Abstract
As the main beta-secretase of the central nervous system, BACE-1 is a key protein in the pathogenesis of Alzheimer’s disease. Excessive expression of the protein might cause an overproduction of the neurotoxic beta-amyloid peptide. Therefore, a tight regulation of BACE-1 expression is expected in vivo. In addition to a possible transcriptional control, the BACE-1 transcript leader contains features that might constitute mechanisms of translational regulation of protein expression. Moreover, recent work has revealed an increase of BACE-1 protein and beta-secretase activity in some Alzheimer’s disease patients, although a corresponding increase of transcript has not been reported. Here we show that BACE-1 translation could be modulated at multiple stages. The presence of several upstream ATGs strongly reduces the translation of the main open reading frame. This inhibition could be overcome with conditions that favour skipping of upstream ATGs. We also report an alternative splicing of the BACE-1 transcript leader that reduces the number of upstream ATGs. Finally, we show that translation driven by the BACE-1 transcript leader is increased in activated astrocytes independently of the splicing event, indicating yet another mechanism of translational control. Our findings might explain why increases in BACE-1 protein or activity are reported in the brain of Alzheimer’s disease patients even in the absence of changes in transcript levels.
INTRODUCTION
The integral membrane protein BACE-1 (Beta Amyloid Cleaving Enzyme 1) is the β-secretase that cleaves the amyloid precursor protein (APP) (1–5), producing a soluble APP β fragment (sAPPβ) and a C-terminal membrane-associated peptide (C99). Further proteolytic cleavage of C99 by a γ-secretase, tentatively identified as a multimeric complex formed by presenilins, nicastrin and other proteins (6–8), produces a cytosolic fragment (APP intracellular domain, AICD) and the amphypatic β-amyloid peptide (A-β). The latter accumulates in the brain of Alzheimer’s disease (AD) patients in the form of amyloid plaques and is thought to be directly or indirectly responsible for the ensuing neurodegeneration (9). Given this premise, BACE-1 expression is expected to play a fundamental role in the control of A-β production, and thus in the etiopathogenesis of AD. Indeed, experimental overexpression of BACE-1 increases the production of A-β in both cell lines (1–3) and transgenic mice (10). Moreover, an increase in β-secretase activity and BACE-1 protein was observed in the brain of AD patients (11–14).
The control of BACE-1 expression could occur at transcriptional, as well as post-transcriptional level. The BACE-1 promoter is currently under investigation and a recent publication reports an initial characterization of the corresponding rat sequence (15). However, there is also evidence that post-transcriptional events might affect expression. For instance, while in neurons the amount of A-β produced is related to BACE-1 expression levels, in astrocytes (16,17), as well as in other tissues (1–3), a poor correlation between BACE-1 transcript and β-secretase activity was found. This is the case for exocrine pancreas, where the extremely high levels of BACE-1 transcript are not reflected by the presence of either the protein or the activity (1,18). A hypothesis to explain this discrepancy is a mechanism involving an inhibition of translation initiation (19). Recently, however, BACE-1 protein has been detected in exocrine pancreas, even though it is cleaved by an unknown protease (20). In any event, it should be noted that, while the amounts of proteolized BACE-1 in pancreas and liver are comparable (20), the levels of mRNA are several-fold higher in pancreatic tissue (1). Another indication in support of a translational control of BACE-1 expression comes from the evidence that changes in BACE-1 mRNA have never been observed in the brains of AD patients, despite the increase in protein and/or activity (21–23).
Translation can be controlled by structures that are present in the mRNA untranslated regions such as internal ribosome entry site (IRES) (24,25) or iron responsive elements (IRE) (26). In addition, other mRNA features can contribute to the translational control of gene expression. Among them upstream AUGs (uAUGs) and upstream open reading frames (uORFs) are thought to down-regulate the efficiency of translation initiation of the main ORF (27,28) and were shown to repress the translation of important proteins such as TPO (29), connexin 41 (30), huntingtin (31), the transcription factors GCN4 (32) and ATF4 (33), as well as other proteins (34–36).
In the present paper we investigate the involvement of these mechanisms in the control of BACE-1 translation and the possibility that this control might operate in astrocytes upon their activation. The possible involvement of this mechanism in the pathogenesis of AD is discussed.
MATERIALS AND METHODS
Materials
Restriction and modification enzymes for DNA cloning were from New England Biolabs; DNA oligonucleotides from Primm; [α-32P]CTP from Amersham; reagents and media for cell culture from Cambrex. RNA extraction was performed with the RNeasy midi kit (Qiagen). Human tissues were provided by F.Bertuzzi (Islet Transplantation Unit) and M.Losa (Neurosurgery Unit), San Raffaele Scientific Institute, Milano, Italy. Human brain total RNA was from Clontech. The EST clone (GenBank accession number BG833894) encoding the full length BACE-1 transcript leader from Sus scrofa cDNA, was obtained from Children’s Hospital Oakland-BACPAC Resources, Oakland, CA. The MVA-T7pol vaccinia virus was provided by Dr Gerd Sutter, Institute of Molecular Virology, GSF-National Research Centre for Environment and Health, Oberschleissheim, Germany.
Cloning of BACE-1 transcript leaders
Reverse transcription (primer, CCG TGG GCA GGC AGC AC) and amplification (sense, AGT CCC ATG GAG CTG CGA GCC GCG AGC T; antisense, CCA TCC ACA GCA GGA GCC AGG GCA) of BACE-1 from total RNA from exocrine pancreas, brain temporal cortex or SK-N-BE cells was performed with the One-Step RT–PCR kit (Qiagen). The same sense oligo was used with a nested antisense oligo (TTG GGC CAT GGT GGG CCC CGG CCT T) for re-amplification. The PCR product was inserted into the pGEM-T vector (Promega) obtaining pGEMT-BACE1-5′y (short BACE-1 transcript leader). To obtain the long BACE-1 transcript leader, the BsrGI fragment from an EST clone (GenBank accession number AL544727, Invitrogen) was cloned into the Acc65I site of pBluescript-KS(+) (Stratagene), to obtain pBS-Bace1-Est, and then amplified by PCR (sense, AGT CCC ATG GAG CTG CGA GCC GCG AGC TGG ATT A; antisense, TTG GGC CAT GGT GGG CCC CGG CCT T), digested with NcoI and cloned into pGEMT-BACE1-5′y after removal of the NcoI fragment. The new plasmid was named pGEMD-BACE1-5′x.
