Abstract
Background
The controversy of CpG island methylator phenotype (CIMP) in gastric cancer persists, despite the fact that many studies have been conducted on its relation with helicobacter pylori (H. pylori), Epstein-Barr virus (EBV), and microsatellite instability (MSI) and prognosis. To drive a more precise estimate of this postulated relationship, a meta-analysis was performed based on existing relevant studies.
Methods
We combined individual patient data from 12 studies which involved 1000 patients with gastric cancer, which met the criteria. We tabulated and analyzed parameters from each study, including H. pylori, EBV, MSI, and clinical information of patients.
Results
The overall OR for H. pylori infection in CIMP positive group vs. negative group revealed that significantly elevated risks of positive H. pylori infection in the former were achieved (OR 2.23 95% CI, 1.25–4.00; P = 0.007, Pheterogeneity = 0.05). Similarly, strong relation between EBV infection and CIMP was achieved by OR 51.27 (95% CI, 9.39–279.86; P<0.00001, Pheterogeneity = 0.39). The overall OR for MSI in CIMP positive group vs. negative group was 4.44 (95% CI, 1.17–16.88; P = 0.03, Pheterogeneity = 0.01). However, there did not appear to be any correlations with clinical parameters such as tumor site, pathological type, cell differentiation, TNM stage, distant metastasis, lymph node metastasis, and 5-year survival.
Conclusions
The meta-analysis highlights the strong relation of CIMP with H. pylori, EBV, and MSI, but CIMP can not be used as a prognostic marker for gastric cancer.
Introduction
Gastric cancer is the fourth most common malignancy and second leading cause of cancer death in the world [1]. Even screening programs with barium photofluorography or endoscopy allow earlier detection in Japan and Korea, the patients with advanced gastric cancer are still worse in 5-year overall survival. Identifying molecular aberrations in gastric cancer may improve our understanding of gastric carcinogenesis, identify strategies for subdividing patients into relevant subgroups, and highlight novel molecular target agents. Although the molecular mechanisms of gastric cancer carcinogenesis remain unclear, both genetic and epigenetic alterations are important. Genetic alterations are responsible for activation of onceogenes and inactivation of tumor-suppressor gene. Epigenetic alteration through DNA methylation is known to play an important role in inhibiting the expression of tumor-related genes.
Aberrant DNA methylation in cancer was summarized as global hypomethylation and regional hypermethylation, which are associated with genomic instability and inactivation of tumor-suppressor genes, respectively [2]. However, regional hypermethylation refers to the aberrant methylation of normally unmethylated sequences, most of which are clusters of CpG sites, denoted CpG island. Because multiple genes are concurrently methylated in the hypermethylated subtype, CIMP concept was first introduced to the molecular pathways of colorectal cancers by Dr. Issa group [3]. After that, Scientists have found that CIMP-positive colorectal cancers have a close association with the molecular and clinicopathological features, and poor prognosis [4]–[6].
Similarly, the presence of CIMP-positive gastric cancer has been reported by many scientists [7]–[24], but controversial data did not confirm the prognostic value of CIMP for gastric cancer. This was possibly due to small sample size or confounding variables. Therefore, we initiated an international collaborative effort which resulted in a meta-analysis of data on individual patient in prospective cohort studies to evaluate the association between CIMP and malignant behavior in gastric cancer.
Methods
Publication Search
Two electronic databases (PubMed and Embase) were searched (last search was updated on 21 November 2012, using the search terms: ‘Gastric cancer’ and ‘CIMP’. All eligible studies were retrieved, and their bibliographies were checked for other relevant publications. Review articles and bibliographies of other relevant studies identified were hand-searched to identify additional eligible studies. Only published studies with full-text articles were included. When the same patient population was included in several publications, only the most recent or complete study was used in this meta-analysis.
Inclusion Criteria
The inclusion criteria were as follows: (a) evaluating the relation between CIMP and H. pylori, EBV, MSI or clinical prognostic parameters; (b) promoter methylation; and (c) sufficient published data to estimate an odds ratio (OR) with 95% confidence interval (CI).
Data Extraction
Information was carefully extracted from all eligible studies by two of the authors (Zong L and Seto Y), according to the inclusion criteria listed above. The following data were collected from each study: first author's surname, publication date, study method, sample size, total number of patients with positive CIMP and negative CIMP, and number of patients divided by age, gender, tumor site, pathological type, cell differentiation, TNM stage, lymph node metastasis, distant metastasis and 5-year overall survival in those with and without CIMP, respectively. We did not define a minimum number of patients for inclusion in our meta-analysis.
