Abstract
DNA microarrays are powerful tools for comparing gene expression profiles from closely related organisms. However, a single microarray design is frequently used in these studies. Therefore, the levels of certain transcripts can be grossly underestimated due to sequence differences between the transcripts and the arrayed DNA probes. Here, we seek to improve the sensitivity and specificity of oligonucleotide microarray-based gene expression analysis by using genomic sequence information to predict the hybridization efficiency of orthologous transcripts to a given microarray. To test our approach, we examine hybridization patterns from three Escherichia coli strains on E.coli K-12 MG1655 gene expression microarrays. We create electronic mask files to discard data from probes predicted to have poor hybridization sensitivity and specificity to cDNA targets from each strain. We increased the accuracy of gene expression analysis and identified genes that cannot be accurately interrogated in each strain using these microarrays. Overall, these studies provide guidelines for designing effective electronic masks for gene expression analysis in organisms where substantial genome sequence information is available.
INTRODUCTION
Differential gene expression is a fundamental mechanism for the development of species-specific traits (1,2). In principle, these traits range from microbial survival in stressful environmental conditions (3) to morphological adaptations in multicellular organisms (1,4,5) to the emergence of human-specific brain functions (6–12). It is a considerable challenge to identify transcriptional changes relevant to these traits since the gene expression profiles of cells and tissues from highly related organisms can vary extensively (3,4,8,13).
One approach to this problem is to systematically catalog inter- as well as intra-species variation in gene expression profiles. Both cDNA and oligonucleotide microarrays have been used to compare expression profiles in cells and tissues from closely related organisms (3,4,8,11,13–15). Typically, DNA probes in these microarrays are specifically designed to interrogate the abundance of transcripts from only one of the organisms examined. However, the majority of RNA transcripts from other highly related organisms, especially ones with over 95% nucleotide identity in orthologous 3′-UTR sequences, should effectively hybridize to the arrayed probes (16–21).
Nevertheless, these cross-species comparative gene expression experiments can yield partially inaccurate data sets with the levels of certain transcripts being underestimated or, more rarely, overestimated. In the former case, mismatches can disrupt binding of specific transcripts to probes designed to interrogate their abundance. These mismatches will affect hybridization to oligonucleotide microarrays much more than cDNA microarrays consisting of PCR products several hundred nucleotides in length. In one commonly used microarray platform, a series of 25mer oligonucleotide probes interrogate the abundance of each transcript (22). The relative abundance of specific transcripts can be underestimated if a significant number of probes interrogating these transcripts are mismatched and thus have weak affinities toward one another (11). In cases where whole genes or 3′-UTR sequences are deleted, hybridization will be compromised for both cDNA and oligonucleotide microarrays (11). Conversely, the relative abundance of specific transcripts can be overestimated due to duplications (23) that increase the potential for the cross-hybridization of highly related sequences to specific probes in the microarray.
We seek to improve the sensitivity and specificity of oligonucleotide microarray-based gene expression analysis of highly related organisms using whole genome sequence information. Here, we use Escherichia coli as a model organism to test our approach. We create electronic mask files to discard data from oligonucleotide probes in commercially available E.coli K-12 MG1655 gene expression microarrays predicted to have poor hybridization sensitivity and specificity to cDNA targets from three different E.coli strains. This allowed us to increase the accuracy of gene expression analysis in each strain and identify genes that cannot be accurately interrogated in different E.coli strains using these microarrays. We validate the effectiveness of these electronic masks on microarray-based gene expression data sets using confirmatory quantitative real-time PCR (qRT–PCR) analysis.
MATERIALS AND METHODS
Growth conditions and RNA isolation
The non-pathogenic E.coli K-12 MG1655 (ATCC 700926) and the pathogenic E.coli O157:H7 EDL933 (ATCC 700927) and E.coli CFT073 (ATCC 700928) strains were obtained from the American Type Culture Collection (Manassas, VA). All strains were maintained on Nutrient Agar (Becton Dickinson, Sparks, MD) at 37°C and stored at –80°C in Nutrient Broth (Becton Dickinson) with 20% glycerol. Strains were initially grown in Nutrient Broth with agitation at 37°C to mid-logarithmic phase and diluted in Nutrient Broth to an OD600 value of 0.04. When cultures again reached mid-logarithmic phase, RNA was harvested using RNAqueous™-4PCR kit (Ambion, Austin, TX) and the manufacturer’s recommended protocols.
Oligonucleotide microarray experiments and data analysis
Escherichia coli total RNA samples (10 µg per sample) were converted into biotin-labeled cDNA using the Enzo™ BioArray™ Terminal Labeling Kit with Biotin-ddUTP and standard protocols recommended by Affymetrix (Santa Clara, CA). For each strain, 2.5 µg of fragmented cDNA was applied to E.coli Antisense Genome Arrays (Affymetrix) which contain probe sets designed to detect the antisense strand of all known E.coli K-12 MG1655 open reading frames. Microarrays were hybridized for 12–16 h at 45°C under standard conditions. Following hybridization, microarrays were washed using the Affymetrix Fluidics Station 400, stained twice with a streptavidin–phycoerythrin conjugate, and then read using a Hewlett Packard GeneArray Scanner. All microarray experiments were performed in duplicate using two different biotin-labeled cDNA targets separately prepared for each sample. This helped us minimize experimental noise associated with cDNA labeling efficiency.
We generated raw gene expression scores for every gene in every sample using Microarray Suite version 5.0 software (Affymetrix) as previously stated (11). In order to minimize noise associated with less robust hybridization, all gene expression scores below 100 were set to 100. Duplicate measurements were averaged to obtain a single gene expression score for each strain. Gene expression data are available in Affymetrix GeneChip (.CEL) format on our web site and Microsoft Excel 2000 (.XLS) format in tables 1 and 2 in the Supplementary Material.
