Abstract
The cytosine base in DNA undergoes hydrolytic deamination at a considerable rate when UV radiation induces formation of a cyclobutane pyrimidine dimer (CPD) with an adjacent pyrimidine base. We have synthesized a phosphoramidite building block of a cis–syn cyclobutane thymine–uracil dimer (T[]U), which is the deaminated form of the CPD at a TC site, and incorporated it into oligodeoxyribonucleotides. The previously reported method for synthesis of the thymine dimer (T[]T) was applied, using partially protected thymidylyl-(3′–5′)-2′-deoxyuridine as the starting material, and after triplet- sensitized irradiation, the configuration of the base moiety in the major product was determined by NMR spectroscopy. Presence of the cis–syn cyclobutane dimer in the obtained oligonucleotides was confirmed by UV photoreversal and reaction with T4 endonuclease V. Using a 30mer containing T[]U, translesion synthesis by human DNA polymerase η was analyzed. There was no difference in the results between the templates containing T[]T and T[]U and pol η bypassed both lesions with the same efficiency, incorporating two adenylates. This enzyme showed fidelity to base pair formation, but this replication causes a C→T transition because the original sequence is TC.
INTRODUCTION
Among the four nucleobases in DNA, cytosine is the most susceptible to hydrolytic deamination. The deamination rate constants for single- and double-stranded DNA at 37°C are reportedly 1 × 10–10 and 7 × 10–13 s–1, respectively (1). When UV radiation causes cytosine to form a cyclobutane pyrimidine dimer (CPD) with an adjacent pyrimidine base, the reaction rate of this deamination is greatly accelerated, as demonstrated using dinucleoside monophosphates (2–4), because stabilization of the amino group by the aromatic resonance is lost due to saturation of the 5,6-double bond. The reported deamination rate constants observed for the cis–syn CPDs of d(TpC) and d(CpT) at 25°C (or room temperature) are 2.5 × 10–5 and 2.8 × 10–5 s–1, respectively (3,4). In DNA, various rate constants have been determined, with reported values of 3.9 × 10–5 and 1.2–1.8 × 10–6 s–1 at 37°C in vitro and 1.5 × 10–4 s–1 at 42°C in Escherichia coli cells (5–7). Besides the first order kinetics, unusual step-function kinetics, in which the deamination occurs rapidly after a latent period, have also been reported (8). Methods for detecting deaminated cytosine within CPDs at specific sequences have been developed, which revealed a sequence context dependence (9).
The hydrolytic deamination of cytosine produces a uracil base, which induces C→T transitions after replication. The cis–syn cyclobutane TC dimer (T[]C) is converted to the TU dimer (T[]U) by this deamination, and C→T mutations of UV-damaged DNA have actually been observed in Escherichia coli cells (7,10,11). Two mechanisms, i.e. bypass and photoreversal mechanisms, were proposed for this mutation (12), but only the bypass mechanism is applicable to human cells, because they lack the photolyases responsible for the photoreversal of UV-induced photoproducts in DNA. Several DNA polymerases that perform translesion synthesis (TLS) have been discovered recently (13–15) and one of them, DNA polymerase η (pol η), which is defective in xeroderma pigmentosum variants (16), efficiently replicates DNA past the CPD (16–18). Both yeast and human pol η insert two adenylates opposite the cis–syn thymine dimer (T[]T) with the same efficiency and accuracy as opposite undamaged thymines (19–22). Templates containing T[]T have been used in all of the biochemical studies on pol η so far. Yu et al. described an error-free bypass of cytosine-containing photodimers in yeast (23), but since the deamination occurs rapidly, as described above, it would be interesting to investigate TLS past T[]U by pol η. If pol η incorporates dGMP opposite the 3′ base of T[]U, then a mutation would not occur, because this damage was originally formed at TC. This may be possible, because the wobble base pair G·U is very common in RNA structures (24,25).
Previously, a decanucleotide containing the T[]U dimer was prepared by triplet-sensitized irradiation of an unmodified oligomer, followed by HPLC purification (12). Since the oligonucleotides prepared by this post-synthetic method have length and sequence context limitations, a better way would involve direct chemical synthesis using a phosphoramidite building block. In this paper, we describe the preparation of a T[]U building block and the synthesis of oligonucleotides containing this deaminated photolesion. Using a 30mer synthesized by this method, TLS by human pol η past this photodimer was investigated.
MATERIALS AND METHODS
General methods
Thin layer chromatography was performed on silica gel 60 F254 plates (Merck, Darmstadt, Germany) using a solvent system of chloroform/methanol, and spots were visualized under a UV lamp. For column chromatography, Wakogel C-200 or C-300 (Wako Pure Chemical Industries, Osaka, Japan) was used. 1H and 31P NMR spectra were recorded on a Varian Unity 600 spectrometer. ROESY spectra were measured with a mixing time of 800 ms. 31P NMR spectra were referenced to an external standard of triphenylphosphine. High resolution mass spectral data were obtained on a JEOL JMS-700 spectrometer. HPLC analysis was performed on a Gilson gradient system, using a µBondasphere C18 5 µm 300 Å column (3.9 × 150 mm) (Waters Corp., Milford, MA) with a linear gradient of acetonitrile in 0.1 M triethylammonium acetate (pH 7.0) or a TSK-GEL DEAE-2SW column (4.6 × 250 mm) (Tosoh Corporation, Tokyo, Japan) with a linear gradient of ammonium formate in 20% aqueous acetonitrile.
