinium nitrogen of Arg690 and the oxygen of Ser740 are 3.13 (gray dotted line), 3.16 (dotted line) and 3.78 Å (solid line), respectively. E.coli counterparts of Arg690 (Arg32) and His736 (His80) are also shown in gray. The drawing was made by using Insight II.
The possibility that His759 is involved in DNA anchoring and drug sensitivity was supported by mapping other amino acid residues that confer drug resistance in the model structure of S.cerevisiae DNA topoisomerase II (Fig. 6). With the Ser740 substitution with Trp, S.cerevisiae DNA topoisomerase II becomes hypersensitive to etoposide and resistant to CP115,953 (14). The Ser740 could make a hydrogen bond between the guanidinium group of Arg690 (Fig. 6), which would not be formed when the side chain is replaced by Trp. Two more mutations have been reported in the α4 helix of the S.cerevisiae DNA topoisomerase II (17,18). These residues, Thr744 and Gly747, are distant from Arg690, but face toward it (Fig. 6). In the presence of drugs, mutations could alter the interaction between the enzyme and 3′-nucleotide of the cleaved DNA terminus. Consistently, the S.cerevisiae Gly747 corresponds to Asn770 in the human enzyme, whose substitution resulted in etoposide resistance (Figs 2, 3 and Table 1).
We also superimposed the α1–α4 helices of E.coli gyrase A subunit on those of the S.cerevisiae topoisomerase II (Fig. 6). In this limited region, the two backbone structures seem to be well overlapped, and like S.cerevisiae counterparts, Arg32 and His80 of gyrase A interact with each other. On the other hand, the α3–α4 loops are located a little distant from each other, and in the primary structures, eukaryotic topoisomerase ‘lacks’ the Pro in the conserved His–Pro–His sequence of prokaryotic gyrase (Fig. 5) (3). Therefore, it could be feasible that the His80 is also involved in the catalytic activity and drug sensitivity in gyrase. In future, it may be interesting to study the possibility that this structural difference underlies the unique drug sensitivity spectra between bacterial gyrase and S.cerevisiae/human DNA topoisomerases II.
Acknowledgments
ACKNOWLEDGEMENTS
The authors are grateful to Ms Tazuko Tomita for technical assistance. This work is supported in a part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Wang J.C. (1985) DNA topoisomerases. Annu. Rev. Biochem., 54, 665–697. [DOI] [PubMed] [Google Scholar]
- 2.Berger J.M., Gamblin,S.J., Harrison,S.C. and Wang,J.C. (1996) Structure and mechanism of DNA topoisomerase II. Nature, 379, 225–232. [DOI] [PubMed] [Google Scholar]
- 3.Caron P.R. (1999) Appendix: Compendium of DNA topoisomerase sequence. In Bjornsti,M.-A. and Osheroff,N. (eds), Protocols in DNA Topology and Topoisomerases, Part I: DNA Topology and Enzymes. Methods in Molecular Biology, vol. 28, Humana Press Inc., Totowa, NJ, pp. 279–316. [Google Scholar]
- 4.Morais Cabral J.H., Jackson,A.P., Smith,C.V., Shikotra,N., Maxwell,A. and Liddington,R.C. (1997) Crystal structure of the breakage-reunion domain of DNA gyrase. Nature, 388, 903–906. [DOI] [PubMed] [Google Scholar]
- 5.Huang W.M. (1996) Bacterial diversity based on type II DNA topoisomerase genes. Annu. Rev. Genet., 30, 79–107. [DOI] [PubMed] [Google Scholar]
- 6.Maxwell A. (1992) The molecular basis of quinolone action. J. Antimicrob. Chemother., 30, 409–416. [DOI] [PubMed] [Google Scholar]