Cloning of expression vectors and site-directed mutagenesis
The dicistronic pBRL-Nco empty vector was prepared from pBATmod2RL-IRES-LUC+ (37) by removal of the EMCV IRES (NcoI–BamHI digestion, followed by T4 DNA polymerase treatment before ligation). For the preparation of the bi-monocistronic pBRm2L-Nco empty vector, pBATmod2-RL-mono-Nco-LUC+ (D.De Pietri Tonelli, unpublished) was opened with BglII and SalI, and a synthetic double-strand oligo with a T7 terminator followed by a T7 promoter was inserted (sense, GAT CTA ACC CCT TGG GGC CTC TAA ACG GGT CTT GAG GGG TTT TTT GCA GAT CTC GAG GCC TTA ATA CGA CTC ACT ATA GGG; antisense, TCG ACC CTA TAG TGA GTC GTA TTA AGG CCT CGA GAT CTG CAA AAA ACC CCT CAA GAC CCG TTT AGA GGC CCC AAG GGG TTA).
BACE-1 fragments were extracted with NcoI from pGEMD-BACE1-5′x and pGEMT-BACE1-5′y and ligated into the NcoI site of pBRL-Nco or pBRm2L-Nco to obtain dicistronic pBRL-B1x, pBRL-B1y, pBRL-B1xas and pBRL-B1yas, and bi-monocistronic pBRm2L-B1x and pBRm2L-B1y. pBRm2L-B1uORF1 and pBRm2L-uPEP were prepared by inserting into pBRm2L-Nco the fragments obtained by PCR amplification of pGEMD-BACE1-5′x (sense, AGT CCC ATG GAG CTG CGA GCC GCG AGC TGG ATT A; antisense for B1uORF, AGG GGC GGC CAT GGC GGG CCG GT; antisense for uPEP, ACG TCC GCG GAG CTG CGA GCC GCG AGC T). The first fragment was inserted using NcoI digestion. The second was cleaved with SmaI and SacII, and inserted into an NcoI- (blunt with Mung Bean Nuclease) and SacII-digested plasmid.
Single point mutations were generated with Pfu turbo polymerase (Stratagene) by PCR-based site directed mutagenesis with overlapping oligos (38). pGEMD-BACE1x-mut1 was generated from pGEMD-BACE1-5′x (sense, ACC GGC CCG CCT TGC CCG CCC CT; antisense, AGG GGC GGG CAA GGC GGG CCG GT); pGEMD-BACE1x-mut2 from pGEMD-BACE1x-mut1 (sense, GTG CCG TTG TAG CGG GCT CCG GA; antisense, TCC GGA GCC CGC TAC AAC GGC AC). pGEMD-BACE1x-mut3 from pGEMD-BACE1x-mut2 (sense, CGA GCT GGA TTT TGG TGG CCT GA; antisense, TCA GGC CAC CAA AAT CCA GCT CG). DNA fragments were digested with NcoI and cloned in pBRm2L-Nco.
RNase protection assay
The full-length 32P-labeled riboprobe was prepared with the T7-SP6 in vitro transcription system (Promega) starting from SpeI-linearized pGEMD-BACE1-5′x and purified over an RNeasy mini spin column (Qiagen) according to the RNA clean-up protocol. Two shorter probes, utilized as size markers, were obtained following the same procedure but starting from BamHI (probe length 316 nt), or XmaI-linearized (probe length 241 nt) pGemT-BACE1-5′y. The RNAse protection assay was performed with the RPA III kit (Ambion) according to the manufacturer’s instructions with the following modifications: 15 µg of total RNA (from either human pancreas or brain) was incubated with the riboprobe (55 000 c.p.m.) overnight at 60°C, and then digested with a 1:100 dilution of the RNAse A/T1 mix. Protected fragments were separated in a 0.75 mm thick and 40 cm long 5% polyacrylamide gel, and revealed by either overnight exposure onto a phosphoimager screen (Molecular Dynamics) or 10 days of exposure onto an X-ray film (Kodak).
Cell culture and transfection
Cells were maintained as follows: U373-MG in Earle’s Minimal Essential Medium (MEM) supplemented with 10% fetal calf serum (FCIII, Hyclone) and 1 mM sodium pyruvate; HeLa and SK-N-BE cells in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 5% FCIII and 10% Donor Horse Serum for HeLa or 15% FCIII, 1 mM sodium pyruvate and 1 mM non-essential amino acids for SK-N-BE. All media were supplemented with 100 U/ml penicillin, 100 µg/ml streptomycin and 2 mM glutamine, and cells were cultured at 37°C in a humidified 5% CO2 atmosphere. Rat cortical astrocytes and hippocampal neurons were prepared according to McCarthy and De Vellis (39) and Ryan and Smith (40), respectively. After 10–15 days in culture, astrocytes were shaken (24 h, 220 r.p.m.) to remove microglia. Proliferating cells were plated the day before an experiment in order to reach about 70% confluence at the time of transfection. Cells were infected with the MVA-T7pol virus in MEM for 30 min at 37°C and then transfected for 6 h with plasmids carrying the DNA construct of interest. The transfection was performed with Superfect (Qiagen) according to the manufacturer’s instructions. Astrocyte activation was obtained by a 24 h treatment with 10 ng/ml recombinant rat IL-1β and 30 ng/ml recombinant rat TNF-α.
Luciferase reporter assay
Firefly luciferase (Fluc) and Renilla luciferase (Rluc) activities were revealed with the Dual-Luciferase reporter assay system (Promega) and measured (20 s readings) using a PlateLumino double injector luminometer (Stratec) or a Victor3 plate reader (Perkin-Elmer). Four independent reactions were carried out for each experimental point.
Polysome analysis
Transfected HeLa cells were lysed with polysomal buffer (100 mM NaCl, 30 mM MgCl2, 10 mM Tris–HCl, 0.1% NP-40, 100 U/ml RNAsin, pH 7.5). Aliquots (100 µl: ∼10 OD) of the cleared lysate were overlayed onto a 7–50% w/v sucrose gradient (containing 50 mM Tris–acetate, 50 mM NH4Cl, 1 mM DTT, pH 7.5) and pelleted by ultracentrifugation in a SW41 rotor (39K, 160 min). Ten fractions (1 ml each) were collected from the top using a gradient collector with continuous monitoring at 254 nm. Total RNA from each fraction was obtained by proteinase K treatment followed by phenol/chloroform extraction. Fluc transcript levels were revealed by RT–PCR (SuperScript and Platinum Taq, Invitrogen) with oligo-dT priming followed by amplification with gene specific oligos (sense, GCC TAA AGG TGT CGC TCT GCC T; antisense, GAA GAT GTT GGG GTG TTG GAG CA).