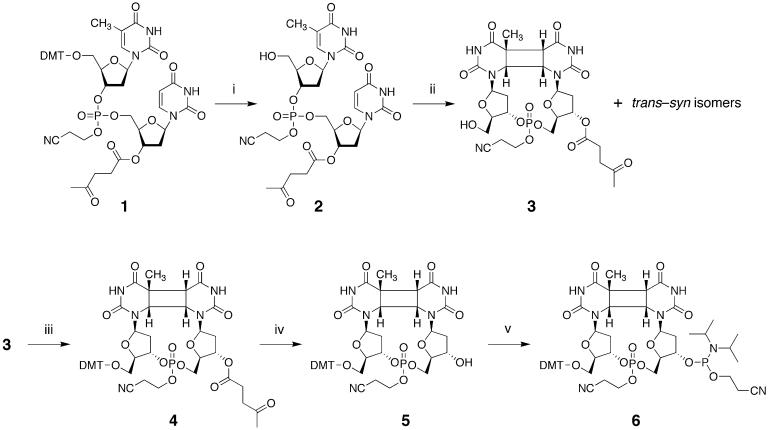
Preparation of the cis–syn cyclobutane TU dimer (3)
A partially protected dinucleoside monophosphate, thymidylyl-(3′–5′)-3′-O-levulinyl-2′-deoxyuridine (2-cyanoethyl)phosphotriester (2), was prepared in a manner similar to that described previously (26), by the coupling of 5′-O-(4,4′-dimethoxytrityl)thymidine 3′-(2-cyanoethyl)-N,N-diisopropylphosphoramidite and 3′-O-levulinyl-2′-deoxyuridine, followed by oxidation with iodine and detritylation with acetic acid. This starting material (2, 683 mg, 1.0 mmol) was dissolved in a mixture of acetonitrile (750 ml) and water (200 ml) and the solution was degassed by a nitrogen purge. After acetone (50 ml) was added, this mixture was irradiated in a Pyrex immersion well apparatus fitted with a 450 W high pressure mercury lamp (UM-452; Ushio, Tokyo, Japan), in a 4°C water bath (EYELA NCB-2200; Tokyo Rikakikai, Tokyo, Japan) for 10 h. After consumption of the starting material was confirmed by TLC and HPLC (Fig. 1A), the solution was evaporated in vacuo and the products were separated by chromatography on silica gel. The major product that eluted most slowly with 5% methanol in chloroform was collected, by analogy with the cis–syn thymine dimer case (26), and a glassy solid was obtained after evaporation. Yield 0.352 g (0.52 mmol, 52%). TLC (CHCl3/MeOH, 5/1) Rf 0.25. 1H NMR (DMSO-d6, the diastereomers are represented with and without an asterisk) δ (p.p.m.) 5.91 (dd, J = 6.2, 8.2Hz, 1H, U-H1′), 5.78 (m, 2H, T-H1′, U*-H1′), 5.47 (dd, J = 5.4, 8.6 Hz, 1H, T*-H1′), 4.92–4.88 (m, 2H, U-H3′, U*-H3′), 4.83 (m, 1H, 5′-OH), 4.79 (m, 2H, T-H3′, T*-H3′), 4.61 (m, 1H, 5′-OH*), 4.50 (dd, J = 5.4, 9.0 Hz, 1H, U*-H6), 4.45 (dd, J = 5.7, 8.5 Hz, 1H, U-H6), 4.39 (m, 1H, U-H5′), 4.23–4.12 (m, 8H, U-H5”, U*-H5′, U*-H5”, T-H6, -OCH2CH2CN, -OCH2CH2CN*), 4.05 (dd, J = 2.4, 5.2 Hz, 1H, T*-H6), 3.94 (dd, J = 2.9, 5.7 Hz, 1H, T*-H4′), 3.91 (m, 2H, U-H4′, U*-H4′), 3.75 (dd, J = 3.8, 7.4 Hz, 1H, T-H4′), 3.43 (m, 4H, T-H5′, T-H5”, T*-H5′, T*-H5”), 3.27 (dd, J = 2.1, 9.1 Hz, 1H, U*-H5), 3.23 (d, J = 8.6 Hz, 1H, U-H5), 2.90 (m, 4H, -OCH2CH2CN, -OCH2CH2CN*), 2.70 (m, 4H, -OCOCH2CH2CO-, -OCOCH2CH2CO-*), 2.63 (m, 1H, T*-H2”), 2.52 (m, 1H, T-H2”), 2.43 (m, 5H, -OCOCH2CH2CO-, -OCOCH2CH2CO-*, U*-H2”), 2.35 (m, 1H, T-H2′), 2.29 (m, 1H, U-H2”), 2.13 (m, 1H, T*-H2′), 2.08 (s, 6H, -CH2COCH3, -CH2COCH3*), 1.98 (m, 2H, U-H2′, U*-H2′), 1.49 (s, 6H, T-CH3, T*-CH3). FABHRMS m/z 684.1919 ([M+H]+, C27H35N5O14P requires 684.1918).
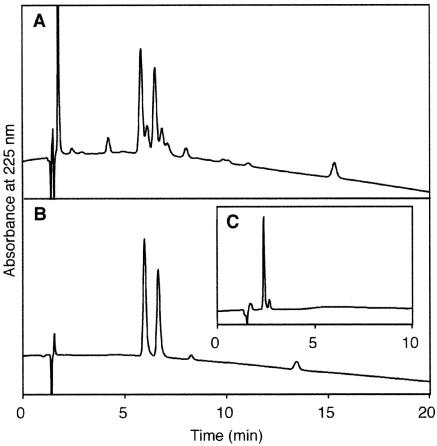
Figure 1.
Reversed phase HPLC analyses of the reaction mixture after the triplet-sensitized photoreaction for 10 h (A), the fractions collected after purification of compound 3 by silica gel chromatography (B) and the same sample as (B) after treatment with ammonia water at room temperature for 2 h (C). The acetonitrile gradient was 11–19% (A and B) or 0–10% (C) for 20 min.
Deprotection of 3
An aliquot of 3 (30 mg, 44 µmol) was dissolved in 28% ammonia water (10 ml). After 2 h at room temperature, the ammonia was removed by evaporation and the residue was passed through a column (1 × 17 cm) of Preparative C18 125 Å (Waters Corp.) on a Bio-Rad Econo System. A short wavelength UV-absorbing substance was eluted out with water immediately after the flow-through fractions. An aliquot was analyzed by HPLC (Fig. 1C) and the rest was used for NMR measurement. 1H NMR (D2O) δ (p.p.m.) 5.93 (dd, J = 5.5, 9.1 Hz, 1H, U-H1′), 5.63 (dd, J = 4.2, 10.0 Hz, 1H, T-H1′), 4.62 (m, 1H, T-H3′), 4.59 (dd, J = 5.7, 8.7 Hz, 1H, U-H6), 4.40 (dd, J = 1.3, 5.6 Hz, 1H, T-H6), 4.33 (m, 1H, U-H3′), 4.13 (m, 2H, U-H5′, T-H4′), 4.00 (m, 1H, U-H5”), 3.83 (m, 1H, U-H4′), 3.65 (d, J = 6.3 Hz, 2H, T-H5′, T-H5”), 3.55 (dd, J = 1.3, 8.6 Hz, 1H, U-H5), 2.53 (m, 1H, T-H2”), 2.32 (m, 1H, T-H2′), 2.19 (m, 1H, U-H2′), 2.00 (m, 1H, U-H2”), 1.61 (s, 3H, T-CH3).