Statistical Analysis
Odd ratios with 95% CI were used to assess the correlation of CIMP with H. pylori, EBV, and MSI and prognosis, according to the method of Woolf. Heterogeneity assumption was confirmed by the X2-based Q-test. A P-value greater than 0.10 for the Q-test indicated a lack of heterogeneity among the studies, therefore, the OR estimate for each study was calculated by the fixed-effects model. Otherwise, the random-effects model was used. The significance of the pooled OR was determined by the Z-test and P>0.05 was considered statistically significant. Sensitivity analyses were carried out to determine if modification of the inclusion criteria for this meta-analysis affected the final results. An estimate of potential publication bias was carried out using the funnel plot, in which the OR for each study was plotted against its log (OR). An asymmetric plot suggested possible publication bias. Funnel plot asymmetry was assessed using Egger's linear regression test, a linear regression approach to measure funnel plot asymmetry on the natural logarithm scale of the OR. The significance of the intercept was determined by the t-test, as suggested by Egger (P<0.05 was considered representative of statistically significant publication bias). All statistical tests were performed with Review Manager Version 5.0 (The Cochrane Collaboration, Oxford, England).
Results
Study Characteristics
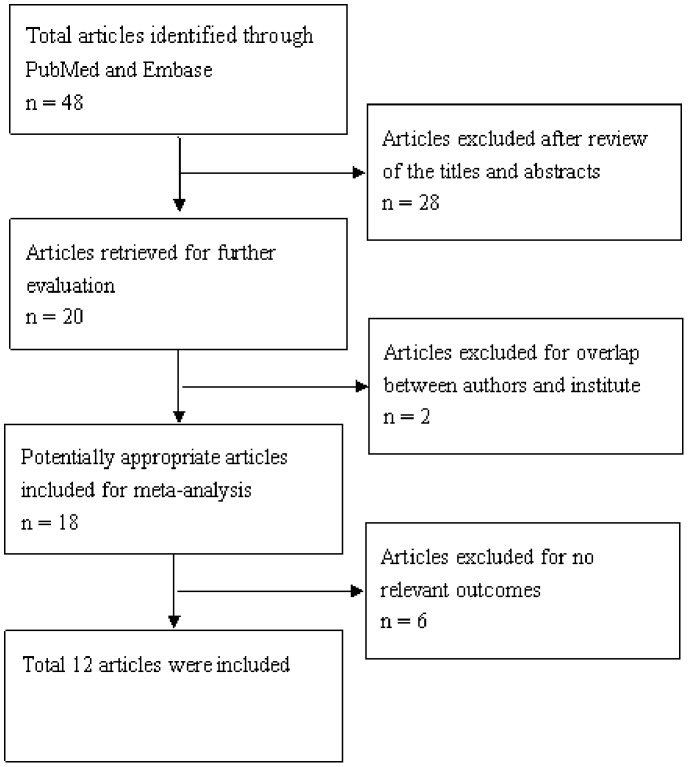
A total of 18 publications met the basic inclusion criteria [7]–[24]. The study by Oue et al was excluded because they did not categorized into CIMP subgroups with methylated gene panel [7]. In addition, the study by Kanai et al was excluded because they focused on DNA methylation of CPG islands and pericentromeric satellite regions in colorectal and stomach cancer [8]. Similarly the study by Oshimo et al was excluded because they mainly analyzed the relation between epigenetic inactivation of RIZ1 and CIMP [9]. The study by Watanabe et al was not included because they tried to prove fidelity in replicating DNA methylation patterns in cancer cells lead to dense methylation of a CpG island [10]. Other studies were excluded due to insufficient information to calculate OR [11], [12]. Hence, a total of 12 studies including 1000 patients were used in the pooled analyses. Table 1 lists the studies identified and their main characteristics. Of the 12 groups, sample size ranged from 40 to 200 (Figure 1).
Table 1. Main characteristics of all studies included in the meta-analysis.