- 7.Froelich-Ammon S.J. and Osheroff,N. (1995) Topoisomerase poisons: harnessing the dark side of enzyme mechanism. J. Biol. Chem., 270, 21429–21432. [DOI] [PubMed] [Google Scholar]
- 8.Kato S. and Kikuchi,A. (1998) DNA topoisomerase: the key enzyme that regulates DNA super structure. Nagoya J. Med. Sci., 61, 11–26. [PubMed] [Google Scholar]
- 9.Everett M.J. and Piddock,L.J.V. (1998) Mechanisms of resistance to fluoroquinolones. In Kuhlman,J., Dalhoff,A. and Zeiler,H.-J. (eds), Handbook of Experimental Pharmacology, vol. 127, Springer, pp. 256–286. [Google Scholar]
- 10.Friedman S.M., Lu,T. and Drlica,K. (2001) Mutation in the DNA gyrase A gene of Escherichia coli that expands the quinolone resistance-determining region. Antimicrob. Agents Chemother., 45, 2378–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel S. and Fisher,L.M. (1993) Novel selection and genetic characterisation of an etoposide-resistant human leukaemic CCRF-CEM cell line. Br. J. Cancer, 67, 456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okada Y., Ito,Y., Kikuchi,A., Nimura,Y., Yoshida,S. and Suzuki,M. (2000) Assignment of functional amino acids around the active site of human DNA topoisomerase IIalpha. J. Biol. Chem., 275, 24630–24638. [DOI] [PubMed] [Google Scholar]
- 13.Mao Y., Yu,C., Hsieh,T.S., Nitiss,J.L., Liu,A.A., Wang,H. and Liu,L.F. (1999) Mutations of human topoisomerase II alpha affecting multidrug resistance and sensitivity. Biochemistry, 38, 10793–10800. [DOI] [PubMed] [Google Scholar]
- 14.Hsiung Y., Elsea,S.H., Osheroff,N. and Nitiss,J.L. (1995) A mutation in yeast TOP2 homologous to a quinolone-resistant mutation in bacteria. Mutation of the amino acid homologous to Ser83 of Escherichia coli gyrA alters sensitivity to eukaryotic topoisomerase inhibitors. J. Biol. Chem., 270, 20359–20364. [DOI] [PubMed] [Google Scholar]
- 15.Hsiung Y., Jannatipour,M., Rose,A., McMahon,J., Duncan,D. and Nitiss,J.L. (1996) Functional expression of human topoisomerase II alpha in yeast: mutations at amino acids 450 or 803 of topoisomerase II alpha result in enzymes that can confer resistance to anti-topoisomerase II agents. Cancer Res., 56, 91–99. [PubMed] [Google Scholar]
- 16.Strumberg D., Nitiss,J.L., Rose,A., Nicklaus,M.C. and Pommier,Y. (1999) Mutation of a conserved serine residue in a quinolone-resistant type II topoisomerase alters the enzyme–DNA and drug interactions. J. Biol. Chem., 274, 7292–7301. [DOI] [PubMed] [Google Scholar]
- 17.Patel S., Sprung,A.U., Keller,B.A., Heaton,V.J. and Fisher,L.M. (1997) Identification of yeast DNA topoisomerase II mutants resistant to the antitumor drug doxorubicin: implications for the mechanisms of doxorubicin action and cytotoxicity. Mol. Pharmacol., 52, 658–666. [DOI] [PubMed] [Google Scholar]
- 18.Dong J., Walker,J. and Nitiss,J.L. (2000) A mutation in yeast topoisomerase II that confers hypersensitivity to multiple classes of topoisomerase II poisons. J. Biol. Chem., 275, 7980–7987. [DOI] [PubMed] [Google Scholar]
- 19.Jensen P.B. and Sehested,M. (1997) DNA topoisomerase II rescue by catalytic inhibitors. Biochem. Pharmacol., 54, 755–759. [DOI] [PubMed] [Google Scholar]
- 20.Strumberg D., Nitiss,J.L., Dong,J., Kohn,K.W. and Pommier,Y. (1999) Molecular analysis of yeast and human type II topoisomerases. Enzyme–DNA and drug interactions. J. Biol. Chem., 274, 28246–28255. [DOI] [PubMed] [Google Scholar]