In vitro translation and mRNA decay
For in vitro translation the plasmids pZac-Bace1x-Luc+ and pZac-Bace1y-Luc+ were prepared by inserting into the SalI–NsiI digested pZac-Luc+ (37) the SalI–NsiI fragments of pBRm2L-B1x and pBRm2L-B1y, respectively. Synthesis of Fluc cRNA from PacI- or PstI-linearized pZac plasmids was obtained with the mMESSAGEmMACHINE kit (Ambion). Translation (0.3 µg of cRNA in 25 µl) was performed with the ReticLysate IVT kit (Ambion). For the decay assay, the radioactive cRNA was prepared with the Riboprobe in vitro transcription system (Promega) and purified with the Rneasy spin column (Qiagen). The cRNA (∼500 000 c.p.m.) was incubated in the translation reaction and purified again at different time points before scintillation counting.
RESULTS
In order to clone the transcript leader of BACE-1 we performed an RT–PCR on total mRNA from human brain and pancreas. In our conditions, we did not get any amplification product from brain. However, in pancreas we obtained a band smaller than expected on the basis of the published sequence (accession number AF190725). Cloning and sequencing of the amplified DNA revealed a putative alternative splice variant of the BACE-1 transcript leader. We submitted this shorter sequence to the GenBank database under the accession number AF324837. The same sequence was also amplified from SK-N-BE cells.
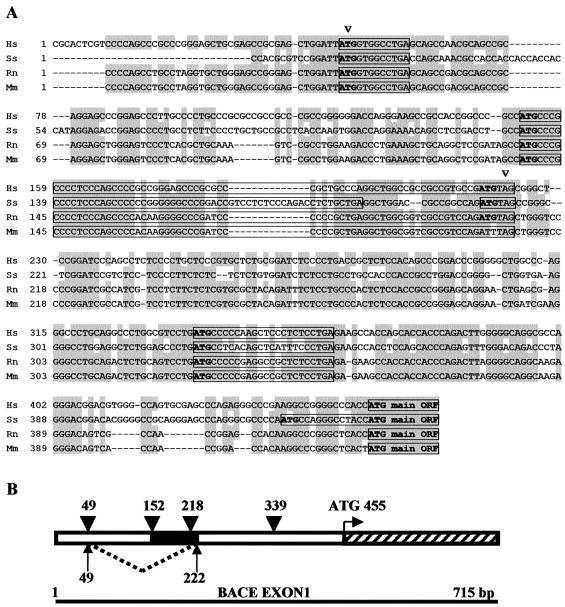
Figure 1 shows the alignment of the cDNA of the BACE-1 transcript leaders from various mammalian species and highlights the putative splicing sites. BACE-1 transcript leader is highly conserved, GC rich and contains several uAUGs and a uORF of 72 nt (60 nt in pig sequence). Although we did not map the actual donor–acceptor splice site (there are three possible combinations: ATTA..GCGG; ATT..AGCGG; AT..TAGCGG), it can be noticed that nucleotides around the splice sites are well conserved, making alternative splicing also likely in the other species analysed. Interestingly, the splicing event removes three out of four uAUGs.
Figure 1.
Scheme of the proposed alternative splicing in BACE-1 transcript leader. (A) Alignment of full-length BACE-1 transcript leaders from human (Hs), pig (Ss), rat (Rn) and mouse (Mm) cDNAs. Arrowheads mark proposed donor–acceptor splice sites for the new variant of the BACE-1 transcript leader. Conserved nucleotides are highlighted in grey. Upstream ATGs are written in bold and boxed sequences represent the putative uORFs. (B) Scheme of the alternative splicing within the first exon of human BACE-1. Dotted line and arrows delimit the sequence missing in the shorter variant. Arrowheads highlight the upstream ATGs, while the black and the striped boxes, the uORF and the first codons of the main ORF, respectively.
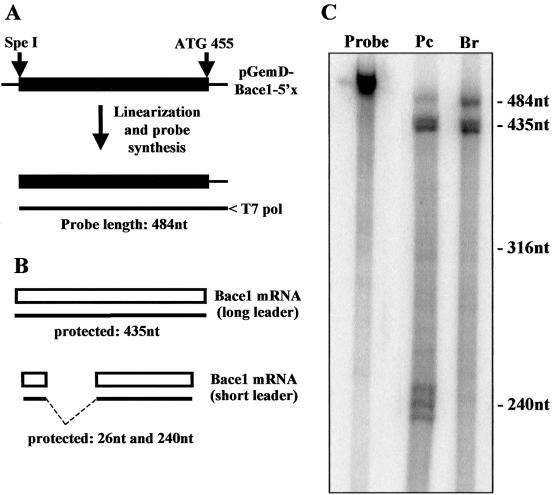
Since we had isolated the short BACE-1 transcript leader from human exocrine pancreas and SK-N-BE cells, we then investigated whether this variant was also expressed in normal human brain. We performed an RNase protection assay with one riboprobe to quantify the relative amount of the two splice variants (Fig. 2A). The long and the short transcript leaders were expected to generate protected fragments of 435 and 240 nt, respectively (Fig. 2B). Total RNA (15 µg) from either human brain or exocrine pancreas were incubated with the same amount of 32P-labelled riboprobe and digested with RNase. In brain tissue only the long form of the BACE-1 transcript leader was expressed, while in pancreas both forms were present at comparable levels (Fig. 2C). The same finding was obtained with an antisense probe synthesized using the short transcript leader as template (data not shown).
Figure 2.
RNase protection assay for the BACE-1 transcript leader. (A) Scheme for the synthesis of the riboprobe for BACE-1. The plasmid pGEMD-BACE1-5′x with the full-length BACE-1 transcript leader was linearized with SpeI and used as template for the synthesis of the 32P-labelled antisense riboprobe performed with the T7 RNA polymerase (probe length 484 nt). (B) Scheme of RNase protection assay. The protected fragments deriving from the full-length and the short BACE-1 leaders are 435 and 240 nt, respectively. The short variant produces an additional fragment of 26 nt that remains undetectable. (C) Result of RNase protection. 15 µg of total RNA from either human exocrine pancreas (Pc) or brain (Br) were incubated with 55 000 c.p.m. of 32P-labelled riboprobe. The longer fragment of 435 nt is visible below the undigested probe, while the shorter fragment of 240 nt is present in the sample from pancreas only. Size markers of 316 and 240 nt were prepared as described in Materials and Methods.
We next decided to test whether the BACE-1 transcript leader was acting as an IRES element, thereby supporting cap-independent initiation of translation. Standard DNA transfection could not be used since a promoterless approach revealed the presence of a cryptic promoter in the transcript leader of BACE-1 (data not shown). For this reason the full-length or the spliced variant of BACE-1 leader, was inserted in the dicistronic plasmid pBRL-Nco between two reporter genes (37). In this way the activity of the 3′ cistron (Fluc) was normalized to the activity of the 5′ cistron (Rluc) to account for variations in the efficiency of transfection. Each dicistronic construct, under the transcriptional control of a single T7 promoter, was transiently transfected in SK-N-BE or HeLa cells. Expression was driven by the MVA-T7pol vaccinia virus system, a strategy that allows the transcription of the gene of interest to occur solely in the cytosol (41). This system was chosen to avoid artefacts arising from nuclear events such as transcription via cryptic promoters or mRNA processing (37). We did not detect any significant IRES activity with either transcript leaders (data not shown). However, our results leave open the possibility that IRES activity might be promoted by a ‘nuclear experience’ of the transcript.
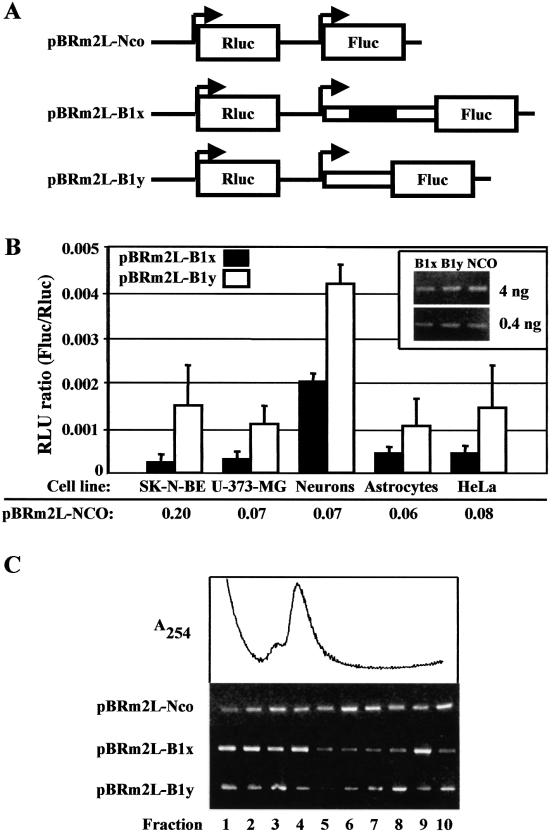
In order to investigate the effect of the BACE-1 transcript leader on cap-dependent translation, we prepared a series of bi-monocistronic plasmids (Fig. 3A). These plasmids contain two independent transcriptional units under the control of two T7 promoters. The first cassette generates a transcript containing the Rluc reporter gene, which was used to normalize for differences in transfection efficiency. The second cassette generates an mRNA with the Fluc gene preceded by either the long (pBRm2L-B1x) or the short (pBRm2L-B1y) BACE-1 transcript leader. These plasmids were transiently transfected by the MVA-T7pol system in the following cells: SK-N-BE (human neuroblastoma); U-373-MG (human astrocytoma); primary hippocampal neurons and cortical astrocytes from newborn rat; HeLa (human adenocarcinoma). Translation was quantified as the ratio of the activities of the two reporter genes (Fluc/Rluc). Both the long and the short BACE-1 transcript leaders were less active (one to two orders of magnitude) than the empty vector pBRm2L-Nco in supporting translation initiation (Fig. 3B) [for quantification, see also De Pietri Tonelli et al. (37)]. The possibility that changes in transcript stability might account for such a difference was ruled out by evaluating the levels of Fluc transcripts by RT–PCR (Fig. 3B, inset). Interestingly, the short form of BACE-1 leader, which lacks three out of four uAUGs, was 2–8-fold more active than the long form. To better evaluate the translation efficiency, we examined by polysome analysis the cytoplasmic distribution of Fluc mRNA in transfected cells. The result in Figure 3C shows that the control mRNA (from pBRm2L-Nco) was mostly present in the polysomal fractions of the gradient. In contrast, the majority of the mRNA from either pBRm2L-B1x or pBRm2L-B1y was detected in the slower sedimenting region of the gradient, thereby indicating repression of translation.
Figure 3.
Translation driven by the BACE-1 transcript leaders. (A) Scheme of bi-monocistronic constructs. The plasmid pBRm2L-Nco drives expression of the two reporter genes, Rluc and Fluc, under the control of separate T7 promoters and in an optimal context for translation initiation. The long and short BACE-1 transcript leaders were cloned in front of the Fluc ORF to generate pBRm2L-B1x and pBRm2L-B1y, respectively. (B) Transient transfection of pBRm2L-B1x or pBRm2L-B1y in SK-N-BE, U-373-MG, neurons, astrocytes and HeLa cells. To account for differences in transient transfection efficiencies, the activity (relative luminescence units, RLU) of Fluc was normalized to that of Rluc (Fluc/Rluc). The corresponding RLU ratio values for the empty vector pBRm2L-Nco are also shown for comparison. All the results are the mean of at least three independent experiments. Measurements of luciferase activity were performed in triplicate as described in Materials and Methods. The inset shows an RT–PCR experiment (with two different total RNA loadings) to evaluate the relative amount of Fluc transcripts in transfected HeLa cells. The efficiency of transfection was monitored by Rluc activity and proved to be comparable in all samples. (C) Cytoplasmic distribution of Fluc mRNA by sucrose gradients. Cytoplasmic extracts of transfected HeLa cells were lysed and resolved on 7–50% w/v sucrose gradients. Fractions were analysed by RT–PCR to reveal the localization of the transcript deriving from the transfected DNA (pBRm2L-Nco, pBRm2L-B1x or pBRm2L-B1y). The trace shows the continuous absorbance profile monitored during the collection of the fractions. Fractions 1–3 contain mainly mRNPs and the ribosomal subunits; the monosome region peaks at fraction 4; fractions 5–10 contain the polyribosomes.
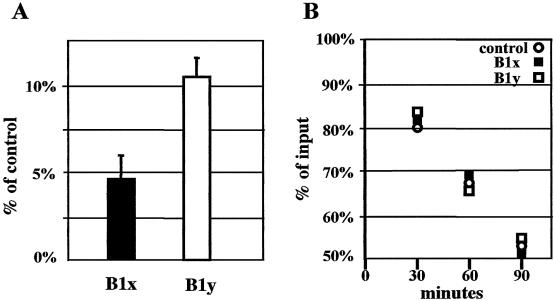
In order to investigate whether the data obtained by transfection were affected by variations in mRNA stability, we performed experiments of in vitro translation also monitoring the mRNA decay rate. Translation driven by either BACE-1 leader in a rabbit reticulocyte translation assay was about two orders of magnitude lower than control (Fig. 4A). Noticeably, the stability of all radioactively labelled transcripts was identical (Fig. 4B).
