Abstract
Here we present vB_BanS-Tsamsa, a novel temperate phage isolated from Bacillus anthracis, the agent responsible for anthrax infections in wildlife, livestock and humans. Tsamsa phage is a giant siphovirus (order Caudovirales), featuring a long, flexible and non-contractile tail of 440 nm (not including baseplate structure) and an isometric head of 82 nm in diameter. We induced Tsamsa phage in samples from two different carcass sites in Etosha National Park, Namibia. The Tsamsa phage genome is the largest sequenced Bacillus siphovirus, containing 168,876 bp and 272 ORFs. The genome features an integrase/recombinase enzyme, indicative of a temperate lifestyle. Among bacterial strains tested, the phage infected only certain members of the Bacillus cereus sensu lato group (B. anthracis, B. cereus and B. thuringiensis) and exhibited moderate specificity for B. anthracis. Tsamsa lysed seven out of 25 B. cereus strains, two out of five B. thuringiensis strains and six out of seven B. anthracis strains tested. It did not lyse B. anthracis PAK-1, an atypical strain that is also resistant to both gamma phage and cherry phage. The Tsamsa endolysin features a broader lytic spectrum than the phage host range, indicating possible use of the enzyme in Bacillus biocontrol.
Introduction
We present a novel temperate phage isolated from Bacillus anthracis, the causative agent of anthrax infections in wildlife, livestock and humans [1]. Bacillus anthracis is a member of the Bacillus cereus sensu lato group of six closely related species: B. cereus, B. anthracis, B. thuringiensis, B. mycoides, B. pseudomycoides and B. weihenstephanensis [2]. This group contains both obligate and opportunistic animal pathogens, including B. anthracis, B. cereus, strains of which cause food poisoning and opportunistic infections in humans [3], and B. thuringiensis, an insect pathogen commonly used as a pesticide in agriculture [4]. Genomic studies have identified a number of putative prophages in the Bacillus cereus sensu lato group (e.g., [5], [6]), including four unique prophage elements in B. anthracis [7].
Lysogeny occurs commonly in B. anthracis [8] and may play an essential role in its life cycle [9]. Isolates of B. anthracis from soil frequently exhibit phage-derived plaques upon subculture [10]. Schuch et al. [9] showed that temperate phage infections of B. anthracis can affect sporulation, induce biofilm formation and promote colonization of earthworms and environmental reservoirs. Furthermore, the lytic activity and high specificity of bacteriophages provide a promising resource for the development of innovative treatments for human pathogens, including B. anthracis.
In this study, we describe the genome and host range of vB_BanS-Tsamsa, a novel temperate phage obtained from B. anthracis isolated in Etosha National Park (Etosha), Namibia. We named the phage Tsamsa, which in Hai||om means “place where the winds blow closed” referring to the endless vista of the Etosha pan and the dust devils that form there. Tsamsa phage is a giant siphovirus capable of infecting some members of Bacillus cereus sensu lato.
Materials and Methods
Phage Isolation and Preparation
We obtained isolates of the siphovirus from two carcass sites in Etosha, a 22,915 km2 national park in northern Namibia with abundant wildlife populations that exhibit regular occurrences of anthrax infections (reviewed in [11]). Field sampling was authorized by the Namibian Ministry of Environment and Tourism under permit number 1448/2009 to HHG. Bacteriophages were isolated from a B. anthracis isolate obtained from a swab of a plains zebra (Equus quagga) carcass from 2006 (Etosha Ecological Institute (EEI) carcass number: EB060318-01WV, GPS coordinates: −18.99736, 15.81584) and from soil collected near another plains zebra carcass in 2010 (EEI carcass number: EB100228-01MK, GPS coordinates: −19.1731, 15.92603). Prior diagnostic testing showed that both carcasses were positive for B. anthracis: isolates from the two carcasses were identified as genotype 6 and genotype 4 in the A cluster of B. anthracis, respectively [12]. Genotypes 6 and 4 are closely related members of a dominant B. anthracis strain that has been causing outbreaks in Etosha for a very long time [12].
Phages were obtained from the two samples by culturing to enrich for bacteria and by exposure to mitomycin C to induce prophages in the host genome to transition into a vegetative state. Methods for enrichment culture and induction are described by Sambrook [13] and Van Twest and Kropinski [14]. We did not obtain phages from either sample without induction. We used two approaches, one for the swab isolate and another for the soil sample, as follows:
For the swab isolate, we inoculated 3 ml of Bovine Heart Infusion medium (BHI, BD Difco, Sparks, MD, USA) with a single B. anthracis colony isolated from the swab and incubated the culture overnight at 37°C with aeration. We diluted the overnight culture 100-fold in 3 ml of BHI and incubated it at 37°C with aeration for one hour. To induce the release of prophages from the genome, mitomycin C was added to achieve a final concentration of 2.5 µg ml−1. The culture was incubated at 37°C with aeration for 20 hrs and pelleted for 15 min at 3000×g. The supernatant was filtered through a 0.22 µm filter unit and stored at 4°C.