| Study | Study Period | N | Study Method | CpG island site | Gene type | Cut off value | Features |
| Kim et al | 2003 | 200 | MSP | Promoter | MINT1, MINT2, MINT12, MINT25, MINT31 | CIMP-high (3–5), CIMP-intermediate (2), CIMP-low (1 or less) | MSI-H GC was related to the high CIMP. |
| An et al | 2005 | 82 | MSP microdissection | Promoter | MINT1, MINT2, MINT25, MINT31, MLH1 | CIMP-high (3 or more), CIMP-low (2 or less), CIMP-negative (0) | CIMP-high GC was associated with MSI and better prognosis. |
| Kim et al | 2005 | 40 | MSP | Promoter | hMLH1, MINT2, TIMP3, THBS1, DAP-K, GST P1, APC | CIMP-high (3–7), CIMP-low(1–2), CIMP-negative (0) | There were no specific findings for CIMP-high GC. |
| Oue et al | 2006 | 75 | COBRA | Promoter | MINT1, MINT2, MINT12, MINT25, MINT31 | CIMP+ (3 or more), CIMP− (2 or less) | There were no specific findings for CIMP+ GC. |
| Chang et al | 2006 | 91 | MSP | Promoter | LOX, HRASLS, FLNc, HAND1, TM | CIMP-high (4–5), CIMP-intermediate (1–3), CIMP-negative (0) | CIMP-high GC was associated with better prognosis. |
| Kusano et al | 2006 | 78 | COBRA | Promoter | MINT1, MINT2, MINT12, MINT25, MINT31 | CIMP-high (4–5), CIMP-intermediate (1–3), CIMP-negative (0) | CIMP-high GC was closely associated with proximal location, diffuse type, less-advanced tumor stage, and better prognosis. |
| Enomoto et al | 2007 | 66 | MSP | Promoter | LOX, HRASLS, FLNc, HAND1,THBD, F2R, NT5E, GREM1, ZNF177, CLDN3, PAX6, CTSL | CIMP-high (5 or more), CIMP-low (1–4), CIMP-negative (0) | CIMP-high GC was closely associated with diffuse type and better prognosis. |
| Zhang et al | 2008 | 47 | MSP | Promoter | hMLH1, MINT1, MINT2, MINT31, p16 | CIMP-high (3–5), CIMP-intermediate (1–2), CIMP-negative (0) | There were no specific findings for CIMP-high GC. |
| Kondo et al | 2009 | 41 | MSP | Promoter | hMLH-1, MINT1, MINT2, MINT31, Kip2, p16, p15, p73, MGMT, DAPK, HCAD | CIMP+ (4 or more), CIMP− (3 or less) | H. pylori infection caused the aberrant DNA hypermethylation of specific genes and induced CIMP. |
| Park et al | 2010 | 150 | MethyLight PCR | Promoter | BCL2, BDNF, CACNA1G, CALCA, CHFR, CYP1B1, DLEC1, GRIN2B, RUNX3, SEZ6L, SFRP4, TERT, THBS1, TIMP3, TP73, TWIST1. | CIMP-high (14–16), CIMP-low (1–3), CIMP-negative (0) | CIMP-high GC was featured with poor prognosis. |
| Chen et al | 2012 | 120 | MethyLight PCR | Promoter | ALX4, TMEFF2, CHCHD10, IGFBP3, NPR1 | CIMP-high (4–5), CIMP-low (1–3), CIMP-negative (0) | CIMP-high GC was associated with more distant lymph node metastasis. |
| Liu et al | 2012 | 72 | MSP | Promoter | APC, WIF1, RUNX3, DLC1, SFRP1, DKK, E-cad | CIMP+ (3 or more), CIMP− (2 or less) | H. pylori+/CIMP+ cases were associated with higher rates of metastasis and recurrence than H. pylori+/CIMP− cases. |
COBRA, combined bisulfite restriction analysis; CIMP, CpG island methylator phenotype; GC, gastric cancer; MSP, methylation-specific PCR; Q-MSP, quantitative MSP.
Figure 1. Flow chart of literature selection.
Correlation with Virus Infection and Molecular Stability
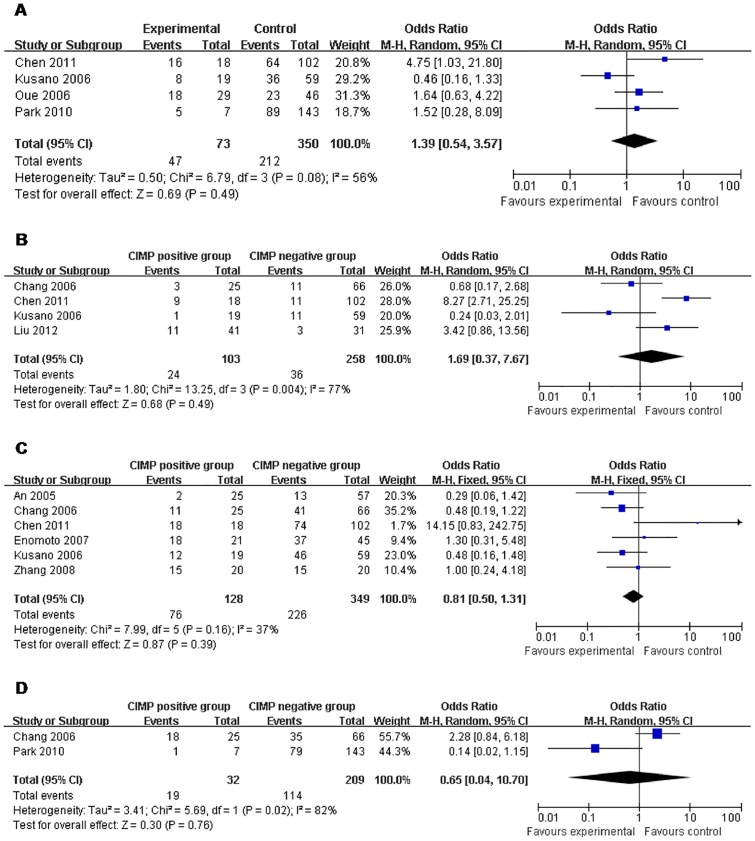
The overall OR for H. pylori infection in CIMP positive group vs. negative group revealed that significantly elevated risks of positive H. pylori infection in the former were achieved (OR 2.23 95% CI, 1.25–4.00;P = 0.007, Pheterogeneity = 0.05). Similarly, strong relation between EBV infection and CIMP was achieved by OR 51.27 (95% CI, 9.39–279.86; P<0.00001, Pheterogeneity = 0.39). The overall OR for MSI in CIMP positive group vs. negative group was 4.44 (95% CI, 1.17–16.88; P = 0.03, Pheterogeneity = 0.01) (Table 2 and Figure 2).