Protection of the 5′-OH to give 4
The partially protected dimer (3, 189 mg, 276 µmol) was dissolved in pyridine (3.5 ml) and mixed with 4,4′-dimethoxytrityl chloride (234 mg, 690 µmol). After stirring at room temperature for 3 h, methanol (0.5 ml) was added and the mixture was concentrated. The residue was dissolved in chloroform (15 ml) and washed with water and, after concentration and co-evaporation with toluene, it was chromatographed on silica gel with a step gradient of 0–3% methanol in chloroform. The fractions containing the tritylated product were collected and 4 was obtained as a foam after evaporation. Yield 219 mg (222 µmol, 80%). TLC (chloroform/methanol, 10/1) Rf 0.40. 1H NMR (CDCl3, the diastereomers are represented with and without an asterisk) δ (p.p.m.) 9.15 (s, 1H, -NH-), 9.12 (s, 1H, -NH-), 8.97 (s, 2H, -NH-), 7.38–7.35 (m, 4H, aromatic), 7.33–7.22 (m, 12H, aromatic), 6.86–6.84 (m, 4H, aromatic), 6.83–6.82 (m, 6H, aromatic), 6.03 (t, J = 5.6 Hz, 1H, T*-H1′), 5.99 (dd, J = 5.3, 7.6 Hz, 1H, T-H1′), 5.93 (dd, J = 5.6, 8.6 Hz, 1H, U*-H1′), 5.92 (dd, J = 5.7, 8.8 Hz, 1H, U-H1′), 5.13 (m, 1H, U-H3′), 5.00 (m, 1H, T-H3′), 4.93 (m, 2H, T*-H3′, U*-H3′), 4.58 (m, 1H, U-H5′), 4.45 (m, 2H, U*-H5′, U*-H5”), 4.40 (dd, 1H, J = 6.0, 8.7 Hz, 1H, U-H6), 4.30 (dd, J = 5.5, 8.3 Hz, 1H, U*-H6), 4.24 (m, 6H, -CH2CH2CN, -CH2CH2CN*, T-H6, U-H5”), 4.16 (d, J = 5.4 Hz, 1H, T*-H6), 4.14 (m, 1H, T-H4′), 4.10 (m, 1H, U-H4′), 4.06 (m, 1H, T*-H4′), 4.02 (m, 1H, U*-H4′), 3.80 (s, 6H, -OCH3 × 2), 3.79 (s, 6H, -OCH3* × 2), 3.66 (dd, J = 2.9, 10.9 Hz, 1H, T*-H5′), 3.50 (dd, J = 3.1, 10.5 Hz, 1H, T-H5′), 3.40 (dd, J = 2.8, 10.7 Hz, 1H, T*-H5”), 3.22 (dd, J = 2.5, 10.5 Hz, T-H5”), 3.03 (d, J = 8.4 Hz, 1H, U*-H5), 2.81 (d, J = 8.6 Hz, 1H, U-H5), 2.76 (m, 4H, -CH2CH2CO-, -CH2CH2CO-*), 2.72 (m, 4H, -OCH2CH2CN, -OCH2 CH2CN*), 2.63–2.50 (m, 8H, T-H2′, T*-H2′, T-H2”, T*-H2”, -CH2CH2CO-, -CH2CH2CO-*), 2.46 (m, 2H, U-H2”, U*-H2”), 2.14 (m, 1H, U-H2′), 2.11 (m, 1H, U*-H2′), 1.22 (s, 3H, T-CH3), 1.20 (s, 3H, T*-CH3). FABHRMS m/z 1008.3029 ([M+Na]+, C48H52N5O16PNa requires 1008.3045).
Removal of the levulinyl group to give 5
The fully protected dimer (4, 354 mg, 359 µmol) was dissolved in pyridine (4.5 ml) and a solution (4.5 ml) of hydrazine monohydrate (175 µl, 3.6 mmol) in pyridine/acetic acid (3:2 v/v) was added. After stirring at room temperature for 5 min, the mixture was cooled in an ice bath and then acetone (3.0 ml) was added. The mixture was diluted with chloroform (30 ml), washed with 2% aqueous NaHCO3 and with water, dried with Na2SO4 and concentrated. After co-evaporation with toluene, the residue was chromatographed on silica gel (6.5 g) with a step gradient of 0–5% methanol in chloroform and a product (5) was obtained as a glassy solid after evaporation. Yield 259 mg (292 µmol, 81%). TLC (chloroform/methanol, 10/1) Rf 0.10. 1H NMR (DMSO-d6, the diastereomers are represented with and without an asterisk) δ (p.p.m.) 10.56 (s, 1H, -NH-), 10.50 (s, 1H, -NH-), 10.47 (s, 1H, -NH-), 10.44 (s, 1H, -NH-), 7.38 (m, 4H, aromatic), 7.30 (m, 4H, aromatic), 7.27–7.21 (m, 10H, aromatic), 6.89–6.86 (m, 8H, aromatic), 5.86 (m, 3H, U*-H1′, U-H1′, T*-H1′), 5.62 (t, J = 6.7 Hz, 1H, T-H1′), 5.35 (d, J = 7.4 Hz, 1H, 3′-OH), 5.34 (d, J = 7.6 Hz, 1H, 3′-OH*), 4.81 (m, 1H, T-H3′), 4.76 (m, 1H, T*-H3′), 4.44 (dd, J = 5.6, 8.9 Hz, 1H, U-H6), 4.32 (m, 3H, U*-H6, U*-H5′, U*-H5”), 4.14 (m, 7H, -CH2CH2CN, -CH2CH2CN*, T*-H6, U-H5′, U-H5”), 4.03 (m, 3H, U-H3′, T-H4′, T-H6), 3.95 (dd, J = 4.2, 6.9 Hz, 1H, T*-H4′), 3.91 (m, 1H, U*-H3′), 3.73 (m, 13H, -OCH3 × 2, -OCH3* × 2, U-H4′), 3.68 (m, 1H, U*-H4′), 3.22 (m, 3H, T*-H5′, U-H5, U*-H5), 3.13 (dd, J = 4.5, 10.2 Hz, 1H, T-H5′), 3.03 (m, 2H, T-H5”, T*-H5”), 2.92 (t, J = 5.8 Hz, 4H, -CH2CH2CN, -CH2 CH2CN*), 2.76 (m, 1H, T-H2”), 2.59 (m, 1H, T*-H2”), 2.43 (m, 1H, T*-H2′), 2.28 (m, 1H, T-H2′), 2.20 (m, 1H, U-H2′), 2.09 (m, 1H, U*-H2′), 1.94 (m, 2H, U-H2”, U*-H2”), 1.32 (s, 3H, T-CH3), 1.24 (s, 3H, T*-CH3). FABHRMS m/z 910.2689 ([M+Na]+, C43H46N5O14PNa requires 910.2677).