For the soil sample, 20 g of soil were added to 15 ml of 1% Nutrient Broth medium (BD Difco, Sparks, MD, USA). After vortexing briefly, the sample was incubated at 35°C with aeration overnight. The resulting culture was centrifuged and the supernatant collected. Then we added 1% Nutrient Broth medium to attain a final volume of 7.5 ml. The sample was incubated in 1 µg ml−1 mitomycin C for 30 minutes with gentle aeration at 30°C. Then the sample was filtered (0.22 µm) and concentrated using a Vivaspin 20 concentrator (Sartorius Stedim, Bohemia, NY, USA) by adding 10 ml of 1% nutrient broth medium and centrifuging the sample at 3000×g for 10 min. The resulting phage extract was stored at 4°C.
Preparation of Plate Stocks of the Two Tsamsa Phage Isolates
Phage preparations were purified and concentrated using standard techniques [13], [15]. Preliminary plaque assays were performed with phage extracts from the two carcass site samples (swab isolate and soil sample) to harvest concentrated plate stocks. Soft agar overlays were performed as described previously by Adams [16]. Briefly, five microliters of a spore preparation of an avirulent (pXO1− pXO2−) B. anthracis strain (6602 R1,[17]) were added to 2.5 ml of LB soft agar (BD Difco, Sparks, MD, USA; containing per liter: 10 g of tryptone, 7 g of agar, 5 g of yeast extract and 5 g of NaCl) and poured over the surface of pre-warmed plates (containing per liter: 8 g of Nutrient Broth, 5 g of NaCl, 15 g of agar, 0.15 g of CaCl2, 0.2 g of MgSO4 and 0.05 g of MnSO4 [18]). After the soft agar solidified, ten microliters of 10−4, 10−6 and 10−8 dilutions of each of the two phage extracts were pipetted on top of each plate and the plates were incubated overnight at 30°C. We selected a single plaque from each of the two phage extract plates and stored it in 0.5 ml of phage buffer (10 mM Tris HCl (pH 8), 10 mM MgCl2 and 200 mM NaCl). Dilutions of the phage plaque buffer were added to B. anthracis strain 6602 R1 [17] spore preparation in 2.5 ml of LB soft agar and soft agar overlays were performed. Plates with complete clearance were harvested by adding 5 ml of phage buffer to each plate and collecting the soft agar overlays with cell scrapers into 50 ml centrifuge tubes. We centrifuged the tubes at 3000×g for 8 min and syringe filtered the supernatant through a 0.22 µm filter unit. The resulting plate stocks were further concentrated using PEG-6000 precipitation [19], followed by cesium chloride density gradient centrifugation to obtain pure phage particles [13] [15] and stored at 4°C for additional analyses.
Phage Host Range
Tsamsa phage was originally propagated on B. anthracis 6602 R1 (an avirulent strain that lacks both pXO1 and pXO2 virulence plasmids, [17]). Using the spot-on-the-lawn method and a 100-fold and a 10,000-fold dilution of the phage stock, 10 µl were spotted on top of plates seeded with a lawn of bacteria. Each plate was tilted to allow the phage solution to run down the plate. If phage titers were higher than 108 pfu/ml, more dilutions were made. A phage was considered positive for infection of a certain strain if single plaques could be observed on one of the spots. We tested a set of 55 strains for susceptibility to Tsamsa phage (Table 1).
Table 1. Host range of Tsamsa phage on 43 different Bacillus strains and 12 non-Bacillus strains and lytic spectrum of Tsamsa endolysin for a subset of the Bacillus strains. Presence of lysis is indicated by+and absence of lysis is indicated by -. n.d.: not determined.
| Strain name | Organism | Notes | Source | Phage lysis | Endolysin lysis |
| 6602 R1 | Bacillus anthracis | pXO1-pXO2 negative | [39] | + | + |
| Sterne | Bacillus anthracis | pXO2 negative | Institut Pasteur #7702 | + | + |
| Weybridge UM44 | Bacillus anthracis | pXO2 negative | [39] | + | + |
| Ames-non reverting | Bacillus anthracis | pXO2 negative | U.S. Dept. of Agriculture, Ames, Iowa | + | + |
| Ames | Bacillus anthracis | U.S. Dept. of Agriculture, Ames, Iowa | + | n.d | |
| Vollum 1b | Bacillus anthracis | Laboratory Strain | + | n.d. | |
| PAK-1 | Bacillus anthracis | Pakistan isolate, M. Hugh-Jones collection | − | n.d. | |
| 569 | Bacillus cereus | [17] | + | n.d. | |
| LA 925 | Bacillus cereus | CHUV | − | + | |
| ATCC 14579 | Bacillus cereus | ATCC | − | + | |
| ATCC 11778 | Bacillus cereus | ATCC | + | + | |
| ATCC 10702 | Bacillus cereus | ATCC | − | + | |
| ATCC 10876 | Bacillus cereus | ATCC | + | n.d. | |
| DSM 2302 | Bacillus cereus | DSM | − | + | |
