Abstract
Oral outbreaks of Chagas disease are increasingly reported in Latin America. The transitory presence of Trypanosoma cruzi parasites within contaminated foods, and the rapid consumption of those foods, precludes precise identification of outbreak origin. We report source attribution for 2 peri-urban oral outbreaks of Chagas disease in Venezuela via high resolution microsatellite typing.
Keywords: Trypanosoma cruzi, oral, orally transmitted, Chagas disease, Venezuela, Caracas, Vargas State, molecular epidemiology, parasites, trypanosome, zoonoses, vector-borne infections
Rapid urbanization presents new challenges for Chagas disease control in Latin America. Foci of disease are now reported in slums surrounding several Andean cities (1–3). Oral transmission is believed responsible for recent outbreaks of Chagas disease, most of which were characterized by atypically severe symptoms (4,5). Many cases have occurred in urban settings (5,6), amplifying the size and effect of the outbreaks.
Sources of orally transmitted disease outbreaks vary, but contaminated food and juices are often blamed. However, after a contaminated food is eaten, it may take weeks for the onset of clinical signs and symptoms, and direct molecular and cytological incrimination of a particular batch of food/beverage has not been possible (5). Thus, evidence pointing to particular foodstuffs is often circumstantial.
Molecular epidemiologic analyses of human and environmental isolates are routinely used to track the source of outbreaks caused by foodborne pathogens. High-resolution molecular markers have been developed and validated for Trypanosoma cruzi, the parasite that causes Chagas disease (7,8). These markers, used in conjunction with careful sampling, can identify the source of foodborne outbreaks.
The Study
We studied 2 outbreaks of orally transmitted Chagas disease (120 cases, 5 deaths). The first occurred in Chichiriviche, Vargas State, a coastal community (population ≈800 persons) ≈50 km northwest of Caracas, Venezuela. The outbreak occurred at a primary school where food was prepared on site. In early April 2009, a total of 71 children (6–13 years of age) who attended the morning school shift and 14 adults became ill. Exposure of these persons to T. cruzi was established by use of IgM and IgG ELISA. Parasitemia was observed in 33 of the patients with serologic results positive for T. cruzi infection (9,10).
The second outbreak occurred in Antimano, a peri-urban slum southwest of central Caracas (Technical Appendix Figure 1). In May 2010, 35 patients with suspected T. cruzi infection were examined at Hospital Miguel Perez Carreno in Caracas. Patients reported that they regularly ate at the same local communal canteen. Among the patients tested, 15 were positive for T. cruzi IgM and IgG (9). Parasitemia in 14 patients was confirmed indirectly by hemoculture. Of the 35 patients, 21 (2 adults, 19 children) were hospitalized.
To enable outbreak source attribution, we undertook intensive additional sampling of contemporary, nonhuman sources local to each outbreak and of human and nonhuman sources from more distant localities throughout Venezuela. In total, 246 T. cruzi strains and clones were typed for 23 microsatellite markers (Technical Appendix 1 Table) (8). A list of the samples and their sites of origin is in Technical Appendix 2).
Individual level sample clustering was defined first by constructing a neighbor joining tree based on pairwise distances between multilocus genotypes (Figure 1). A second analysis used K-means clustering and discriminant analysis of principal components (Figure 2) (11). To assess connectivity between human and nonhuman outbreak cases, pairwise genetic differentiation (FST) was calculated (Table 1). Population-level genetic diversity was assessed first by calculating allelic richness then private allele frequency over loci between each human–nonhuman population pair (Table 2). Geographic sampling distribution is shown in Technical Appendix 1 Figure 2.