Table 2. Outcomes of the meta-analysis.
| Parameters | No. Studies | Sample Size | Heterogeneity | OR | 95% CI of Overall Effect | P | |
| CIMP+ | CIMP− | ||||||
| H. pylori | 5 | 137 | 243 | P = 0.05, I2 = 58% | 2.23 | 1.25–4.00 | P = 0.007 |
| EBV | 2 | 40 | 104 | P = 0.39, I2 = 0% | 51.27 | 9.39–279.86 | P<0.00001 |
| MSI | 4 | 136 | 306 | P = 0.01, I2 = 71% | 4.44 | 1.17–16.88 | P = 0.03 |
| Age | 4 | 87 | 195 | P = 0.52, I2 = 0% | 1.10 | 0.63–1.93 | P = 0.74 |
| Gender | 9 | 184 | 565 | P = 0.91, I2 = 0% | 0.69 | 0.48–1.00 | P = 0.05 |
| Tumor site | 4 | 85 | 206 | P = 0.17, I2 = 40% | 0.85 | 0.51–1.44 | P = 0.55 |
| Pathological type | 7 | 142 | 440 | P = 0.03, I2 = 56% | 0.63 | 0.31–1.28 | P = 0.20 |
| Cell differentiation | 3 | 45 | 264 | P = 0.68, I2 = 0% | 0.64 | 0.29–1.42 | P = 0.27 |
| TNM stage | 4 | 47 | 212 | P = 0.08, I2 = 56% | 1.39 | 0.54–3.57 | P = 0.49 |
| Distant metastasis | 4 | 103 | 258 | P = 0.004, I2 = 77% | 1.69 | 0.37–7.67 | P = 0.49 |
| Lymph node metastasis | 6 | 128 | 349 | P = 0.16, I2 = 37% | 0.81 | 0.50–1.31 | P = 0.39 |
| 5-year survival | 2 | 32 | 114 | P = 0.02, I2 = 82% | 0.65 | 0.04–10.70 | P = 0.76 |
CIMP, CpG island methylator phenotype; CI, confidence interval; EBV, Epstein-Barr virus; H. pylori, Helicobacter pylori; MSI, microsatellite instability; OR, odds ratio.
Figure 2. CIMP+ vs. CIMP−: a) H. pylori; b) EBV; c) MSI.