| BO 366 | Bacillus cereus | This study | − | − | |
| BO 372 | Bacillus cereus | This study | − | − | |
| BO 493 | Bacillus cereus | This study | − | − | |
| DSM 4218 | Bacillus cereus | DSM | − | + | |
| ATCC 33019 | Bacillus cereus | ATCC | + | + | |
| ATCC 14737 | Bacillus cereus | ATCC | + | + | |
| DSM1274 | Bacillus cereus | DSM | − | + | |
| ATCC 27522 | Bacillus cereus | ATCC | − | + | |
| NCTC 11143 | Bacillus cereus | NCTC | − | + | |
| NCIMB 8705 | Bacillus cereus | NCIMB | + | + | |
| ATCC 6464 | Bacillus cereus | ATCC | − | + | |
| B346 | Bacillus cereus | Mouse isolate | − | + | |
| DSM360 | Bacillus cereus | DSM | − | − | |
| HER1399 | Bacillus cereus | HER | + | + | |
| WSBC 10530 | Bacillus cereus | WSBC | − | − | |
| WSBC 10556 | Bacillus cereus | WSBC | − | + | |
| WSBC 10566 | Bacillus cereus | WSBC | − | − | |
| WSBC 10583 | Bacillus cereus | WSBC | − | − | |
| DSM4421 | Bacillus thuringiensis | DSM | − | + | |
| WSBC 10204 | Bacillus thuringiensis | WSBC | − | − | |
| HER1211 | Bacillus thuringiensis | HER | + | + | |
| Kurstaki | Bacillus thuringiensis | Industry isolate | − | − | |
| ATCC 10792 | Bacillus thuringiensis | ATCC | + | + | |
| DSM168 | Bacillus subtilis | DSM | − | − | |
| DSM675 | Bacillus subtilis | DSM | − | − | |
| ATCC 23059 | Bacillus subtilis | ATCC | − | n.d. | |
| DSM395 | Bacillus sphaericus | DSM | − | − | |
| DSM90 | Bacillus megaterium | DSM | − | − | |
| WSBC 10550 | Bacillus weihenstephanensis | WSBC | − | + | |
| WSLC 3009 | Listeria ivanovii | WSLC | − | ||
| ATCC BAA-679 | Listeria monocytogenes | ATCC | − | ||
| PSK | Staphylococcus aureus | Laboratory Stock | − | ||
| Twort | Staphylococcus aureus | Laboratory Stock | − | ||
| 414 | Staphylococcus epidermidis | Laboratory Stock | − | ||
| 100655 | Staphylococcus epidermidis | Laboratory Stock | − | ||
| 602 | Staphylococcus epidermidis | Laboratory Stock | − | ||
| DT7155 | Salmonella Typhimurium | Laboratory Stock | − | ||
| CGSC 4401 | Escherichia coli | CGSC | − | ||
| DSM 20560 | Streptococcus salivarius | DSM | − | ||
| NZ9000 | Lactococcus lactis | Laboratory Stock | − | ||
| ATCC 19433 | Enterococcus faecalis | ATCC | − |
Source abbreviations: CHUV: Strain collection of the Centre Hospitalier universitaire Vaudois, Switzerland; HER: Félix d’Hérelle Reference Center for bacterial viruses, Laval, Canada; ATCC: American Type Culture Collection; NCTC: National Collection of Type Cultures; DSM: Deutsche Sammlung von Mikroorganismen; NCIMB: National Collection of Industrial Bacteria; WSBC: Weihenstephan Bacillus Collection; WSLC = Weihenstephan Listeria Collection; CGSC: Coli Genetic Stock Center, Yale, USA.
Phage Morphology
The Tsamsa phage was negatively stained with 2% uranyl acetate on carbon-coated copper grids (Carbongrids, Quantifoil, Jena, Germany) and observed in a Philips CM12 TEM microscope at 120 kV acceleration voltage with a Gatan Orius 1 k camera.
Genome Sequencing and Genetic Characteristics
We performed a standard DNA extraction using phenol-chloroform-isoamyl-alcohol of the cesium chloride purified stock [13]. After washing with 70% ethanol and drying, the DNA pellet was resuspended in sterile ultrapure water. We sequenced DNA from the two isolates using a SMRT sequencing approach (Pacific Biosciences RS) with 10 kb and 800 bp insert libraries (C2 chemistry) and one SMRT sequencing cell for each library. We used the standard error-correction workflow and SMRT portal software 1.3.1 for assembly of 36166 post-filter reads (with 2582 bp average read length). Open reading frames were predicted by RAST [20] and edited manually. The genome sequence was deposited at GenBank under accession number KC481682. The unassembled reads for both sequencing runs are available in the DNA Databank of Japan Sequence Read Archive under accession number DRA001229.
Phylogenetic Analysis
Comparisons were made between sequences of the large terminase subunit of Tsamsa phage and 17 other Bacillus phages (GenBank accession numbers: 955214, 955254, 7070024, 12980149, 13164871, 14697218, 14697335, 14697413, 14697831, NC_001884, NC_006557, NC_007457, NC_007458, NC_007734, NC_007814, NC_011167, NC_011421). Alignments and phylogenetic tree construction were performed in Geneious version 6.1 (Biomatters Ltd., http://www.geneious.com). Muscle [21] was used to align the terminase gene (with 16 iterations). Then MrBayes 3.1.2 [22], [23] was used to build the tree and determine Bayesian posterior probabilities (with the Monte Carlo Markov Chain run for 1.1×106 generations).