Thermodynamic calculations of Gibbs free energies
Nearest-neighbor rules were used to calculate the Gibbs free energy for the longest continuous duplex (LCD) and second longest continuous duplex (SLCD) of each perfectly matched (PM) probe and LCD of each mismatched (MM) probe sequence with genomic sequences from each E.coli strain (24,25). For example, the ΔG for a non-self-complementary 25 bp duplex was calculated as the sum of the ΔG values of all 24 nearest neighbor pairs (25–28) plus the free energy of helix initiation (ΔGinit).
Criteria for masking probes
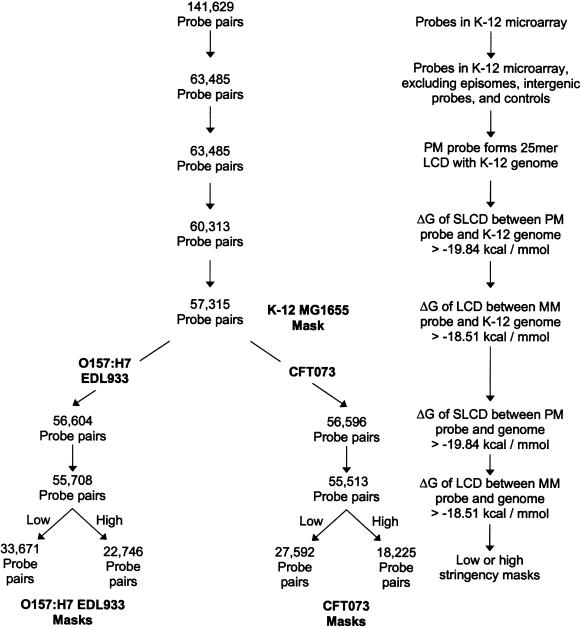
All probe sequences on the E.coli Antisense Genome Arrays (Affymetrix) were compared against the K-12 MG1655, O157:H7 EDL933, or CFT073 genome sequences (available through NCBI) using the BLAST algorithm (29). We used extremely strict gap and mismatch penalties (‘F’ and –1000, respectively) in order to identify the LCD of a probe that matches the genome sequence without mismatches or gaps. In designing a series of masks for each E.coli strain, we excluded PM/MM probe pairs according to the criteria given in Figure 1.
Figure 1.
Flowchart for the construction of strain-specific electronic masks. The number of PM/MM probe pairs remaining at each stage of constructing electronic masks for three E.coli strains are given. The criteria for selecting ΔG values for the LCD and SLCD that could be formed between the arrayed probes and genomic sequences of the three E.coli strains are described in the text.
Mask file structure
A mask file is written in a specified text format that (a) lists the probe Id (gene identifier) in the first column and (b) probe numbers in the second column. The mask files are available as Supplementary Material.
Masking protocol
Affymetrix Microarray Suite v5.0 software utilizes the electronic mask file to exclude probes from consideration when calculating the final expression scores for that gene. This mask file is applied when the .CEL files, that assign a hybridization signal to each probe, are being processed into .CHP files, that assign hybridization scores for each probe tiling path. Alternatively, the mask can also be applied when the .DAT file is being processed into the .CEL file, in which case, the .CEL file will contain the information of the masked probes. Although we use the Affymetrix GeneChip v5.0 program, our strategy is also applicable to other software (30).
qRT–PCR analysis
First, total RNA (500 ng) was converted to cDNA using iScript cDNA synthesis kit (Bio-Rad Laboratories). RT–PCR reactions were assembled in triplicate using the iQ SYBR Green Supermix Kit (Bio-Rad Laboratories). The amount of PCR product synthesized at each cycle was quantified by measuring the fluorescent signal generated by SYBR Green binding to the amplified cDNA. The relative abundance of each transcript was calculated based on PCR efficiency and cycle number at which the fluorescence crosses a threshold for the GAPDH internal reference and the gene tested using iCycler iQ optical system software (Bio-Rad Laboratories).
RESULTS
Choice of model organism and microarray platform
We sought to identify a group of closely related organisms with sequenced genomes that share significant nucleotide identity with oligonucleotide probes in an established microarray platform. Three E.coli strains (K-12 MG1655, O157:H7 EDL933 and CFT073) fulfill these criteria (31–34). The E.coli K-12 microarrays from Affymetrix (Santa Clara, CA), designed to interrogate the abundance of all currently known E.coli K-12 MG1655 open reading frames, were chosen for these studies (35). The E.coli K-12 microarrays consist of a series of 25mer probes complementary to segments of E.coli K-12 MG1655 transcripts (35–38). Each PM probe to these transcripts has a cognate mismatch probe (MM probe) identical in sequence except for a single nucleotide change near the center of the probe. The latter are often used to compensate for non-specific hybridization to PM probes (39). Hybridization data from a set of PM/MM probe pairs interrogating a specific transcript, referred to as a probe tiling path, are considered when assigning individual gene expression scores.
To obtain a global estimate for how well the K-12 MG1655 microarray probes match sequences from these strains, we aligned the E.coli K-12 MG1655 genomic segments from which PM probes sequences were selected to the orthologous genomic segments in the other E.coli strains. In doing so, we excluded probe pairs that interrogate episomal, plasmid and phage sequences that are not present in the E.coli K-12 MG1655 genome. As expected, the genomic segments from which the oligonucleotide probes were designed were 100% identical to K-12 MG1655 genome sequences. These segments also shared 97.7 and 96.5% identity with the O157:H7 EDL933 and CFT073 genomic sequences, respectively.
Masking criteria
As an initial step in creating strain-specific electronic masks for the K-12 microarrays, we focused on setting uniform standards for E.coli K-12 MG1655 RNA sample hybridization (Fig. 1). Since the K-12 microarray was designed to evaluate the abundance of transcripts from this strain, we expected minimal probe masking would be needed to optimize microarray performance. First, we exclude 1580 PM/MM probe pairs that interrogate sequences that are not present in the E.coli K-12 MG1655 genome. Second, we excluded 76 405 PM/MM probe pairs that interrogate intergenic regions. Many of these probes are present in non-transcribed regions and thus can confound data interpretation. We also excluded 159 probes that contained homopolymeric sequence tracts greater than six bases in length. Such probes provide poor hybridization specificity and/or affinity (40). After these initial masking steps, we found that 4345 known or putative E.coli K-12 MG1655 transcripts are interrogated by these microarrays. Over 95.7% of these tiling paths consist of 14–15 PM/MM probe pairs.
