Abstract
Double stranded, short interfering RNAs (siRNA) of 21–22 nt length initiate a sequence-specific, post-trancriptional gene silencing in animals and plants known as RNA interference (RNAi). Here we show that RNAi can block a pathophysiological pain response and provide relief from neuropathic pain in a rat disease model by down regulating an endogenous, neuronally expressed gene. Rats, intrathecally infused with a 21 nt siRNA perfectly complementary to the pain-related cation-channel P2X3, showed diminished pain responses compared to missense (MS) siRNA-treated and untreated controls in models of both agonist-evoked pain and chronic neuropathic pain. This form of delivery caused no adverse effects in any of the animals receiving P2X3 siRNA, MS siRNA or vehicle. Molecular analysis of tissues revealed that P2X3 mRNA expressed in dorsal root ganglia, and P2X3 protein translocated into the dorsal horn of the spinal cord, were significantly diminished. These observations open a path toward use of siRNA as a genetic tool for drug target validation in the mammalian central nervous system, as well as for proof of concept studies and as therapeutic agents in man.
INTRODUCTION
RNAi is a naturally occurring mechanism for regulating gene expression which has been observed in several model organisms and is mediated by double stranded RNA (dsRNA) (1–3). Previously, RNAi has been observed and applied in plants (4), in invertebrates (1,2), in mammalian cell culture (5,6) and in early mouse development (7–9). Recently, siRNAs, either administered directly or provided via a variety of expression constructs including virus vectors, have been used in vivo to demonstrate efficacy in inhibition of expression of several disease-related targets including VEGF (10,11), Caspase-8 (12), hepatitis B (13,14) and C viruses (15), β-catenin (16), TNF-α (17), AGRP (18), β-glucuronidase (19) and the endogenous Fas gene expressed in adult mouse liver (20).
The intracellular presence of dsRNA homologous to a gene results in post-transcriptional gene silencing via induced sequence-specific degradation of the corresponding mRNA. Mechanistic studies have shown that Dicer, a ribonuclease III, specifically cleaves long dsRNA into short 21–22-nucleotide (nt) dsRNA (siRNA) carrying 2–3-nt 3′-overhangs on both sense and antisense strands (21–23). These siRNAs subsequently serve as guide sequences that instruct a multicomponent nuclease (the RISC complex) to destroy complementary mRNA (23,24). Unlike plants and invertebrates, mammalian cells respond to dsRNAs that are greater than 30 bp in length. These responses include interferon production (25) and activation of two pathways: PKR (26) and 2′-5′ polyadenylic acid (27). The former phosphorylates eIF2α, leading to a global shutdown of protein synthesis, and the latter activates RNaseL, mediating non-specific degradation of cellular mRNA. Activation of such mechanisms has been circumvented in mammalian cell culture by using siRNAs directly (5).
These observations indicate that it will now be possible to down regulate adult mammalian genes on a broader scale in vivo with RNAi, as is already done systematically in Caenorhabditis elegans (28,29), and enable the comprehensive study, proof of concept and validation of the roles of putative disease genes in whole mammalian organisms. Although the field is advancing rapidly, inhibition of a pathophysiological condition upon treatment with an siRNA has, to date, only been reported for concavalin A-induced liver damage (20) to prevent sepsis (17). siRNAs have been applied successfully to down regulate gene expression in the central nervous system (CNS) (18); however, no experiments have yet been reported demonstrating a direct effect of an siRNA on a pathophysiological disease phenotype in the CNS. In the work described here, we have examined the efficacy of a 21-nt siRNA bearing 3′-overhangs of two phosphorothioated (PS) 2′-O-(2-methoxyethyl)-ribonucleotides (MOE) (30), targeting P2X3 in vivo, using a system we had previously established for administration of P2X3 antisense oligonucleotides (ASO) (31) in the CNS, in which we measured effects of specific intrathecally administered oligonucleotides in a chronic neuropathic pain model. The siRNA sequence used in this study was chosen based upon the optimal ASO sequence selected in previous studies (32) (indicating that the site is available for oligonucleotide targeting) and which, after thorough testing in vitro, was shown to be even more potent than the original ASO (33). The purpose of our study was two-fold. Firstly, our goal was to evaluate the roles of putative pain genes in pain models in order to identify novel drug targets. Secondly, our plan was to test to see if siRNAs could be used therapeutically to alleviate a disease phenotype. We chose P2X3 as our first target for testing the principle of siRNA since we had already proven its role in pain via ASO treatment in vivo and had already established all the conditions for optimal delivery of oligonucleotides. Studies on P2X3 knockout mice also indicated that this gene plays a role in inflammatory pain signalling (34,35). This system, therefore, provided the ideal means for testing the efficacy of siRNAs in vivo in an adult animal disease model in the CNS. Administration of these potent P2X3 siRNAs in vivo resulted in a dramatic relief of pain as indicated by a significant inhibition in the neuropathic pain response, while exhibiting no apparent toxicity or non-specific side effects. Furthermore, in addition to being safer, siRNA was also more efficacious when compared with the administration of equivalent doses of ASO.
