Abstract
Physical clusters of co-regulated, but apparently functionally unrelated, genes are present in many genomes. Despite the important implication that the genomic environment contributes appreciably to the regulation of gene expression, no simple statistical method has been described to identify physical clusters of co-regulated genes. Here we report the development of a model that allows the direct calculation of the significance of such clusters. We have implemented the derived statistical relation in a software program, Pyxis, and have analyzed a selection of Saccharomyces cerevisiae gene expression microarray data sets. We have identified many gene clusters where constituent genes exhibited a regulatory dependence on proteins previously implicated in chromatin structure. Specifically, we found that Tup1p-dependent gene domains were enriched close to telomeres, which suggested a new role for Tup1p in telomere silencing. In addition, we identified Sir2p-, Sir3p- and Sir4p-dependent clusters, which suggested the presence of Sir-mediated heterochromatin in previously unidentified regions of the yeast genome. We also showed the presence of Sir4p-dependent gene clusters bordering the HMRa heterothallic locus, which suggested leaky termination of the heterochromatin by the boundary elements. These results demonstrate the utility of Pyxis in identifying possible higher order genomic features that may contribute to gene regulation in extended domains.
INTRODUCTION
There is considerable evidence that many eukaryotic genes are not isolated regulatory units responding only to the presence of regulatory proteins bound to the local promoter, but are responsive to regulatory mechanisms that affect tens or hundreds of kilobases (1–5). Examples of such extended regulatory domains include the bithorax complex in Drosophila (6), the silent mating type loci in Saccharomyces cerevisiae (7), extensive regions of the inactivated mammalian X chromosome (8) and the β-globin gene locus in chicken erythrocytes (9). Although telomere-proximal regions in S.cerevisiae are not, strictly speaking, regulatory, genes that are located within regions close to chromosome ends are silenced in a coordinated fashion (10). In most cases, these domains are related to extensive specialized chromatin structures encompassing the domain.
The effect of chromatin on transcriptional regulation is well documented (11). Repressive chromatin structures were shown to contain core histones that were covalently modified at specific amino acid residues (9). The precise pattern of modifications, including acetylation, methylation and phosphorylation (12), facilitated the structural stability of the repressive heterochromatin and were recognized by non-chromatin proteins, including HP1 in Drosophila (13) and the Sir proteins or Tup1p in S.cerevisiae (12), which were then recruited to local regions of the chromatin and contributed to a state of transcriptional silencing in a domain (14).
The widespread application of whole-genome technologies, particularly gene expression microarrays, has allowed the identification of physical groups or clusters of genes that were co-regulated. Two approaches have been reported to calculate the statistical significance of such physical clusters of co-regulated genes. The first involved determination of the frequency of appearance of clusters of genes in randomly generated sample sets composed of the same total number of genes as the experimental set (3–5). The definition of what constituted a putative cluster, and therefore the property of the cluster that was compared to the random data set, differed among studies. Kim and colleagues defined clusters as groups of at least two genes with translation start positions within 10 kb and identified numerous clusters of genes that were co-regulated in the muscle tissue of L1 Caenorhabditis elegans larvae (3). In a similar approach, clusters were defined by grouping genes within a given maximum distance and extensive clustering of up-regulated genes was shown in senescent human mammary epithelial cells and in human fibroblasts (5). In a slight modification to this approach, Spellman and Rubin identified clusters by comparing the average pair-wise Pearson correlation of gene expression in a sliding 10 gene window with that obtained in a random data set (4) and reported the presence of extensive clusters of functionally unrelated genes in Drosophila.
The second type of approach to determine cluster significance made use of a binomial distribution. In an analysis of the median level of gene expression along each of the 23 human chromosomes, clusters were defined by groups of genes with an average expression level four times that of the genomic average in consecutive settings of a window. The numerical significance of such clusters was approximated from a binomial distribution and revealed the presence of distinct regions of highly expressed genes on all human chromosomes (15). A cumulative binomial distribution was used in a study by Church and colleagues who showed that correlated levels of expression occurred more often for adjacent genes during the S.cerevisiae cell cycle than expected by random chance (1). These pairs of co-regulated genes, which often belonged to the same functional category, infrequently contained identical promoter elements, which suggested a regulatory domain effect, possibly an extended influence of activators within a region of open chromatin (1). Genes that were associated with hematopoietic stem cell proliferation in mice were present on chromosome 11 more often than expected from a random distribution of such genes and appeared in three clusters of between 7 and 22 cM containing from 6 to 11 genes (2). The statistical significance of these clusters was, however, not reported (2).
These studies clearly illustrate that eukaryotic genes are often not strictly regulated as independent, isolated units, but that the expression of one gene may influence the level of expression of its neighbors. It is therefore clear that an analysis of regulatory pathways of gene expression cannot simply concentrate on the distribution and presence of DNA cis-elements bound by transcriptional activators and repressors at local promoters, but must also consider less discriminate regulatory mechanisms. The identification of clusters of co-regulated genes in genomes is therefore a very useful way to locate domains of linked regulation and will allow the study of the mechanistic aspects of extended, general transcriptional control within such domains.
No bioinformatics tool is generally available that allows the analysis of whole genomes for physical gene clustering and that can calculate the statistical significance of any identified putative clusters. In this study, we develop a rigorous method from the first principles of probability theory that allowed the direct calculation of the statistical significance of observed physical gene clusters. We report the implementation of our method in Pyxis, a web-based software program that allowed the identification of putative gene clusters at a chosen stringency and that calculated the significance of the identified gene clusters. We finally show the application of Pyxis to public microarray data sets and demonstrate the presence of previously unidentified Tup1p- and Sir2p/Sir3p/Sir4p- dependent physical clusters of co-regulated genes in the S.cerevisiae genome.