Endolysin Production and Determination of Lytic Spectrum
Purified Tsamsa phage endolysin was recombinantly produced [24] and characterized. Lysin activity against different bacterial strains was tested with a turbidity reduction assay or plate lysis. Turbidity reduction experiments were performed by harvesting and washing an overnight bacterial culture (grown in half-strength BHI medium) in PBS (120 mM NaCl, 50 mM NaH2PO4, pH 8.0) buffer. Cell density was adjusted to OD600nm of 1±0.05. 100 µM of lysin were added to the wells of a 96-well plate and 200 µl bacterial suspension was added. Wells were measured at a wavelength of 600 nm in 10 second intervals until clear turbidity reduction was observed relative to the control.
Lysis activity by plate lysis assay was tested as follows. The Bacillus strains were grown in half-strength BHI medium to an OD600nm of 0.4–0.6 and diluted 1/100 in PBS immediately before plating. The freshly spread lawns of Bacillus cells on ½ BHI agar plates were air dried for 30 min. Ten microliters of a 10-fold serial dilution (50, 5 and 0.5 µM) of the purified endolysin were spotted onto the plates. Spots were air-dried and plates incubated at 30°C for 16 hours. Cleared spots indicating cell lysis were assessed visually.
Results
We obtained two phage isolates from two different carcass sites (one from soil and one carcass swab). Both phages were morphologically very similar and unusually large compared with previously described Siphoviridae of Bacillus. We found by sequencing the genomes of these two isolates that both phages were 100% identical. The representative phage was named Tsamsa and its characteristics were investigated further.
Tsamsa Features a Broad Host Range in the B. anthracis Subgroup
A set of 55 bacterial strains was analyzed for susceptibility to Tsamsa. The phage lysed seven out of 25 B. cereus strains as well as two out of five B. thuringiensis strains and six out of seven B. anthracis strains (6602 R1, Sterne, Weybridge UM44, Ames-non-reverting and Vollum 1b but not PAK-1) (Table 1). It should be noted that PAK-1 is an atypical member of B. anthracis, belonging to the A2 branch that contains very few isolates [25], and is resistant to both Cherry phage and Gamma phage [26]. Tsamsa phage is unable to infect B. subtilis 168 and DSM675, B. megaterium DSM90, B. sphaericus DSM395, B. weihenstephanensis WSBC10550 as well as all tested strains from other bacterial genera (Listeria ivanovii and L. monocytogenes, Staphylococcus aureus and S. epidermidis, Salmonella Typhimurium, Escherichia coli, Streptococcus salivarius, Lactococcus lactis or Enterococcus faecalis) (Table 1). Thus it is a narrow host-range virus that infects some members of the Bacillus cereus sensu lato group and exhibits moderate specificity for B. anthracis.
Tsamsa is a Giant Siphovirus
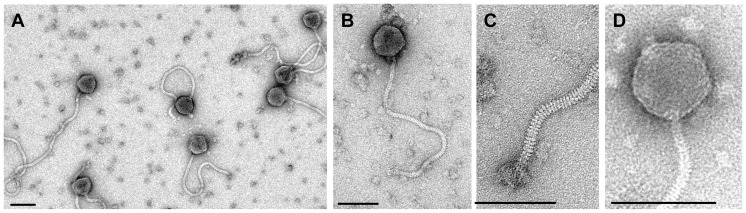
Most bacteriophages belong to the Order Caudovirales, which contains three families: Myoviridae with a contractile tail, Siphoviridae with a non-contractile flexible tail and Podoviridae with a short, non-contractile tail. Tsamsa exhibits typical siphovirus morphology, featuring a long, flexible and non-contractile tail of 440 nm (not including baseplate structure) and an isometric head of 82 nm in diameter (Figure 1 A and B). Individual tail striations (disk-like structure) and a baseplate structure with appendages are visible (Figure 1C). The head features visible individual capsomers (Figure 1D), an observation previously made for a different class of large virulent phages belonging to the Spounavirinae subfamily within the family Myoviridae [27]. Because of the large head dimensions and our experience from similarly sized myoviruses, we anticipate a triangulation number of 16 or higher but experimental proof is lacking.
Figure 1. Electron microscopy of Tsamsa phage.
TEM images were acquired from a preparation of pure phage particles negatively stained with 2% uranyl acetate on carbon-coated copper grids (Quantifoil, Jena, Germany) and observed using a Philips CM12 microscope at 120 kV acceleration voltage with a Gatan Orius digital camera. A. Preparation overview. B. Close-up of single phage particle. C. Details of the phage tail distal end. D. Details of the phage head structure. Individual capsomers are visible, an observation previously made for SPO1-related phages ([27]). Scale bars represent 100 nm.