MATERIALS AND METHODS
siRNA and ASO compounds
The ASO compound has been previously published (31). P2X3 siRNA sequences were synthesized (5′-UCACUCGGCUG GAUGGAGUtt and 5′-ACUCCAUCCAGCCGAGUGAag), together with a corresponding 4-nt missense (MS) control siRNA sequence (5′-UCACUGCGCUCGAUGCAGUtt and 5′-ACUGCAUCGAGCGCAGUGAag) where lower case letters represent phosphorothioated ribonucleotides carrying a 2′-O-(2-methoxyethyl)-group (30).
Details of siRNA synthesis
Synthetic oligoribonucleotides were prepared using standard TOM-phosphoramidite chemistry (Xeragon AG) on an OligoPilot II synthesizer (Amersham Pharmacia Biotech) at 180 µmol scale. Phosphoramidites were dissolved in acetonitrile at 0.2 M concentration, mixed in a 1:1 ratio with a 0.2 M solution of benzimidazolium triflate in acetonitrile for coupling over 5 min. A first capping was made using standard capping reagents. Sulfurization was made by using a 0.05 M solution of N-ethyl,N-phenyl-5-amino-1,2,4-dithiazol-3-thione for 2 min. Oxidation was made by a 0.5 M t-butylhydroxyperoxide in dichloromethane for 2 min. A second capping was performed after oxidation or sulfurization. Oligonucleotides were detritylated for the following coupling by 2% dichloroacetic acid in dichloroethane. Upon completion of the sequences, the support-bound compounds were cleaved and base and phosphodiester deprotected as ‘Trityl-on’ material by a Methylamine solution (41% aqueous methylamine/33% ethanolic methylamine 1:1 v/v) at 35°C for 6 h. Resulting suspensions were lyophilized to dryness. 2′-O-silyl groups were removed upon treatment with 1 M tetrabutylammonium fluoride (10 min at 50°C and 6 h at 35°C). The obtained crude solutions were directly purified by RP-HPLC. The purified detritylated compounds were analysed by capillary gel electrophoresis for purity and quantified by UV according to their extinction coefficient at 260 nM. Identity was checked by electrospray mass spectrometry (5′-UCA CUCGGCUGGAUGGAGUtst: Mcalc = 6866.5 Da, Mmeas = 6869.1 Da; 5′-ACUCCAUCCAGCCGAGUGAasg: Mcalc = 6826.5 Da, Mmeas = 6830.5 Da; 5′-UCACUGCGCUC GAUGCAGUtst: Mcalc = 6784.4 Da, Mmeas = 6784.0 Da; 5′-ACUGCAUCGAGCGCAGUGAasg: Mcalc = 6904.5 Da, Mmeas = 6903.5 Da).
siRNA annealing
For annealing of siRNA, 1 mM single strands were incubated in isotonic buffer (100 mM potassium acetate, 30 mM HEPES–KOH, 2 mM magnesium acetate, 26 mM NaCl, pH 7.4 at 37°C) for 5 min at 90°C followed by 1 h at 37°C. These conditions differ slightly from the original conditions described by Elbashir et al. (5): higher concentrations of oligonucleotides were used for annealing, which was confirmed by gel analysis; denaturation at 90°C was performed for 5 min instead of 1 min; NaCl was added to assure a non-irritating, isotonic solution for in vivo testing. At the time of annealing, both strands were fully deprotected.