Correlation with Clinical Information
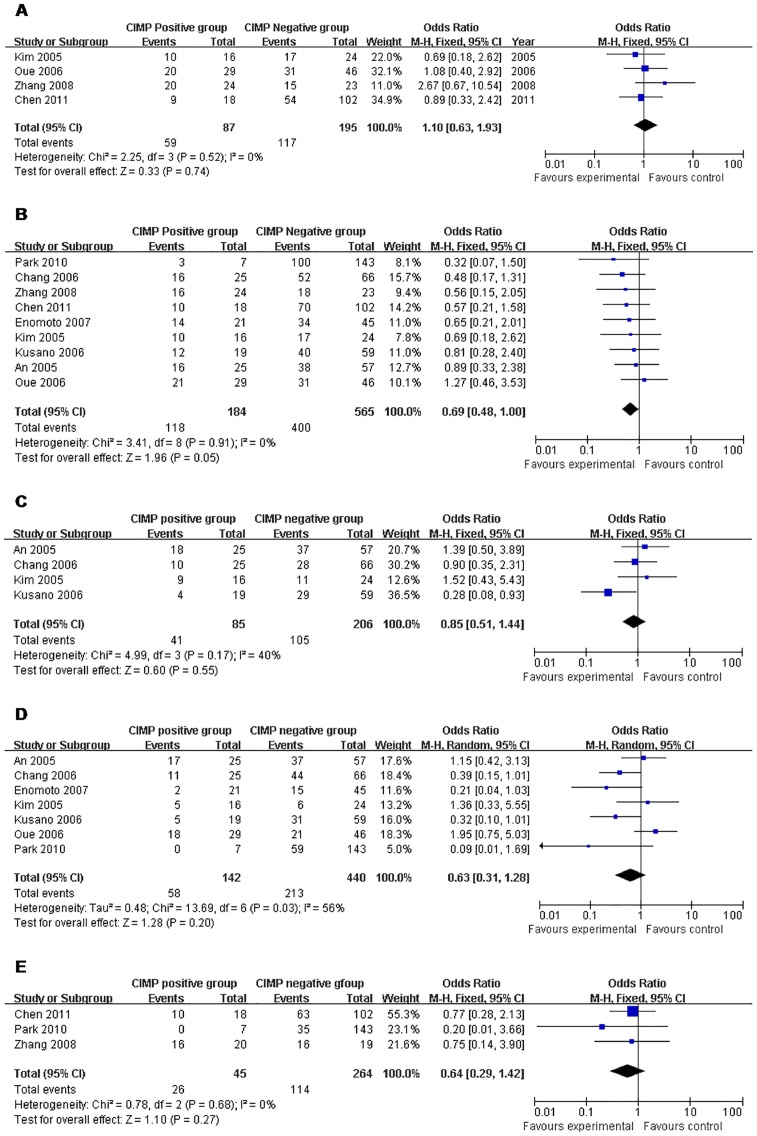
The meta-analysis of both age distribution and gender in the CIMP-positive vs. -negative groups did not attain statistical significance (OR 1.10 95% CI, 0.63–1.93; P = 0.74, Pheterogeneity = 0.52) and (OR 0.69 95% CI, 0.48–1.00; P = 0.05, Pheterogeneity = 0.91). The overall OR for tumor site in the CIMP-positive vs. -negative subgroups was 0.85 (95% CI, 0.51–1.44; P = 0.55, Pheterogeneity = 0.17). The overall OR for either pathological type or cell differentiation in the CIMP-positive vs. -negative subgroups did not show any positive finding (OR 0.63 95% CI, 0.31–1.28; P = 0.20, Pheterogeneity = 0.03 and OR 0.64 95% CI, 0.29–1.42; P = 0.027, Pheterogeneity = 0.68, respectively) (Table 2 and Figure 3).
Figure 3. CIMP+ vs. CIMP−: a) Age; b) Gender; c) Tumor site; d) Pathological type; e) Cell differentiation.
Correlation with Prognostic Parameters
Although some studies reported that CIMP may correlate with a better prognosis in gastric cancer, no matter on TNM stage, distant metastasis, lymph node metastasis, or even 5-year survival rate, CIMP did not have any significant correlation with one of them in our analysis (Table 2 and Figure 4).
Figure 4. CIMP+ vs. CIMP−: a) TNM stage; b) Distant metastasis; c) Lymph node metastasis; d) 5-year survival.
Publication Bias
Begg's funnel plot was performed to assess publication bias. The heterogeneity tests for comparing the 12 combined studies showed heterogeneity in some analyses such as MSI, pathological type, distant metastasis and 5-year survival. However, no single study influenced the pooled OR qualitatively as indicated by the sensitivity analyses (data not shown).
Discussion
Epigenetic alterations have been suggested to be significant initiating events in cancerization [25]. However, with the deep involvement of aberrant DNA methylation in human cancers becoming clear, the occasional presence of aberrant DNA methylation in non-cancerous tissues was recognized in the Barrett's esophagus [26], stomach [27], colon [28], [29], and liver [30], which suggested the involvement of the former in the field for cancerization. Chronic inflammation, possibly specific types is likely to induce aberrant DNA methylation in normal tissues and thus form an “epigenetic field for cancerization”. To supply the evidence, it is well known that chronic inflammation plays an important role in ulcerative colitis for colon cancers, chronic hepatitis for liver cancers, and Barrett's esophagus for esophagus cancers. Infection by H. pylori is known to induce severe chronic inflammation, which were involved in the induction of the field for gastric cancers. In addition, aberrant DNA methylation in gastric biopsies from H. pylori + patients was found to be correlated with a greater gastric cancer risk in several studies [31], [32]. Therefore, aberrant DNA methylation might be the key event in tumor genesis of gastric cancer.