Phosphoramidite building block of T[]U (6)
An aliquot of 5 (143 mg, 161 µmol) was dissolved in tetrahydrofuran (1.8 ml) and mixed with N,N-diisopropylethylamine (112 µl, 644 µmol). To this solution, 2-cyanoethyl N,N-diisopropylchlorophosphoramidite (72 µl, 322 µmol) was added. This mixture was stirred at room temperature for 30 min and diluted with ethyl acetate (20 ml). The resulting solution was washed with 2% aqueous NaHCO3 and with water, dried with Na2SO4, concentrated to a gum and co-evaporated with toluene. The residue was chromatographed on silica gel (3 g) with a step gradient of 0–2% methanol in chloroform containing 0.1% pyridine. Prior to the chromatography, the column and the solvents were cooled with ice to avoid degradation of the phosphoramidite on the silica gel. The appropriate fractions were collected and, after concentration, the residue was dissolved in chloroform (1 ml) and precipitated in n-pentane (20 ml). Yield 104 mg (95 µmol, 59%). TLC (chloroform/methanol, 10/1) Rf 0.55. 31P NMR (pyridine-d5) δ 166.62, 166.49, 166.37, 166.21, 15.47, 15.41, 13.04, 12.92 p.p.m. FABHRMS m/z 1110.3739 ([M+Na]+, C52H63N7O15P2Na requires 1110.3755).
Synthesis of oligonucleotides
The phosphoramidite building block of T[]U was dissolved in anhydrous acetonitrile at a concentration of 0.12 M and installed on an Applied Biosystems Model 394 DNA/RNA synthesizer. Nucleoside phosphoramidites for ultramild DNA synthesis (Glen Research, Sterling, VA), as well as the base-unprotected thymidine phosphoramidite, were also dissolved in acetonitrile to make 0.1 M solutions and installed on the synthesizer. An 8mer, d(GTAT[]UATG), and a 30mer, d(CTCGTCAGCATCT[]UCATCATACAGTCAGTG), were synthesized using a reaction column containing 0.2 µmol of the 3′-terminal deoxyguanosine. The 0.2 µmol synthesis cycle supplied by the manufacturer was changed to prolong the reaction time for coupling of 6 to 20 min. A solution of phenoxyacetic anhydride (obtained from Glen Research) was used as a capping agent. After chain assembly on the synthesizer, the solid support containing the oligonucleotide was treated with 28% aqueous ammonia at room temperature for 2 h. The resulting ammoniac solution was concentrated on a rotary evaporator equipped with a vacuum pump and the residue was dissolved in distilled water (1.0 ml). Aliquots were analyzed by HPLC, as shown in Figure 2A, and the remaining portions were purified with the same HPLC system. The yields of the 8mer and the 30mer were 3.6 and 6.9 A260 units (53 and 26 nmol), respectively. The 30mer containing the T[]T dimer, d(CTCGTCAGCATCT[]TCATCATACAG TCAGTG), was synthesized as described previously (26). For the photoreversal of T[]U, a solution (10 µl) of the T[]U 8mer (∼0.1 nmol) was subjected to UV irradiation on a Spectrolinker XL-1500 UV Crosslinker (Spectronics Corp., Westbury, NY) for 2 min and the product was analyzed by HPLC, as shown in Figure 3B.
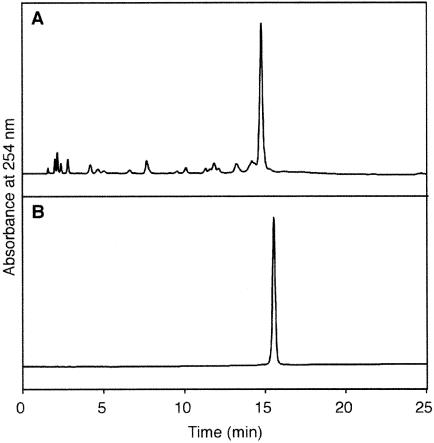
Figure 2.
(A) Reversed phase HPLC analysis of a crude sample of the T[]U-containing 30mer, using a gradient of 7–13% acetonitrile for 20 min. (B) Anion exchange HPLC analysis of the 30mer after purification by reversed phase HPLC, using a gradient of 0.4–1.2 M ammonium formate for 20 min.
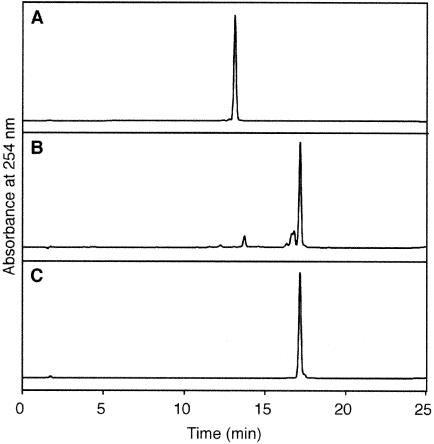
Figure 3.
Reversed phase HPLC analysis to confirm the presence of T[]U in the synthesized oligonucleotide. The purified 8mer, d(GTAT[]UATG) (A), a UV-irradiated sample of the T[]U 8mer (B) and the undamaged, uracil-containing 8mer, d(GTATUATG) (C) were analyzed using a 5–13% acetonitrile gradient for 20 min.
T4 endonuclease V assays
T4 endonuclease V was kindly provided by Dr Eiko Ohtsuka. The 5′-32P-labeled 30mer containing T[]T or T[]U (3.3 pmol) was mixed with the complementary oligonucleotide (10 pmol) in a buffer (33 µl) containing 10 mM Tris–HCl (pH 7.5), 1 mM EDTA and 0.1 mM NaCl. The mixtures were incubated at 65°C for 5 min and cooled down slowly to room temperature. The T4 endonuclease V reactions contained 25 mM Tris–HCl (pH 7.5), 2.5 mM EDTA, 200 µg/ml BSA, 50 mM NaCl, 5 nM 32P-labeled double-stranded DNA and 2.5 ng/ml T4 endonuclease V. The reaction was performed at 37°C and was terminated by the addition of formamide, followed by boiling. The products were electrophoresed on a 20% polyacrylamide gel containing 7 M urea and the bands were detected by autoradiography.
Translesion synthesis (TLS) assays
Recombinant human pol η, tagged with (His)6 at its C-terminal end, was prepared as described previously (19). Pol α was purified from mouse FM3A cells, as described previously (27). Standard reactions (10 µl) contained 25 mM potassium phosphate (pH 7.5), 5 mM MgCl2, 10 mM DTT, 250 µg/ml BSA, 60 mM KCl, 2.5% glycerol, 40 nM 5′-32P-labeled primer–template DNA, 100 µM dNTPs and pol η or α. The reaction was performed at 37°C for 15 min and was terminated by the addition of formamide (10 µl), followed by boiling. The products were electrophoresed on a 20% polyacrylamide gel containing 7 M urea and the bands were detected by autoradiography.