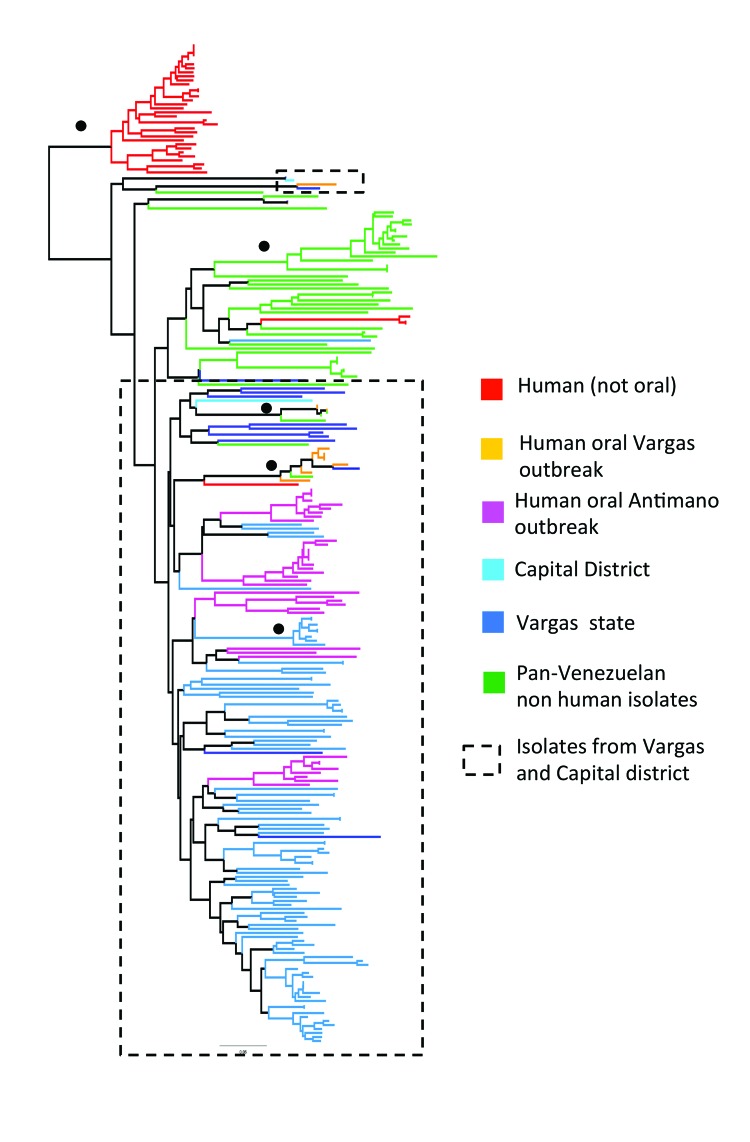
Figure 1.
Unrooted neighbor joining tree showing genetic clustering among Trypanosoma cruzi isolates from 2 outbreaks of oral disease in northern Venezuela. Based on pairwise genetic distances (1 – proportion of shared alleles) between multilocus microsatellite profiles (23 loci) generated from 246 isolates and clones. Black circles indicate nodes with >60% bootstrap support. Branch color key is shown. Dashed boxes indicate isolates associated with the outbreaks.
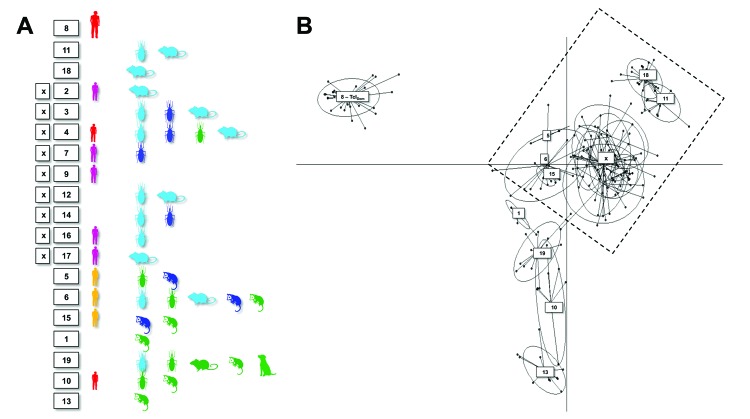
Figure 2.