Predicting hybridization characteristics
In principle, it is possible to identify other PM/MM probe pairs with poor hybridization specificity and selectivity based upon the thermodynamic properties of nucleic acid hybridization. Since the thermodynamic properties of perfectly matched DNA duplexes are much better understood than those containing internal mismatches (24–28,41,42), we focused on the predicted stabilities of the longest continuous duplexes (LCDs) that could form between E.coli genomic sequences and the PM/MM probe pairs. By definition, these LCDs contain no internal mismatches. In order to maximize the stringency of our analysis, we used whole genome sequences to ensure that all possible sense and antisense transcripts (43) are represented. The sequence and lengths of LCDs between PM and MM probes with each of the three E.coli strains can be identified by BLAST analysis (29) using strict gap and mismatch penalties (Tables 1 and 2). This analysis also provides the sequence and lengths of second longest continuous duplex (SLCD) that forms between these probes and genomic sequences (Table 3). This can be used to predict the cross-hybridization potential of each probe.
Table 1. Length of LCD with PM probes in E.coli K-12 MG1655 microarray.
| Strain | Length in base pairs | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 25 | 24 | 23 | 22 | 21 | 20 | 19 | 18 | 17 | 16 | 15 | 14 | 13 | 12 | |
| K-12 MG1655 | 63 485 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| O157:H7 EDL933 | 40 668 | 1086 | 881 | 899 | 1123 | 1039 | 1037 | 1306 | 1405 | 1679 | 2831 | 4332 | 3968 | 1195 |
| CFT073 | 32 050 | 1312 | 1212 | 1183 | 1510 | 1398 | 1345 | 1751 | 1986 | 2201 | 4065 | 6134 | 5680 | 1592 |
Table 2. Length of LCD with MM probes in E.coli K-12 MG1655 microarray.
| Strain | Length in base pairs | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 25 | 24 | 23 | 22 | 21 | 20 | 19 | 18 | 17 | 16 | 15 | 14 | 13 | 12 | |
| K-12 MG1655 | 0 | 0 | 0 | 0 | 4 | 11 | 31 | 156 | 555 | 1918 | 6636 | 17 357 | 25 195 | 11 604 |
| O157:H7 EDL933 | 57 | 2 | 2 | 3 | 9 | 13 | 41 | 171 | 562 | 2143 | 7210 | 18 351 | 24 942 | 9883 |
| CFT073 | 55 | 7 | 1 | 1 | 15 | 20 | 32 | 173 | 584 | 2104 | 7158 | 18 378 | 25 007 | 9884 |
Table 3. Lengths of SLCDs formed by PM probes.
| Strain | First LCDa | Length of second LCD in base pairs | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 25 | 24 | 23 | 22 | 21 | 20 | 19 | 18 | 17 | 16 | 15 | 14 | 13 | 12 | ||
| K-12 | 25 | 977 | 4 | 11 | 4 | 5 | 20 | 93 | 248 | 908 | 2886 | 9029 | 19 982 | 22 210 | 6864 |
| O157:H7 EDL933 | 25 | 943 | 8 | 17 | 17 | 14 | 24 | 82 | 167 | 612 | 2087 | 6242 | 13 347 | 13 603 | 3420 |
| 24 | 4 | 4 | 0 | 0 | 0 | 0 | 4 | 28 | 59 | 182 | 375 | 340 | 89 | ||
| 23 | 8 | 0 | 0 | 0 | 1 | 4 | 18 | 45 | 144 | 320 | 281 | 58 | |||
| 22 | 9 | 1 | 1 | 1 | 8 | 11 | 50 | 144 | 304 | 277 | 91 | ||||
| 21 | 12 | 2 | 0 | 4 | 28 | 60 | 169 | 378 | 390 | 78 | |||||
| 20 | 12 | 0 | 4 | 19 | 61 | 173 | 343 | 342 | 82 | ||||||
| 19 | 20 | 9 | 14 | 57 | 158 | 342 | 321 | 113 | |||||||
| 18 | 27 | 25 | 75 | 229 | 442 | 402 | 105 | ||||||||
| CFT073 | 25 | 771 | 4 | 19 | 6 | 19 | 24 | 53 | 125 | 512 | 1620 | 4896 | 10 614 | 10 627 | 2685 |
| 24 | 6 | 3 | 2 | 0 | 3 | 2 | 4 | 26 | 82 | 204 | 434 | 443 | 97 | ||
| 23 | 8 | 1 | 0 | 1 | 3 | 3 | 21 | 65 | 229 | 394 | 399 | 85 | |||
| 22 | 6 | 0 | 2 | 1 | 6 | 20 | 59 | 193 | 407 | 419 | 69 | ||||
| 21 | 6 | 5 | 4 | 7 | 21 | 82 | 254 | 505 | 508 | 116 | |||||
| 20 | 3 | 1 | 8 | 17 | 96 | 220 | 464 | 478 | 108 | ||||||
| 19 | 12 | 13 | 34 | 78 | 176 | 445 | 474 | 109 | |||||||
| 18 | 15 | 35 | 102 | 284 | 600 | 571 | 138 | ||||||||
aLength of first LCD in base pairs formed with indicated strain.