In last decade, careful quantitative evaluation showed that many genes that are highly methylated in carcinoma also show a low but measurable degree of methylation in normal mucosa [33]. Furthermore, aberrant methylated region focusing in the promoter rich in CpG islands suggested the key step in epigenetic gene silencing. It is therefore necessary to elucidate the methylation statuses of a panel of representative genes in an individual disease. To achieve this goal, CpG island methylator phenotype was introduced by Toyota et al [3]. Till now, it was well evidenced that CIMP is associated with poor prognosis in colorectal cancer, lung cancer and neuroblastoma. Although the term “CIMP” has been used in a variety of ways in the context of gastric cancer [18], its prognostic value in gastric cancer is still controversial. For example, An et al. showed that CIMP, which correlated with malignant features on histopathology, was an independent prognostic factor for overall and cause-specific survival in patients with gastric cancer [14], whereas Kim et al. and Zhang et al. failed to observe such an association [15], [20]. Park et al. suggested that CIMP-high GCs were featured with characteristic clinicopathological parameters, including poor prognosis [22]. However, in contrast, Chang et al. and Kusano et al. suggested that CIMP-high showing better prognosis [17], [18]. Some scientists supposed this discrepancy might come from using different CIMP marker panels as the determination of CIMP status; however, there was a common point of these studies in which the methylated cite of CIMP marker genes lied in promoter. Therefore, it still represents a trend of methylation level in promoter, by which the CIMP is meaningful in tumorigenesis of gastric cancer. In our opinion, prospective data was less convincing mainly due to small sample size and the lack of statistical power to integrate sporadic individual studies.
With the goal to explore the potential value of CIMP in gastric cancer, we performed this meta-analysis of published studies to derive an overall pooled estimation. Since some studies have divided patients into three groups, CIMP-high, CIMP-immediate, CIMP-low, we combine the latter two into CIMP-negative subtype in comparison with no changed CIMP-high as CIMP-positive subtype. From table 2, our findings strongly suggested that H. pylori and EBV infections cause the aberrant DNA hypermethylation of specific genes and induce CIMP, an important epigenetic mechanism of the tumorigenesis. However, the mechanism for aberrant DNA hypermethylation induced by H. pylori might be different from that by EBV. Recent study by Huang et al. supported that H. pylori infection causes gastric mucosal inflammatory responses, resulting in up-regulation of interleukin-1b (IL-1b) and overproduction of mutagenic nitric oxide (NO), by which aberrant DNA methylation was induced [34]. As for EBV infection, it was suggested that the methylation mechanism in host cells might be primarily for defense against foreign DNA and that the host-driven extensive methylation of viral genome may also trigger host genome methylation [35]. It was also demonstrated repeatedly that direct interaction of viral latent proteins with DNA methyl transferases (DNMT), up-regulation of DNMT genes by viral latent proteins, and increased expression of polycomb group proteins may contribute to alternations in DNA methylation and histone modifications [35]–[38].
Furthermore, strong relation of CIMP with MSI reveals that CIMP may have a potential relation with gene mutations, which may cooperate with each other in development and progression of gastric cancer. However, the meta-analysis did not show any correlations with clinical parameters such as age, gender, tumor site, pathological type, cell differentiation, TNM stage, distant metastasis, lymph node metastasis, and 5-year survival.
If CIMP was a key incidence in gastric cancer, the reason why the CIMP in promoter could not be used as a prognostic marker is not clear. It is possible that gene methylation in promoter lead to the primary tumor genesis but not progressing in gastric cancer. However, another possible reason is that limited methylated CpG island sites do not represent the true trend of CIMP. Regardless of the above analysis, heterogeneity is also one of the important sources that limited us to make more precise conclusion. Therefore, it is essential to develop more extensive large-scale study with bead-array technology in future.
Supporting Information
(DOC)
(DOC)
Funding Statement
No current external funding sources for this study.