Animal models
All experiments were carried out according to Home Office (UK) guidelines and with approval of the local Novartis Animal Welfare and Ethics Committee. The partial ligation model and infusion of siRNA, MS siRNA and ASO into naïve and neuropathic rats via minipump as well as behavioural testing for mechanical hyperalgesia and allodynia were performed as described previously (31,36). The intrathecal dose of 400 µg/day P2X3 siRNA and MS siRNA or 180 µg/day P2X3 ASO was continuously infused over the course of 6–7 days. Each animal was tested in a blind random order. Statistical significance of mechanical hyperalgesia and allodynia data were analysed with ANOVA followed by Tukey’s HSD test, *P < 0.05, **P < 0.005, ***P < 0.001.
Molecular analysis
Fixed, cryopreserved spinal cord was dissected and treated with an immunohistochemistry protocol described previously (31) with one difference: as a control, biotinylated IB4 (Sigma, Buchs, Switzerland; 1:35) was used instead of anti-CGRP followed by streptavidin-TRITC (Jackson ImmunoResearch, West Grove, PA; 1:200). Dorsal root ganglia (DRG) from non-perfused rats were snap-frozen and RNA was isolated separately for each animal by crushing the frozen DRG with mortar and pestle and syringing 10 times to fragment genomic DNA. The RNA was then purified with RNeasy Columns (Qiagen, Basel, Switzerland). 50 ng of each sample was analysed three times with the ABI PRISM™ 7700 Sequence Detector (PE Applied Biosystems, Foster City, CA) as described previously (31). Values were then normalized to β-actin analysis done in parallel on the same samples. Statistics: ANOVA followed by Tukey’s HSD test, *P < 0.05.
RESULTS AND DISCUSSION
PS-MOE modifications are known to offer superior stability against nucleases when incorporated into ASO and have proven particularly useful in in vivo studies (31). Used as 3′-overhangs in siRNAs, these RNA analogues have been shown not to adversely affect the RNAi mechanism in cell culture experiments compared to standard siRNAs carrying deoxythymidine dimers (dTdT) (33). Furthermore, this chemical modification, 2′-MOE at two 3′ terminal nucleotides of each strand linked via a phosphorothioate, has been shown to be just as potent in vitro as native dsRNA (33) and was included as a chemical protection against possible in vivo nuclease degradation. P2X3 is an ATP-gated cation channel, expressed in a restricted fashion in the cell bodies of sensory neurons and their projections. Evidence for its involvement in pain is based on the regulation of P2X3 message expression in animal models of chronic pain and altered pain-related responses in knockout mice (34,35). More direct proof was provided by our demonstration of an inhibition of hyperalgesia in a model of neuropathic pain by ASO delivery, a finding which has been independently confirmed (37) and is further supported by the recent report of analgesic activity of a small molecule antagonist of P2X3 in a variety of animal models of pain (38,39). Since we have already established a role for P2X3 in a rat model of neuropathic pain, and have delineated the optimal conditions for delivery of oligonucleotides in this model, this target was chosen to test the principle of the utility of siRNA in validating potential targets in disease states in vivo.
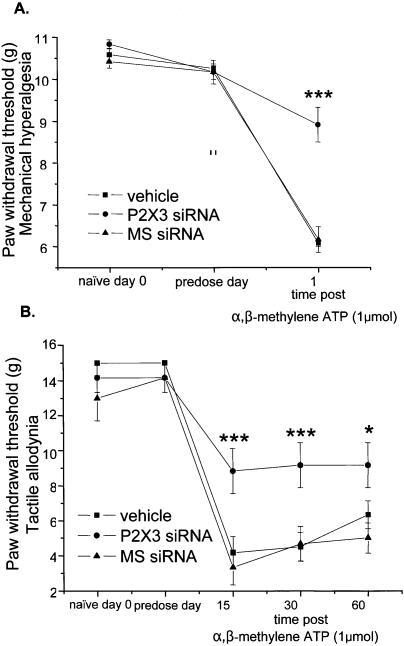