RESULTS
Synthesis of the T[]U building block
The synthesis of the phosphoramidite building block of the T[]T dimer was originally described by Taylor et al. (28). One of the authors of the present work improved the method, for use in current oligonucleotide synthesis protocols (26), and applied it to the study of bacteriophage T4 endonuclease V (29–31), which recognizes this damaged base and initiates its repair. For the T[]U building block, we used a combination of the protecting groups used in our previous study (26). The 4,4′-dimethoxytrityl (DMT) and 2-cyanoethyl groups were used to protect the 5′-hydroxyl function and the internucleoside phosphate, respectively, and the 3′-hydroxyl function of the intermediates was protected by the levulinyl group (Scheme 1). The DMT group in the protected thymidylyl-(3′–5′)-2′-deoxyuridine (1), which was prepared using a general procedure, was removed prior to the photoreaction, as originally described by Taylor et al. (28). The detritylated dinucleoside monophosphate (2) was irradiated in a Pyrex immersion well apparatus fitted with a 450 W high pressure mercury lamp, using acetone as a sensitizer. The reaction was monitored by reversed phase HPLC. After 7 h, the starting material still remained (data not shown), but as shown in Figure 1A, the product peaks were detected almost exclusively in the sample irradiated for 10 h. The small peak with a retention time of 15.3 min in Figure 1A was the starting material, although its diastereomers caused by the chiral phosphorus were not separated. In this analysis, two major and several minor products were observed, but the peak separation was not sufficient for the isolation of each product on a preparative scale using this system. Silica gel chromatography using a solvent system of chloroform/methanol provided better separation. The two major products were isolated from the minor ones, but they could not be separated from each other on silica gel, as shown in Figure 1B. Since these two compounds were presumed to be diastereomers, due to the chirality of the phosphorus atom, a mixture of these two products was used for analysis of the base structure and for further synthesis.
Scheme 1. Synthesis of a phosphoramidite building block of the cis–syn cyclobutane thymine–uracil dimer. (i) 80% AcOH, room temperature, 2 h; (ii) hν, CH3CN–H2O–acetone (15/4/1 v/v/v), 4°C, 10 h; (iii) DMT-Cl, pyridine, room temperature, 3 h; (iv) NH2NH2·H2O, pyridine–AcOH (4/1 v/v), room temperature, 5 min; (v) [(CH3)2CH]2NP(Cl)OCH2CH2CN, [(CH3)2CH]2NC2H5, THF, room temperature, 30 min.
The stereochemistry of the cyclobutane dimer structure was analyzed by NMR spectroscopy. Since specificity is often lost due to spin diffusion in nuclear Overhauser effect spectroscopy (NOESY) of intermediate size molecules, rotating frame Overhauser effect spectroscopy (ROESY) spectra of the mixture of the two products were measured, after successful signal assignment by correlated spectroscopy (COSY). For both of the compounds, ROE crosspeaks were observed between the following protons: T CH3–U H6, T CH3–U H5, T CH3–T H6, U H6–U H5 and U H6–T H6. This result indicates that all of these protons exist on the same side of the cyclobutane ring in both of the compounds. The base structure was analyzed further after deprotection of the products, with the expectation that removal of the cyanoethyl group would simplify the NMR spectra. A single product was obtained by treatment of the mixed products with ammonia water at room temperature for 2 h, as shown in Figure 1C. This result confirmed that the two major products were the diastereomers of the phosphotriester with the same base moiety. The 1H NMR spectrum of this deprotected compound, for which all of the signals were assigned by the COSY measurement, was completely identical with that reported previously for the unprotected cis–syn cyclobutane TU dimer (2). The ROESY crosspeaks described above were observed more distinctly for this deprotected dimer. The T CH3–U H6 crosspeak was weaker than that of T H6–U H5, which suggests that the T H6 and U H5 are in axial positions in the conformation of the cyclobutane ring. From these results, we concluded that the desired compound (3), which had the cis–syn T[]U dimer, was obtained, although the diastereomers arising from the chirality of the phosphorus atom could not be separated.
As shown in Scheme 1, the 5′-hydroxyl group of 3 was protected again with the DMT group. Next, the levulinyl group was removed from 4 and, finally, the phosphoramidite building block (6) was obtained by phosphitylation of the 3′-hydroxyl function of 5. This procedure is the same as those for the T[]T (26) and (6–4) photoproduct (32) building blocks.
Synthesis of oligonucleotides containing T[]U
Using the T[]U building block (6) described in the above section, an 8mer, d(GTAT[]UATG), and a 30mer, d(CTC GTCAGCATCT[]UCATCATACAGTCAGTG), were synthe sized. Although the conventional protecting groups for the exocyclic amino functions could be used, as described previously for the T[]T dimer (26), nucleoside 3′-phosphoramidites bearing base-labile protecting groups, i.e. phenoxyacetyl, (4-isopropylphenoxy)acetyl and acetyl groups for adenine, guanine and cytosine, respectively, were used to reduce the reaction time for deprotection. The coupling reaction of 6 was prolonged to 20 min, according to the previous study (26), and chain elongation on the DNA synthesizer proceeded almost quantitatively. After the DMT group at the 5′-end was removed on the synthesizer, the solid support containing each oligonucleotide was treated with ammonia water at room temperature for 2 h for cleavage and deprotection and the products were analyzed by reversed phase HPLC. The elution profile of the 30mer is shown in Figure 2A. A single main peak was detected, and this product was isolated by the same HPLC system. The purified sample gave a single peak in an anion exchange HPLC analysis, as shown in Figure 2B.
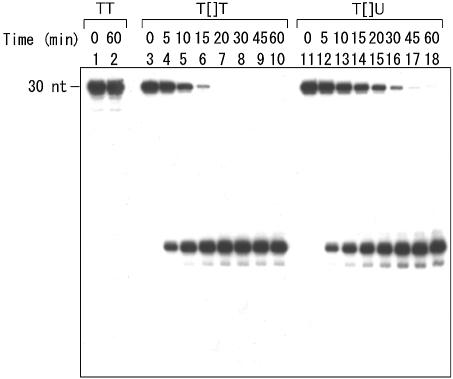
Presence of the T[]U dimer in the obtained oligonucleotides was confirmed by photoreversal. The CPDs are reversed to the original pyrimidine bases by UV irradiation, and this is a general property of this photolesion (33). An aliquot of the purified 8mer was irradiated on a UV crosslinker, at a UV dose of ∼16 mJ, and was analyzed by reversed phase HPLC. As shown in Figure 3, the peak of the synthesized 8mer decreased to a great extent and a new peak appeared. This peak had the same retention time as that of the undamaged, uracil-containing 8mer, d(GTATUATG), and the identity was confirmed by co-injection. The cis–syn configuration was confirmed by the reaction of T4 endonuclease V. After 5′-32P-labeling, the T[]U-containing 30mer was hybridized to the complementary strand and this duplex was treated with the enzyme. The reaction was compared to that with the substrate containing T[]T, which was chemically synthesized in the same way (26). As shown in Figure 4, T4 endonuclease V cleaved the substrate containing T[]U effectively, although the reaction rate was slightly lower than that for the T[]T substrate.