We next sought to predict the thermodynamic stability of each LCD and SLCD formed by the arrayed probes and E.coli genomes. Predicted Gibbs free energy (ΔG) values are more informative than simple calculations of LCD length since the former takes both duplex length and probe sequence composition into consideration (25–28,41,42). First, we identified PM/MM probe pairs with the greatest potential to cross-hybridize with the K-12 MG1655 genome (Fig. 1). We define these as being PM/MM probe pairs whose PM probes can form very stable SLCDs with the K-12 MG1655 genome. The top 5% most stable SLCDs formed by PM probes and the K-12 MG1655 genome had a maximum Gibbs free energy of –19.84 kcal/mmol. To set stringent criteria for PM probe hybridization, we exclude 3172 PM/MM probe pairs that have a SLCD with ΔG < –19.84 kcal/mmol. Second, we identified PM/MM probe pairs whose MM probes have the greatest potential for significant hybridization to the K-12 MG1655 genome and thus are not optimal controls for cross-hybridization. The top 5% most stable LCDs formed between MM probes and the K-12 MG1655 genome had a maximum ΔG of –18.51 kcal/mmol. To set stringent criteria for MM probe hybridization, we exclude 2998 PM/MM probe pairs having MM LCDs with ΔG < –18.51 kcal/mmol. Collectively, these three criteria exclude 14 935 PM/MM probe pairs (10.6% of the initial design devoid of non-K12 MG1655 sequences) and comprise the basic E.coli K-12 MG1655 electronic mask.
Next, we focused on creating O157:H7 EDL933 and CFT073 strain-specific electronic masks. To simplify the analysis, the probe pairs masked for the K-12 MG1655 strain were also excluded in these strain-specific masks. In addition, we masked probe pairs whose PM probes were predicted to have the greatest potential to cross-hybridize to the genomic sequences of a given strain or whose MM probes were not optimal controls using the same ΔG criteria as stated above (Fig. 1).
Although only 64.1 and 50.5% of the PM probes form fully complementary duplexes with the O157:H7 EDL933 and CFT073 genomic sequences, respectively (Table 1), a significant subset of PM probes that are not fully complementary to these genomes can still provide high quality data (18). To begin to identify such probes, we first calculated the average ΔG of LCDs formed by PM probes within a given probe tiling path and the K-12 MG1655 genome. Next, we used these average ΔG values as baseline parameters upon which to decide whether a probe pair should be included in the analysis. To do this, we compared the ΔG of the LCD formed between each PM probe and the O157:H7 EDL933 or CFT073 genomes to the previously determined average ΔG of its probe tiling path. For ‘low stringency’ electronic masks, the ΔG for the LCD formed between the PM probe and genomic DNA from the appropriate strain must be at most two standard deviations higher (less stable) than the mean ΔG of its tiling path in order to be retained. For ‘high stringency’ electronic masks, the ΔG for the LCD of the PM probe with genomic DNA from the appropriate strain must be at most one standard deviation higher (less stable) than the mean ΔG of the tiling path in order to be retained. Overall, this strategy provides flexible criteria for masking probe pairs.
Evaluation of probe tiling paths
We sought to minimize ascertainment biases caused by significantly different numbers of PM/MM probe pairs interrogating a given transcript in each strain after applying electronic masks. Here, we define acceptable quality probe tiling paths as containing five or more probes (i.e. at least one-third of all possible PM/MM probe pairs for more than 99% of the tiling paths). Nevertheless, we realize that some of the unacceptable quality probe tiling paths could provide useful information, as discussed below. By definition, genes not represented in the K-12 microarray (i.e. genes found in pathogenicity islands in the E.coli O157:H7 EDL933 and CFT073 strains) cannot be interrogated by the microarrays (31–34,44). Table 4 lists the number of probe tiling paths in each of these categories for each E.coli strain after applying electronic masks. We will only consider genes interrogated by acceptable quality probe tiling paths when discussing the results from applying electronic masks.
Table 4. Effect of electronic masks on the number of PM/MM probe pairs in different probe tiling paths.
| Strain | Mask stringency | Number of probe tiling paths with the indicated percentage of possible PM/MM probe pairsa | |||
|---|---|---|---|---|---|
| >67%a | 33–67%a | 1–33%a | 0%a | ||
| K-12 MG1655 | Basic | 4187 | 65 | 11 | 3049 |
| O157:H7 EDL933 | High | 283 | 2308 | 1172 | 3549 |
| Low | 1327 | 2169 | 334 | 3482 | |
| CFT073 | High | 140 | 1804 | 1666 | 3802 |
| Low | 694 | 2367 | 663 | 3588 | |
aRelative to number of PM/MM probe pairs in the original probe tiling paths in the K-12 MG1655 microarray design.
Comparison of gene expression profiles using electronic masks
We compared gene expression profiles between K-12 MG1655 and O157:H7 EDL933 strains grown to mid-logarithmic phase before masking and after applying the O157:H7 EDL933 electronic masks (Methods). In order to ensure the cross-strain comparisons were as consistent as possible, we applied the O157:H7 EDL933 electronic masks to both the K-12 MG1655 and O157:H7 EDL933 data sets when conducting masking analysis. Therefore, the number and identity of the probes evaluating both strains are identical. A total of 494 genes (disregarding those encoded by episomes, plasmids, phage genomes and intergenic regions) were predicted to be a least 2-fold differentially expressed (P < 0.05) between these strains prior to electronic masking (table 3 in the Supplementary Material). Only 157 (31.8%) and 264 (53.4%) of these 494 genes were still predicted to be least 1.8-fold differentially expressed (P < 0.05) in the same direction after applying the high and low stringency O157:H7 EDL933 masks, respectively (table 3 in the Supplementary Material). In part, this is due to the fact that 205 (41.5%) and 90 (18.2%) of the original 494 genes had unacceptable tiling paths after applying the high and low stringency O157:H7 EDL933 masks, respectively.
Likewise, we compared gene expression profiles between K-12 MG1655 and CFT073 strains before masking and after applying the CFT073 electronic masks. Prior to masking, 551 genes were predicted to be a least 2-fold differentially expressed (P < 0.05) among these strains (table 4 in the Supplementary Material). Of these, a total of 161 (29.2%) and 245 (44.5%) genes were still predicted to be at least 1.8-fold differentially expressed in the same direction (P < 0.05) using the high and low stringency CFT073 masks, respectively. Again, this is mainly due to the fact that 278 (50.5%) and 136 (24.7%) of the original 551 genes had unacceptable tiling paths after applying these high and low stringency masks, respectively.