P2X3 siRNA sequences, complementary to the same region of the gene targeted in our earlier ASO study (31), together with an appropriate mismatch control were synthesized. The reagents that we employed in this study were thoroughly characterized in vitro for their ability to inhibit P2X3 at mRNA, protein and functional levels (33). In preliminary experiments, varying doses of siRNAs were delivered intrathecally into the spinal fluid of naïve rats (n/group = 3) via an indwelling cannula attached to an osmotic minipump (data not shown). This work established that doses up to 400 µg/day did not elicit any signs of neurotoxicity (hind limb paralysis, vocalization or anatomical damage to the spinal cord). This dose, approximately equivalent to the ASO dose evaluated in our previous study (31), was used in all subsequent studies, resulting in significant and specific behavioural effects. Thus, following intrathecal delivery of P2X3 siRNA for 6 days in naïve rats, an intraplantar injection of the P2X3 agonist α,β-methylene ATP (1 µmol) into the left hindpaw resulted in a diminished level of mechanical hyperalgesia (hypersensitivity to a noxious pressure stimulus of ∼100 g) and tactile allodynia (hypersensitivity to a normally non-noxious pressure stimulus of ∼15–20 g), as measured by a reduction in paw withdrawal thresholds (PWTs) (P < 0.001, Fig. 1) compared to vehicle- and MS siRNA-treated rats. Molecular analysis using tissues isolated from the rats revealed that the down regulation of P2X3 mRNA levels in DRG and protein levels in the dorsal horn of the spinal cord correlated with the decrease in pain response in P2X3 siRNA-treated rats (Fig. 2). Quantification of P2X3 mRNA from DRG using quantitative RT–PCR, the most sensitive method of mRNA quantitation, demonstrated 40% down regulation of the target for P2X3 siRNA-treated animals compared to MS siRNA and vehicle controls. The incomplete knock-down of target mRNA may be a result of residual mRNA in the cell nucleus, or may simply reflect an incomplete transfection of DRGs. P2X3 protein was analysed immunohistochemically at its site of action in the entire dorsal horn of lumbar spinal cord tissue slice and appeared visibly down regulated in lamina II in the P2X3 siRNA group compared to MS siRNA and vehicle-treated controls (Fig. 2B).
Figure 1.
P2X3 siRNA reduced agonist-induced pain in vivo. Significant inhibition of α,β-methylene-ATP evoked (A) mechanical hyperalgesia and (B) tactile allodynia in the hind paw of naïve rats after 6-day intrathecal treatment with P2X3 siRNA compared to MS siRNA or vehicle. PWTs were measured prior to cannulation on day 0, and on day 6 before and after intraplantar injection of 1 µmol α,β-methylene-ATP (n/group = 6).
Figure 2.
Molecular down regulation of P2X3 RNA and protein upon treatment with siRNA. (A) P2X3 mRNA levels in lumbar L4/L5 DRG, normalized to β-actin, were significantly reduced in P2X3 siRNA- versus MS siRNA- and vehicle-treated rats (n/group = 3). (B) P2X3 immunoreactivity in lamina II of the spinal cord was visibly reduced in siRNA-treated animals (top panel). Co-immunostaining with neuron-specific isolectin B4 (green) showed that equivalent regions of each spinal cord were analysed (n/group = 3).
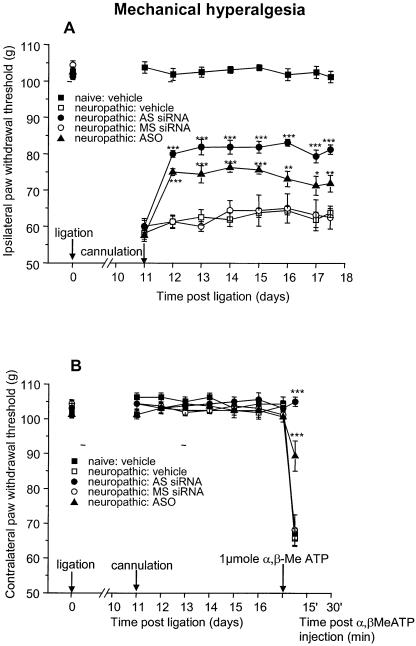
The significant reduction of mechanical hyperalgesia and tactile allodynia observed in the agonist-induced pain model described above was subsequently confirmed in the partial sciatic ligation model of neuropathic pain (40). Within 2 days of initiation of treatment, P2X3 siRNA, but not MS siRNA or vehicle, produced a significant reduction (P < 0.001) in tactile allodynia which was maintained for the duration of the siRNA administration (Fig. 3A). In the same experiment, administration of P2X3 ASO at a dose of 180 µg/day, in contrast, did not reduce allodynia, confirming results from our earlier study (31). The cannulation process itself does not cause any change in nociceptive indices as indicated by the PWTs of the naïve vehicle group. In the contralateral paws, where the nerve was not ligated (Fig. 3B), all groups exhibited similar PWTs to the naïve controls. However, intraplantar application of α,β-methylene ATP on the final day of the experiment evoked tactile allodynia, measurable 15, 30 and 60 min post-agonist injection in vehicle-, ASO- and MS siRNA-treated groups. In contrast, the allodynic response was significantly (P < 0.001) diminished in the P2X3 