- 21.Strumberg D., Nitiss,J.L., Dong,J., Walker,J., Nicklaus,M.C., Kohn,K.W., Heddle,J.G., Maxwell,A., Seeber,S. and Pommier,Y. (2002) Importance of the fourth alpha-helix within the CAP homology domain of type II topoisomerase for DNA cleavage site recognition and quinolone action. Antimicrob. Agents Chemother., 46, 2735–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richardson J.S. and Richardson,D.C. (1988) Amino acid preferences for specific locations at the ends of alpha helices. Science, 240, 1648–1652. [DOI] [PubMed] [Google Scholar]
- 23.Visiers I., Braunheim,B.B. and Weinstein,H. (2000) Pro kink: a protocol for numerical evaluation of helix distortions by proline. Protein Eng., 13, 603–606. [DOI] [PubMed] [Google Scholar]
- 24.Kim M.K. and Kang,Y.K. (1999) Positional preference of proline in alpha-helices. Protein Sci., 8, 1492–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe K., Masuda,T., Ohashi,H., Mihara,H. and Suzuki,Y. (1994) Multiple proline substitutions cumulatively thermostabilize Bacillus cereus ATCC7064 oligo-1,6-glucosidase. Irrefragable proof supporting the proline rule. Eur. J. Biochem., 226, 277–283. [DOI] [PubMed] [Google Scholar]
- 26.Tosaka A., Ogasa,M., Yoshida,S. and Suzuki,M. (2001) O-helix mutant T664P of Thermus aquaticus DNA polymerase I. J. Biol. Chem., 276, 27562–27567. [DOI] [PubMed] [Google Scholar]
- 27.Ogawa M., Limsirichaikul,S., Niimi,A., Iwai,S., Yoshida,S. and Suzuki,M. (2003) Distinct function of conserved amino acids in the fingers of Saccharomyces cerevisiae DNA polymerase alpha. J. Biol. Chem., 278, 19071–19078. [DOI] [PubMed] [Google Scholar]
- 28.Okada Y., Tosaka,A., Nimura,Y., Kikuchi,A., Yoshida,S. and Suzuki,M. (2001) Atypical multidrug resistance may be associated with catalytically active mutants of human DNA topoisomerase II alpha. Gene, 272, 141–148. [DOI] [PubMed] [Google Scholar]
- 29.Suda N., Nakagawa,Y., Kikuchi,A., Sawada,M., Takami,Y., Funahashi,H., Nakao,A., Yoshida,S. and Suzuki,M. (2003) Function of the loop residue Thr792 in human DNA topoisomerase IIα. Biochem. Biophys. Res. Commun., 303, 46–51. [DOI] [PubMed] [Google Scholar]
- 30.Adachi N., Ikeda,H. and Kikuchi,A. (1994) Mutant isolation of mouse DNA topoisomerase II alpha in yeast. Nucleic Acids Res., 22, 4229–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wasserman R.A., Austin,C.A., Fisher,L.M. and Wang,J.C. (1993) Use of yeast in the study of anticancer drugs targeting DNA topoisomerases: expression of a functional recombinant human DNA topoisomerase II alpha in yeast. Cancer Res., 53, 3591–3596. [PubMed] [Google Scholar]
- 32.Liu Q. and Wang,J.C. (1998) Identification of active site residues in the ‘GyrA’ half of yeast DNA topoisomerase II. J. Biol. Chem., 273, 20252–20260. [DOI] [PubMed] [Google Scholar]
- 33.Wilstermann A.M. and Osheroff,N. (2001) Positioning the 3′-DNA terminus for topoisomerase II-mediated religation. J. Biol. Chem., 276, 17727–17731. [DOI] [PubMed] [Google Scholar]
- 34.Lu T., Zhao,X. and Drlica,K. (1999) Gatifloxacin activity against quinolone-resistant gyrase:allele-specific enhancement of bacteriostatic and bactericidal activities by the C-8-methoxy group. Antimicrob. Agents Chemother., 43, 2969–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]