References
- 1. Brenner H, Rothenbacher D, Arndt V (2009) Epidemiology of stomach cancer. Methods Mol Biol 472: 467–477. [DOI] [PubMed] [Google Scholar]
- 2. Park SY, Yoo EJ, Cho NY, Kim N, Kang GH (2009) Comparison of CpG island hypermethylation and repetitive DNA hypomethylation in premalignant stages of gastric cancer, stratified for Helicobacter pylori infection. J Pathol 219: 410–416. [DOI] [PubMed] [Google Scholar]
- 3. Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, et al. (1999) CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A 96: 8681–8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jo P, Jung K, Grade M, Conradi LC, Wolff HA, et al. (2012) CpG island methylator phenotype infers a poor disease-free survival in locally advanced rectal cancer. Surgery 151: 564–570. [DOI] [PubMed] [Google Scholar]
- 5. La Rosa S, Marando A, Furlan D, Sahnane N, Capella C (2012) Colorectal poorly differentiated neuroendocrine carcinomas and mixed adenoneuroendocrine carcinomas: insights into the diagnostic immunophenotype, assessment of methylation profile, and search for prognostic markers. Am J Surg Pathol 36: 601–611. [DOI] [PubMed] [Google Scholar]
- 6. Pai RK, Jayachandran P, Koong AC, Chang DT, Kwok S, et al. (2012) BRAF-mutated, microsatellite-stable adenocarcinoma of the proximal colon: an aggressive adenocarcinoma with poor survival, mucinous differentiation, and adverse morphologic features. Am J Surg Pathol 36: 744–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oue N, Motoshita J, Yokozaki H, Hayashi K, Tahara E, et al. (2002) Distinct promoter hypermethylation of p16INK4a, CDH1, and RAR-beta in intestinal, diffuse-adherent, and diffuse-scattered type gastric carcinomas. J Pathol 198: 55–59. [DOI] [PubMed] [Google Scholar]
- 8. Kanai Y, Ushijima S, Kondo Y, Nakanishi Y, Hirohashi S (2001) DNA methyltransferase expression and DNA methylation of CPG islands and peri-centromeric satellite regions in human colorectal and stomach cancers. Int J Cancer 91: 205–212. [DOI] [PubMed] [Google Scholar]
- 9. Oshimo Y, Oue N, Mitani Y, Nakayama H, Kitadai Y, et al. (2004) Frequent epigenetic inactivation of RIZ1 by promoter hypermethylation in human gastric carcinoma. Int J Cancer 110: 212–218. [DOI] [PubMed] [Google Scholar]
- 10. Watanabe N, Okochi-Takada E, Yagi Y, Furuta JI, Ushijima T (2006) Decreased fidelity in replicating DNA methylation patterns in cancer cells leads to dense methylation of a CpG island. Curr Top Microbiol Immunol 310: 199–210. [DOI] [PubMed] [Google Scholar]
- 11. Toyota M, Ahuja N, Suzuki H, Itoh F, Ohe-Toyota M, et al. (1999) Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Res 59: 5438–5442. [PubMed] [Google Scholar]
- 12. Zouridis H, Deng N, Ivanova T, Zhu Y, Wong B, et al. (2012) Methylation subtypes and large-scale epigenetic alterations in gastric cancer. Sci Transl Med 4: 156ra140. [DOI] [PubMed] [Google Scholar]
- 13. Kim H, Kim YH, Kim SE, Kim NG, Noh SH, et al. (2003) Concerted promoter hypermethylation of hMLH1, p16INK4A, and E-cadherin in gastric carcinomas with microsatellite instability. J Pathol 200: 23–31. [DOI] [PubMed] [Google Scholar]
- 14. An C, Choi IS, Yao JC, Worah S, Xie K, et al. (2005) Prognostic significance of CpG island methylator phenotype and microsatellite instability in gastric carcinoma. Clin Cancer Res 11: 656–663. [PubMed] [Google Scholar]
- 15. Kim HC, Kim JC, Roh SA, Yu CS, Yook JH, et al. (2005) Aberrant CpG island methylation in early-onset sporadic gastric carcinoma. J Cancer Res Clin Oncol 131: 733–740. [DOI] [PubMed] [Google Scholar]
- 16. Oue N, Mitani Y, Motoshita J, Matsumura S, Yoshida K, et al. (2006) Accumulation of DNA methylation is associated with tumor stage in gastric cancer. Cancer 106: 1250–1259. [DOI] [PubMed] [Google Scholar]
- 17. Chang MS, Uozaki H, Chong JM, Ushiku T, Sakuma K, et al. (2006) CpG island methylation status in gastric carcinoma with and without infection of Epstein-Barr virus. Clin Cancer Res 12: 2995–3002. [DOI] [PubMed] [Google Scholar]
- 18. Kusano M, Toyota M, Suzuki H, Akino K, Aoki F, et al. (2006) Genetic, epigenetic, and clinicopathologic features of gastric carcinomas with the CpG island methylator phenotype and an association with Epstein-Barr virus. Cancer 106: 1467–1479. [DOI] [PubMed] [Google Scholar]