MATERIALS AND METHODS
Software development
Pyxis was developed in Java and compiled with JBuilder version 9.0 (Borland Software Corp., Scotts Valley, CA), making use of the Java 2 runtime environment version 1.4.0_01 (http://java.sun.com) and utilizing libraries of the Java 2 Software Development Kit Standard Edition version 1.4.0_01 (http://java.sun.com) on the Windows XP operating system platform (Microsoft Corp., Redmond, WA). The mySQL server version 4.0 (http://www.mysql.org) was used to manage database queries. The dynamic web pages used in the Java Server Page framework were developed in Dreamweaver version MX (Macromedia Inc., San Francisco, CA). The web application was tested using the Apache Tomcat version 4.1.27 web server (http://jakarta.apache.org/tomcat). All binaries and program code files are available freely for academic use from http://www.bioinformatics.uct.ac.za/pyxis.
Statistical analyses of telomeric enrichment
The statistical significance of the enrichment of Tup1p-dependent gene clusters overlapping with the terminal 20 kb of each chromosome was calculated from the product of the probabilities of finding a telomeric location for the observed e number of clusters from a total of t clusters for each chromosome, n, from a total of N chromosomes. This was given by
∏Nn = 1(peqt – etCe),
where p represents the probability of a telomeric location for a cluster and is given by R/(L/2 – R), where R is the maximum allowed distance from the telomere end (20 kb) and L is the length of the chromosome. The parameter q represents the probability of a non-telomeric location for a cluster (= 1 – p). The term tCe is the binomial coefficient, with e ≤ 2, since each chromosome has a maximum of two telomeric ends, and t is the total number of observed gene clusters on chromosome n.
Removal of recently duplicated genes from cluster sets
A homology matrix of the genomic sequences of all S.cerevisiae ORFs was calculated using the BLAST algorithm (16). Possible homologs or recently duplicated copies of every ORF in an assigned cluster were identified in this matrix by entries with E values of 10–10 or less (17). Any such identified homologous pair or group in a cluster was removed from the cluster and the cluster significance recalculated. The homology matrix of E values can be downloaded from http://www.bioinformatics.uct.ac.za. Where noted, promoter sequences were analyzed for known sequence motifs or transcription factor binding sites with AlignAce (http://atlas.med.harvard.edu) and DNAssist (http://www.dnassist.org), respectively.
RESULTS
A statistical model to calculate the significance of physical gene clustering
In order to derive a rigorous model to describe the statistical significance of physical clusters of genes on a chromosome, we started from the first principles of probability theory. When performing a gene expression microarray study, only genes conforming to a minimum significance requirement are used in the subsequent analysis of the data set. If we specified that r genes from such a data set appeared on a chromosome and that the chromosomal location was independent of expression, then there are nCr possible combinations in which the r genes can appear on a chromosome composed of n genes, as given by equation 1.
nCr = n!/[(n – r)!r!] 1
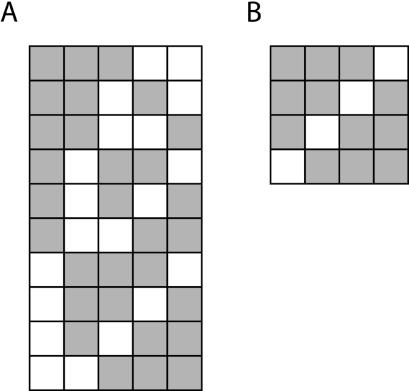
Solving equation 1 in an illustrative case where r = 3 and n = 5, shows that there are 10 possible combinations, representing the entire sample space, of three induced genes on a chromosome composed of five genes, as shown in Figure 1A. A physical cluster of genes is generally understood to mean that genes within a limited range or window conform to a test condition. If we are interested in identifying clusters of co-regulated genes, we need to compute the chance of seeing a given number of up-regulated (or down-regulated) genes within a window composed of a chosen number of genes. For instance, if we are interested in determining the significance of a cluster composed of three induced genes in a window that is four genes wide (a 3,4 cluster), a visual inspection of Figure 1A reveals that such a cluster will be present 40% of the time in each of the two possible settings of a four gene window. In any one setting, the four instances of three genes appearing in a four gene window is composed of all possible combinations, which, in the general case, is given by wCg, where w represents the width of the window and g the number of genes within the window (Fig. 1B). Although it immediately appears that the probability of finding three genes in a four gene window [p(3,4)] is simply
Figure 1.
Matrix of combinations. (A) The full sample space, consisting of all possible combinations, for the distribution of three objects among five possible settings. (B) All possible combinations of three objects among four settings. The objects are represented by the grey squares.
p(3,4) = (wCg)/(nCr), 2
this relation is only true in the special case where g = r. This is readily demonstrated by considering the probability of finding exactly two genes in a three gene window. A visual inspection of Figure 1A shows that 2,3 clusters occurred six times (P = 0.6) in the first setting of the window and not three times (P = 0.3), as suggested by equation 2. This is due to the duplicated presence of each possible combination of two genes in a three gene window, brought about by the combinations of the remaining genes (r – g) outside the window (see Fig. 1A). To incorporate the contribution of the genes outside the window, the number of combinations of g genes in a w gene window is multiplied by the number of combinations of the genes outside the window, given by (n – w)C(r – g). Therefore, in the general case, the number of occurrences of g genes in a w gene window on a chromosome where r genes from a total of n genes are randomly distributed is given by wCg × (n – w)C(r – g). The probability p(g,w) is finally given by
p(g,w) = wCg × (n – w)C(r – g)/nCr = {w!/[(w – g)!g!]} × 〈(n – w)!/{[(n – w) – (r – g)]!(r – g)!}〉/{n!/[(n – r)!r!]} 3
Equation 3 represents a hypergeometric distribution (18) and states that the probability of observing a g,w cluster is the number of combinations of g among w genes multiplied by the number of combinations of (r – g) among (n – w) genes, normalized for the total number of combinations of r among n genes on the chromosome. This probability is independent of the genomic location of the putative cluster.