Figure 4.
T4 endonuclease V reaction of a 30mer duplex containing T[]U. The 5′-32P-labeled undamaged (lanes 1 and 2) and T[]T- (lanes 3–10) and T[]U-containing (lanes 11–18) 30mers were hybridized to a 3-fold excess of the complementary 30mer and incubated with T4 endonuclease V at 37°C. The products after the indicated times were subjected to PAGE under denaturing conditions.
Translesion synthesis past T[]U by DNA polymerase η
Previously, we showed that human pol η, the product of the XPV gene that is mutated in xeroderma pigmentosum variant, can catalyze TLS past the cis–syn T[]T dimer by efficiently incorporating two adenylates opposite this damaged base (16,19). Among the translesion polymerases found in eukaryotes, only pol η replicates CPD-containing DNA accurately (34,35). TLS past the cis–syn T[]U dimer by pol η is interesting, because accurate replication, i.e. the incorporation of dAMP opposite the U component of T[]U, results in a C→T mutation. In this study, we analyzed TLS by pol η using the aforementioned 30mer as a template.
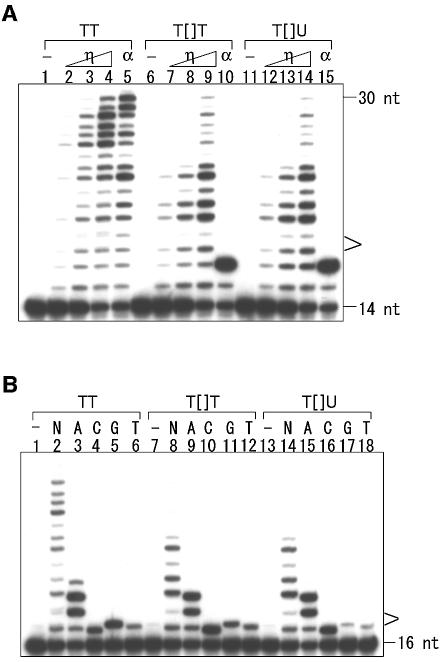
Running- and standing-start experiments were carried out using a 32P-labeled 14mer, 32P-d(CACTGACTGTATGA), and a 16mer, 32P-d(CACTGACTGTATGATG), respectively, as primer, according to previous studies (18,22). Only the results of the former are shown in Figure 5A, because DNA polymerase stalling immediately before the damaged base can be observed in this set of experiments. Whereas mouse pol α stalled at the lesion (Fig. 5A, lanes 10 and 15), human pol η bypassed both types of CPDs efficiently (Fig. 5A, lanes 7–9 and 12–14) and, basically, there was no difference between T[]T and T[]U. The results for the T[]U-containing template were also undistinguishable from those for the T[]T-containing one in the standing-start experiments (data not shown). To determine the nucleotide selectivity for incorporation opposite the CPD, chain elongation of the 16mer primer was analyzed in the presence of only one of the four nucleoside triphosphates. For all three templates, i.e. the undamaged one and those containing T[]T and T[]U, pol η incorporated three adenylates (Fig. 5B, lanes 3, 9 and 15), whereas it stopped after incorporating one nucleotide in the presence of dCTP, dGTP or dTTP (Fig. 5B, lanes 4–6, 10–12 and 16–18). There were no obvious differences between the three data sets. These results indicate that pol η efficiently incorporates two adenylates opposite the cis–syn T[]U dimer, in the same way as opposite T[]T.
Figure 5.
Comparison of translesion synthesis past T[]T and T[]U. (A) The 5′-32P-labeled 14mer was hybridized to the undamaged TT 30mer template (lanes 1–5) or the template containing T[]T (lanes 6–10) or T[]U (lanes 11–15) and incubated with increasing amounts of pol η or pol α in the presence of four dNTPs. The amounts of pol η were 0.56 (lanes 2, 7 and 12), 1.7 (lanes 3, 8 and 13) and 5.0 fmol (lanes 4, 9 and 14) and the amount of pol α was 5.0 fmol (lanes 5, 10 and 15). Lanes 1, 6 and 11 contained no enzyme. The products were subjected to PAGE under denaturing conditions. (B) The 5′-32P labeled 16mer primer was hybridized to the undamaged 30mer template (lanes 1–6) or the template containing T[]T (lanes 7–12) or T[]U (lanes 13–18) and incubated with pol η (1.0 fmol), in the presence of four dNTPs (lanes 2, 8 and 14) or one of the indicated dNTPs (lanes 3–6, 9–12 and 15–18) or in the absence of dNTP (lanes 1, 7 and 13).
DISCUSSION
The cis–syn T[]C dimer is formed in DNA in a yield comparable to T[]T. It was reported that when cellular DNA was irradiated with UVB at a dose of 1 kJ/m2, the yields of T[]T and T[]C were 3.15 and 1.29 per 104 bases, respectively (36). The T[]C dimer undergoes hydrolytic deamination, as described in the Introduction, and becomes T[]U. In this study, we have synthesized oligonucleotides containing the cis–syn T[]U dimer and analyzed TLS past this damaged base by human pol η, using one of the synthesized oligomers as a template. The synthetic method, shown in Scheme 1, was basically the same as that previously described for the synthesis of oligonucleotides containing the T[]T dimer (26), but the structure of the damaged base in the intermediates and the oligonucleotides was analyzed thoroughly by NMR spectroscopy and biochemical experiments, respectively. After the triplet-sensitized photoreaction, two major products, which were assumed to be diastereoisomers caused by the chirality of the phosphorus atom, were eluted from the silica gel column more slowly than the minor products. This property is the same as that of the cis–syn T[]T dimer, and NMR studies of both the protected and deprotected compounds showed that these products were the diastereomers of cis–syn T[]U (compound 3). The spectra of the deprotected dimer can be analyzed more easily than those of the protected compound. Among the observed ROE crosspeaks, the T H6–U H5 crosspeak was stronger than the T CH3–U H6 one. This result suggests that T H6 and U H5 are in axial positions, which agrees with the cyclobutane ring conformation in a T[]T-containing duplex reported by Taylor et al. (37). After the determination of this stereochemistry, a phosphoramidite building block (compound 6) was prepared in three steps, and was used for oligonucleotide synthesis.