We conducted quantitative RT–PCR (qRT–PCR) analyses to test the effectiveness of applying the high stringency electronic masks to improve the accuracy of data analysis. First, we selected 12 genes (dnaJ, lgt, lldP, oppB, pssR, slpA, cutF, eco, lspA, sodA, pspE and yaeC) predicted to be at least 2-fold up-regulated in K-12 MG1655 relative to the O157:H7 EDL933 or CFT073 strains prior to masking and not differentially expressed (at most 1.8-fold change) after applying the high stringency mask (table 5 in the Supplementary Material). We confirmed by qRT–PCR that 12/12 of these genes were not up-regulated in K-12 MG1655 relative to either the O157:H7 EDL933 or CFT073 strains. One of these genes (sodA) was only analyzed by four probe pairs after high stringency masking yet was still called correctly. Surprisingly, two (pspE and yaeC) of the 12 genes were actually significantly down-regulated in K-12 MG1655 relative to O157:H7 EDL933. In these cases, both the original and masking analyses failed to detect this change. Next, we examined seven genes (accC, hisB, sucD, mopA, rplW, thiL and yfiD) that were predicted to be over 2-fold differentially expressed in K-12 MG1655 relative to the O157:H7 EDL933 or CFT073 strains only after using the high stringency electronic masks. qRT–PCR confirmed 5/7 of these cases (all except sucD and yfiD).
Next, we compared the accuracy of both the high and low stringency electronic masks. There was a conflict in predicting significant (at least 1.8-fold) differential gene expression for only two (lldP and rplW) genes listed in Table 5. For lldP, only the high stringency mask correctly predicted no significant differential gene expression between K-12 MG1655 and CFT073. For rplW, only the high stringency mask correctly predicted differential gene expression between K-12 MG1655 and O157:H7 EDL933. Although the high stringency mask may be of preferential use, the low stringency mask analysis will allow more genes to be interrogated by acceptable quality probe tiling path (Table 4).
Table 5. Evaluation of gene expression data by qRT–PCR.
| Gene | o maska | HS maskb | LS maskc | qRT–PCRd | ||||
|---|---|---|---|---|---|---|---|---|
| Ratio | Probes | Ratio | Probes | Ratio | Probes | |||
| CFT073 | accC | –1.3 | 15 | –2.0 | 5 | –1.8 | 8 | –2.1 |
| dnaJ | 2.0 | 15 | 1.4 | 6 | 1.4 | 9 | 1.0 | |
| hisB | –1.4 | 15 | –2.2 | 6 | –2.2 | 6 | –2.4 | |
| lgt | 2.7 | 14 | 1.3 | 6 | 1.6 | 8 | 1.3 | |
| lldP | 5.3 | 15 | 1.3 | 5 | 2.0 | 7 | –1.1 | |
| oppB | 3.2 | 15 | 1.0 | 5 | 1.2 | 6 | –1.2 | |
| pssR | 2.5 | 11 | 1.0 | 5 | 1.3 | 6 | 1.1 | |
| slpA | 2.6 | 15 | 1.5 | 6 | 1.6 | 7 | 1.3 | |
| sucD | –1.4 | 15 | –2.1 | 8 | –1.9 | 11 | –1.5 | |
| O157:H7 EDL933 | cutF | 2.6 | 15 | 1.1 | 5 | 1.2 | 6 | 1.3 |
| eco | 2.1 | 15 | 1.2 | 8 | 1.2 | 9 | –1.2 | |
| lspA | 3.2 | 15 | 1.5 | 8 | 1.6 | 10 | 1.8 | |
| mopA | –1.5 | 15 | –3.0 | 8 | –3.1 | 11 | –3.6 | |
| rplW | –1.3 | 14 | –2.0 | 5 | –1.3 | 13 | –1.8 | |
| sodA | 2.6 | 15 | 1.3 | 4 | 1.3 | 7 | 1.2 | |
| thiL | –1.5 | 14 | –2.2 | 7 | –1.9 | 8 | –5.0 | |
| pspE | 2.0 | 15 | –1.1 | 8 | 1.4 | 11 | –2.1 | |
| yaeC | 2.6 | 15 | 1.4 | 7 | 1.1 | 8 | –3.1 | |
| yfiD | 1.1 | 14 | –3.9 | 7 | –4.0 | 7 | –1.3 | |
aRatio of gene expression level of K12 MG1655 and the indicated strain before masking and number of probe pairs used for this calculation.
bRatio of gene expression level of K12 MG1655 and the indicated strain after application of the high stringency electronic mask and the number of probe pairs used for this calculation.
cRatio of gene expression level of K12 MG1655 and the indicated strain after application of the low stringency electronic mask and the number of probe pairs used for this calculation.
dRatio of gene expression level of K12 MG1655 and the indicated strain as determined by quantitative RT–PCR.
DISCUSSION
Using E.coli as a model system, we use whole genome sequence information to create electronic masks aimed at increasing the sensitivity and specificity of oligonucleotide microarray-based gene expression analysis of closely related organisms. The electronic masks showed a high efficiency (12/12 cases) in removing genes that were falsely called to be differentially expressed in a given direction using hybridization data from all probe sets. Nevertheless, the electronic masks do not completely compensate for all errors in the initial analysis. The discordances between the electronic mask-based microarray data and qRT–PCR analysis could be due to differences in the inter- and intra-molecular structures of cDNA targets from each strain that affect their hybridization properties. For example, fixed sequence differences among the strains will affect the folding of the cDNA targets. Although chemical and enzymatic fragmentation minimizes these structures, it does not completely eliminate them (40). Thus, binding sites might be differentially exposed for hybridization.
This limited comparison indicates that the high stringency mask may be of preferential use for acceptable quality tiling paths than the low stringency mask. Nevertheless, the low stringency mask analysis will allow more genes to be interrogated by acceptable quality probe tiling paths (Table 4). This is especially important in cases where one is interested in the expression levels of specific genes. Therefore, each electronic mask has its own specific advantages depending on the nature of the data set and genes of interest.