The Phage Tsamsa Genome is Large and Unique
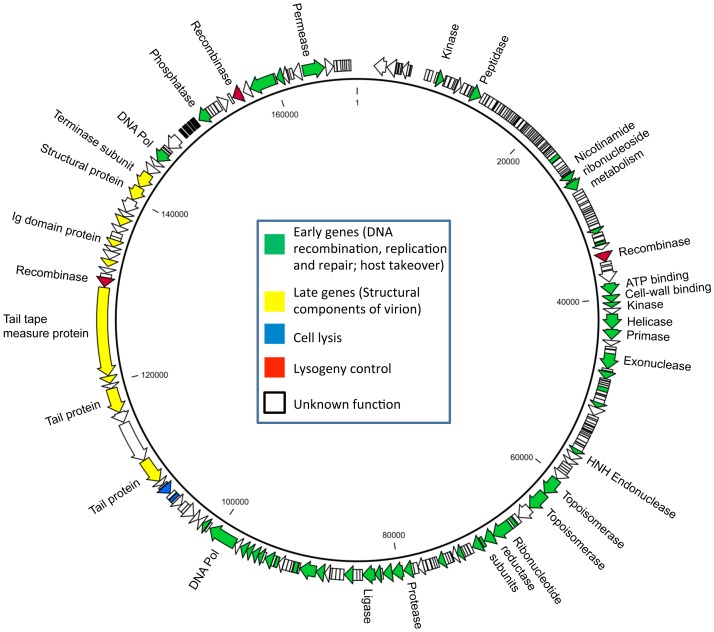
Genome sequencing and assembly resulted in a single large contig with an average coverage of 550-fold of error-corrected SMRT reads (Figure 2). Both phage isolates were identical. A repeat structure of 284 bp at both genome ends was identified during assembly and confirmed in restriction profiles (Figure 3). Methylome analysis revealed no base modifications in the genome. The genome sequence is 168,876 bp in length. Tsamsa features 272 open reading frames, 17 tRNA and 2 pseudo-tRNA genes. Database matches of predicted proteins encoded by Bacillus anthracis phage Tsamsa are provided in Table S1. The GC content is 34%, similar to published genome sequences of B. anthracis.
Figure 2. Genome map of Tsamsa phage.
Open reading frames are drawn to scale and transcription direction is indicated by arrows. Selected proteins with putative function are labeled. Genetic modules (i.e. structural genes, early genes) are indicated by coloring.
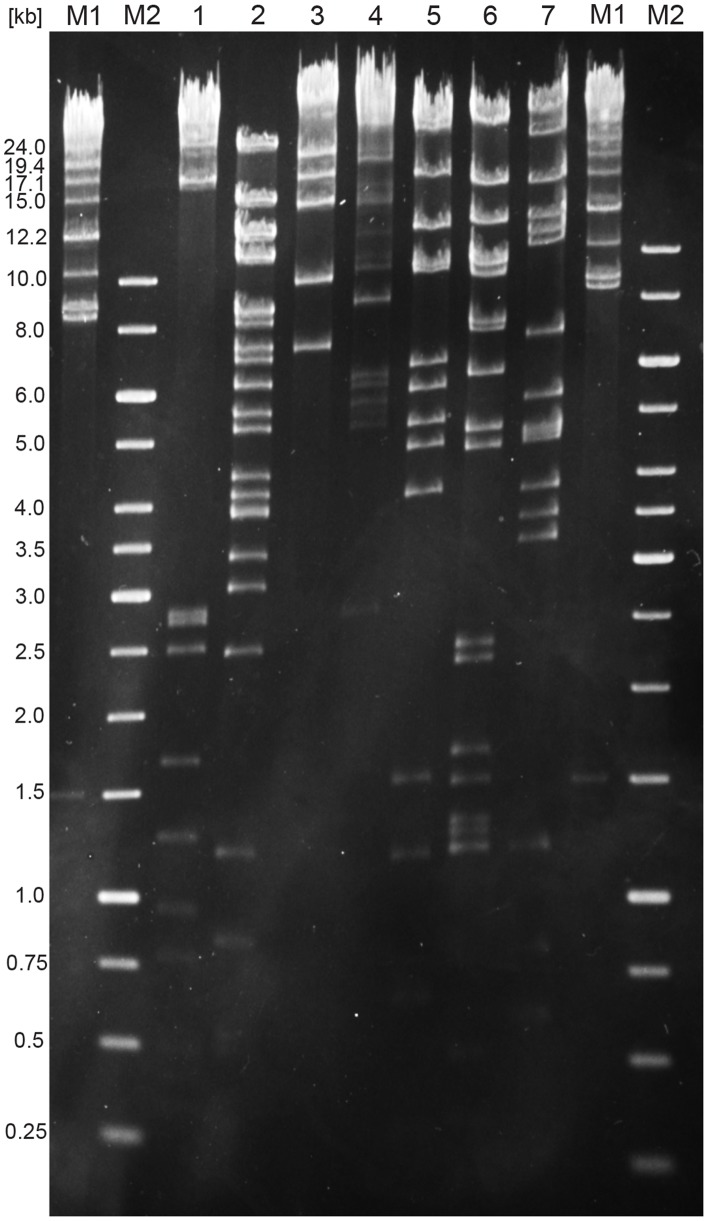
Figure 3. Restriction profile of Tsamsa phage.
500 ng DNA were digested with different restriction enzymes for 2 h at 37°C and electrophoresed. Clear band separation up to 24 kb in size was achieved and the restriction profiles matched with the sequenced genome size. The terminal redundancy location and size was determined from the fragment sizes as previously described [13], [15]. Enzymes used: 1: Alw44I (NEB); 2: Eco91I (Fermentas); 3: NheI (NEB); 4: PacI (NEB); 5: SwaI (NEB); 6: Van91I (Fermentas); 7: XcmI (NEB). M1: Lambda 19 Mix Size standard (Fermentas); M2: 1 kb size standard (Fermentas). Numbers to the left indicate band size in kb.