Verifying the accuracy of the model
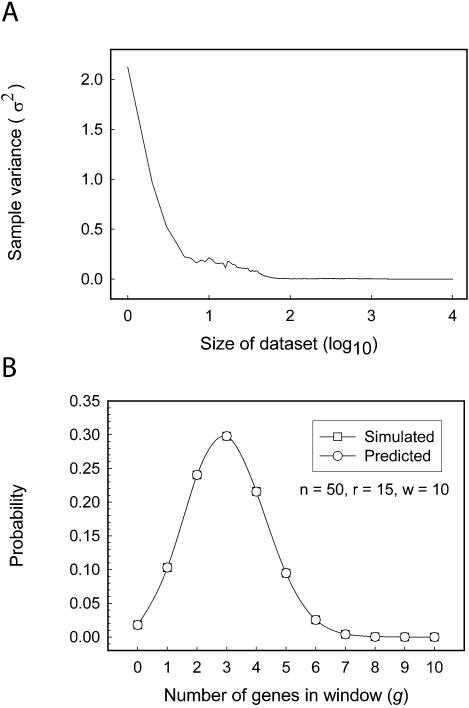
We verified the accuracy of the derived hypergeometric distribution that describes cluster probability by comparing the value calculated from equation 3 to the observed frequency of clusters in a randomly generated data set (Fig. 2). Since the variance in the average number of g,w clusters observed in different sized randomly generated data sets reached a plateau for data sets larger than ∼1000 chromosomes (see Fig. 2A), we determined the frequency of appearance of g,w clusters in a dataset composed of random distributions of r genes on each of 105 chromosomes, where error due to random variability is expected to be negligible (Fig. 2A). There was little difference between the calculated probability and the observed frequency of gene clusters (normalized sum of the squares of the differences ≈ 10–7) in a random data set (see Fig. 2B), which demonstrated that equation 3 could be applied to accurately calculate the probability and hence significance of gene clusters in genomic microarray data sets.
Figure 2.
The accuracy of the hypergeometric distribution to calculate the probability of physical clustering. (A) The dependence of the variance in cluster frequency on the size of the randomly generated population. The variance in the occurrence of clusters of defined size (y-axis) is shown as a function of the number of random distributions of 15 objects among 50 settings (x-axis). (B) The probability of the occurrence of a cluster calculated with the hypergeometric distribution is indistinguishable from the frequency observed in randomly generated in silico data. The frequency of occurrence of clusters that contained the indicated number of objects within a 10 setting wide window was calculated using equation 3 or determined in a population of 105 randomly generated distributions of 15 objects among 50 settings. The calculated (circles) and determined (squares) probabilities are shown (y-axis) as a function of the number of objects in the cluster window (x-axis).
Implementing the model in a computer program
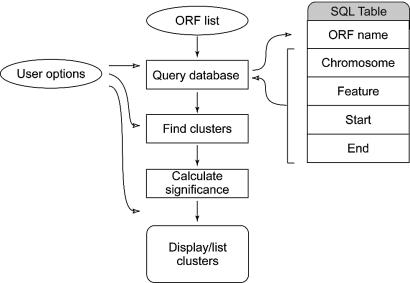
To allow the determination of the statistical significance of physical gene clusters in microarray data sets, we implemented the hypergeometric distribution represented by equation 3 in a computer program titled Pyxis. The program was developed in the Java programming language and a web interface created in the Java Server Page framework. The flow of the program logic in Pyxis is summarized in Figure 3. Briefly, a list of ORF names, either entered individually or as a text file, is provided by the user. Pyxis queries the appropriate SQL format database selected by the user and retrieves information on the chromosomal location of each of the entered ORFs. Putative genes clusters are identified by locating groups of genes on each chromosome that are within the maximum number of ORFs stipulated by the user. In this study we have used a maximum setting of five, which is appropriate in the general case. The statistical significance of each putative cluster is calculated and clusters that fall within the significance range selected by the user are displayed graphically at their chromosomal positions. The user can also select to save a text file that lists all putative clusters and the statistical significance and ORFs that formed each. Pyxis is freely accessible at http://www.bioinformatics.uct.ac.za/pyxis.
Figure 3.
Summary of the program logic of Pyxis. The ovals represent user selections or data supplied by the user, the rectangles show the main groups of programmatic actions, and the rounded rectangle represents the generated result.