- 19. Enomoto S, Maekita T, Tsukamoto T, Nakajima T, Nakazawa K, et al. (2007) Lack of association between CpG island methylator phenotype in human gastric cancers and methylation in their background non-cancerous gastric mucosae. Cancer Sci 98: 1853–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang KL, Sun Y, Li Y, Liu M, Qu B, et al. (2008) Increased frequency of CpG island methylator phenotype and CDH1 methylation in a gastric cancer high-risk region of china. Transl Oncol 1: 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kondo T, Oka T, Sato H, Shinnou Y, Washio K, et al. (2009) Accumulation of aberrant CpG hypermethylation by Helicobacter pylori infection promotes development and progression of gastric MALT lymphoma. Int J Oncol 35: 547–557. [DOI] [PubMed] [Google Scholar]
- 22. Park SY, Kook MC, Kim YW, Cho NY, Jung N, et al. (2010) CpG island hypermethylator phenotype in gastric carcinoma and its clinicopathological features. Virchows Arch 457: 415–422. [DOI] [PubMed] [Google Scholar]
- 23. Chen HY, Zhu BH, Zhang CH, Yang DJ, Peng JJ, et al. (2012) High CpG island methylator phenotype is associated with lymph node metastasis and prognosis in gastric cancer. Cancer Sci 103: 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu JB, Wu XM, Cai J, Zhang JY, Zhang JL, et al. (2012) CpG island methylator phenotype and Helicobacter pylori infection associated with gastric cancer. World J Gastroenterol 18: 5129–5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Feinberg AP, Ohlsson R, Henikoff S (2006) The epigenetic progenitor origin of human cancer. Nat Rev Genet 7: 21–33. [DOI] [PubMed] [Google Scholar]
- 26. Eads CA, Lord RV, Wickramasinghe K, Long TI, Kurumboor SK, et al. (2001) Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res 61: 3410–3418. [PubMed] [Google Scholar]
- 27. Waki T, Tamura G, Tsuchiya T, Sato K, Nishizuka S, et al. (2002) Promoter methylation status of E-cadherin, hMLH1, and p16 genes in nonneoplastic gastric epithelia. Am J Pathol 161: 399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, et al. (1994) Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet 7: 536–540. [DOI] [PubMed] [Google Scholar]
- 29. Hsieh CJ, Klump B, Holzmann K, Borchard F, Gregor M, et al. (1998) Hypermethylation of the p16INK4a promoter in colectomy specimens of patients with long-standing and extensive ulcerative colitis. Cancer Res 58: 3942–3945. [PubMed] [Google Scholar]
- 30. Kondo Y, Kanai Y, Sakamoto M, Mizokami M, Ueda R, et al. (2000) Genetic instability and aberrant DNA methylation in chronic hepatitis and cirrhosis – A comprehensive study of loss of heterozygosity and microsatellite instability at 39 loci and DNA hypermethylation on 8 CpG islands in microdissected specimens from patients with hepatocellular carcinoma. Hepatology 32: 970–979. [DOI] [PubMed] [Google Scholar]
- 31. Niwa T, Tsukamoto T, Toyoda T, Mori A, Tanaka H, et al. (2010) Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res 70: 1430–1432. [DOI] [PubMed] [Google Scholar]
- 32. Nakajima T, Enomoto S, Yamashita S, Ando T, Nakanishi Y, et al. (2010) Persistence of a component of DNA methylation in gastric mucosae after Helicobacter pylori eradication. J Gastroenterol 45: 37–44. [DOI] [PubMed] [Google Scholar]
- 33. Rashid A, Issa JP (2004) CpG island methylation in gastroenterologic neoplasia: a maturing field. Gastroenterology 127: 1578–1588. [DOI] [PubMed] [Google Scholar]
- 34. Huang FY, Chan AO, Rashid A, Wong DK, Cho CH, et al. (2012) Helicobacter pylori induces promoter methylation of E-cadherin via interleukin-1β activation of nitric oxide production in gastric cancer cells. Cancer 118: 4969–4980. [DOI] [PubMed] [Google Scholar]
- 35. Kaneda A, Matsusaka K, Aburatani H, Fukayama M (2012) Epstein-Barr Virus Infection as an Epigenetic Driver of Tumorigenesis. Cancer Res 72: 3445–3450. [DOI] [PubMed] [Google Scholar]
- 36. Tsai CL, Li HP, Lu YJ, Hsueh C, Liang Y, et al. (2006) Activation of DNA methyltransferase 1 by EBV LMP1 Involves c-Jun NH(2)-terminal kinase signaling. Cancer Res 66: 11668–11676. [DOI] [PubMed] [Google Scholar]
- 37. Seo SY, Kim EO, Jang KL (2008) Epstein-Barr virus latent membrane protein 1 suppresses the growth-inhibitory effect of retinoic acid by inhibiting retinoic acid receptor-h2 expression via DNA methylation. Cancer Lett 270: 66–76. [DOI] [PubMed] [Google Scholar]
- 38. Fukayama M, Hino R, Uozaki H (2008) Epstein-Barr virus and gastric carcinoma: virus-host interactions leading to carcinoma. Cancer Sci 99: 1726–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOC)