Abstract
Most cases of BCR-ABL1-negative myeloproliferative neoplasms (MPNs), essential thrombocythemia, polycythemia vera and primary myelofibrosis are associated with JAK2 V617F mutations. The outcomes of these cases are critically influenced by the transition from JAK2 V617F heterozygosity to homozygosity. Therefore, a technique providing an unbiased assessment of the critical allele burden, 50% JAK2 V617F, is highly desirable. In this study, we present an approach to assess the JAK2 V617F burden from genomic DNA (gDNA) and complementary DNA (cDNA) using one-plus-one template references for allele-specific quantitative-real-time-PCR (qPCR). Plasmidic gDNA and cDNA constructs encompassing one PCR template for JAK2 V617F spaced from one template for JAK2Wild Type were constructed by multiple fusion PCR amplifications. Repeated assessments of the 50% JAK2V617F burden within the dynamic range of serial dilutions of gDNA and cDNA constructs resulted in 52.53±4.2% and 51.46±4.21%, respectively. The mutation-positive cutoff was estimated to be 3.65% (mean +2 standard deviation) using 20 samples from a healthy population. This qPCR approach was compared with the qualitative ARMS-PCR technique and with two standard methods based on qPCR, and highly significant correlations were obtained in all cases. qPCR assays were performed on paired gDNA/cDNA samples from 20 MPN patients, and the JAK2 V617F expression showed a significant correlation with the allele burden. Our data demonstrate that the qPCR method using one-plus-one template references provides an improved assessment of the clinically relevant transition of JAK2 V617F from heterozygosity to homozygosity.
Introduction
The discovery of a mutation in the Janus kinase 2 (JAK2) gene opened a new era in the understanding of BCR-ABL- negative myeloproliferative neoplasms (MPNs) [1]–[5]. An acquired transversion in JAK2 exon 14 (c.1849G>T) that is confined to hematopoietic cells and results in p.Val617Phe (JAK2 V617F) is observed in approximately 90% of patients with polycythemia vera (PV), 50% of essential thrombocythemia (ET) cases and 50% of primary myelofibrosis (PMF) cases [1], [3]. JAK2 V617F impacts the function of the pseudokinase JH2 domain, which normally plays a role in the auto-inhibition of JAK2 kinase activity [4]. In vitro studies have demonstrated that JAK2 V617F leads to a specific phosphorylation associated with the constitutive activation of the tyrosine kinase function [3].
Primarily involved in myeloid development, the JAK2 protein is a non-receptor tyrosine kinase associated with the cytoplasmic regions of several cytokine membrane receptors [6]. JAK2 is activated when these receptors bind to hematopoietic growth factors, and it acts as a molecular intermediary through the constitutive activation of STAT5-, AKT- and ERK-dependent pathways [7], [8].
After the acquisition of JAK2 V617F, loss of heterozygosity (LOH) may occur by the duplication of the mutant allele via mitotic recombination of the short arm of chromosome 9, resulting in homozygosity. Consequently, the quantity of mutant versus wild-type JAK2 may vary significantly, introducing the concept of allele burden. The term homozygosity is employed to indicate patients in whom the level of mutant allele in the test sample is greater than 50% of the total JAK2 (mutant [MT] plus wild type [WT]). The JAK2 V617F burden has been correlated with changes in clinical phenotype and disease complications, such as thrombosis and myelofibrosis [9]–[11]. Homozygosity is associated with a significantly longer duration of disease, treatment with cytoreductive therapy and a higher rate of complications [3]. JAK2 V617F LOH has been observed in approximately 30% of patients with PV and PMF, compared to only 2–4% of patients with ET [12], [13].
Therefore, the accurate estimation of the V617F allele burden and (in particular) the unbiased assessment of the 50% allele burden has gained major clinical relevance in patients with PV, ET and PMF because values significantly greater than 50% guarantee the presence of at least some cells exhibiting LOH and the prognostic consequences associated with this condition. The current methods to analyze the JAK2 V617F allele burden are based on the absolute or disconnected quantification of standards for the MT and WT alleles. Hence, a practical approach to measure the V617F allele burden with a special focus on the accurate assessment of the one-plus-one MT:WT allelic ratio and the associated experimental error is highly desirable in this field.
This work presents a new approach to assess the JAK2 V617F allele burden in gDNA (genomic DNA) and cDNA (complementary DNA from total RNA) samples using one-plus-one template references in a general strategy of allele-specific quantitative real time-PCR (qPCR).
Materials and Methods
Studied Population and Samples
Peripheral blood samples were obtained from a total of 53 patients with MPNs and 20 healthy donors (control group). Twenty of the MPN patients were diagnosed according to the current hematological criteria established by the World Health Organization (WHO) as six PV, five ET and nine PMF cases; these patients were used to test the allele burden and transcript expression of JAK2 V617F for correlation analysis. Another group of 33 cases was used to validate the above method by comparing it with ARMS-PCR, and with two other standard qPCR assays. This study was approved by the local Institutional Ethics Committee (Academia Nacional de Medicina de Buenos Aires). Written informed consent was obtained in all cases. The patients’ characteristics are listed in Table 1.
Table 1. Patient characteristics.
| PV (n = 6) | ET (n = 5) | MF (n = 9) | |
| Males/females | 3/3 | 2/3 | 3/6 |
| Median age (years) | 64 | 58 | 55 |
| Range age (years) | 42–90 | 50–90 | 50–68 |
| Characteristics at diagnosis: | |||
| Hematocrit values (%) | 57.2±2.3 | 42.2±2.3 | 33±0.9 |
| White blood cells, ×109/L | 11.5±2 | 8.9±1.2 | 10.5±2 |
| Neutrophils (%) | 65.8±6,2 | 59±5 | 62.3±7.2 |
| Platelets, x 109/L | 354.2±73.9 | 2943±2100 | 234.1±50.4 |
| Splenomegaly | 1/6 | 0/6 | 4/9 |
| Patients on cytoreductivetreatment | 4/6 | 3/5 | 6/9 |
gDNA and total RNA were extracted from total leukocytes by standard procedures after 3 cycles of lysing red cells from peripheral blood samples. Leukocyte pellets were either treated with TRIzol (Invitrogen, Argentina) for total RNA extraction or with phenol/Tris-HCl (pH: 8) for gDNA extraction. One microgram of total RNA was reverse transcribed into cDNA using random hexamer primers and reverse transcriptase M-MLV (Promega, Biodynamics Argentina). In addition, a gDNA sample from SET-2, a cell line derived from a MPN patient with JAK2V617F heterocigosity, was used to confirm the inaccurateness of using JAK2V617F positive cell lines as standards.
Construction of JAK2 V617F-JAK2 Wild Type (JAK2 MT-JAK2 WT) One-plus-one Template Reference Plasmids
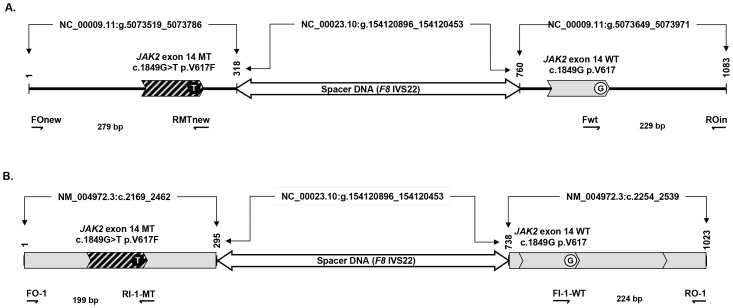
The JAK2 gDNA-MT::WT 1::1 and JAK2 cDNA-MT::WT 1::1 reference constructs consisted of a tripartite structure (i.e., an MT-left arm, a spacer and a WT-right arm) (Figure 1A and 1B). Each construct provided two templates for qPCR amplification: one for JAK2V617F and one for JAK2 WT. These constructs were assembled following a strategy of multiple fusion PCR amplifications with conventional primers and specially designed fusion oligonucleotides (Table 2), as described in detail in Methods S1 and Figure S1.
Figure 1. JAK2 MT:WT 1∶1 reference constructs.
Upper diagram: reference construct from genomic DNA. Bottom diagram: reference construct from complementary DNA. These diagrams show the allele-specific primers used to amplify the mutated allele (MT) and the wild-type allele (WT). Indicated are the GenBank files and the molecular sizes of each segment and of each amplification product.
Table 2. Oligonucleotide primers.
| Name | Sequence (5′->3′) | Reference |
| Construction of cDNA MT:WT 1::1 template reference | ||
| FO-1 | ATTTTTAAAGGCGTACGAAGAGAAGTAG | (14) |
| RO-1 | ATAAGCAGAATATTTTTGGCACATACAT | (14) |
| Up-Sp-cDNA | GAAGTTGCTAAACAGaagaaaccccaggaaacaga | This study. |
| Lo-Sp-cDNA | CTGAATAGTTTCTGTctcagcccctaagtcgtatc | This study. |
| Construction of gDNA MT:WT 1::1 template reference | ||
| FOnew | CATATAAAGGGACCAAAGCACA | This study. |
| FOin | TCCTCAGAACGTTGATGGCAG | (15) |
| ROin | ATTGCTTTCCTTTTTCACAAGAT | (15) |
| Up-Sp-gDNA | CAGAGCATCTGTTTTaagaaaccccaggaaacaga | This study. |
| Lo-Sp-gDNA | GACTGTTGTCCATAActcagcccctaagtcgtatc | This study. |
| Allele specific cDNA primers | ||
| RI-1 | ACCAGAATATTCTCGTCTCCACAaAA | (14) |
| FI-1 | GCATTTGGTTTTAAATTATGGAGTATaTG | (14) |
| Allele specific gDNA primers | ||
| Rmt | GTTTTACTTACTCTCGTCTCCACAaAA | (15) |
| Fwt | GCATTTGGTTTTAAATTATGGAGTATaTG | (15) |
| RMTnew | TTACTTACTCTCGTCTCCACAaAA | This study. |
For the amplification and storage of the qPCR amplification references, the cDNA and gDNA MT-WT one-plus-one template PCR products were cloned into plasmid vector pCR2.1-TOPO (Invitrogen SRL, Argentina) (details of the procedure are provided in the last section of Methods S1).
The cDNA and gDNA JAK2 V617F -JAK2WT one-plus-one template reference plasmids are available for research use only after a Material Transfer Agreement (MTA) form is signed.
Confirmation of the Uniqueness of JAK2 V617F in both the gDNA and cDNA Constructs by BsaXI Restriction Analysis and DNA Sequencing
The JAK2 V617F mutation (c.1849G>T) introduces a single BsaXI restriction site in both gDNA and cDNA constructs. To investigate the presence of a single copy of mutated JAK2 in each construct, BsaXI restriction analysis was performed. Three microliters of PCR products obtained from an aliquot of a 10−3 dilution of the gDNA plasmid with primers FOin and ROin, as well as 3 µL of PCR products from a 10−7 dilution of the cDNA plasmid with primers FO-1 and RO-1, were subjected to BsaXI restriction with 20 units of enzyme in a total volume of 20 µl under the conditions recommended by the manufacturer (New England Biolabs, USA). The restriction products were analyzed using EtBr-stained agarose gel electrophoresis (2%), Figure S2 (E).
In addition, the JAK2 constructs (gDNA and cDNA MT::WT 1::1) were bidirectionally sequenced (with FOin and ROin for the gDNA construct and with FO-1 and RO-1 for the cDNA construct) using the fluorescently labeled chain termination approach (BigDye ABI, Argentina) and an ABI 3130 XL apparatus (Genetic Analyzer from Applied Biosystems). The DNA sequences of MT-arm and WT-arm from the gDNA and cDNA constructs are shown in Figure S2 C and D, respectively.
Primer Specificity and Structures of JAK2 gDNA and cDNA Reference Plasmids
The molecular structures of the gDNA and cDNA reference plasmids were studied using PCR amplification experiments with multiple primer pair combinations (Table 2). Two different annealing temperatures (58°C and 60°C) were evaluated, and 2 µl from a 10−7 dilution of the gDNA and cDNA plasmids was amplified. The following optimized PCR thermocycling protocol was applied: an initial step of 94°C for 2 min; 25 cycles of 94°C for 30 sec, 58°/60°C for 45 sec and 72°C for 1 min, and a final extension step at 72°C for 5 min.
The desired specific structures of the gDNA and cDNA constructs (Figure 1A and 1B) were positively confirmed by the results shown in Figure S3 A and B, respectively. The results demonstrated that only the properly oriented primers produced size-specific PCR amplifications: FOn/RMTn, UpSp-g/LoSp-g and Fwt/ROin for the gDNA plasmid; and FO-1/RI-1, UpSp-c/LoSp-c and FI-1/RO-1 for the cDNA plasmid.
Quantitative Real-time PCR
Quantitative real-time PCR (qPCR) was performed using the LightCycler 2.0 (Roche Diagnostics, Mannheim, Germany), which is based on SYBR Green chemistry. The 20-µl qPCR reaction mixtures contained 5 µl of sample cDNA or 40 ng of gDNA, 1X PCR Mix (LC FastStart DNA Master SYBR Green I, Roche Diagnostics, Argentina), 3.5 mM MgCl2 and 0.25 µM of each primer.
The optimal reaction conditions for amplifying JAK2 V617F and JAK2WT from cDNA templates were 50 cycles of a 4-step PCR (95°C for 5 sec, 58°C for 3 sec, 72°C for 20 sec and 75°C for 1 sec). The optimal conditions for gDNA templates were 45 cycles of a 4-step PCR (95°C for 5 sec, 62°C for 6 sec and 72°C for 12 sec) after an initial denaturation (95°C for 10 min). The allele-specific primer sets used in this study to perform the relative quantification of JAK2 V617F and JAK2WT from the patient cDNA samples were previously published by Vannucchi et al. [14], and the allele-specific primer sets for quantification from patient gDNA samples were modified from a qualitative ARMS-PCR strategy published by Jones et al. [15] (Table 2).
Calibration curves were generated using serial dilutions of the cDNA and gDNA JAK2 V617F::JAK2WT 1::1 reference plasmids to estimate the qPCR amplification efficiencies and to quantify the JAK2 V617F and JAK2WT alleles on gDNA and transcripts within the dynamic range.
Quantification Strategy, Formulas and Error Estimation
The allele burden (AB) magnitude was calculated with the formula MT/(MT+WT) and expressed in arbitrary units related to the dilutions of the MT::WT 1::1 template reference curves. The errors associated with the MT and WT measurements were estimated using a linear regression (data not shown) between the mean and the standard deviation (SD) of each reference template dilution triplicate: 10−5, 10−6, 10−7 and 10−8 (Figure S4). To calculate the AB error (ΔAB) (in SD), the estimated MT and WT SDs were propagated using Gauss’ method of partial derivatives, i.e., ΔAB2 = |δAB/δMT|2ΔMT2+|δAB/δWT|2ΔWT2, which resulted in ΔAB (MT; ΔMT; WT; ΔWT) = [(WT * ΔMT)2+(MT*ΔWT)2]½/(MT+WT)2.
JAK2 V617F Genotyping by the Amplification Refractory Mutation System (ARMS)
Genomic DNA was extracted from total peripheral blood leucocytes obtained from 20 patients with suspected diagnoses of MPNs using phenol-chloroform according to standard procedures. The JAK2 V617F ARMS (amplification refractory mutation system) analysis was performed using a multiplex PCR strategy, as described by Jones et al. [15]. The allele-specific primers contained a mismatch three bases from the 3′ end to maximize allele discrimination. The ARMS-PCR assay was performed using Taq DNA polymerase (Promega, Argentina), 25 ng of genomic DNA substrate and 30 amplification cycles (including a critical annealing temperature of 60°C) under standard amplification conditions. The results were analyzed by agarose gel electrophoresis (3%).
Independent JAK2 V617F Quantification Methods for Validation of the One-plus-one Reference System
Two independent methods were applied to validate our one-plus-one plasmid-based reference system by use of the Pearson correlation statistics. First, a qPCR system based on allele specific Taqman-probe quantification was performed as described Bousquet et al [16]. A second qPCR system based on allele specific amplification, which take advantage of human gDNA samples from a MPN patient with JAK2V617F homozygosity and a healthy blood donor with JAK2WT genotype to achieve the standard curves for qPCR, was performed as it is applied in a number of laboratories worldwide [8]. In order provide accurate standard curves the amount of JAK2 PCR template copy number in both gDNA samples (i.e., JAK2V617F and JAK2WT) was equaled by experiments of PCR amplification analysis on a common reference region in ABL1 exon 3.
Results
Strategy to Assess the JAK2 V617F Allele Burden Using One-plus-one Template References
The JAK2 V617F allele burden (AB) percentage (i.e., AB% = 100×MT/[MT+WT]) represents a weighted average of cells with zero, one or two copies of JAK2 V617F in a given gDNA sample (ABg). The ABg% is largely similar for cDNA samples (ABc) but may be modified by differential JAK2 allele mRNA expression, which is either produced by differential transcription rates of MT and WT or differential mRNA stability. In addition, the eventual contribution of allelic mRNA from enucleated elements in the whole blood samples (e.g., platelets) may introduce another source of variation to the ABc measurements.