It was previously shown that clusters of co-regulated genes were present in several genomes, including in the S.cerevisiae, C.elegans, Drosophila and human genomes (1,3,4,15). Church and colleagues suggested that the frequency with which adjacent genes were co-regulated in S.cerevisiae suggested that an upstream regulatory element present in the promoter of one gene may influence the expression of adjacent genes due to the open chromatin structure of the region (1). Accessibility was similarly suggested by Cohen and co-workers as the reason for the observed clustering of up-regulated genes in senescent human mammary epithelial cells (5). To investigate this possibility, we used Pyxis to analyze physical clustering in the transcription profiles of S.cerevisiae mutants lacking proteins previously implicated in chromatin structure. It was expected that the absence of a protein complex that was involved in either the formation of heterochromatic structures or alleviating the repressive effect of chromatin was likely to cause a similar transcriptional response in a domain of genes. Although SWI/SNF is the canonical ATP-dependent chromatin remodeling activity (19), the Winston laboratory have previously shown that there was no correlation between transcriptional response and promoter distance in a S.cerevisiae strain that lacked Snf2p, the catalytic subunit of SWI/SNF (20). This result suggested that SWI/SNF acted at the level of single genes and was not involved in the remodeling of extended domains of chromatin (20). For this reason, we chose to investigate physical gene clustering in Tup1p and Sir2/3/4p mutant strains, where these proteins have previously been shown to be involved in the formation of extensive heterochromatic regions (21,22). Since it was a concern that some genes may be co-regulated due to evolutionary recent duplication, we removed all ORFs in homologous pairs or groups in a cluster with BLAST E values ≤ 10–10 before calculating the statistical significance of all clusters reported in this study (17).
Tup1p influences the expression of extended gene domains
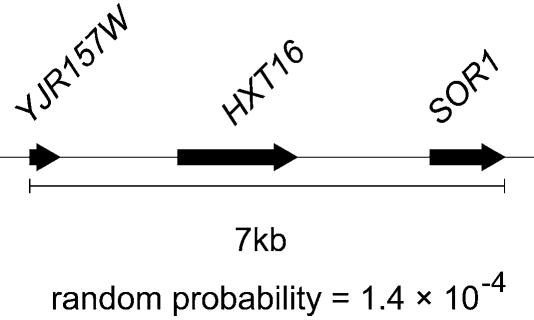
Genes with expression levels that increased at least 2-fold in the absence of Tup1p in comparison with the wild-type yeast strain, and were therefore co-regulated with respect to the absence of Tup1p, were selected from a database (23). An analysis of the distribution of co-regulated gene clusters in a S.cerevisiae strain lacking the general transcriptional repressor Tup1p showed clear evidence of statistically significant (P < 0.001) groups of adjacent genes in the genome that displayed a regulatory dependence on Tup1p (Fig. 4). There was also evidence of extensive regions, ranging from 1 kb containing two adjacent genes to 13 kb containing six adjacent genes, which appeared to share a common regulatory mechanism. The constituent genes in one such cluster on chromosome 10, for which a genomic map is shown (Fig. 5), did not exhibit significant sequence homology and an analysis of the promoter regions of these genes did not reveal any common sequence motifs or regulatory sites. This result suggested that Tup1p was involved in the establishment of extended regulatory domains and may indicate a processive association and contribution to repressive chromatin structures, as was shown for mini-chromosomes containing the STE6 gene in MATα yeast cells (22). The appearance of clusters of transcribed genes that exactly matched genomic regions where chromatin was previously shown to contribute to gene silencing in extended domains, such as at the HMRa locus in the sir4– strain (Fig. 4), provides strong support to the idea that the clusters represented extended regions where altered chromatin structure allowed gene expression. In addition, few significant clusters were observed in mutant strains lacking the ANP1 and CUP5 genes that encoded an endoplasmic reticulum protein and a vacuolar ATPase, respectively, proteins that were unlikely to influence chromatin structure directly (data not shown).
Figure 4.
Physical clusters of genes in the genome of S.cerevisiae in the absence of Tup1p, Sir2p, Sir3p or Sir4p. The positions of genes that were induced by at least 2-fold in a tup1–, sir2–, sir3– or a sir4– strain compared to the wild-type strain are shown for each of the 16 chromosomes. Gene clusters, composed of genes within five ORFs of its closest neighbor and where the probability of a similar grouping arising randomly was <0.1%, are identified by rectangles. Homologous gene pairs or groups were removed from clusters before calculation of the cluster significance. The positions of the MFA1 and BAR1 genes, the HMLα and HMRa heterothallic loci and the MAT locus are indicated. The 28 kb gene cluster on chromosome XIII in the sir2– strain is indicated by the asterisk. The microarray data for the tup1– strain was obtained from the Brown study (23) and the sir2–, sir3– and sir4– data from the Young study (39).
Figure 5.
ORF map of a statistically significant (P < 0.001) cluster composed of directly adjacent ORFs that were induced by at least 2-fold in a tup1– S.cerevisiae strain. The analysis was performed on the data from the Brown study (23). The black arrows represent ORFs with the direction of transcription indicated. The length and spacing of ORFs are shown to scale. The random probability for the appearance of the cluster is indicated.
Some individual genes, such as BAR1 and MFA1, that were previously shown to be repressed in a Tup1p-dependent manner, and where clear extended nucleosomal arrays were mapped on the repressed genes in vivo (22), did not appear in clustered groups (Fig. 4). This result suggested that the expansion of Tup1p-directed heterochromatic structures were efficiently terminated at some genomic loci.
Tup1p is involved in the regulation of telomere-proximal genes
A visual inspection of the distribution of the statistically significant gene clusters (P < 0.001) showed that the clusters often appeared close to the telomeric ends of the chromosomes (see Fig. 4). We determined the frequency of appearance of genes clusters within a 20 kb region at the ends of chromosomes and compared that to the frequency expected from a random distribution of gene clusters. The result suggested that genes that were co-regulated in the absence of Tup1p appeared more often in proximity to telomeres than expected for a random distribution (Table 1). This was most clearly seen for chromosome 10, where the likelihood that a random distribution resulted in the observed telomeric location of the two gene clusters was <1%. Assuming that the distribution of clusters on each chromosome was an independent event, the probability of the distribution seen on all chromosomes can be calculated from the product of the individual probabilities shown in Table 1, and is <10–11. This clearly demonstrated the significance of the telomeric enrichment observed for clusters on all the chromosomes and suggested that Tup1p was involved in the regulation of transcription in extended telomeric gene loci. This involvement can occur either by a direct participation as a structural component of telomeric heterochromatin or indirectly, by changing the expression of a structural or regulatory component of telomeric chromatin.
Table 1. Statistical significance of the observed telomeric versus non-telomeric distribution of gene clusters in the tup1– strain.
| Chromosome | No. of clusters | Random probability for telomeric | |
|---|---|---|---|
| Total | Telomeric | ||
| 1 | 0 | ||
| 2 | 1 | 1 | 0.08 |
| 3 | 0 | ||
| 4 | 2 | 1 | 0.08 |
| 5 | 1 | 1 | 0.11 |
| 6 | 0 | ||
| 7 | 1 | 1 | 0.07 |
| 8 | 0 | ||
| 9 | 0 | ||
| 10 | 2 | 2 | 0.009 |
| 11 | 1 | 1 | 0.08 |
| 12 | 1 | 0 | |
| 13 | 0 | ||
| 14 | 1 | 1 | 0.08 |
| 15 | 2 | 2 | 0.01 |
| 16 | 1 | 0 | |
The probability that a random distribution of the identified gene clusters would result in the partitioning observed in the tup1– strain was calculated from the fraction of the total number of positions where a cluster would overlap with the terminal 20 kb of a chromosome, as described in Materials and Methods.
Common clusters of genes are repressed by Sir2p and Sir3p
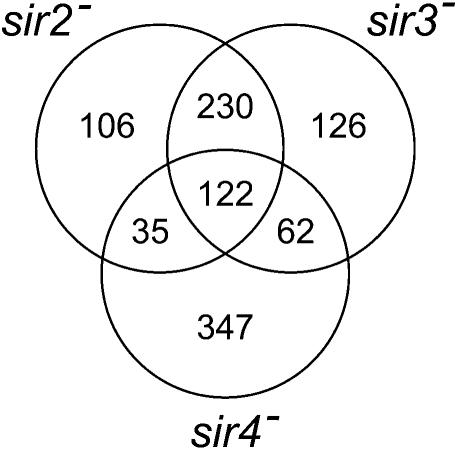
We next investigated whether genes that were markedly up-regulated in the absence of the Sir proteins appeared in physical groupings in the genome. It was previously shown that transcriptional repression of telomeric genes and repression of recombination at the rRNA locus, as well as silencing of the heterothallic mating-type loci, were dependent on the Sir proteins (24). An analysis of the distribution of genes that were induced at least 2-fold in the absence of Sir2p, Sir3p or Sir4p revealed several statistically significant (P < 0.001) groupings of genes (Fig. 4). A cursory examination of the genomic distribution of these clusters showed that clusters frequently appeared at similar locations on chromosomes, particularly in the sir2– and the sir3– strains (Fig. 4). We therefore asked which genes were commonly induced in the absence of the Sir proteins. The results, shown in Figure 6, revealed that a total of 352 common genes were induced by at least 2-fold in the absence of Sir2p or Sir3p. This represented 71% of the genes repressed by Sir2p and 65% of the genes repressed by Sir3p and was a significantly larger pool of common genes than that shared by Sir2p and Sir4p or by Sir3p and Sir4p (see Fig. 6). This was not unexpected, since Sir2p is a NAD-dependent histone deacetylase specific for K9 and K14 of histone H3 and K16 of histone H4 (25). The C-terminal tail of Sir3p interacted preferentially with the hypoacetylated N-terminal tail of histone H4 (26). The dependence of Sir3p histone binding on Sir2p activity therefore suggested a functional co-dependency, as was indeed observed (Figs 4 and 6). The gene clusters observed at identical positions in the chromosomes of the sir2– and sir3– strains probably reflected this functional dependency.
Figure 6.
Genes that are regulated in common by the Sir proteins. Genes that were up-regulated by 2-fold or more in the sir2–, sir3– or sir4– strain (39) were identified and common genes in each data set pair selected from the SQL database. The number of genes that displayed a regulatory dependence on one or more of the Sir proteins is shown in the Venn diagram.
Extensive Sir-dependent gene domains are present at locations other than the silent mating-type loci and the telomeres
The gene clusters in the genomes of sir2–, sir3– or sir4– yeast strains were often located at positions close to the telomeric ends (Fig. 4), in agreement with the established role of the Sir proteins in telomeric silencing. However, there were also several examples of internal locations of gene groupings. In some instances the gene clusters were very extensive (Fig. 4), in one case spanning ∼28 kb from YMR168C to YMR182C on chromosomes XIII of the sir2– strain. This cluster did not contain any genes of related sequence, demonstrating the presence of extended gene domains that exhibited a regulatory dependence on one or more of the members of the Sir protein family. Genes that fell within this cluster were functionally diverse and encoded proteins that participated in processes ranging from centromere binding (YMR168C/CEP3) to cytoplasmic polyamine degradation (YMR169C/ALD3). This result suggested that the regulation of gene expression by the Sir proteins was not limited to simple functional classes, such as the silent mating type loci or the rRNA gene repeat, but included gene groupings without an obvious functional commonality. This result also suggested that Sir-dependent heterochromatic regions were present at previously unidentified loci on the chromosomes of S.cerevisiae.
The repressive effects of Sir4p extends beyond the E and I elements of HMRa
The involvement of Sir2p, Sir3p and Sir4p in initiating and maintaining a silenced state of the silent mating type loci on chromosome III is well established (27). The appearance of the genes located at the transcriptionally active MAT locus in a Sir-dependent cluster (Fig. 4) was likely due to an increase in the HMLα1 and HMLα2 mRNA and corresponding microarray probe, following derepression of the HMLα locus in the absence of the Sir proteins in the MATα yeast strain. Removal of the HMLα and MATα genes from the microarray data set used in the physical clustering analysis resulted in the disappearance of gene clusters at the HMLα and at the MATα loci. This result suggested, as expected, that the Sir proteins were not involved in extended repressive structures at the active MAT locus. The cross-hybridization of the MATα and HMLα genes precluded an analysis of the HMLα locus. However, a statistically significant gene cluster was present in the absence of Sir4p at the HMRa locus (Fig. 4). The cluster, spanning 29 kb, included regions of ∼19 kb on the centromere-proximal and 9 kb on the telomere-proximal sides, beyond the silenced locus demarcated by the E and I boundary elements of the HMRa locus. These extended regions that displayed a Sir4p regulatory dependence included the KIN82 and CDC39 genes (centromere-proximal) and the GIT1, YCR100C and YCR101C genes (telomere-proximal). It was recently shown that the histone H2A.Z isotype was an antagonist to Sir2p-mediated repression in yeast (28) and influenced the expression of genes in a group that extended in both the centromere- and telomere-proximal directions from the HMRa locus. This Sir2p-dependent grouping overlapped with the Sir4p-dependent cluster identified above (see Fig. 4) and confirmed that the repressive effects of the HMRa silent mating type locus extended beyond the boundary E and I elements.
DISCUSSION
In this study we have developed a relation from first principles that described the statistical significance of physical gene clustering in a genome. We have implemented this relation in a computer program that allowed the easy identification of statistically significant clusters of genes in a data set of specified ORF names. We have applied this program to publicly available microarray data sets and have demonstrated its utility in identifying new domains of co-regulated genes.
The use of a hypergeometric distribution
In most published studies on the physical grouping of genes, one of two approaches was used to determine the statistical significance of putative physical gene clusters. The first involved calculating the frequency with which an observed cluster appeared in a randomly generated data set composed of the same number of genes as the experimental data set (3–5). The second approach made use of a binomial distribution (1,15).
There are several different distributions that can be used to estimate the likelihood of a certain number of events occurring within a Bernoulli trial. These distributions have different properties and although the limits of each approach are mathematically rigorous, the suitability of its application to biology is subtle. Where a small number of probability trials are carried out, such as selecting a ball from a bag that contained five red and five white balls, the random selection of a red ball diminished the subsequent probability of choosing another red ball. To ensure that successive events in a Bernoulli trial are independent, a ball must be replaced in the bag following its random selection. A distribution that described the probability p of an event in such a system is referred to as a distribution ‘with replacement’. The hypergeometric distribution is an example of a distribution ‘with replacement’. Generally, for a large number of events (n→∞) the law of large numbers applies and the effect of a single event (ni) on p is negligible (ni/n→p) (18). Therefore, distributions ‘without replacement’, such as the binomial distribution, are more accurate for large n. For this reason, the use of a hypergeometric distribution may be more accurate to calculate statistical significance in the relatively small data sets often found in microarray analyses.
The applicability of a hypergeometric distribution to real genes
The hypergeometric function used in Pyxis calculates the probability of a distribution composed of a number of equi-probable events. This can be represented graphically by the distribution of balls among several holes separated by a constant distance on a wooden beam. This type of arrangement is unlike the distribution of genes in a genome, where individual genes are of variable length, adjacent pairs of proximal promoters are spaced at variable distances and some genes respond to distal enhancer elements. The question therefore arises whether a simple statistical model that described a pattern of equi-probable events was applicable to the complex geometric arrangement of genes in the genome. It may be envisioned, for instance, that two divergently transcribed genes may respond to a distributed regulatory effect and appear in a cluster, whereas two convergently transcribed genes of identical length may place their promoters beyond the range of the distributed effect and not be part of the same cluster. It is stressed that Pyxis calculates the statistical significance of clusters based on gene groupings identified by independent criteria, such as a 2-fold induction or repression. Pyxis does not provide any information on why adjacent genes were or were not included in a cluster. The geometry of gene arrangements may have an effect on the mechanism that was responsible for the co-regulation of adjacent genes, but is irrelevant to the calculation of the statistical significance of any putative clusters.
The role of Tup1p in telomeric repression
Tup1p is a broad spectrum transcriptional repressor involved in the regulated shutdown of the mating type, glucose- and oxygen-responsive genes in yeast (29). This repression is thought to occur by a combination of chromatin binding (30), where Tup1p associated with the hypoacetylated N-terminal tail of histone H3, forming specialized repressive chromatin structures (22), and by interacting with Srb11p (31) and with Srb7p (32), which formed part of the polymerase II holoenzyme.
We have shown in this study that there is a statistically significant enrichment of clusters of Tup1p-repressible genes at the telomeres, suggesting a role for Tup1p in telomeric silencing. This involvement can be direct, with Tup1p acting as a component of telomeric heterochromatin, or indirect, where Tup1p, for instance, exerted an influence on the level of other telomere-associated proteins. Rap1p was shown to associate with the telomere repeat sequence and recruited the Sir2/3/4p proteins, allowing Sir3p and Sir4p to serially propagate along the telomeric chromatin by binding to the hypoacetylated N-terminal tails of histones H3 and H4 (26). This association may render the histone tails unavailable for interactions with other proteins, although the presence of two copies of each core histone in the nucleosome represents two possible binding sites that may allow simultaneous interaction of a nucleosome with Tup1p and with the Sir proteins. Alternatively, it is also possible that Tup1p was recruited to the telomeric regions by associating with another protein. An in vivo interaction between Tup1p and the RNA polymerase II-associated elongation factor Cdc73p was, for instance, reported and disruption of the CDC73 gene was shown to result in telomeric deprepression (33), a result that potentially places Tup1p at the telomeres. However, an in vitro interaction between Cdc73p and Tup1p has not been demonstrated and it was pointed out that the in vivo interaction, determined by dimerization of ubiquitin fusion fragments, could have been due to independent localization of the two proteins to a promoter region (33).
To consider a possible indirect effect, we looked at the transcriptional response of genes in the tup1– strain (23) that were also implicated in telomeric silencing in the gene ontology database (34,35). Of the 33 genes that fitted this category, only the SET1 gene exhibited a significant change and was down-regulated by ∼2-fold in the tup1– strain. We and others have previously shown that Set1p was a histone H3 methyltransferase specific for lysine 4 (36,37). It was also shown that the absence of Set1p resulted in telomeric derepression (38). It is therefore possible that the Tup1p-dependence of telomeric gene clusters were due to a decrease in the Set1p level, the concomitant decrease in H3 K4 methylation and the resulting derepression of telomere-proximal genes in the tup1– strain. However, we cannot exclude alternative mechanisms with the data available to us.
Biological significance of clusters
We have used the web-based program Pyxis to identify numerous extended regions in the S.cerevisiae genome containing genes that appeared to be co-regulated. In some cases the co-regulation was due to a clear functional link that had previously been identified. Examples were also seen of co-regulated genes without any clear functional relation, which suggested the presence of an indiscriminate or relaxed regulatory mechanism at some genomic locations. Recent studies have suggested that the co-regulation of genes in large regions may be due to localized changes in the chromatin structure of the genomic region (1,3–5). Spellman and Rubin proposed that such structural changes could be induced by the transcriptional activity of a gene which caused the decondensation of chromatin in the surrounding areas, facilitating the binding of transcriptional factors in the whole region (4). Alternatively, it was suggested that a UAS-bound transcriptional activator could more easily contact distal promoters and facilitate transcription of neighboring genes in an extended region of decondensed chromatin (4). It is possible that the recruitment of histone modification enzymes such as methyltransferases and acetyltransferases to a promoter results in a transient local increase in enzyme activity and gratuitous modification of histones in the immediate genomic vicinity. This, in turn, may facilitate the local decondensation of chromatin and subsequent transcriptional activation of genes close to the original recruitment point. The appearance of clusters of co-regulated genes at only a limited number of locations in the genome (see Fig. 4) suggests that such a relaxed regulatory mechanism does not apply generally. However, it may also not pose an evolutionary disadvantage at the limited number of locations where co-regulation was observed (see Fig. 4).
In this study we have demonstrated that the absence of proteins previously shown to influence chromatin structure, and specifically the absence of proteins known to be structural components of repressive heterochromatin, resulted in derepression of gene domains. This provides the first direct link between factors that influence chromatin structure and the co-regulation of domains of genes, defined by physical clusters of genes with similar transcriptional behaviors.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to express their gratitude to Cathal Seoighe for generating the all-against-all BLAST homology matrix, Charles Ducker for helpful comments on Tup1p binding, Rodger Duffett for his assistance with the web server and Janet Kelso for helpful suggestions on the manuscript. This work was funded by a grant from the National Bioinformatics Network (to H.G.P.). C.-F.C. was the recipient of a NBN bursary. H.G.P. is a Wellcome Trust International Senior Research Fellow in Biomedical Science in South Africa.
REFERENCES
- 1.Cohen B.A., Mitra,R.D., Hughes,J.D. and Church,G.M. (2000) A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nature Genet., 26, 183–186. [DOI] [PubMed] [Google Scholar]
- 2.de Haan G., Bystrykh,L.V., Weersing,E., Dontje,B., Geiger,H., Ivanova,N., Lemischka,I.R., Vellenga,E. and Van Zant,G. (2002) A genetic and genomic analysis identifies a cluster of genes associated with hematopoietic cell turnover. Blood, 100, 2056–2062. [DOI] [PubMed] [Google Scholar]
- 3.Roy P.J., Stuart,J.M., Lund,J. and Kim,S.K. (2002) Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature, 418, 975–979. [DOI] [PubMed] [Google Scholar]
- 4.Spellman P.T. and Rubin,G.M. (2002) Evidence for large domains of similarly expressed genes in the Drosophila genome. J. Biol., 1, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H., Pan,K.H. and Cohen,S.N. (2003) Senescence-specific gene expression fingerprints reveal cell-type-dependent physical clustering of up-regulated chromosomal loci. Proc. Natl Acad. Sci. USA, 100, 3251–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orlando V., Jane,E.P., Chinwalla,V., Harte,P.J. and Paro,R. (1998) Binding of trithorax and Polycomb proteins to the bithorax complex: dynamic changes during early Drosophila embryogenesis. EMBO J., 17, 5141–5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bi X., Braunstein,M., Shei,G.J. and Broach,J.R. (1999) The yeast HML I silencer defines a heterochromatin domain boundary by directional establishment of silencing. Proc. Natl Acad. Sci. USA, 96, 11934–11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plath K., Mlynarczyk-Evans,S., Nusinow,D.A. and Panning,B. (2002) Xist RNA and the mechanism of X chromosome inactivation. Annu. Rev. Genet., 36, 233–278. [DOI] [PubMed] [Google Scholar]