Figure 4.
In vitro translation driven by the BACE-1 transcript leader. (A) Activity of Fluc translated under the control of the long (B1x) or the short (B1y) BACE-1 transcript leaders. The cRNA produced from the empty vector pZac-Luc+ gives the 100% value. (B) Decay of the corresponding cRNAs during the translation reactions. cRNA from pZac-Luc+ is the control and radioactive input is considered to be 100%.
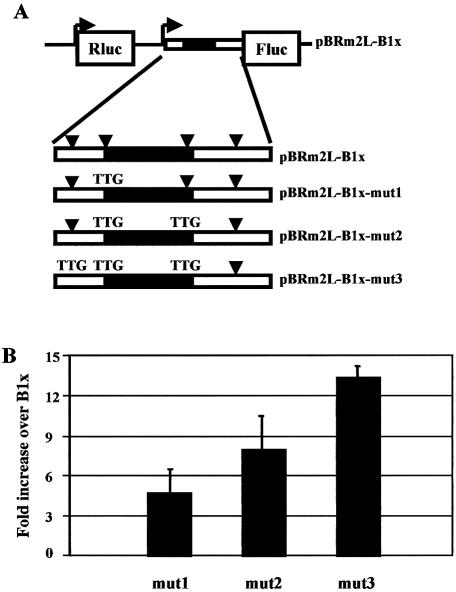
To understand the relative contribution of the four uATGs in the translation repression, we performed an analysis of the context for translation initiation surrounding each uATG (27) with the NetStart program (42). The prediction showed that the second uATG, which is positioned in a good Kozak context, is recognized by the ribosome with at least the same efficiency as the main ATG, while the other three uATGs are not in good contexts for translation initiation. To test this prediction, we mutated the second uATG of the long BACE-1 transcript leader (pBRm2L-B1x-mut1 plasmid) (Fig. 5A). The transfection in SK-N-BE cells revealed that the activity of the mutant was ∼5-fold higher than the long BACE-1 leader (pBRm2L-B1x), but still lower than the short form (pBRm2L-B1y). Then, we mutated sequentially the other uATGs as described in Figure 5A, generating pBRm2L-B1x-mut2 and pBRm2L-B1x-mut3. The effect of these mutations was additive in overcoming the translational hurdle (Fig. 5B).
Figure 5.
Effect of upstream ATGs in BACE-1 transcript leader on translation efficiency. (A) Scheme of the long BACE-1 transcript leader in the bi-monocistronic plasmid. The four upstream ATGs are marked with arrowheads. The black box represents the upstream ORF. The upstream ATGs were mutated to TTG, generating pBRm2L-B1x-mut1, pBRm2L-B1x-mut2, pBRm2L-B1x-mut3. (B) Transient transfection of bi-monocistronic plasmids in SK-N-BE cells. Results are normalized to the value of the long BACE-1 leader (pBRm2L-B1x).
According to this result, the second uATG exerts a prominent repression on translation. This might be accomplished by recruiting the ribosomal machinery for an abortive cycle of translation. To test this hypothesis, we generated the bi-monocistronic plasmid pBRm2L-B1x-uORF1, in which the second reporter gene (Fluc) is positioned immediately after the second uATG. As predicted, transfection in SK-N-BE and U-373-MG cells confirmed that this uATG is efficiently recognized by the translation machinery (data not shown).
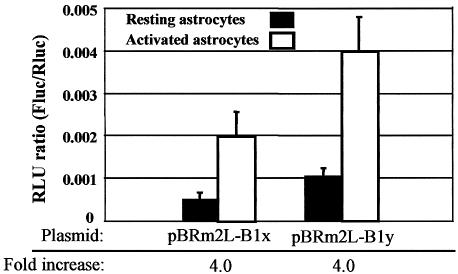
To see whether translational control of BACE-1 has relevance in physiopathological conditions, we used the activated astrocyte model. Astrocytes pretreated for 24 h with IL-1β (10 ng/ml) and TNF-α (30 ng/ml) or nothing were transfected for 4 h with bi-monocistronic plasmids using the MVA-T7pol system. In this way differences between expressions of the reporter genes can be attributed to variations in translation efficiency. Interestingly, Fluc translation driven by BACE-1 transcript leader, either long or short, was 4-fold higher in activated astrocytes, while translation of the empty construct pBRm2L-Nco was unaffected (Fig. 6). In contrast, no changes in translation were observed when astrocytes were exposed to acute (2 h) stimulation with maximal concentration of physiological agonists such as glutamate, ATP and PGE2 (data not shown).
Figure 6.
Translation driven by BACE-1 transcript leader in resting and activated astrocytes. Astrocytes were transfected in normal culture conditions (resting) or after activation with IL-1β and TNF-α (activated). Transfection was performed with bi-monocistronic plasmids containing either the long (pBRm2L-B1x) or short (pBRm2L-B1y) BACE-1 transcript leader. The increase in Fluc translation between resting and activated astrocytes is also reported (fold increase).
DISCUSSION
The BACE-1 transcript leader is long and highly conserved among various mammalian species, two features that are strongly suggestive of a translational control of protein expression. By working with reporter genes, we demonstrate that BACE-1 transcript leader acts as a ‘silencer’ of translation. Since we had previously reported that the level of a toxic protein, called Scamper, is maintained at a low level by an IRES (37), we explored whether this was also the case for BACE-1. The use of the dicistronic approach, an experimental strategy conventionally used to assess the presence of an IRES, did not provide confirmation for the presence of such a mechanism in the BACE-1 transcript. Rather, we obtained evidence that the inhibition of translation is, at least partially, due to the presence of uATGs. It is known that uATGs can inhibit translation in several ways. When they are recognized by the translation machinery, a futile cycle can occur, so that only the ribosomes skipping the uATG (leaky-scanning) can reach the main ORF. Accordingly, the level of inhibition is directly related to the context for translation initiation: the better an uATG is recognized, the higher is the resulting inhibition (27). Even in conditions of optimal uATG recognition, a small percentage of ribosomes, after translation of the uORF, can continue scanning for start sites (re-initiation), eventually reaching the ATG of the main ORF (43,44). Moreover, some peptides produced by translation of a uORF have been reported to interact with the ribosome, further diminishing the efficiency of protein translation (45–48). In the case of BACE-1, the second uATG is efficiently recognized by the ribosome. However, the presence of another ATG as the last codon before termination in the 72 nt uORF made it impossible to discriminate between leaky scanning and a reinitiation mechanism. We cannot provide conclusive evidence whether complete translation of this uORF occurs and has relevance to the process. Since the recognition of start sites can be modulated, the presence of uATGs might provide a way to regulate protein expression. For instance, phosphorylation of the eukaryotic initiation factor eIF2-α is known to decrease the efficiency of ATG recognition with ensuing inhibition of the translation of most transcripts (49). However, translation of uATG-containing transcripts is paradoxically increased because more ribosomes overcome uATGs and become available for the translation of the main ORF (50). According to this view, BACE-1 translation might increase in conditions that favour eIF2-α phosphorylation.