The damaged base in the obtained oligonucleotides was characterized firstly by photoreversal. UV irradiation of the synthesized 8mer resulted in its conversion to the undamaged, uracil-containing 8mer (Fig. 3), which demonstrated the presence of a cyclobutane-type dimer in the oligonucleotide. The cis–syn configuration was confirmed by the reaction of T4 endonuclease V. The rate of the T4 endonuclease V reaction with a substrate containing the trans–syn thymine dimer was reportedly at most 1% of that with the cis–syn substrate (38). As shown in Figure 4, the 30 bp substrate containing T[]U was cleaved by this enzyme efficiently, although the reaction rate was slightly lower than that for the T[]T substrate. These results indicate that the oligonucleotides synthesized with the obtained building block contained the cis–syn T[]U dimer, as expected.
Pol η efficiently incorporates two adenylates opposite the cis–syn T[]T dimer (19,20). This is accurate and error-free TLS. In the case of T[]U, accurate TLS means the incorporation of dAMP opposite the U component, considering the base pairing complementarity, but this is error prone replication since this type of lesion originates from a TC sequence. To characterize the TLS past the deaminated form of the T[]C dimer by pol η, TLS assays using a template containing T[]U were carried out and the results were compared to those with a T[]T-containing template. As shown in Figure 5, there was no difference between the two damaged templates. Compared to the undamaged template, incorporation of dGMP opposite the 3′ component of the CPD, which was expected at the beginning, was even suppressed (Fig. 5B, lanes 5 and 17). Thus, we concluded that pol η efficiently catalyzes accurate TLS past T[]U, which will induce a C→T mutation. This probably occurs by making a Watson–Crick A·U pair, by analogy with the crystal structure of an archaeal pol η homolog complexed with a T[]T-containing DNA (39).
Among the other TLS polymerases, human pol ι is the next target. Although the preference of nucleotide incorporation opposite the cis–syn T[]T dimer depends on the sequence context (40,41), this enzyme misincorporates G opposite T (42,43) and U (44) efficiently, and G is incorporated opposite C (42). If dGMP is specifically incorporated opposite the U component of T[]U, then error-free replication past T[]C can be achieved, regardless of the deamination. Our method for the synthesis of T[]U-containing oligonucleotides, described in this paper, can be used for these future studies.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Takeshi Imanishi and Satoshi Obika for the analysis of the compounds.
REFERENCES
- 1.Frederico L.A., Kunkel,T.A. and Shaw,B.R. (1990) A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry, 29, 2532–2537. [DOI] [PubMed] [Google Scholar]
- 2.Liu F.-T. and Yang,N.C. (1978) Photochemistry of cytosine derivatives. 1. Photochemistry of thymidylyl-(3′→5′)-deoxycytidine. Biochemistry, 17, 4865–4876. [DOI] [PubMed] [Google Scholar]
- 3.Douki T. and Cadet,J. (1992) Far-UV photochemistry and photosensitization of 2′-deoxycytidylyl-(3′–5′)-thymidine: isolation and characterization of the main photoproducts. J. Photochem. Photobiol. B., 15, 199–213. [DOI] [PubMed] [Google Scholar]
- 4.Lemaire D.G.E. and Ruzsicska,B.P. (1993) Kinetic analysis of the deamination reactions of cyclobutane dimers of thymidylyl-3′,5′-2′-deoxycytidine and 2′-deoxycytidylyl-3′,5′-thymidine. Biochemistry, 32, 2525–2533. [DOI] [PubMed] [Google Scholar]
- 5.Barak Y., Cohen-Fix,O. and Livneh,Z. (1995) Deamination of cytosine-containing pyrimidine photodimers in UV-irradiated DNA. Significance for UV light mutagenesis. J. Biol. Chem., 270, 24174–24179. [DOI] [PubMed] [Google Scholar]
- 6.Peng W. and Shaw,B.R. (1996) Accelerated deamination of cytosine residues in UV-induced cyclobutane pyrimidine dimers leads to CC→TT transitions. Biochemistry, 35, 10172–10181. [DOI] [PubMed] [Google Scholar]
- 7.Burger A., Fix,D., Liu,H., Hays,J. and Bockrath,R. (2003) In vivo deamination of cytosine-containing cyclobutane pyrimidine dimers in E. coli: a feasible part of UV-mutagenesis. Mutat. Res., 522, 145–156. [DOI] [PubMed] [Google Scholar]
- 8.Tessman I., Kennedy,M.A. and Liu,S.-K. (1994) Unusual kinetics of uracil formation in single and double-stranded DNA by deamination of cytosine in cyclobutane pyrimidine dimers. J. Mol. Biol., 235, 807–812. [DOI] [PubMed] [Google Scholar]
- 9.Tu Y., Dammann,R. and Pfeifer,G.P. (1998) Sequence and time-dependent deamination of cytosine bases in UVB-induced cyclobutane pyrimidine dimers in vivo. J. Mol. Biol., 284, 297–311. [DOI] [PubMed] [Google Scholar]
- 10.Tessman I., Liu,S.-K. and Kennedy,M.A. (1992) Mechanism of SOS mutagenesis of UV-irradiated DNA: mostly error-free processing of deaminated cytosine. Proc. Natl Acad. Sci. USA, 89, 1159–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horsfall M.J., Borden,A. and Lawrence,C.W. (1997) Mutagenic properties of the T–C cyclobutane dimer. J. Bacteriol., 179, 2835–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang N. and Taylor,J.-S. (1993) In vivo evidence that UV-induced C→T mutations at dipyrimidine sites could result from the replicative bypass of cis–syn cyclobutane dimers or their deamination products. Biochemistry, 32, 472–481. [DOI] [PubMed] [Google Scholar]
- 13.Lehmann A.R. (2002) Replication of damaged DNA in mammalian cells: new solutions to an old problem. Mutat. Res., 509, 23–34. [DOI] [PubMed] [Google Scholar]
- 14.Prakash S. and Prakash,L. (2002) Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev., 16, 1872–1883. [DOI] [PubMed] [Google Scholar]
- 15.Friedberg E.C., Wagner,R. and Radman,M. (2002) Specialized DNA polymerases, cellular survival and the genesis of mutations. Science, 296, 1627–1630. [DOI] [PubMed] [Google Scholar]
- 16.Masutani C., Kusumoto,R., Yamada,A., Dohmae,N., Yokoi,M., Yuasa,M., Araki,M., Iwai,S., Takio,K. and Hanaoka,F. (1999) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature, 399, 700–704. [DOI] [PubMed] [Google Scholar]