The number of electronic masking applications will continue to increase due to the rapid expansion of publicly available genomic sequence information from highly related organisms (45–47). These include microbial strains that may differ in pathogenicity such as the ones in this study as well as other prokaryotic and eukaryotic organisms. Of special interest are comparisons between the human and African great ape (chimpanzee, bonobo and gorilla) gene expression patterns (8,11,12,48). Their genomes are over 98.5% identical at the nucleotide level (49,50) and thus are similar to the percent nucleotide identities of the strains analyzed in this study. It has been shown that mismatches between bonobo transcripts and human probes were responsible for approximately half of the genes predicted to be up-regulated in human relative to bonobo cultured fibroblasts (11). Electronic masks will increase the value of these data sets by minimizing aberrant readings causing these mismatches. Nevertheless, it should be noted that the value of these electronic masks is dependent upon the quantity and quality of genomic sequence information for the organism of interest. In the absence of such sequence information, our strategy cannot be employed. In such cases, one may consider using microarrays consisting of longer DNA fragments that more effectively cross-hybridize to orthologous transcripts and also confirm the expression levels of genes of interest by northern blot or qRT–PCR analysis.
In theory, it is possible to use whole genome sequence information to design oligonucleotide microarrays that can analyze gene expression patterns in multiple organisms with comparable specificity and sensitivity (51). One strategy involves designing probes that share complete sequence identity to RNAs from both organisms and have equivalent specific as well as non-specific hybridization potential. Another strategy involves designing sets of probes to specifically bind to RNAs from a given species (species-specific probes) and using appropriate electronic masks to analyze only those probes for a given species. In the latter case, electronic masks provide a computational means to tailor the microarray to the organism being examined. A combination of both strategies is likely to provide the most economical use of the microarray surface area. One would first design probes with equivalent hybridization properties among the organisms of interest. For some genes, this may be difficult due to sequence differences among the organisms. Therefore, additional species-specific probes would be included in the microarray to ensure that an adequate number of probes are present in each tiling path.
Electronic masks can be used in several other contexts to increase the accuracy of gene expression analysis. For example, it is desirable to selectively retain or eliminate probes containing common polymorphisms within a species that are sensitive to the genotype of the individual. Likewise, it would be useful to retain or eliminate probes that overlap splice junctions and thus selectively evaluate the abundance of differentially spliced transcripts.
Electronic masks could also increase the accuracy of microarray-based resequencing analysis. Oligonucleotide microarrays designed to resequence human DNA have been used to identify fixed sequence differences and polymorphisms in non-human primates and other mammals (17–19,52). However, mismatches between the orthologous targets and oligonucleotide probes cause loss of hybridization signal in certain sequence tracts that could falsely indicate or obscure the presence of polymorphism (18). By eliminating sequence tracts with poor hybridization specificity and sensitivity from analysis, more stringent hybridization conditions can be applied in order to increase the sensitivity and specificity of sequence variation.
Our studies are meant to provide guidelines for designing electronic masks that can be used for organisms for which genomic sequence information is available. Nevertheless, the first generation electronic masks described here are based upon predictions of hybridization sensitivity and specificity using several simplifying assumptions. For example, the secondary structure and the heterogeneity of the complex library of the randomly fragmented nucleic acid targets containing repetitive sequences cannot be readily modeled (53). In principle, one can begin to address this challenging problem using nucleic acid structure predictions and other algorithms for predicting microarray hybridization (54–60). All these considerations can be implemented in future versions of electronic masks in order to further improve the analysis of microarray data.
SUPPLEMENTARY MATERIAL
Additional information about the data reported in this manuscript, including raw and formatted gene expression data and the code for the custom software developed in these studies, is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
This manuscript is dedicated to Dr Masayori Inouye at the University of Medicine and Dentistry of New Jersey. We thank Juergen Reichardt and Susan Groshen at USC for thoughtful discussion and Michael Apicella at the University of Iowa and Garry Miyada at Affymetrix for their support of this project. This work was partially funded by National Institutes of Health Grant P50-HG002790.