The discovery that BACE-1 transcript leader can be alternatively spliced provides an additional possibility for the regulation of BACE-1 expression. In fact, the alternative splicing we describe here reduces the number of uATGs (from 4 to 1) and, as expected, increases the efficiency of translation of reporter genes. By RNase protection and RT–PCR we show that this splicing occurs in exocrine pancreas and neuroblastoma cells, but not in normal human brain. Therefore, BACE-1 expression is likely to be affected by changes in the efficiency of this splicing event and this might play a role under different physiological as well as pathological conditions.
Although it has long been assumed that the toxic A-β peptide is produced by neurons, the possibility that astrocytes can contribute to this process is gaining momentum (51,52). Noticeably, in astrocytes from either AD patients or the Tg2576 transgenic mouse model, BACE-1 protein expression was found only around amyloid plaques, where reactive astrocytes are likely to be present (17,51). The current view is that astrocytes do not express BACE-1—and thus do not produce A-β—unless they are brought to a state of chronic activation or they are put in culture (51). Nothing is known about the switch to promote BACE-1 expression. Transcriptional mechanisms are likely to be involved, but we provide evidence that translational regulation can contribute as well. Our data clearly show that when astrocytes are activated by chronic exposure to cytokines in culture, a marked reduction in the inhibition of translation driven by the BACE-1 transcript leader is recorded. This mechanism is independent of the splicing event described above, since it was observed with both splice variants when transient transfection of reporter genes was performed with the RNA-T7pol vaccinia system, i.e. under conditions that bypass nuclear processing of the transcripts. Interestingly, translation driven by the BACE-1 transcript leader in activated astrocytes reaches an efficiency that is similar to that obtained in neurons. This increase has been observed during a process of chronic activation, which also occurs in vivo, while acute activation with physiological stimuli failed to reproduce the effect. In our conditions, the most likely explanation of this effect resides in a variation in translation efficiency. We cannot exclude that variations in transcript stability may contribute. However, this clearly shows that after astrocyte activation a non-transcriptional effect on BACE-1 protein production operates.
In conclusion, we provide evidence that BACE-1 expression is inhibited at the translational level by the presence of uATGs. Furthermore, we report that splicing of the transcript leader can change the number of uATGs that are present, thereby providing an additional mechanism for the control. Finally, we show that a release of the inhibition of translation occurs in activated astrocytes, potentially contributing to A-β production. Therefore, even though it is known that BACE-1 can also act on substrates other than APP (53), our findings provide a possible explanation of why in brain tissue of AD patients an increase in BACE-1 protein or activity does not require an increase in BACE-1 mRNA or in the ratio between α- and β-secretase mRNAs (21,22,23), and why changes in BACE-1 mRNA have never been observed around amyloid plaques in transgenic Tg2576 mice (16).
Acknowledgments
ACKNOWLEDGEMENTS
We thank S.Durante for excellent technical help, L.Sala for help with polysome analysis, D.Dunlap for critical reading of the manuscript and M.Losa and F.Bertuzzi for the gift of human temporal cortex and exocrine pancreas, respectively. We also thank G.Sutter and the Institute of Molecular Virology of the GSF—Forschungszentrum fuer Umwelt und Gesundheit GmbH for the MVA-T7pol virus. This work was carried out within the framework of the Italian Ministry of Research Centre of Excellence in Physiopathology of Cell Differentiation. Financial support was also from the Armenise-Harvard Foundation, the Italian Telethon (grant E.0888 to FG), the CNR target project in Biotechnology (to FG), and grants from the EU Commission (DECG project CLG3-CT-2001-02004 to FG; APOPIS project LSHM-CT-2003-503330 to DZ).
DDBJ/EMBL/GenBank accession no. AF324837
REFERENCES
- 1.Vassar R., Bennett,B.D., Babu-Khan,S., Kahn,S., Mendiaz,E.A., Denis,P., Teplow,D.B., Ross,S., Amarante,P., Loeloff,R. et al. (1999) Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science, 286, 735–741. [DOI] [PubMed] [Google Scholar]
- 2.Yan R., Bienkowski,M.J., Shuck,M.E., Miao,H., Tory,M.C., Pauley,A.M., Brashier,J.R., Stratman,N.C., Mathews,W.R., Buhl,A.E. et al. (1999) Membrane-anchored aspartyl protease with Alzheimer’s disease beta-secretase activity. Nature, 402, 533–537. [DOI] [PubMed] [Google Scholar]
- 3.Sinha S., Anderson,J.P., Barbour,R., Basi,G.S., Caccavello,R., Davis,D., Doan,M., Dovey,H.F., Frigon,N., Hong,J. et al. (1999) Purification and cloning of amyloid precursor protein beta-secretase from human brain. Nature, 402, 537–540. [DOI] [PubMed] [Google Scholar]
- 4.Hussain I., Powell,D., Howlett,D.R., Tew,D.G., Meek,T.D., Chapman,C., Gloger,I.S., Murphy,K.E., Southan,C.D., Ryan,D.M. et al. (1999) Identification of a novel aspartic protease (Asp 2) as beta-secretase. Mol. Cell. Neurosci., 14, 419–427. [DOI] [PubMed] [Google Scholar]