- 9.Litt M.D., Simpson,M., Gaszner,M., Allis,C.D. and Felsenfeld,G. (2001) Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science, 293, 2453–2455. [DOI] [PubMed] [Google Scholar]
- 10.Moretti P. and Shore,D. (2001) Multiple interactions in Sir protein recruitment by Rap1p at silencers and telomeres in yeast. Mol. Cell. Biol., 21, 8082–8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolffe A.P. (1998) Chromatin: Structure and Function. Academic Press, New York, NY. [Google Scholar]
- 12.Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- 13.Bannister A.J., Zegerman,P., Partridge,J.F., Miska,E.A., Thomas,J.O., Allshire,R.C. and Kouzarides,T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–124. [DOI] [PubMed] [Google Scholar]
- 14.Braunstein M., Sobel,R.E., Allis,C.D., Turner,B.M. and Broach,J.R. (1996) Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol. Cell. Biol., 16, 4349–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caron H., van Schaik,B., van der Mee,M., Baas,F., Riggins,G., van Sluis,P., Hermus,M.C., van Asperen,R., Boon,K., Voute,P.A. et al. (2001) The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science, 291, 1289–1292. [DOI] [PubMed] [Google Scholar]
- 16.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 17.Seoighe C., Federspiel,N., Jones,T., Hansen,N., Bivolarovic,V., Surzycki,R., Tamse,R., Komp,C., Huizar,L., Davis,R.W. et al. (2000) Prevalence of small inversions in yeast gene order evolution. Proc. Natl Acad. Sci. USA, 97, 14433–14437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feller W. (1961) An Introduction to Probability Theory and its Applications. John Wiley & Sons, New York, NY. [Google Scholar]
- 19.Peterson C.L. and Tamkun,J.W. (1995) The SWI-SNF complex: a chromatin remodeling machine? Trends Biochem. Sci., 20, 143–146. [DOI] [PubMed] [Google Scholar]
- 20.Sudarsanam P., Iyer,V.R., Brown,P.O. and Winston,F. (2000) Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 97, 3364–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hecht A., Laroche,T., Strahl-Bolsinger,S., Gasser,S.M. and Grunstein,M. (1995) Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell, 80, 583–592. [DOI] [PubMed] [Google Scholar]
- 22.Ducker C.E. and Simpson,R.T. (2000) The organized chromatin domain of the repressed yeast a cell-specific gene STE6 contains two molecules of the corepressor Tup1p per nucleosome. EMBO J., 19, 400–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeRisi J.L., Iyer,V.R. and Brown,P.O. (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science, 278, 680–686. [DOI] [PubMed] [Google Scholar]
- 24.Fritze C.E., Verschueren,K., Strich,R. and Easton,E.R. (1997) Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J., 16, 6495–6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imai S., Armstrong,C.M., Kaeberlein,M. and Guarente,L. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature, 403, 795–800. [DOI] [PubMed] [Google Scholar]
- 26.Carmen A.A., Milne,L. and Grunstein,M. (2002) Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J. Biol. Chem., 277, 4778–4781. [DOI] [PubMed] [Google Scholar]
- 27.Rusche L.N., Kirchmaier,A.L. and Rine,J. (2002) Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell, 13, 2207–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meneghini M., Wu,M. and Madhani,H. (2003) Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell, 112, 725–736. [DOI] [PubMed] [Google Scholar]
- 29.Tzamarias D. and Struhl,K. (1994) Functional dissection of the yeast Cyc8-Tup1 transcriptional co-repressor complex. Nature, 369, 758–761. [DOI] [PubMed] [Google Scholar]
- 30.Edmondson D.G., Smith,M.M. and Roth,S.Y. (1996) Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev., 10, 1247–1259. [DOI] [PubMed] [Google Scholar]
- 31.Schuller J. and Lehming,N. (2003) The cyclin in the RNA polymerase holoenzyme is a target for the transcriptional repressor Tup1p in Saccharomyces cerevisiae. J. Mol. Microbiol. Biotechnol., 5, 199–205. [DOI] [PubMed] [Google Scholar]
- 32.Gromoller A. and Lehming,N. (2000) Srb7p is a physical and physiological target of Tup1p. EMBO J., 19, 6845–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kerkmann K. and Lehming,N. (2001) Genome-wide expression analysis of a Saccharomyces cerevisiae strain deleted for the Tup1p-interacting protein Cdc73p. Curr. Genet., 39, 284–290. [DOI] [PubMed] [Google Scholar]
- 34.Cherry J.M., Adler,C., Ball,C., Chervitz,S.A., Dwight,S.S., Hester,E.T., Jia,Y., Juvik,G., Roe,T., Schroeder,M. et al. (1998) SGD: Saccharomyces Genome Database. Nucleic Acids Res., 26, 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T. et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boa S., Coert,C. and Patterton,H.G. (2003) Saccharomyces cerevisiae Set1p is a methyltransferase specific for lysine 4 of histone H3 and is required for efficient gene expression. Yeast, 20, 827–835. [DOI] [PubMed] [Google Scholar]
- 37.Briggs S.D., Bryk,M., Strahl,B.D., Cheung,W.L., Davie,J.K., Dent,S.Y., Winston,F. and Allis,C.D. (2001) Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev., 15, 3286–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nislow C., Ray,E. and Pillus,L. (1997) SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol. Biol. Cell, 8, 2421–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wyrick J.J., Holstege,F.C., Jennings,E.G., Causton,H.C., Shore,D., Grunstein,M., Lander,E.S. and Young,R.A. (1999) Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature, 402, 418–421. [DOI] [PubMed] [Google Scholar]