Virus Proteome
Virion proteins of Tsamsa were separated on a 10–20% SDS gradient PAGE. Resulting bands were extracted and protein content identified by mass spectrometry [28]. Six bands were allocated to gene products. The tape measure protein is present in two protein bands of 280 and 100 kDa in size, presumably because of instability of the large protein or post-translational modification. gp206 was identified in a band with an estimated mass of 38 kDa and gp199 and 207 were identified in bands of 26.5 and 19 kDa, respectively.
Comparison with Other Sequenced Bacillus Phages
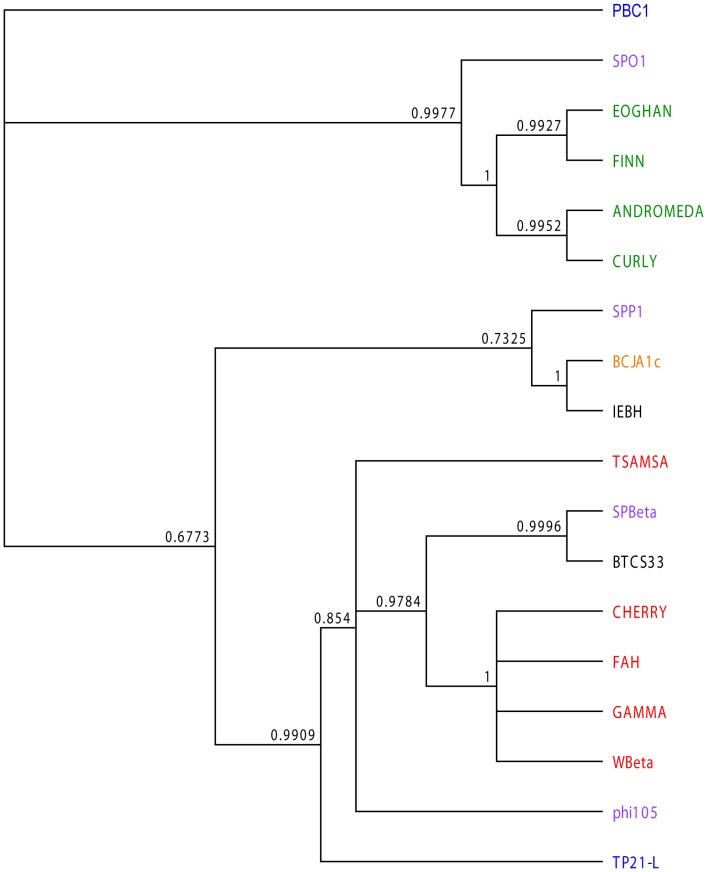
The sequence of the large terminase subunit of Tsamsa phage was compared with 17 previously sequenced Bacillus phages: four B. anthracis phages (Fah, Cherry, WBeta, Gamma), two B. thuringiensis phages (IEBH, BTCS33), four B. subtilis phages (SPBeta, SPO1, SPP1, phi105), one B. clarkii phage (BCJA1c), two B. cereus phages (PBC1, TP21-L) and four B. pumilus phages (Andromeda, Curly, Eoghan, Finn). The Tsamsa phage terminase clearly differs from previously described phages isolated from B. anthracis (Figure 4), which were shown to be derived from a single B. anthracis prophage named W [29], [30].
Figure 4. Phylogenetic relationship of the large terminase subunit gene in 18 Bacillus phages.
Phages are color coded according to their host bacterium: B. anthracis in red, B. cereus in blue, B. clarkii in orange, B. pumilus in green, B. subtilis in purple and B. thuringiensis in black. The tree and posterior probabilities were determined from 1.1x106 generations with MrBayes 3.1.2 [22], [23].
The Tsamsa Endolysin Shows Broad Activity
The endolysin of phage Tsamsa (gp217) was cloned and recombinantly produced in E. coli. The 35.78 kDa protein features an isoelectric point of 9.01 and contains three domains, one Amidase_2 enzymatic active domain (PF01510) and two SH3_3 (PF08239) cell-wall binding domains. Lytic activity of the protein was assessed with either turbidity reduction or plate lysis assay and activity against a set of Bacillus strains is outlined in Table 1. The Tsamsa endolysin features a broad spectrum of lytic activity and is able to lyse more Bacillus cereus sensu lato strains than the phage can infect.
Discussion
We obtained identical phage isolates of an unusually large siphovirus induced from B. anthracis in two different environmental samples. Such a large siphovirus is rarely isolated; only nine of the 539 Siphovirus genomes in the NCBI Genome database have a larger genome than Tsamsa phage (six Caulobacter phages, two Synechococcus phages and one Erwinia phage). To our knowledge, Tsamsa is the largest sequenced siphovirus infecting Bacillus. Two larger Bacillus siphoviruses are known but not characterized: B. mycoides phage N5 and B. thuringiensis phage II, both of which feature approximately 20% larger dimensions than Tsamsa and are speculated to be identical (H.-W. Ackermann, personal communication). Large siphoviruses may be isolated infrequently due to a bias in phage isolation procedures towards smaller phages. Consequently, the distribution and abundance of large siphoviruses are poorly understood. Such a sampling bias has been reported for the so-called Jumbo-Phages [31].