Discriminant analysis of principal components showing genetic clustering among Trypanosoma cruzi isolates from 2 outbreaks of oral disease in northern Venezuela. Six principal components were retained, explaining 80% of the diversity. Ellipses correspond to the optimal (as defined by the Bayesian information criterion minimum) number of population clusters among the genotypes analyzed. Images indicate sample host origin (human, rodent, marsupial, or triatomine), while colors correspond to the key in Figure 1. A full list of samples and population assignments (numbered boxes) is included in Technical Appendix 2. Dashed box indicates the isolates associated with the outbreaks.
Table 1. FST values in a 4-way comparison for differentiation between Trypanosoma cruzi isolates derived from humans and the local environment during an outbreak of orally transmitted Chagas disease in 2 areas of Venezuela*.
| Isolate, location | Human isolates from |
Nonhuman isolates from |
||
|---|---|---|---|---|
| Antimano | Chichiriviche | Antimano | Chichiriviche | |
| Human | ||||
| Antimano | 0.000 | 0.000 | 0.000 | |
| Chichiriviche |
0.201 |
|
0.000 |
0.004 |
| Nonhuman | ||||
| Antimano | 0.093 | 0.170 | 0.000 | |
| Chichiriviche | 0.088 | 0.053 | 0.079 | |
*Lower left shows linearized FST (genetic differentiation) values; upper right shows associated p values.
Table 2. Sample size corrected diversity between Trypanosoma cruzi isolates derived from humans and the local environment during an outbreak of orally transmitted Chagas disease in 2 areas of Venezuela*.
| Isolate, location | No. isolates/no. genotypes | Sample size corrected allelic richness ± SE | Mean no. private alleles/locus ± SE |
|---|---|---|---|
| Human | |||
| Antimano | 30/26 | 2.735 ± 0.291 | 0.32 ± 0.113 |
| Chichiriviche |
12/9 |
3.459 ± 0.412 |
0 |
| Nonhuman | |||
| Antimano | 107/91 | 2.946 ± 0.320 | 0.86 ± 0.203 |
| Chichiriviche | 13/13 | 3.443 ± 0.409 | 0 |
Clustering results determined by discriminant analysis of principal components and neighbor joining were broadly congruent. In the former, 19 clusters were defined; sample allocations are included in Technical Appendix 2. Substantial overlap existed between some clusters, especially those from Capital and Vargas States (e.g., those labeled “x” in Figure 2), while others were highly distinct (e.g., cluster 8 in Figure 2). Human isolates from both oral outbreaks are extremely distinct from non-orally transmitted isolates collected from humans throughout Venezuela. Almost all these presumably vector-transmitted strains are closely related to one another, despite their geographic dispersal (cluster 8 in Figure 2). By comparison, oral outbreak strains that were isolated a mere 50 km apart (clusters 2, 5, 7, 9, 15–17) are far more globally diverse. Unlike most human isolates in Venezuela, which are distinct from nonhuman strains, oral outbreak isolates are interspersed among nonhuman strains from Venezuela. Furthermore, samples from both outbreaks clustered among nonhuman strains local to that outbreak, clearly indicating a local origin. Oral samples from each outbreak are polyphyletic with respect to strains from their immediate environment, a finding consistent with multiple contamination events or multiclonal infection sources.
FST values further support connectivity between outbreak and local environmental samples in both Antimano and Chichiriviche (Table 1). A lack of private alleles between human and nonhuman isolates also supports a local source for the Chichiriviche outbreak (Table 2). FST values in the 4-way comparison between outbreak and local environmental strains are, however, somewhat equivocal with respect to the entire dataset (Table 1). Cluster analysis showed that the human and nonhuman strains from Chichiriviche interspersed with strains from other states in Venezuela (Figure 2). Thus, we cannot confirm a uniquely local origin for the Chichiriviche outbreak, despite a low value for FST, and it is possible that some contaminating strains originated elsewhere.