- 5.Lin X., Koelsch,G., Wu,S., Downs,D., Dashti,A. and Tang,J. (2000) Human aspartic protease memapsin 2 cleaves the beta-secretase site of beta-amyloid precursor protein. Proc. Natl Acad. Sci. USA, 97, 1456–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu G., Nishimura,M., Arawaka,S., Levitan,D., Zhang,L., Tandon,A., Song,Y.Q., Rogaeva,E., Chen,F., Kawarai,T. et al. (2000) Nicastrin modulates presenilin mediated notch/glp-1signal transduction and βAPP processing. Nature, 407, 48–54. [DOI] [PubMed] [Google Scholar]
- 7.Esler W.P., Kimberly,W.T., Ostaszewski,B.L., Ye,W., Diehl,T.S., Selkoe,D.J. and Wolfe,M.S. (2002) Activity-dependent isolation of the presenilin-gamma-secretase complex reveals nicastrin and a gamma substrate. Proc. Natl Acad. Sci. USA, 99, 2720–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farmery M.R., Tjernberg,L.O., Pursglove,S.E., Bergman,A., Winblad,B. and Naslund,J. (2003) Partial purification and characterization of γ-secretase from post-mortem human brain. J. Biol. Chem., 278, 24277–24284. [DOI] [PubMed] [Google Scholar]
- 9.Selkoe D.J. (2001) Alzheimer’s disease: genes, proteins, and therapy. Physiol. Rev., 81, 741–766. [DOI] [PubMed] [Google Scholar]
- 10.Bodendorf U., Danner,S., Fischer,F., Stefani,M., Sturchler-Pierrat,C., Wiederhold,K.H., Staufenbiel,M. and Paganetti,P. (2002) Expression of human β-secretase in the mouse brain increases the steady-state level of β-amyloid. J. Neurochem., 80, 799–806. [DOI] [PubMed] [Google Scholar]
- 11.Holsinger R.M., McLean,C.A., Beyreuther,K., Masters,C.L. and Evin,G. (2002) Increased expression of the amyloid precursor beta-secretase in Alzheimer’s disease. Ann. Neurol., 51, 783–786. [DOI] [PubMed] [Google Scholar]
- 12.Tyler S.J., Dawbarn,D., Wilcock,G.K. and Allen,S.J. (2002) alpha- and beta-secretase: profound changes in Alzheimer’s disease. Biochem. Biophys. Res. Commun., 299, 373–376. [DOI] [PubMed] [Google Scholar]
- 13.Fukumoto H., Cheung,B.S., Hyman,B.T. and Irizarry,M.C. (2002) Beta-secretase protein and activity are increased in the neocortex in Alzheimer disease. Arch. Neurol., 59, 1381–1389. [DOI] [PubMed] [Google Scholar]
- 14.Yang L.B., Lindholm,K., Yan,R., Citron,M., Xia,W., Yang,X.L., Beach,T., Sue,L., Wong,P., Price,D. et al. (2003) Elevated beta-secretase expression and enzymatic activity detected in sporadic Alzheimer disease. Nat. Med., 9, 3–4. [DOI] [PubMed] [Google Scholar]
- 15.Lange-Dohna C., Zeitschel,U., Gaunitz,F., Perez-Polo,J.R., Bigl,V. and Rossner,S. (2003) Cloning and expression of the rat BACE1 promoter. J. Neurosci. Res., 73, 73–80. [DOI] [PubMed] [Google Scholar]
- 16.Bigl M., Apelt,J., Luschekina,E.A., Lange-Dohna,C., Rossner,S. and Schliebs,R. (2000) Expression of β-secretase mRNA in transgenic Tg2576 mouse brain with Alzheimer plaque pathology. Neurosci. Lett., 292, 107–110. [DOI] [PubMed] [Google Scholar]
- 17.Rossner S., Apelt,J., Schliebs,R., Perez-Polo,J.R. and Bigl,V. (2001) Neuronal and glial beta-secretase (BACE) protein expression in transgenic Tg2576 mice with amyloid plaque pathology. J. Neurosci. Res., 64, 437–446. [DOI] [PubMed] [Google Scholar]
- 18.Bodendorf U., Fischer,F., Bodian,D., Multhaup,G. and Paganetti,P. (2001) A splice variant of beta-secretase deficient in the amyloidogenic processing of the amyloid precursor protein. J. Biol. Chem., 276, 12019–12023. [DOI] [PubMed] [Google Scholar]
- 19.Ehehalt R., Michel,B., De Pietri Tonelli,D., Zacchetti,D., Simons,K. and Keller,P. (2002) Splice variants of the beta-site APP-cleaving enzyme BACE1 in human brain and pancreas. Biochem. Biophys. Res. Commun. 293, 30–37. [DOI] [PubMed] [Google Scholar]
- 20.Huse J.T., Byant,D., Yang,Y., Pijak,D.S., D’Souza,I., Lah,J.J., Lee,V.M., Doms,R.W. and Cook,D.G. (2003) Endoproteolysis of beta-secretase (beta-site amyloid precursor protein-cleaving enzyme) within its catalytic domain. A potential mechanism for regulation. J. Biol. Chem., 278, 17141–17149. [DOI] [PubMed] [Google Scholar]
- 21.Marcinkiewicz M. and Seidah,N.G. (2000) Coordinated expression of beta-amyloid precursor protein and the putative beta-secretase BACE and alpha-secretase ADAM10 in mouse and human brain. J. Neurochem., 75, 2133–2143. [DOI] [PubMed] [Google Scholar]
- 22.Gatta L.B., Albertini,A., Ravid,R. and Finazzi,D. (2002) Levels of beta-secretase BACE and alpha-secretase ADAM10 mRNAs in Alzheimer hippocampus. Neuroreport, 13, 2031–2033. [DOI] [PubMed] [Google Scholar]
- 23.Preece P., Virley,D.J., Costandi,M., Coombes,R., Moss,S.J., Mudge,A.W., Jazin,E. and Cairns,N.J. (2003) beta-Secretase (BACE) and GSK-3 mRNA levels in Alzheimer’s disease. Brain Res. Mol. Brain Res., 116, 155–158. [DOI] [PubMed] [Google Scholar]
- 24.Hellen C.U. and Sarnow,P. (2001) Internal ribosome entry site in eukaryotic mRNA molecules. Genes Dev., 15, 1593–1612. [DOI] [PubMed] [Google Scholar]
- 25.Vagner S., Galy,B. and Pyronnet,S. (2001) Irresistible IRES: attracting the translation machinery to internal ribosome entry sites. EMBO Rep., 2, 893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hentze M.W. (1994) Translational control by iron-responsive elements. Adv. Exp. Med. Biol., 356, 119–126. [DOI] [PubMed] [Google Scholar]
- 27.Kozak M. (1999) Initiation of translation in prokaryotes and eukaryotes. Gene, 234, 187–208. [DOI] [PubMed] [Google Scholar]
- 28.Morris D.R. and Geballe,A.P. (2000) Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol., 20, 8635–8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghilardi N., Wiestner,A. and Skoda,R.C. (1998) Thrombopoietin production is inhibited by a translational mechanism. Blood, 92, 4023–4030. [PubMed] [Google Scholar]
- 30.Meijer H.A., Dictus,W.J., Keuning,E.D. and Thomas,A.A. (2000) Translational control of the Xenopus laevis connexin-41 5′-untranslated region by three upstream open reading frames. J. Biol. Chem., 275, 30787–30793. [DOI] [PubMed] [Google Scholar]
- 31.Lee J., Park,E.H., Couture,G., Harvey,I., Garneau,P. and Pelletier,J. (2002) An upstream open reading frame impedes translation of the huntingtin gene. Nucleic Acids Res., 30, 5110–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaba A., Wang,Z., Krishnamoorthy,T., Hinnebusch,A.G. and Sachs,M.S. (2001) Physical evidence for distinct mechanisms of translational control by upstream open reading frames. EMBO J., 20, 6453–6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harding H.P., Novoa,I., Zhang,Y., Zeng,H., Wek,R., Schapira,M. and Ron,D. (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell, 6, 1099–1108. [DOI] [PubMed] [Google Scholar]
- 34.Schluter G., Boinska,D. and Nieman-Seyde,S.C. (2000) Evidence for translational repression of the SOCS-1 major open reading frame by an upstream open reading frame. Biochem. Biophys. Res. Commun., 268, 255–261. [DOI] [PubMed] [Google Scholar]
- 35.Holzmann K., Ambrosch,I., Elbling,L., Micksche,M. and Berger,W. (2001) A small upstream open reading frame causes inhibition of human major vault protein expression from a ubiquitous mRNA splice variant. FEBS Lett., 494, 99–104. [DOI] [PubMed] [Google Scholar]
- 36.Guyot B., Arnaud,S., Phothirath,P., Rigal,D. and Mouchiroud,G. (2002) Upstream open reading frames regulate translation of Mona/Gads adapter mRNA in the megakaryocytic lineage. Platelets, 13, 459–464. [DOI] [PubMed] [Google Scholar]
- 37.DePietriTonelli D., Mihailovich,M., Schnurbus,R., Pesole,G., Grohovaz,F. and Zacchetti,D. (2003) Translational control of Scamper expression via a cell-specific internal ribosome entry site. Nucleic Acids Res., 31, 2508–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fisher C.L. and Pei,G.K. (1997) Modification of a PCR-based site-directed mutagenesis method. Biotechniques, 23, 570–571. [DOI] [PubMed] [Google Scholar]
- 39.McCarthy K.D. and De Vellis,J. (1980) Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J. Cell Biol., 85, 890–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan T.A. and Smith,S.J. (1995) Vesicle pool mobilization during action potential firing at hippocampal synapses. Neuron, 14, 983–989. [DOI] [PubMed] [Google Scholar]
- 41.Sutter G., Ohlmann,M. and Erfle,V. (1995) Non replicating vaccinia virus vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett., 371, 9–12. [DOI] [PubMed] [Google Scholar]
- 42.Pedersen A.G. and Nielsen,H. (1997) Neural network prediction of translation initiation sites in eukaryotes: perspectives for EST and genome analysis. ISMB, 5, 226–233. [PubMed] [Google Scholar]
- 43.Child S.J., Miller,M.K. and Geballe,A.P. (1999) Translational control by an upstream open reading frame in the HER-2/neu transcript. J. Biol. Chem., 274, 24335–24341. [DOI] [PubMed] [Google Scholar]
- 44.Griffin E., Re,A., Hamel,N., Fu,C., Bush,H., McCaffrey,T. and Asch,A.S. (2001) A link between diabetes and atherosclerosis: Glucose regulates expression of CD36 at the level of translation. Nat. Med. 7, 840–846. [DOI] [PubMed] [Google Scholar]
- 45.Fang P., Wang,Z. and Sachs,M.S. (2000) Evolutionarily conserved features of the arginine attenuator peptide provide the necessary requirements for its function in translational regulation. J. Biol. Chem., 275, 26710–26719. [DOI] [PubMed] [Google Scholar]
- 46.Jousse C., Bruhat,A., Carraro,V., Urano,F., Ferrara,M., Ron,D. and Fafournoux,P. (2001) Inhibition of CHOP translation by a peptide encoded by an open reading frame localized in the chop 5′UTR. Nucleic Acids Res., 29, 4341–4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gong F. and Yanofsky,C. (2002) Instruction of translating ribosome by nascent peptide. Science, 297, 1864–1867. [DOI] [PubMed] [Google Scholar]
- 48.Raney A., Law,G.L., Mize,G.J. and Morris D.R. (2002) Regulated translation termination at the upstream open reading frame in S-adenosylmethionine decarboxylase mRNA. J. Biol. Chem., 277, 5988–5994. [DOI] [PubMed] [Google Scholar]
- 49.Harding H.P., Calfon,M., Urano,F., Novoa,I. and Ron,D. (2002) Transcriptional and translational control in the Mammalian unfolded protein response. Annu. Rev. Cell. Dev. Biol., 18, 575–599. [DOI] [PubMed] [Google Scholar]
- 50.Dever T.E. (2002) Gene-specific regulation by general translation factors. Cell, 108, 545–556. [DOI] [PubMed] [Google Scholar]
- 51.Hartlage-Rubsamen M., Zeitschel,U., Apelt,J., Gartner,U., Franke,H., Stahl,T., Gunther,A., Schliebs,R., Penkowa,M., Bigl,V. et al. (2003) Astrocytic expression of the Alzheimer’s disease beta-secretase (BACE1) is stimulus-dependent. Glia, 41, 169–179. [DOI] [PubMed] [Google Scholar]
- 52.Lesne S., Docagne,F., Gabriel,C., Liot,G., Lahiri,D.K., Buee,L., Plawinski,L., Delacourte,A., MacKenzie,E.T., Buisson,A. et al. (2003) Transforming growth factor-beta 1 potentiates amyloid-beta generation in astrocytes and in transgenic mice. J. Biol. Chem., 278, 18408–18418. [DOI] [PubMed] [Google Scholar]
- 53.Kitazume S., Tachida,Y., Oka,R., Shirotani,K., Saido,T.C. and Hashimoto,Y. (2001) Alzheimer’s beta-secretase, beta-site amyloid precursor protein-cleaving enzyme, is responsible for cleavage secretion of a Golgi-resident sialyltransferase. Proc. Natl Acad. Sci. USA, 98, 13554–13559. [DOI] [PMC free article] [PubMed] [Google Scholar]