Like many siphoviruses, the genome is structured in functional modules. The early gene cluster (genes for DNA replication, modification and repair, host takeover and nucleotide metabolism) spans roughly 70% of the genome, indicating active participation of virus-encoded genes in the metabolic processes associated with replication in the host cell. It is notable that the Tsamsa genome encodes three tyrosine integrase/recombinase enzymes of the Cre/XERD type (gp94, gp227, gp255; [32], [33], which exhibit no homology to each other. Tsamsa features a temperate lifestyle and these three enzymes may serve as means to integrate into different attB sites and ensure a large host range for lysogeny. Further work will elucidate the specificity and activity of the three recombinases. We also note the presence of two Ig-domain containing proteins, gp233 and gp213 [33], [34], which may play accessory roles during infection [35]. Ig-like domains in structural proteins were recently shown to play a potential role in phage attachment to mucosa of humans and animals by interaction of the Ig-like domain with the mucosa glycan residues, providing a non-host derived immunity [36].
Tsamsa features distributed homologies in its structural proteins to SPO1-related phages: A511, A9, LP65 and SPO1 [27]. This finding is very unusual because SPO1-related phages belong to the Spounavirinae subfamily within an unrelated family of bacteriophages (Myoviridae) [27]. In addition, Tsamsa is a temperate phage and phages within the Spounavirinae are strictly virulent. Tsamsa also displays individual capsomers thought to be a hallmark of the Spounavirinae (Figure 1) [27]. An assessment of the virus particle proteome identified six structural protein bands in Tsamsa, namely the tape measure protein in two bands, and one band each for gp199, 206, 207 and 222. Tsamsa features an unusually long tail of 440 nm, which corresponds with the large size of the tape measure protein (3123 aa) [37], [38] and the protein is disproportionately large in comparison to other sequenced bacteriophages. The large unknown gene 221 likely encodes for a tail fiber component, with the C-terminus featuring significant homologies to Cellobiosidase, S-layer associated endoglucanase or glycoside hydrolase domains. The Tsamsa endolysin (gp217) is accompanied by a holin (gp215) and is active when produced recombinantly. The enzyme shows a broad lytic spectrum and lysed more B. cereus and B. thuringiensis strains than the phage infects. Thus the Tsamsa lysin might be useful as an antimicrobial agent against some Bacillus cereus sensu lato organisms.
In conclusion, we present vB_BanS-Tsamsa, a novel temperate phage obtained from B. anthracis that is specific to some members of the Bacillus cereus sensu lato group. To our knowledge Tsamsa is the largest sequenced siphovirus infecting Bacillus organisms.
Supporting Information
Database matches of predicted proteins encoded by Bacillus anthracis phage Tsamsa.
(DOCX)
Acknowledgments
We thank Margaret Smith for help with recombinase identification, Matthew Boitano and Tyson A. Clark for genome assembly and initial analysis, and Martina Kusters, Wendy C. Turner and Wilferd Versfeld for field support. In addition, we thank Brian Souza, Paul Jackson and Raymond Schuch for advice on phage isolation methods. We are grateful to the scientific staff at the Etosha Ecological Institute for facility resources and logistical support. Finally, we thank Ted Hadfield, John Houston, David Malcolm, Raymond Schuch and two anonymous reviewers for their helpful comments on the manuscript.
Funding Statement
This research was funded by a Pre-Graduate Pathway Research Grant from the University of California, Berkeley (UCB), Biology Scholars Program, the UCB College of Natural Resources-Biology Scholars Program, a Rosberg-Geist Fellowship from the UCB Center for African Studies and an Undergraduate Merit Scholarship from the UCB Institute of International Studies to CL, and NIH Grant GM083863 to WMG. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Carter KC (1988) The Koch-Pasteur dispute on establishing the cause of anthrax. Bull Hist Med 62: 42–57. [PubMed] [Google Scholar]
- 2.Okstad ØA, Kolstø A-B (2011) Genomics of Bacillus Species. In: Wiedmann M, Zhang W, editors. Genomics of Foodborne Pathogens. New Yorl: Springer. pp. 29–55.
- 3. Drobniewski FA (1993) Bacillus cereus and related species. Clin Microbiol Rev 6: 324–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Soberon M, Pardo-Lopez L, Lopez I, Gomez I, Tabashnik BE, et al. (2007) Engineering modified Bt toxins to counter insect resistance. Science 318: 1640–1642. [DOI] [PubMed] [Google Scholar]
- 5. Stromsten NJ, Benson SD, Burnett RM, Bamford DH, Bamford JK (2003) The Bacillus thuringiensis linear double-stranded DNA phage Bam35, which is highly similar to the Bacillus cereus linear plasmid pBClin15, has a prophage state. J Bacteriol 185: 6985–6989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rasko DA, Altherr MR, Han CS, Ravel J (2005) Genomics of the Bacillus cereus group of organisms. FEMS Microbiol Rev 29: 303–329. [DOI] [PubMed] [Google Scholar]
- 7. Read TD, Peterson SN, Tourasse N, Baillie LW, Paulsen IT, et al. (2003) The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423: 81–86. [DOI] [PubMed] [Google Scholar]
- 8. Buck CA, Anacker RL, Newman FS, Eisenstark A (1963) Phage Isolated from Lysogenic Bacillus anthracis . J Bacteriol 85: 1423–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schuch R, Fischetti VA (2009) The secret life of the anthrax agent Bacillus anthracis: bacteriophage-mediated ecological adaptations. PLoS One 4: e6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Saile E, Koehler TM (2006) Bacillus anthracis multiplication, persistence, and genetic exchange in the rhizosphere of grass plants. Appl Environ Microbiol 72: 3168–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Turner WC, Imologhome P, Havarua Z, Kaaya GP, Mfune JK, et al. (2013) Soil ingestion, nutrition and the seasonality of anthrax in herbivores of Etosha National Park. Ecosphere 4: art13. [Google Scholar]
- 12. Beyer W, Bellan S, Eberle G, Ganz HH, Getz WM, et al. (2012) Distribution and molecular evolution of bacillus anthracis genotypes in Namibia. PLoS Negl Trop Dis 6: e1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook J, Russell DW (2001) Molecular Cloning - A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press.