siRNA-treated group, demonstrating a functional down regulation of the P2X3 channel (Fig. 3B). Taken together with the effects observed with the match and mismatch siRNAs (sequences of which do not exist in any other P2X family member (e.g. P2X1 or P2X2) and the fact that the P2X3 isoform is the primary α,β-methylene-ATP sensitive P2X target expressed in DRG neurons (36), confines the functional effects of this siRNA to inhibition of P2X3. When the same animals were tested for mechanical hyperalgesia, a significant reversal of PWTs was observed in neuropathic paws 1 day after initiating treatment, and this was maintained for the duration of the application of P2X3 siRNA and ASO. However, the double-stranded P2X3 siRNA (400 µg/day) was more effective than the ASO at approximately equimolar concentrations (180 µg/day). Reversal of mechanical hyperalgesia was not seen in the control groups (Fig. 4A), supporting a specific down regulation of P2X3 receptors. Mechanical hyperalgesia was not observed in the contralateral paws of any group until the agonist α,β-methylene ATP was injected on the last day of the experiment, whereupon vehicle- and MS siRNA-treated groups exhibited a considerable decrease in PWTs (Fig. 4B). Remarkably, P2X3 siRNA treatment completely suppressed the hyperalgesic response of α,β-methylene ATP, whereas P2X3 ASO diminished it by more than 50%, both confirming functional down regulation of the channel (Fig. 4B). Both measurements of the hyperalgesic response (Fig. 4A and B) confirm that the P2X3 siRNA reagent is considerably more effective at reducing pain than the analogous ASO and this difference may be one possible explanation as to why the P2X3 siRNA is more potent than the analogous ASO in the allodynia model measurements.
Figure 3.
P2X3 siRNA reduced tactile allodynia in a neuropathic pain model. (A) Tactile allodynia was significantly inhibited in the neuropathic paw (partially ligated sciatic nerve) in P2X3 siRNA- versus MS siRNA-, P2X3 ASO- and vehicle-treated rats. An additional naïve (non-ligated) group that received vehicle only controlled for the cannulation process. (B) PWTs in the contralateral paws of all groups were the same as naïve animals. Upon agonist injection (1 µmol α,β-methylene-ATP) on the last day of the experiment (day 17), all control groups and the ASO group showed agonist-induced tactile allodynia when assayed at 15, 30 and 60 min post injection. P2X3 siRNA significantly diminished agonist-induced tactile allodynia (n/group = 8).
Figure 4.
P2X3 siRNA reduced mechanical hyperalgesia in a neuropathic pain model. (A) Mechanical hyperalgesia was significantly inhibited in the neuropathic paw (partially ligated sciatic nerve) in P2X3 siRNA- and in P2X3 ASO- versus MS siRNA- and vehicle-treated rats. As a control, an additional naïve vehicle group had not been ligated and showed normal pain perception. (B) PWTs in the contralateral paws of all groups was the same as normal animals. Upon agonist injection (1 µmol α,β-methylene-ATP) on the last day of the experiment (day 17), all groups showed agonist-induced mechanical hyperalgesia at 15 min after injection, except for the P2X3 siRNA- and the ASO-treated groups. P2X3 siRNA completely suppressed agonist-induced mechanical hyperalgesia (n/group = 8).
An important observation was the absence of any apparent toxicity due to intrathecal administration of siRNAs at the doses described in these studies. There were no animals lost; no hind leg paralysis was observed, and no signs of discomfort were exhibited throughout the experiments. The rats were carefully observed during the experiments, both during the behavioural studies and during dissection of the DRG and spinal cord tissue. Although the siRNA was administered intrathecally directly into the spinal fluid, there were no observable behavioural changes and no obvious signs of spinal inflammation upon dissection.