Conclusions
This study demonstrates the value of rigorous molecular epidemiologic analysis of orally transmitted T. cruzi outbreaks, including the importance of appropriate sampling to identify the origin of the infecting strains. The foodstuff that propagated the peri-urban outbreak in Antimano was certainly contaminated locally. An active nonhuman transmission cycle in the slums of Caracas, maintained by Rattus rattus rodents and Panstrongylus geniculatus triatomines, is the likely source. The Chichiriviche outbreak, however, has potential sources both in and outside the immediate area. As found in Chagas disease outbreaks linked to açaí palm fruit in Brazil (12), the T. cruzi parasite can survive for several days in some foodstuffs (13). Also, triatomines can survive for months in harvested crops; thus, multiple hygiene interventions are potentially necessary along the food production line (14). Nonetheless, if the foodstuff implicated was prepared locally, local contamination represents the most likely source of the outbreak. Study of additional nonhuman strains from Chichiriviche is necessary to support this assertion.
Crucial to understanding parasite transmission in general, we believe, are genetic differences between strains from orally and non-orally transmitted human cases. All TcI strains appear to be infective to humans and adapted to long-term carriage (8). However, the presence of a common, reduced-diversity TcI genotype cluster (TcIDOM) among a high proportion of human Chagas disease cases in South America is also well established (7,8). We originally hypothesized that TcIDOM was maintained, despite the presence of sympatric and infective sylvatic strains, because of low parasite transmission efficiency by invasive sylvatic vectors (8). Oral transmission is likely to be much more efficient. Thus, unlike TcIDOM strains, those from orally transmitted T. cruzi cases demonstrated high genetic diversity and clearly originated from local nonhuman T. cruzi populations. However, it is also true that all TcIDOM strains we isolated originated from patients with chronic infection, and all orally transmitted cases were in the acute phase. We cannot, therefore, rule out a role for immune selection in driving the frequency of TcIDOM infections among humans; such selection represents an intriguing topic for future enquiry.
Molecular tools and reference datasets are now available to determine the source of acute Chagas disease outbreaks within days of their occurrence. The plummeting cost of such analyses means it is time to apply population genetic techniques and markers developed for trypanosomes as genuine epidemiologic tools.
Photo of Antimano, a suburban slum of Caracas, map showing the geographic distribution of Trypanosoma cruzi I isolates analyzed, and primers and chromosomal positions for microsatellite loci used in a study of the molecular epidemiologic source tracking of 2 outbreaks of oral Chagas disease, Venezuela.
Trypanosoma cruzi isolates and clones analyzed in a study of the molecular epidemiologic source tracking of 2 outbreaks of oral Chagas disease, Venezuela.
Acknowledgments
Sincere thanks to Maria Dora Feliciangeli for her unconditional support. We also thank M. Rodriguez, L. Briceño, J. Ortegoza, M. Torres. F. Alfonso, H. Montañez, G. de Valenzuela, M. Sanchez, J. Lara, C. Viloria, J. Bracho, and L. Barrios for their support in the clinic, laboratory, and field.
This work was supported by FONACIT (grant G-2005000827) and the European Union (Fp7 grant 223034).
Mr. Segovia is a researcher at the Laboratorio de Biología Molecular de Protozoarios, Universidad Central de Venezuela, in Caracas, and he works in several clinical laboratories in the area. He has a keen practical interest in disease epidemiology and pathogen population genetics.
Footnotes
Suggested citation for this article: Segovia M, Carrasco HJ, Martínez CE, Messenger LA, Nessi A, Londoño JC, et al. Molecular epidemiologic source tracking of orally transmitted Chagas disease, Venezuela. Emerg Infect Dis [Internet]. 2013 Jul [date cited]. http://dx.doi.org/10.3201/eid1907.121576
References
- 1.Levy MZ, Bowman NM, Kawai V, Waller LA, Cornejo del Carpio JG, Cordova Benzaquen E, et al. Periurban Trypanosoma cruzi–infected Triatoma infestans, Arequipa, Peru. Emerg Infect Dis. 2006;12:1345–52 . 10.3201/eid1209.051662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medrano-Mercado N, Ugarte-Fernandez R, Butron V, Uber-Busek S, Guerra HL, Araujo-Jorge TC, et al. Urban transmission of Chagas disease in Cochabamba, Bolivia. Mem Inst Oswaldo Cruz. 2008;103:423–30. 10.1590/S0074-02762008000500003 [DOI] [PubMed] [Google Scholar]
- 3.Carrasco HJ, Segovia M, Llewellyn MS, Morocoima A, Urdaneta-Morales S, Martinez C, et al. Geographical distribution of Trypanosoma cruzi genotypes in Venezuela. PLoS Negl Trop Dis. 2012;6:e1707. PMID: 22745843 [DOI] [PMC free article] [PubMed]
- 4.Rassi A Jr, Rassi A, Marin-Neto JA. Chagas disease. Lancet. 2010;375:1388–402. 10.1016/S0140-6736(10)60061-X [DOI] [PubMed] [Google Scholar]
- 5.Shikanai-Yasuda MA, Carvalho NB. Oral transmission of Chagas disease. Clin Infect Dis. 2012;54:845–52 . 10.1093/cid/cir956 [DOI] [PubMed] [Google Scholar]
- 6.Alarcón de Noya B, Diaz-Bello Z, Colmenares C, Ruiz-Guevara R, Mauriello L, Zavala-Jaspe R, et al. Large urban outbreak of orally acquired acute Chagas disease at a school in Caracas, Venezuela. J Infect Dis. 2010;201:1308–15 . 10.1086/651608 [DOI] [PubMed] [Google Scholar]
- 7.Ramírez JD, Guhl F, Messenger LA, Lewis MD, Montilla M, Cucunuba Z, et al. Contemporary cryptic sexuality in Trypanosoma cruzi. Mol Ecol. 2012;21:4216–26 . 10.1111/j.1365-294X.2012.05699.x [DOI] [PubMed] [Google Scholar]
- 8.Llewellyn MS, Miles MA, Carrasco HJ, Lewis MD, Yeo M, Vargas J, et al. Genome-scale multilocus microsatellite typing of Trypanosoma cruzi discrete typing unit I reveals phylogeographic structure and specific genotypes linked to human infection. PLoS Pathog. 2009;5:e1000410 . 10.1371/journal.ppat.1000410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suárez J, de Suárez D, Alarcón de Noya B, Espinosa R, Chiurillo M, Villaroel PA, et al. Enfermedad de Chagas sistémico en fase aguda por transmisión oral: diagnóstico integral de un caso autopsiado. Gac Med Caracas. 2010;118:212–22. [Google Scholar]
- 10.Alarcón de Noya B, Martínez J. Transmisión oral de la enfermedad de Chagas en Venezuela: un segundo brote escolar. Salus. 2009;13:9–10. [Google Scholar]
- 11.Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 2010;11:94 . 10.1186/1471-2156-11-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nóbrega AA, Garcia MH, Tatto E, Obara MT, Costa E, Sobel J, et al. Oral transmission of Chagas disease by consumption of acai palm fruit, Brazil. Emerg Infect Dis. 2009;15:653–5. 10.3201/eid1504.081450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbosa RL, Dias VL, Pereira KS, Schmidt FL, Franco RM, Guaraldo AM, et al. Survival in vitro and virulence of Trypanosoma cruzi in acai pulp in experimental acute Chagas disease. J Food Prot. 2012;75:601–6 . 10.4315/0362-028X.JFP-11-233 [DOI] [PubMed] [Google Scholar]
- 14.Miles MA. Orally acquired Chagas disease: lessons from an urban school outbreak. J Infect Dis. 2010;201:1282–4 . 10.1086/651609 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Photo of Antimano, a suburban slum of Caracas, map showing the geographic distribution of Trypanosoma cruzi I isolates analyzed, and primers and chromosomal positions for microsatellite loci used in a study of the molecular epidemiologic source tracking of 2 outbreaks of oral Chagas disease, Venezuela.
Trypanosoma cruzi isolates and clones analyzed in a study of the molecular epidemiologic source tracking of 2 outbreaks of oral Chagas disease, Venezuela.