- 14.Van Twest R, Kropinski AM (2009) Bacteriophage enrichment from water and soil. Bacteriophages: Springer. 15–21. [DOI] [PubMed]
- 15. Klumpp J, Dorscht J, Lurz R, Bielmann R, Wieland M, et al. (2008) The Terminally Redundant, Nonpermuted Genome of Listeria Bacteriophage A511: a Model for the SPO1-Like Myoviruses of Gram-Positive Bacteria. J Bacteriol 190: 5753–5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adams MH (1959) Methods of study of bacterial viruses. Bacteriophages. New York: Interscience publishers, Inc. 443–457.
- 17. Battisti L, Green BD, Thorne CB (1985) Mating System for Transfer of Plasmids among Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis . Journal of Bacteriology 162: 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thorne CB (1968) Transducing bacteriophage for Bacillus cereus . Journal of virology 2: 657–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yamamoto KR, Alberts BM, Benzinger R, Lawhorne L, Treiber G (1970) Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology 40: 734–744. [DOI] [PubMed] [Google Scholar]
- 20. Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, et al. (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- 23. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 24. Pastagia M, Schuch R, Fischetti VA, Huang DB (2013) Lysins: the arrival of pathogen-directed anti-infectives. Journal of medical microbiology 62: 1506–1516. [DOI] [PubMed] [Google Scholar]
- 25. Keim P, Price L, Klevytska A, Smith K, Schupp J, et al. (2000) Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis . Journal of Bacteriology 182: 2928–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fulmer P (2006) Bacillus anthracis: Antibiotic and Phage Sensitivity and Resistances: Louisiana State University.
- 27. Klumpp J, Lavigne R, Loessner MJ, Ackermann HW (2010) The SPO1-related bacteriophages. Arch Virol 155: 1547–1561. [DOI] [PubMed] [Google Scholar]
- 28.Marti R, Zurfluh K, Hagens S, Pianezzi J, Klumpp J, et al.. (2013) Long tail fibers of the novel broad host range T-even bacteriophage S16 specifically recognize Salmonella OmpC. Mol Microbiol. [DOI] [PubMed]
- 29. Fouts DE, Rasko DA, Cer RZ, Jiang L, Fedorova NB, et al. (2006) Sequencing Bacillus anthracis typing phages gamma and cherry reveals a common ancestry. J Bacteriol 188: 3402–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Minakhin L, Semenova E, Liu J, Vasilov A, Severinova E, et al. (2005) Genome sequence and gene expression of Bacillus anthracis bacteriophage Fah. J Mol Biol 354: 1–15. [DOI] [PubMed] [Google Scholar]
- 31. Hendrix RW (2009) Jumbo bacteriophages. Curr Top Microbiol Immunol 328: 229–240. [DOI] [PubMed] [Google Scholar]
- 32. Smith MC, Thorpe HM (2002) Diversity in the serine recombinases. Mol Microbiol 44: 299–307. [DOI] [PubMed] [Google Scholar]
- 33. Kilcher S, Loessner MJ, Klumpp J (2010) Brochothrix thermosphacta bacteriophages feature heterogeneous and highly mosaic genomes and utilize unique prophage insertion sites. J Bacteriol 192: 5441–5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kelly G, Prasannan S, Daniell S, Fleming K, Frankel G, et al. (1999) Structure of the cell-adhesion fragment of intimin from enteropathogenic Escherichia coli . Nat Struct Biol 6: 313–318. [DOI] [PubMed] [Google Scholar]
- 35. Fraser JS, Yu Z, Maxwell KL, Davidson AR (2006) Ig-like domains on bacteriophages: a tale of promiscuity and deceit. J Mol Biol 359: 496–507. [DOI] [PubMed] [Google Scholar]
- 36. Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, et al. (2013) Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci U S A 110: 10771–10776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Katsura I (1987) Determination of bacteriophage lambda tail length by a protein ruler. Nature 327: 73–75. [DOI] [PubMed] [Google Scholar]
- 38. Katsura I (1990) Mechanism of length determination in bacteriophage lambda tails. Adv Biophys 26: 1–18. [DOI] [PubMed] [Google Scholar]
- 39. Green BD, Battisti L, Koehler TM, Thorne CB, Ivins BE (1985) Demonstration of a capsule plasmid in Bacillus anthracis . Infect Immun 49: 291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Database matches of predicted proteins encoded by Bacillus anthracis phage Tsamsa.
(DOCX)