In summary, we have shown in an agonist-induced pain model, that siRNA produced a significant behavioural reduction of mechanical hyperalgesia and allodynia, which correlated with a molecular down regulation of P2X3 in DRG and spinal cord. This behavioural effect was subsequently reproduced in a disease model of chronic neuropathic pain. The reversal of pain was not complete, possibly due to the partial down regulation of P2X3, or more likely, the involvement of proteins other than P2X3 in neuropathic pain.
However, it has been already demonstrated that partial inhibition of P2X3 with ASO treatment produces significant behavioural effects (31,37). In general, the behavioural effects produced by application of P2X3 siRNA were similar to those observed after administration of an equivalent dose of P2X3 ASO; however, an important difference was the reversal of allodynia by the former, an effect that we did not observe upon ASO treatment. Both the knock-out animals and the siRNA-treated animals clearly show an important role of P2X3 in pain behaviour. The experiments are complementary, as P2X3 knock-out animals were tested for sensitivity to warming stimuli and bladder response, whereas the siRNA-treated animals were assessed in agonist-induced and neuropathic pain models. Concerning neuropathic pain, the effects observed using these siRNAs correlate very well with those from two recently published studies characterizing a low molecular weight inhibitor of P2X3 and P2X2/3 (38,39).
In comparison to previous reports, our results are novel in three important respects: firstly, an endogenous disease-related neuronal gene in the nervous system has been targeted. Secondly, siRNA was delivered intrathecally to the spinal fluid, a delivery method which is commonly used for clinical treatment of severe pain. Thirdly, and most importantly, the readout was a measurable pathophysiological response in a relevant and well-defined disease model. These data have immediate implications in the study of gene function in that the mechanism is sequence specific, rapid and scalable, allowing one to envisage multiparallel analyses on collections of genes for in vivo target validation in drug discovery.
Finally, considering its effectiveness in vivo and relative lack of toxicity even at a high concentration, these data indicate that siRNA could be used extensively to test the involvement of potential novel targets in disease processes, both in animal models and in man. This is particularly attractive where drugs or small molecule compounds directed at these targets are not available. On a more speculative level there is the additional possibility that RNAi can be used itself as a therapy for certain disease states where delivery and target protein knockdown can be optimized.
Acknowledgments
ACKNOWLEDGEMENT
We would like to thank Dr Fred Asselbergs for critical review of this manuscript.
REFERENCES
- 1.Sharp P.A. (2001) RNA interference–2001. Genes Dev., 15, 485–490. [DOI] [PubMed] [Google Scholar]
- 2.Tuschl T. (2001) RNA interference and small interfering RNAs. ChemBioChem, 2, 239–245. [DOI] [PubMed] [Google Scholar]
- 3.Hammond S.M., Caudy,A.A. and Hannon,G.J. (2001) Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. Genet., 2, 110–119. [DOI] [PubMed] [Google Scholar]
- 4.Waterhouse P.M., Wang,M.B. and Lough,T. (2001) Gene silencing as an adaptive defence against viruses. Nature, 411, 834–842. [DOI] [PubMed] [Google Scholar]
- 5.Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 6.Harborth J., Elbashir,S.M., Bechert,K., Tuschl,T. and Weber,K. (2001) Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci., 114, 4557–4565. [DOI] [PubMed] [Google Scholar]
- 7.Svoboda P., Stein,P., Hayashi,H. and Schultz,R.M. (2000) Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development, 127, 4147–4156. [DOI] [PubMed] [Google Scholar]