- 17.Johnson R.E., Prakash,S. and Prakash,L. (1999) Efficient bypass of a thymine–thymine dimer by yeast DNA polymerase, Polη. Science, 283, 1001–1004. [DOI] [PubMed] [Google Scholar]
- 18.Masutani C., Araki,M., Yamada,A., Kusumoto,R., Nogimori,T., Maekawa,T., Iwai,S. and Hanaoka,F. (1999) Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J., 18, 3491–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masutani C., Kusumoto,R., Iwai,S. and Hanaoka,F. (2000) Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J., 19, 3100–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Washington M.T., Johnson,R.E., Prakash,S. and Prakash,L. (2000) Accuracy of thymine–thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc. Natl Acad. Sci. USA, 97, 3094–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson R.E., Washington,M.T., Prakash,S. and Prakash,L. (2000) Fidelity of human DNA polymerase η. J. Biol. Chem., 275, 7447–7450. [DOI] [PubMed] [Google Scholar]
- 22.Kusumoto R., Masutani,C., Iwai,S. and Hanaoka,F. (2002) Translesion synthesis by human DNA polymerase η across thymine glycol lesions. Biochemistry, 41, 6090–6099. [DOI] [PubMed] [Google Scholar]
- 23.Yu S.-L., Johnson,R.E., Prakash,S. and Prakash,L. (2001) Requirement of DNA polymerase η for error-free bypass of UV-induced CC and TC photoproducts. Mol. Cell. Biol., 21, 185–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masquida B. and Westhof,E. (2000) On the wobble GoU and related pairs. RNA, 6, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varani G. and McClain,W.H. (2000) The G·U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep., 1, 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murata T., Iwai,S. and Ohtsuka,E. (1990) Synthesis and characterization of a substrate for T4 endonuclease V containing a phosphorodithioate linkage at the thymine dimer site. Nucleic Acids Res., 18, 7279–7286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eki T., Enomoto,T., Masutani,C., Miyajima,A., Takada,R., Murakami,Y., Ohno,T., Hanaoka,F. and Ui,M. (1991) Mouse DNA primase plays the principal role in determination of permissiveness for polyomavirus DNA replication. J. Virol., 65, 4874–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor J.-S., Brockie,I.R. and O’Day,C.L. (1987) A building block for the sequence-specific introduction of cis–syn thymine dimers into oligonucleotides. Solid-phase synthesis of TpT[c,s]pTpT. J. Am. Chem. Soc., 109, 6735–6742. [Google Scholar]
- 29.Iwai S., Maeda,M., Shimada,Y., Hori,N., Murata,T., Morioka,H. and Ohtsuka,E. (1994) Endonuclease V from bacteriophage T4 interacts with its substrate in the minor groove. Biochemistry, 33, 5581–5588. [DOI] [PubMed] [Google Scholar]
- 30.Iwai S., Maeda,M., Shirai,M., Shimada,Y., Osafune,T., Murata,T. and Ohtsuka,E. (1995) Reaction mechanism of T4 endonuclease V determined by analysis using modified oligonucleotide duplexes. Biochemistry, 34, 4601–4609. [DOI] [PubMed] [Google Scholar]
- 31.Vassylyev D.G., Kashiwagi,T., Mikami,Y., Ariyoshi,M., Iwai,S., Ohtsuka,E. and Morikawa,K. (1995) Atomic model of a pyrimidine dimer excision repair enzyme complexed with a DNA substrate: structural basis for damaged DNA recognition. Cell, 83, 773–782. [DOI] [PubMed] [Google Scholar]
- 32.Iwai S., Shimizu,M., Kamiya,H. and Ohtsuka,E. (1996) Synthesis of a phosphoramidite coupling unit of the pyrimidine (6–4) pyrimidone photoproduct and its incorporation into oligodeoxynucleotides. J. Am. Chem. Soc., 118, 7642–7643. [Google Scholar]
- 33.Herbert M.A., LeBlanc,J.C., Weinblum,D. and Johns,H.E. (1969) Properties of thymine dimers. Photochem. Photobiol., 9, 33–43. [DOI] [PubMed] [Google Scholar]
- 34.Goodman M.F. (2002) Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem., 71, 17–50. [DOI] [PubMed] [Google Scholar]
- 35.Kannouche P. and Stary,A. (2003) Xeroderma pigmentosum variant and error-prone DNA polymerases. Biochimie, 85, 1123–1132. [DOI] [PubMed] [Google Scholar]
- 36.Douki T. and Cadet,J. (2001) Individual determination of the yield of the main UV-induced dimeric pyrimidine photoproducts in DNA suggests a high mutagenicity of CC photolesions. Biochemistry, 40, 2495–2501. [DOI] [PubMed] [Google Scholar]
- 37.Taylor J.-S., Garrett,D.S., Brockie,I.R., Svoboda,D.L. and Telser,J. (1990) 1H NMR assignment and melting temperature study of cis–syn and trans–syn thymine dimer containing duplexes of d(CGTATTATGC)·d(GCATAATACG). Biochemistry, 29, 8858–8866. [DOI] [PubMed] [Google Scholar]
- 38.Smith C.A. and Taylor,J.-S. (1993) Preparation and characterization of a set of deoxyoligonucleotide 49-mers containing site-specific cis–syn, trans–syn-I, (6–4) and Dewar photoproducts of thymidylyl(3′→5′)-thymidine. J. Biol. Chem., 268, 11143–11151. [PubMed] [Google Scholar]
- 39.Ling H., Boudsocq,F., Plosky,B.S., Woodgate,R. and Yang,W. (2003) Replication of a cis–syn thymine dimer at atomic resolution. Nature, 424, 1083–1087. [DOI] [PubMed] [Google Scholar]
- 40.Tissier A., Frank,E.G., McDonald,J.P., Iwai,S., Hanaoka,F. and Woodgate,R. (2000) Misincorporation and bypass of thymine–thymine dimers by human DNA polymerase ι. EMBO J., 19, 5259–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaisman A., Frank,E.G., Iwai,S., Ohashi,E., Ohmori,H., Hanaoka,F. and Woodgate,R. (2003) Sequence context-dependent replication of DNA templates containing UV-induced lesions by human DNA polymerase ι. DNA Repair, 2, 991–1006. [DOI] [PubMed] [Google Scholar]
- 42.Tissier A., McDonald,J.P., Frank,E.G. and Woodgate,R. (2000) polι, a remarkably error-prone human DNA polymerase. Genes Dev., 14, 1642–1650. [PMC free article] [PubMed] [Google Scholar]
- 43.Washington M.T., Johnson,R.E., Prakash,L. and Prakash,S. (2004) Human DNA polymerase ι utilizes different nucleotide incorporation mechanisms dependent upon the template base. Mol. Cell. Biol., 24, 936–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaisman A. and Woodgate,R. (2001) Unique misinsertion specificity of polι may decrease the mutagenic potential of deaminated cytosines. EMBO J., 20, 6520–6529. [DOI] [PMC free article] [PubMed] [Google Scholar]