REFERENCES
- 1.Brunetti C.R., Selegue,J.E., Monteiro,A., French,V., Brakefield,P.M. and Carroll,S.B. (2001) The generation and diversification of butterfly eyespot color patterns. Curr. Biol., 11, 1578–1585. [DOI] [PubMed] [Google Scholar]
- 2.Tautz D. (2000) Evolution of transcriptional regulation. Curr. Opin. Genet. Dev., 10, 575–579. [DOI] [PubMed] [Google Scholar]
- 3.Cavalieri D., Townsend,J.P. and Hartl,D.L. (2000) Manifold anomalies in gene expression in a vineyard isolate of Saccharomyces cerevisiae revealed by DNA microarray analysis. Proc. Natl Acad. Sci. USA, 97, 12369–12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rifkin S.A., Kim,J. and White,K.P. (2003) Evolution of gene expression in the Drosophila melanogaster subgroup. Nature Genet., 33, 138–144. [DOI] [PubMed] [Google Scholar]
- 5.Wang R.L., Stec,A., Hey,J., Lukens,L. and Doebley,J. (1999) The limits of selection during maize domestication. Nature, 398, 236–239. [DOI] [PubMed] [Google Scholar]
- 6.King M.C. and Wilson,A.C. (1975) Evolution at two levels in humans and chimpanzees. Science, 188, 107–116. [DOI] [PubMed] [Google Scholar]
- 7.Hacia J.G. (2001) Genome of the apes. Trends Genet., 17, 637–645. [DOI] [PubMed] [Google Scholar]
- 8.Enard W., Khaitovich,P., Klose,J., Zollner,S., Heissig,F., Giavalisco,P., Nieselt-Struwe,K., Muchmore,E., Varki,A., Ravid,R. et al. (2002) Intra- and interspecific variation in primate gene expression patterns. Science, 296, 340–343. [DOI] [PubMed] [Google Scholar]
- 9.Olson M.V. and Varki,A. (2003) Sequencing the chimpanzee genome: insights into human evolution and disease. Nat. Rev. Genet., 4, 20–28. [DOI] [PubMed] [Google Scholar]
- 10.Carroll S.B. (2003) Genetics and the making of Homo sapiens. Nature, 422, 849–857. [DOI] [PubMed] [Google Scholar]
- 11.Karaman M.W., Houck,M.L., Chemnick,L.G., Nagpal,S., Chawannakul,D., Sudano,D., Pike,B.L., Ho,V.V., Ryder,O.A. and Hacia,J.G. (2003) Comparative analysis of gene-expression patterns in human and African great ape cultured fibroblasts. Genome Res., 13, 1619–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caceres M., Lachuer,J., Zapala,M.A., Redmond,J.C., Kudo,L., Geschwind,D.H., Lockhart,D.J., Preuss,T.M. and Barlow,C. (2003) Elevated gene expression levels distinguish human from non-human primate brains. Proc. Natl Acad. Sci. USA, 100, 13030–13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su A.I., Cooke,M.P., Ching,K.A., Hakak,Y., Walker,J.R., Wiltshire,T., Orth,A.P., Vega,R.G., Sapinoso,L.M., Moqrich,A. et al. (2002) Large-scale analysis of the human and mouse transcriptomes. Proc. Natl Acad. Sci. USA, 99, 4465–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandberg R., Yasuda,R., Pankratz,D.G., Carter,T.A., Del Rio,J.A., Wodicka,L., Mayford,M., Lockhart,D.J. and Barlow,C. (2000) Regional and strain-specific gene expression mapping in the adult mouse brain. Proc. Natl Acad. Sci. USA, 97, 11038–11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marvanova M., Menager,J., Bezard,E., Bontrop,R.E., Pradier,L. and Wong,G. (2003) Microarray analysis of nonhuman primates: validation of experimental models in neurological disorders. FASEB J., 17, 929–931. [DOI] [PubMed] [Google Scholar]
- 16.Kayo T., Allison,D.B., Weindruch,R. and Prolla,T.A. (2001) Influences of aging and caloric restriction on the transcriptional profile of skeletal muscle from rhesus monkeys. Proc. Natl Acad. Sci. USA, 98, 5093–5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frazer K.A., Sheehan,J.B., Stokowski,R.P., Chen,X., Hosseini,R., Cheng,J.F., Fodor,S.P., Cox,D.R. and Patil,N. (2001) Evolutionarily conserved sequences on human chromosome 21. Genome Res., 11, 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hacia J.G., Makalowski,W., Edgemon,K., Erdos,M.R., Robbins,C.M., Fodor,S.P., Brody,L.C. and Collins,F.S. (1998) Evolutionary sequence comparisons using high-density oligonucleotide arrays. Nature Genet., 18, 155–158. [DOI] [PubMed] [Google Scholar]
- 19.Hacia J.G., Fan,J.B., Ryder,O., Jin,L., Edgemon,K., Ghandour,G., Mayer,R.A., Sun,B., Hsie,L., Robbins,C.M. et al. (1999) Determination of ancestral alleles for human single-nucleotide polymorphisms using high-density oligonucleotide arrays. Nature Genet., 22, 164–167. [DOI] [PubMed] [Google Scholar]
- 20.Chismar J.D., Mondala,T., Fox,H.S., Roberts,E., Langford,D., Masliah,E., Salomon,D.R. and Head,S.R. (2002) Analysis of result variability from high-density oligonucleotide arrays comparing same-species and cross-species hybridizations. Biotechniques, 33, 516–518, 520,, 522 passim. [DOI] [PubMed] [Google Scholar]
- 21.Bigger C.B., Brasky,K.M. and Lanford,R.E. (2001) DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J. Virol., 75, 7059–7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGall G.H. and Christians,F.C. (2002) High-density genechip oligonucleotide probe arrays. Adv. Biochem. Eng. Biotechnol., 77, 21–42. [DOI] [PubMed] [Google Scholar]
- 23.Samonte R.V. and Eichler,E.E. (2002) Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet., 3, 65–72. [DOI] [PubMed] [Google Scholar]
- 24.Breslauer K.J., Frank,R., Blocker,H. and Marky,L.A. (1986) Predicting DNA duplex stability from the base sequence. Proc. Natl Acad. Sci. USA, 83, 3746–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.SantaLucia J. Jr, Allawi,H.T. and Seneviratne,P.A. (1996) Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry, 35, 3555–3562. [DOI] [PubMed] [Google Scholar]
- 26.Allawi H.T. and SantaLucia,J.,Jr (1997) Thermodynamics and NMR of internal G.T mismatches in DNA. Biochemistry, 36, 10581–10594. [DOI] [PubMed] [Google Scholar]
- 27.Allawi H.T. and SantaLucia,J.,Jr (1998) Nearest-neighbor thermodynamics of internal A.C mismatches in DNA: sequence dependence and pH effects. Biochemistry, 37, 9435–9444. [DOI] [PubMed] [Google Scholar]
- 28.Peyret N., Seneviratne,P.A., Allawi,H.T. and SantaLucia,J.,Jr (1999) Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A,C.C,G.G and T.T mismatches. Biochemistry, 38, 3468–3477. [DOI] [PubMed] [Google Scholar]
- 29.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 30.Dudoit S., Gentleman,R.C. and Quackenbush,J. (2003) Open source software for the analysis of microarray data. Biotechniques (Suppl.), 45–51. [PubMed] [Google Scholar]