- 8.Wianny F. and Zernicka-Goetz,M. (2000) Specific interference with gene function by double-stranded RNA in early mouse development. Nat. Cell Biol., 2, 70–75. [DOI] [PubMed] [Google Scholar]
- 9.McCaffrey A.P., Meuse,L., Pham,T.T., Conklin,D.S., Hannon,G.J. and Kay,M.A. (2002) RNA interference in adult mice. Nature, 418, 38–39. [DOI] [PubMed] [Google Scholar]
- 10.Reich S.J., Fosnot,J., Kuroki,A., Tang,W., Yang,X., Maguire,A.M., Bennett,J. and Tolentino,M.J. (2003) Small interfering RNA (siRNA) targeting VEGF effectively inhibits ocular neovascularization in a mouse model. Mol. Vis., 9, 210–216. [PubMed] [Google Scholar]
- 11.Filleur S., Courtin,A., Ait-Si-Ali,S., Guglielmi,J., Merle,C., Harel-Bellan,A., Clezardin,P. and Cabon,F. (2003) SiRNA-mediated inhibition of vascular endothelial growth factor severely limits tumor resistance to antiangiogenic thrombospondin-1 and slows tumor vascularization and growth. Cancer Res., 63, 3919–3922. [PubMed] [Google Scholar]
- 12.Zender L., Hutker,S., Liedtke,C., Tillmann,H.L., Zender,S., Mundt,B., Waltemathe,M., Gosling,T., Flemming,P., Malek,N.P. et al. (2003) Caspase 8 small interfering RNA prevents acute liver failure in mice. Proc. Natl Acad. Sci. USA, 100, 7797–7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giladi H., Ketzinel-Gilad,M., Rivkin,L., Felig,Y., Nussbaum,O. and Galun,E. (2003) Small interfering RNA inhibits hepatitis B virus replication in mice. Mol. Ther., 8, 769–776. [DOI] [PubMed] [Google Scholar]
- 14.McCaffrey A.P., Nakai,H., Pandey,K., Huang,Z., Salazar,F.H., Xu,H., Wieland,S.F., Marion,P.L. and Kay,M.A. (2003) Inhibition of hepatitis B virus in mice by RNA interference. Nat. Biotechnol., 21, 639–644. [DOI] [PubMed] [Google Scholar]
- 15.Yokota T., Sakamoto,N., Enomoto,N., Tanabe,Y., Miyagishi,M., Maekawa,S., Yi,L., Kurosaki,M., Taira,K., Watanabe,M. and Mizusawa,H. (2003) Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. EMBO Rep., 4, 602–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verma U.N., Surabhi,R.M., Schmaltieg,A., Becerra,C. and Gaynor,R.B. (2003) Small interfering RNAs directed against beta-catenin inhibit the in vitro and in vivo growth of colon cancer cells. Clin. Cancer Res., 9, 1291–1300. [PubMed] [Google Scholar]
- 17.Sorensen D.R., Leirdal,M. and Sioud,M. (2003) Gene silencing by systemic delivery of synthetic siRNAs in adult mice. J. Mol. Biol., 327, 761–766. [DOI] [PubMed] [Google Scholar]
- 18.Makimura H., Mizuno,T.M., Mastaitis,J.W., Agami,R. and Mobbs,C.V. (2002) Reducing hypothalamic AGRP by RNA interference increases metabolic rate and decreases body weight without influencing food intake. BMC Neurosci., 3, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia H., Mao,Q., Paulson,H.L. and Davidson,B.L. (2002) siRNA-mediated gene silencing in vitro and in vivo. Nat. Biotechnol., 20, 1006–1010. [DOI] [PubMed] [Google Scholar]
- 20.Song E., Lee,S.K., Wang,J., Ince,N., Ouyang,N., Min,J., Chen,J., Shankar,P. and Lieberman,J. (2003) RNA interference targeting Fas protects mice from fulminant hepatitis. Nature Med., 9, 347–351. [DOI] [PubMed] [Google Scholar]
- 21.Elbashir S.M., Lendeckel,W. and Tuschl,T. (2001) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev., 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. [DOI] [PubMed] [Google Scholar]
- 23.Zamore P.D., Tuschl,T., Sharp,P.A. and Bartel,D.P. (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell, 101, 25–33. [DOI] [PubMed] [Google Scholar]
- 24.Hammond S.M., Bernstein,E., Beach,D. and Hannon,G.J. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature, 404, 293–296. [DOI] [PubMed] [Google Scholar]
- 25.Stark G.R., Kerr,I.M., Williams,B.R., Silverman,R.H. and Schreiber,R.D. (1998) How cells respond to interferons. Annu. Rev. Biochem., 67, 227–264. [DOI] [PubMed] [Google Scholar]
- 26.Clemens M.J. and Elia,A. (1997) The double-stranded RNA-dependent protein kinase PKR: structure and function. J. Interferon Cytokine Res., 17, 503–524. [DOI] [PubMed] [Google Scholar]
- 27.Player M.R. and Torrence,P.F. (1998) The 2–5A system: modulation of viral and cellular processes through acceleration of RNA degradation. Pharmacol. Ther., 78, 55–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fraser A.G., Kamath,R.S., Zipperlen,P., Martinez-Campos,M., Sohrmann,M. and Ahringer,J. (2000) Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature, 408, 325–330. [DOI] [PubMed] [Google Scholar]
- 29.Gonczy P., Echeverri,C., Oegema,K., Coulson,A., Jones,S.J., Copley,R.R., Duperon,J., Oegema,J., Brehm,M., Cassin,E. et al. (2000) Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature, 408, 331–336. [DOI] [PubMed] [Google Scholar]
- 30.Martin P. (1995) New access to 2′-O-alkylated ribonucleosides and properties of 2′-O-alkylated oligoribonucleotides. Helv. Chem. Acta, 78, 486–504. [Google Scholar]
- 31.Barclay J., Patel,S., Dorn,G., Wotherspoon,G., Moffatt,S., Eunson,L., Abdel’al,S., Natt,F., Hall,J., Winter,J. et al. (2002) Functional downregulation of P2X3 receptor subunit in rat sensory neurons reveals a significant role in chronic neuropathic and inflammatory pain. J. Neurosci., 22, 8139–8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dorn G., Abdel’al,S., Natt,F.J., Weiler,J., Hall,J., Meigel,I., Mosbacher,J. and Wishart,W. (2001) Specific inhibition of the rat ligand-gated ion channel P2X3 function via methoxyethoxy-modified phosphorothioated antisense oligonucleotides. Antisense Nucleic Acid Drug Dev., 11, 165–174. [DOI] [PubMed] [Google Scholar]
- 33.Hemmings-Mieszczak M., Dorn,G., Natt,F.J., Hall,J. and Wishart,W.L. (2003) Independent combinatorial effect of antisense oligonucleotides and RNAi-mediated specific inhibition of the recombinant rat P2X3 receptor. Nucleic Acids Res., 31, 2117–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cockayne D.A., Hamilton,S.G., Zhu,Q.M., Dunn,P.M., Zhong,Y., Novakovic,S., Malmberg,A.B., Cain,G., Berson,A., Kassotakis,L. et al. (2000) Urinary bladder hyporeflexia and reduced pain-related behaviour in P2X3-deficient mice. Nature, 407, 1011–1015. [DOI] [PubMed] [Google Scholar]
- 35.Souslova V., Cesare,P., Ding,Y., Akopian,A.N., Stanfa,L., Suzuki,R., Carpenter,K., Dickenson,A., Boyce,S., Hill,R. et al. (2000) Warm-coding deficits and aberrant inflammatory pain in mice lacking P2X3 receptors. Nature, 407, 1015–1017. [DOI] [PubMed] [Google Scholar]
- 36.Dunn P.M., Zhong,Y. and Burnstock,G. (2001) P2X receptors in peripheral neurons. Prog. Neurobiol., 65, 107–134. [DOI] [PubMed] [Google Scholar]
- 37.Honore P., Kage,K., Mikusa,J., Watt,A.T., Johnston,J.F., Wyatt,J.R., Faltynek,C.R., Jarvis,M.F. and Lynch,K. (2002) Analgesic profile of intrathecal P2X(3) antisense oligonucleotide treatment in chronic inflammatory and neuropathic pain states in rats. Pain, 99, 11–19. [DOI] [PubMed] [Google Scholar]
- 38.Jarvis M.F., Burgard,E.C., McGaraughty,S., Honore,P., Lynch,K., Brennan,T.J., Subieta,A., Van Biesen,T., Cartmell,J., Bianchi,B. et al. (2002) A-317491, a novel potent and selective non-nucleotide antagonist of P2X3 and P2X2/3 receptors, reduces chronic inflammatory and neuropathic pain in the rat. Proc. Natl Acad. Sci. USA, 99, 17179–17184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGaraughty S., Wismer,C.T., Zhu,C.Z., Mikusa,J., Honore,P., Chu,K.L., Lee,C.H., Faltynek,C.R. and Jarvis,M.F. (2003) Effects of A-317491, a novel and selective P2X3/P2X2/3 receptor antagonist, on neuropathic, inflammatory and chemogenic nociception following intrathecal and intraplantar administration. Br. J. Pharmacol., 140, 1381–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seltzer Z., Dubner,R. and Shir,Y. (1990) A novel behavioral model of neuropathic pain disorders produced in rats by partial sciatic nerve injury. Pain, 43, 205–218. [DOI] [PubMed] [Google Scholar]