- 31.Blattner F.R., Plunkett,G.,3rd, Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 32.Perna N.T., Plunkett,G.,3rd, Burland,V., Mau,B., Glasner,J.D., Rose,D.J., Mayhew,G.F., Evans,P.S., Gregor,J., Kirkpatrick,H.A. et al. (2001) Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature, 409, 529–533. [DOI] [PubMed] [Google Scholar]
- 33.Kudva I.T., Evans,P.S., Perna,N.T., Barrett,T.J., Ausubel,F.M., Blattner,F.R. and Calderwood,S.B. (2002) Strains of Escherichia coli O157:H7 differ primarily by insertions or deletions, not single-nucleotide polymorphisms. J. Bacteriol., 184, 1873–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Welch R.A., Burland,V., Plunkett,G.,3rd, Redford,P., Roesch,P., Rasko,D., Buckles,E.L., Liou,S.R., Boutin,A., Hackett,J. et al. (2002) Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl Acad. Sci. USA, 99, 17020–17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenow C., Saxena,R.M., Durst,M. and Gingeras,T.R. (2001) Prokaryotic RNA preparation methods useful for high density array analysis: comparison of two approaches. Nucleic Acids Res., 29, e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tjaden B., Saxena,R.M., Stolyar,S., Haynor,D.R., Kolker,E. and Rosenow,C. (2002) Transcriptome analysis of Escherichia coli using high-density oligonucleotide probe arrays. Nucleic Acids Res., 30, 3732–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tjaden B., Haynor,D.R., Stolyar,S., Rosenow,C. and Kolker,E. (2002) Identifying operons and untranslated regions of transcripts using Escherichia coli RNA expression analysis. Bioinformatics, 18 (Suppl. 1), S337–S344. [DOI] [PubMed] [Google Scholar]
- 38.Selinger D.W., Saxena,R.M., Cheung,K.J., Church,G.M. and Rosenow,C. (2003) Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res., 13, 216–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irizarry R.A., Bolstad,B.M., Collin,F., Cope,L.M., Hobbs,B. and Speed,T.P. (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res., 31, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hacia J.G., Edgemon,K., Fang,N., Mayer,R.A., Sudano,D., Hunt,N. and Collins,F.S. (2000) Oligonucleotide microarray based detection of repetitive sequence changes. Hum. Mutat., 16, 354–363. [DOI] [PubMed] [Google Scholar]
- 41.Allawi H.T. and SantaLucia,J.,Jr (1998) Thermodynamics of internal C.T mismatches in DNA. Nucleic Acids Res., 26, 2694–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allawi H.T. and SantaLucia,J.,Jr (1998) Nearest neighbor thermodynamic parameters for internal G.A mismatches in DNA. Biochemistry, 37, 2170–2179. [DOI] [PubMed] [Google Scholar]
- 43.Delihas N. and Forst,S. (2001) MicF: an antisense RNA gene involved in response of Escherichia coli to global stress factors. J. Mol. Biol., 313, 1–12. [DOI] [PubMed] [Google Scholar]
- 44.Kolisnychenko V., Plunkett,G.,3rd, Herring,C.D., Feher,T., Posfai,J., Blattner,F.R. and Posfai,G. (2002) Engineering a reduced Escherichia coli genome. Genome Res., 12, 640–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kellis M., Patterson,N., Endrizzi,M., Birren,B. and Lander,E.S. (2003) Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature, 423, 241–254. [DOI] [PubMed] [Google Scholar]
- 46.Frazer K.A., Elnitski,L., Church,D.M., Dubchak,I. and Hardison,R.C. (2003) Cross-species sequence comparisons: a review of methods and available resources. Genome Res., 13, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cooper G.M., Brudno,M., Green,E.D., Batzoglou,S. and Sidow,A. (2003) Quantitative estimates of sequence divergence for comparative analyses of mammalian genomes. Genome Res., 13, 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gu J. and Gu,X. (2003) Induced gene expression in human brain after the split from chimpanzee. Trends Genet., 19, 63–65. [DOI] [PubMed] [Google Scholar]
- 49.Chen F.C. and Li,W.H. (2001) Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet., 68, 444–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Britten R.J. (2002) Divergence between samples of chimpanzee and human DNA sequences is 5%, counting indels. Proc. Natl Acad. Sci. USA, 99, 13633–13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.vanDam R.M. and Quake,S.R. (2002) Gene expression analysis with universal n-mer arrays. Genome Res., 12, 145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frazer K.A., Chen,X., Hinds,D.A., Pant,P.V., Patil,N. and Cox,D.R. (2003) Genomic DNA insertions and deletions occur frequently between humans and nonhuman primates. Genome Res., 13, 341–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hacia J.G., Woski,S.A., Fidanza,J., Edgemon,K., Hunt,N., McGall,G., Fodor,S.P. and Collins,F.S. (1998) Enhanced high density oligonucleotide array-based sequence analysis using modified nucleoside triphosphates. Nucleic Acids Res., 26, 4975–4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res., 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luebke K.J., Balog,R.P. and Garner,H.R. (2003) Prioritized selection of oligodeoxyribonucleotide probes for efficient hybridization to RNA transcripts. Nucleic Acids Res., 31, 750–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matveeva O.V., Shabalina,S.A., Nemtsov,V.A., Tsodikov,A.D., Gesteland,R.F. and Atkins,J.F. (2003) Thermodynamic calculations and statistical correlations for oligo-probes design. Nucleic Acids Res., 31, 4211–4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rouillard J.M., Zuker,M. and Gulari,E. (2003) OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res., 31, 3057–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang L., Miles,M.F. and Aldape,K.D. (2003) A model of molecular interactions on short oligonucleotide microarrays. Nat. Biotechnol., 21, 818–821. [DOI] [PubMed] [Google Scholar]
- 59.Mei R., Hubbell,E., Bekiranov,S., Mittmann,M., Christians,F.C., Shen,M.M., Lu,G., Fang,J., Liu,W.M., Ryder,T. et al. (2003) Probe selection for high-density oligonucleotide arrays. Proc. Natl Acad. Sci. USA, 100, 11237–11242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Held G.A., Grinstein,G. and Tu,Y. (2003) Modeling of DNA microarray data by using physical properties of hybridization. Proc. Natl Acad. Sci. USA, 100, 7575–7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.