ABg and ABc have no units because the units of MT and WT are equal (arbitrary units associated with the copies of the 1∶1 reference plasmid) and, therefore, cancelled. This is not the case when using two independent reference plasmids (MT and WT), whose accuracy in assessing the relative ABg relies upon two independent absolute copy number estimations. Hence, the main objective behind applying a one-plus-one template reference strategy is to reduce the inevitable biases associated with assessing JAK2 V617F AB to approximately 50%, considering that this value is of major clinical significance.
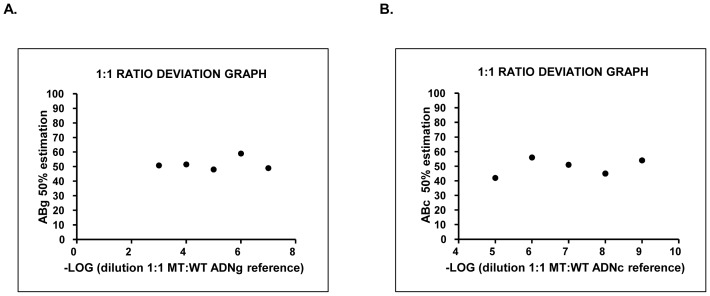
The capacity of the gDNA and cDNA reference plasmids to assess ABg and ABc was investigated by repeated measurements of the same reference plasmid dilution within the dynamic range (defined as the range in which the fluctuation of 50% AB was minimal) (Figure 2). The ABg reference plasmid exhibited a mean of 52.53% and a standard deviation of 4.20% in the range 10−3–10−7 dilution (Figure 2A). Therefore, a limit value of JAK2 V617F ABg of 56.73% (mean+SD) was predetermined to indicate a reliable transition to homozygosity. ABc exhibited a mean of 51.46% and a standard deviation of 4.21% in the range 10−6–10−9 (Figure 2B).
Figure 2. Graphics of deviation from the 1∶1 ratio in the dynamic range of dilutions of the reference plasmids.
Left panel, gAB values for each plasmid dilution of gDNA; right panel, gAB values for plasmid cDNA.
The dynamic range of the ABg analysis, reference plasmid dilutions with minimal errors (10−3–10−7) that corresponded to approximately 1.2×106–1.2 copies, included the average JAK2 copy number in gDNA inputs of 20 ng (6×103 copies), which was used in our qPCR system. Although the dynamic range of ABc (10−6–10−9) corresponded to 9.65×103–9.65 JAK2 template copies, the difficulty in estimating the absolute JAK2 template copies in the cDNA samples prevented a determination of whether the cDNA dynamic range actually contained the absolute template copy number.
Individual values of MT and WT were associated with an intrinsic operative error (SD), which was obtained by interpolating these MT and WT values by a linear regression performed with the reference plasmid dilution triplicates (plasmid dilutions versus SD triplicates). The propagation of these MT and WT errors in the allele burden formula (Materials and Methods) allowed the provision of each AB measurement with its corresponding experimental SD.
Experimental Cutoff for Detecting JAK2 V617F Positive Samples
In addition, to determine the experimental cutoff for discriminating JAK2 V617F-positive from -negative samples using qPCR, we assessed the ABg values from 20 healthy donors and obtained a mean value of 1.04% and an SD of 1.3%. A reliable JAK2 V617F cutoff was based on an ABg threshold of 3.65%, which resulted from the mean plus two SD of the control population.
Experimental Correlation between ARMS-PCR and qPCR Using One-plus-one Template References
To analyze the qPCR method based on one-plus-one references against the widely used qualitative method based on ARMS-PCR [15], 20 DNA samples from patients with a suspected diagnosis of MPNs (10 positive cases and 10 determined to be negative for JAK2 V617F using ARMS-PCR) were analyzed by qPCR in a blind experiment. The negative samples (according to ARMS-PCR) showed ABg values estimated by qPCR (mean ± standard error (SE)) of 1.9±0.6%; the ABg values of the positive samples were 55±9% (Figure 3).
Figure 3. Comparison of ARMS-PCR and qPCR assays.

Ten cases shown to be positive for the JAK2V617F mutation using ARMS-PCR exhibited an allele burden of 55% ±9% (mean ± SE) according to qPCR. Ten negative cases according to ARMS-PCR showed an allele burden of 1.9% ±0.6%, including two cases that were negative by ARMS-PCR and positive by qPCR with a value above our cutoff (>3.65%, estimated from a healthy population).
Using a cutoff value of 3.65%, 18 out of 20 cases showed coincident results by both approaches. Interestingly, 2 of the 10 cases that were negative according to ARMS-PCR were positive according to qPCR, with ABg values of 5.1% and 6.7%. The most likely explanation is that these values scored below the detection limit of ARMS-PCR, which can be estimated on ABg values greater than 6.7%. Therefore, this discrepancy between the two methods may be ascribed to the greater sensitivity of qPCR. Quantitative PCR using one-plus-one template references, as a qualitative tool with a cutoff of 3.65%, allowed the identification of two false negatives by ARMS-PCR and produced no false positives.
Two Independent Correlations Analyzes between Quantitative JAK2 V617F ABg Determinations and the One-plus-one Reference System
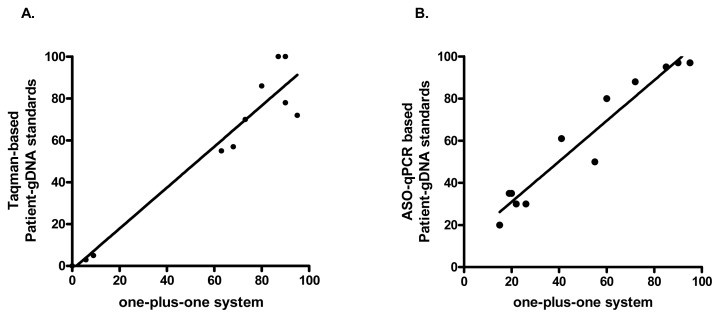
To validate the qPCR method based on one-plus-one reference, 12 gDNA samples from patients with diagnosis of MPNs and healthy controls were subjected to a blind JAK2V617F ABg quantification using the Taqman-based method described by Bousquet et al [16]. Figure 4A shows the highly significant correlation (p<0.0001); Pearson r = 0.968 between both methods.
Figure 4. Validation of the one-plus-one reference system qPCR assay.
A. Twelve MPN cases (n = 10) and negative controls (n = 2) shown highly significant (p<0.0001) correlation (r = 0.967 95%CI = 0.88–0.99) between JAK2V617F ABg measured by an allele-specific Taqman probe-based qPCR method (16) and by the one-plus-one reference system. B. Other group of 12 MPN cases shown highly significant (p<0.0001) correlation (r = 0.968 95%CI = 0.88–0.99) between JAK2V617F ABg measured by a MPN-patient/healthy-control DNA-standardized allele-specific qPCR amplification method (17) and by the one-plus-one reference system. Linear regressions are shown in both graphs.
In addition, gDNA samples from 12 patients with MPNs were used for a second validation of the one-plus-one system against an allele-specific qPCR amplification method using a selected JAK2V617F+/+ patient and a healthy control DNA samples as standards. Equally to the other validation, Figure 4B shows a highly significant (p<0.0001) correlation (Pearson r = 0.967) between both quantitative methods.
In addition, a JAK2-V617F heterozygous patient-derived cell line, SET-2, was evaluated giving a JAK2-V617F allelic burden of 80%.
Allele Burden and the Expression of JAK2 V617F in Patients with MPNs
The application and performance of these new strategies of allele-specific qPCR using one-plus-one template references were tested in 19 cases with JAK2 V617F-positive MPNs: 6 PV, 5 ET and 8 PMF cases. The JAK2 V617F allele burden and RNA expression (mean±SD) were as follows: 62.8±32.1 and 71±32.6 for PV; 53±20.6 and 53.6±21.3 for ET; and 80±14 and 97±3.4 for PMF, respectively (Table 3). This series represents preliminary results from our population and indicates a higher JAK2 V617F allele burden and RNA expression in patients with PMF than in those with PV or ET.
Table 3. Allele burden and expression of JAK2 V617F mutation.
| Patient (N°) | MNP | JAK2 V617F gAB[%]* | JAK2 V617F cAB [%]* |
| 1 | PV | 34.8 | 99.9 |
| 2 | PV | 92.6 | 83.4 |
| 3 | PV | 53.08 | 57.3 |
| 4 | PV | 19.3 | 12.8 |
| 5 | PV | 97.27 | 97.3 |
| 6 | PV | 80.3 | 78.6 |
| 7 | ET | 39.5 | 45.7 |
| 8 | ET | 67.7 | 45.7 |
| 9 | ET | 45.1 | 53.3 |
| 10 | ET | 31.5 | 35.02 |
| 11 | ET | 81.1 | 89.9 |
| 12 | MF | 86.2 | 99.8 |
| 13 | MF | 93.05 | 90.1 |
| 14 | MF | 62.3 | 94.1 |
| 15 | MF | 60.1 | 99.9 |
| 16 | MF | 98.6 | 99.8 |
| 17 | MF | 67.18 | 99.3 |
| 18† | MF | 0.54 | 5.21E-04 |
| 19 | MF | 83.4 | 99.9 |
| 20 | MF | 91.2 | 95.4 |
The propagated error (SD) of the AB from individual values of MT and WT measurements was negligible; therefore, it was not considered (range 6.37×10−8–1.5310−5).
Case N° 18 was negative for the JAK2 V617F mutation.
The patient-paired assessment of the JAK2 V617F allele burden (ABg) and the RNA expression level (ABc) from 19 positive MPNs (Table 3) allowed us to perform a correlation analysis. A positive correlation was observed (Spearman r = 0.53, p = 0.02) even with the inclusion of four cases with increased JAK2 V617F RNA expression levels (outliers) (Figure S5). Interestingly, all four patients in this group of outliers exhibited splenomegaly, increased white blood cell counts and bone marrow fibrosis. Although the small number of cases exhibiting JAK2 V617F overexpression suggests that caution should be exercised concerning reaching general conclusions, this result encourages the performance of further studies.
Discussion
The discovery of mutations in JAK2 has allowed crucial advances in the understanding of the pathophysiology of typical BCR-ABL1-negative MPNs. The mutation JAK2 V617F is a useful molecular marker that has improved and simplified the diagnosis of these disorders. The JAK2 V617F mutation is found in more than 90% of patients with PV and in nearly one-half of those with PMF or ET [17], [18]. Consequently, all the recommended diagnostic algorithms for these entities include qualitative molecular information regarding JAK2 mutations [19]. However, a quantitative study stratifying patients into different quartiles according to their allele burden at diagnosis may be even more appropriate for evaluating the clinical implications of JAK2 V617F load.
A multicenter study demonstrated large discrepancies between the different methods used to quantify the JAK2 V617F mutation [20]. Hence, it is extremely important to employ suitable reference standards to allow an exact quantification of the JAK2 V617F allele burden.
Considering that a blood leukocyte sample represents a potential mixture of cells that are homo/heterozygous for JAK2 V617F, homozygosity cannot be determined when the allele burden is lower than 50% and it can only be warranted when the proportion of the JAK2 V617F allele is significantly greater than 50%. Because the presence of a JAK2 V617F homozygous clone is associated with major clinical consequences, it is crucial to determine the AB turning point (i.e., 50%) without bias.
In addition, a method that permits the exact and reproducible quantification of JAK2 V617F is extremely valuable for the evaluation of patients with MPNs, particularly for the follow-up of patients treated with JAK2 inhibitors.
There is a growing interest in assessing the JAK2 V617F allele burden and in its potential influence on disease phenotype, disease complications and evolution [11]; raising the possibility that homozygosity for the mutant allele is a time-dependent clonal evolution event [4]. The use of different reference standards for quantitative assays may generate discrepancies between AB values.
We provided two independent validations comparing the one-plus-one plasmid-based method with an allele-specific Taqman-probe based qPCR method [16]; and with a method based on curves made from patient samples, using a V617F JAK2 homozygous patient and a JAK2 non-mutated control, as has been used in a number of laboratories worldwide [20]. Recently, the European Leukemia Net (ELN) performed a study for establishing optimal quantitative-polymerase chain reaction assays for routine diagnosis of JAK2-V617F by comparing 12 laboratories: three of them using unpublished ‘in-house’ developed assays and nine of them applying published standard curves using either independently measured plasmid DNA for JAK2-WT and JAK2-V617F or, alternatively, DNA samples from a homozygous JAK2-V617 patient and a healthy donor [21]. Quentmeier et al revealed an active mitotic recombination on JAK2-V617F positive cell lines such as MB-02, MUTZ8, HEL or SET-2 using FISH (fluorescent in situ hybridization) and microsatellite analysis, which associates with genetic imbalances on JAK2 locus and may cause quantification inconsistencies when these cell lines are used for standard curves [22]. In agree with this evidence, we measured an allelic burden of 80% from SET-2, a JAK2V617F heterozygous patient-derived cell line, reflecting an active mitotic recombination in vitro and the lack of reliability to use it for standard curves.
The quantification method presented in this paper would be most appropriate for assessing ABs of approximately 50% because the molecular structure of the construct (one-plus-one) warrants a fixed 1∶1 ratio between the mutated and wild-type JAK2 PCR templates. To the best of our knowledge, no standard for real-time PCR-based quantitative approaches has used the one-plus-one template structure thus far.
As a qualitative tool, our approach using a threshold value (obtained in healthy donors) of 3.65% (mean +2×SD) allowed the positive molecular detection of JAK2 V617F in 19 cases with MPNs and demonstrated a more sensitive detection limit than ARMS-PCR (≥6.7%).
This qPCR-based approach using one-plus-one template references allowed the rapid estimation of the allele burden and RNA expression of JAK2 V617F in 19 positive cases with classical MPNs and detected 13 cases associated with homozygous clones (ABg≥56.73% [mean+SD]).
Although the sample size prevents general conclusions about Argentinian patients with MPNs, a similar trend to those reported in the literature for the JAK2 V617F allele was observed in our group: higher ABg and ABc expression in patients with PMF or PV than in patients with ET.
Although the relative expression level of JAK2 V617F was variable, this depends mainly on the percentage of ABg in the majority of cases. We observed correlations between the levels of JAK2 V617F ABg and ABc in patients with PV, ET and PMF, in agreement with the results reported by Lippert et al. [23] and Tiedt et al. [24]. In contrast to the general trend, we found four outliers (i.e., patients with relatively increased levels of JAK2 V617F transcripts) (Figure S5) who exhibited splenomegaly, high white blood counts and bone marrow fibrosis. The possibility of JAK2 V617F allele overexpression or differential RNA stability in MPNs and the possible clinical consequences are extremely interesting points that merit further investigation.
In conclusion, the qPCR method using one-plus-one template references reported here for JAK2 V617F allele quantification represents a cost-effective tool that is particularly appropriate for measuring the critical AB associated with the transition to the homozygosity state, which is of prognostic value in classical MPN cases.
Supporting Information
JAK2 V617F MT::WT 1::1 reference PCR construction strategy. A. gDNA. The first series of PCR amplifications was performed to obtain products (i), (ii) and (iii) from genomic DNA substrates (references in the gDNA-construct section of the main text). The second series produced (iv) and (v) from PCR substrates (i) plus (ii) and (ii) plus (iii), respectively. The third series produced the full-length gDNA JAK2 V617F MT::WT 1::1 reference construct. The primers and DNA substrates for PCR amplification are indicated. B. cDNA. The first series of PCR amplifications was performed to obtain products (i’), (ii’) and (iii’) from complementary DNA (randomly primed, reverse-transcribed total RNA) substrates (references in the cDNA construct section of the main text). The second series produced (iv’) from substrates (i’) plus (ii’) and (v’) from substrates (ii’) plus (iii’). The third series produced the full-length cDNA JAK2 V617F MT::WT 1::1 reference construct by fusing PCR products (iv’) and (v’). The primers and DNA substrates for each PCR amplification are indicated.
(PPT)
cDNA and gDNA JAK2 V617F MT::WT 1::1 PCR construction steps and MT/WT analysis by DNA sequencing and BsaXI restriction enzyme digestion. A. Agarose gel electrophoresis of all the PCR products and the final product, the JAK2 MT/WT 1::1 gDNA construct. (1) amplimer of JAK2 gDNA V617F MT-arm (453 bp), (2) amplimer of JAK2 gDNA V617 WT-arm (453 bp), (3) DNA-spacer (473 bp, F8 gene part of IVS22), (4) fusion amplimer of (1) (MT-arm), plus (3) (spacer) (775 bp), (5) fusion amplimer of (2) (WT-arm), plus (3) (spacer) (781 bp), (6) final fusion amplimer of (4) (MT-arm+spacer) plus (5) (WT-arm+spacer) (1083 bp). B. Agarose gel electrophoresis of all the PCR products and the final product, the JAK2 MT/WT 1::1 cDNA construct. M indicates 100-bp ladder molecular marker. (1) amplimer of JAK2 cDNA V617F MT-arm (371 bp), (2) amplimer of JAK2 cDNA V617 WT-arm (371 bp), (3) DNA-spacer (473 bp, F8 gene part of IVS22), (4) fusion amplimer of (1) (MT-arm) plus (3) (spacer) (752 bp) and (5) final fusion amplimer of (4) (MT-arm+spacer) plus (spacer+WT-arm) (1023 bp). The final cDNA and gDNA constructs (i.e., A. [6] and B. [5]) were cloned and DNA sequenced. C and D show the relevant DNA sequences of the WT-arm (upper panel) and MT-arm (lower panel) of the gDNA and cDNA recombinant plasmids, respectively. E. Agarose gel electrophoresis showing the BsaXI restriction analysis of both constructs: (1) undigested gDNA, (2) BsaXI-digested gDNA, (3) undigested cDNA and (4) BsaXI-digested cDNA.
(PPT)
Experiments to check the structural specificity of the gDNA (A) and cDNA (B) reference plasmids. The experimental results are shown by the agarose gel electrophoresis analysis of the PCR products. The annealing temperatures used in the PCR amplification and the combined primer pairs are indicated at the top and bottom of each gel image, respectively.
(PPT)
JAK2 V617F and JAK2 WT DNA standard curves. A. cDNA. The upper curves show the PCR amplification cycle versus the fluorescence (530 nm) from triplicates of serial dilutions (i.e., 10−5, 10−6, 10−7 and 10−8) of the JAK2 cDNA MT:WT 1∶1 plasmid. The lower graphs show the corresponding log-transformed standard curves of the cDNA-plasmid concentration (arbitrary units, AUc associated with a specific dilution of the same plasmid) versus the crossing points for the JAK2 V617F mutation (left) and JAK2 WT (right), as indicated. Eff. indicates the efficiency of the real-time PCR amplification. Note that standard curves share the same cDNA-plasmid concentration units (AUc); therefore, these units may be added or canceled in relative quantification equations. B. gDNA. The upper curves show the PCR amplification cycle versus the fluorescence (530 nm) from triplicates of serial dilutions (i.e., 10−3, 10−4, 10−5, 10−6 and 10−7) of the JAK2 gDNA MT:WT 1∶1 plasmid. The lower graphs show the corresponding log-transformed standard curves of the gDNA-plasmid concentration (arbitrary units, AUg associated with a specific dilution of the same plasmid) versus the crossing point for the JAK2 V617F mutation (left) and JAK2WT (right), as indicated. Eff. indicates the efficiency of the real-time PCR amplification. Again, the standard curves share the same plasmid concentration units (AUg); therefore, these may be added or canceled in relative quantification equations.
(PPT)
Correlation analysis between qPCR results using gDNA and cDNA as substrates. There was a significant correlation between the allelic burden and expression levels of the JAK2V617F mutation (Spearman P<0.0002). Four cases with increased JAK2 V617F RNA expression levels (outliers) are indicated.
(PPT)
(DOC)
Acknowledgments
We thank Evangelina Agrielo and Lorena Zanella (Laboratorio de Especialidades Bioquímicas, LEB, Bahía Blanca, Argentina) for their contribution comparing our results with those obtained using the method described by Bousquet et al (2006). We also thank the hematologists Beatriz Moiraghi, Raquel Bengió and Federico Sackman Muriel for providing the patient samples.
Funding Statement
This study was supported by grants from the National Research Council (CONICET) and the National Agency for Promotion of Science and Technology (ANPCyT). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, et al. (2005) Acquired mutations of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 365: 1054–1061. [DOI] [PubMed] [Google Scholar]
- 2. James C, Ugo V, Le Couédic JP, Staerk J, Delhommeau F, et al. (2005) A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 434: 1144–1148. [DOI] [PubMed] [Google Scholar]
- 3. Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, et al. (2005) A gain of function mutation of JAK2 in Myeloproliferative Disorders. N Engl J Med 352: 1779–1790. [DOI] [PubMed] [Google Scholar]
- 4. Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, et al. (2005) Acting mutation in the tyrosine kinase JAK2 in polycithemia vera, essential thrombocythemia and myeloid metaplasia with myelofibrosis. Cancer Cell 7: 387–397. [DOI] [PubMed] [Google Scholar]
- 5. Zhao R, Xing S, Li Z, Fu X, Li Q, et al. (2005) Identification of an acquired JAK2 mutation in polycythemia vera. J. Biol. Chem 280: 22788–22792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Parganas E, Wang D, Stravopodis D, Topham DJ, Marine JC, et al. (1998) JAK2 is essential for signaling through a variety of cytokine receptors. Cell 93: 385–395. [DOI] [PubMed] [Google Scholar]
- 7.Vainchenker W, Constantinescu S (2005) A unique activating mutation in JAK2 (V617) is at the Origin of polycythemia vera and allows a new classification of myeloproliferative diseases. Hematology Am Soc Hematol Educ Program 195–200. [DOI] [PubMed]
- 8. Levine RL, Pardanani A, Tefferi A, Gilliland DG (2007) Role of JAK2 in the pathogenesis and therapy of myeloproliferative disorders. Nat Rev Cancer 7: 673–683. [DOI] [PubMed] [Google Scholar]
- 9. Vannucchi AM, Antonioli E, Guglielmelli P, Longo G, Pancrazzi A, et al. (2007) MPD Research Consortium: Prospective identification of high-risk polycythemia vera patients based on JAK2 V617F allele burden. Leukemia 21: 1952–1959. [DOI] [PubMed] [Google Scholar]
- 10. Carobbio A, Finazzi G, Antonioli E, Guglielmelli P, Vannucchi AM, et al. (2009) JAK2V617F allele burden and thrombosis: A direct comparison in essential thrombocythemia and polycythemia vera. Exp Hematol 37: 1016–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Passamonti F, Rumi E (2009) Clinical relevance of JAK2 (V617F) mutant allele burden. Haematologica 94: 7–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen G, Prchal J (2006) Polycythemia vera and its molecular basis: An update. Best Pract Res Clin Haematol. 19: 387–397. [DOI] [PubMed] [Google Scholar]
- 13. Vannucchi A, Antonioli E, Guglielmelli P, Pardanani A, Tefferi A (2008) Clinical correlates of JAK2V617F presence or allele burden in myeloproliferative neoplasms: a critical reappraisal. Leukemia 22: 1299–1307. [DOI] [PubMed] [Google Scholar]
- 14. Vannucchi AM, Pancrazzi A, Bogani C, Antonioli E, Guglielmelli P (2006) A quantitative assay for JAK2V617F mutation in myeloproliferative disorders by ARMS-PCR and capillary electrophoresis. Leukemia 20: 1055–1060. [DOI] [PubMed] [Google Scholar]
- 15. Jones AV, Kreil S, Zoi K, Waghorn K, Curtis C, et al. (2005) Widespread occurrence of the JAK2 V617F mutation in chronic myeloproliferative disorders. Blood 106: 2162–2168. [DOI] [PubMed] [Google Scholar]
- 16. Bousquet M, Le Guellec S, Quelen C, Rigal-Huguet F, Delsol G, et al. (2006) Frequent detection of the JAK2 V617F mutation in bone marrow core biopsy specimens from chronic myeloproliferative disorders using the TaqMan polymerase chain reaction single nucleotide polymorphism genotyping assay. A retrospective study with pathologic correlations. Hum Pathol 37: 1458–1464. [DOI] [PubMed] [Google Scholar]
- 17. Tefferi A, Terra M, Lasho L, Gilliland G (2005) JAK2 Mutations in Myeloproliferative Disorders. N Engl J Med 353: 1416–1417. [DOI] [PubMed] [Google Scholar]
- 18. Campbell PJ, Green AR (2006) The Myeloproliferative Disorders. N Engl J Med 355: 2452–2466. [DOI] [PubMed] [Google Scholar]
- 19. Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, et al. (2009) The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood 114: 937–951. [DOI] [PubMed] [Google Scholar]
- 20. Lippert E, Girodon F, Hammond E, Jelinek J, Reading NS, et al. (2009) Concordance of assays designed for the quantification of JAK2V617F: a multicenter study. Haematologica 94: 38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jovanovic J, Ivey A, Vannuchi A, Lippert E, Oppliger Leibundgut E, et al. (2013) Establishing optimal quantitative-polymerase chain reaction assays for routine diagnosis and tracking of minimal residual disease in JAK2-V617F-associated myeloproliferative neoplasms: a joint European LeukemiaNet/MPN&MPNr-EuroNet (COST action BM0902) study. Leukemia 27: 2032–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Quentmeier H, MacLeod R, Zaborski M, Drexler H (2006) JAK2 V617F tyrosine kinase mutation in cell lines derived from myeloproliferative disorders. Leukemia 20: 471–476. [DOI] [PubMed] [Google Scholar]
- 23. Lippert E, Boissinot M, Kralovics R, Girodon F, Dobo I, et al. (2006) The JAK2-V617F mutation is frequently present at diagnosis in patients with essential thrombocythemia and polycythemia vera. Blood 108: 1865–1867. [DOI] [PubMed] [Google Scholar]
- 24. Tiedt R, Hao-Shen H, Sobas MA, Looser R, Dirnhofer S, et al. (2008) Ratio of mutant JAK2V617F to wild type Jak2 determines the MPD phenotypes in transgenic mice. Blood 111: 3931–3940. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
JAK2 V617F MT::WT 1::1 reference PCR construction strategy. A. gDNA. The first series of PCR amplifications was performed to obtain products (i), (ii) and (iii) from genomic DNA substrates (references in the gDNA-construct section of the main text). The second series produced (iv) and (v) from PCR substrates (i) plus (ii) and (ii) plus (iii), respectively. The third series produced the full-length gDNA JAK2 V617F MT::WT 1::1 reference construct. The primers and DNA substrates for PCR amplification are indicated. B. cDNA. The first series of PCR amplifications was performed to obtain products (i’), (ii’) and (iii’) from complementary DNA (randomly primed, reverse-transcribed total RNA) substrates (references in the cDNA construct section of the main text). The second series produced (iv’) from substrates (i’) plus (ii’) and (v’) from substrates (ii’) plus (iii’). The third series produced the full-length cDNA JAK2 V617F MT::WT 1::1 reference construct by fusing PCR products (iv’) and (v’). The primers and DNA substrates for each PCR amplification are indicated.
(PPT)
cDNA and gDNA JAK2 V617F MT::WT 1::1 PCR construction steps and MT/WT analysis by DNA sequencing and BsaXI restriction enzyme digestion. A. Agarose gel electrophoresis of all the PCR products and the final product, the JAK2 MT/WT 1::1 gDNA construct. (1) amplimer of JAK2 gDNA V617F MT-arm (453 bp), (2) amplimer of JAK2 gDNA V617 WT-arm (453 bp), (3) DNA-spacer (473 bp, F8 gene part of IVS22), (4) fusion amplimer of (1) (MT-arm), plus (3) (spacer) (775 bp), (5) fusion amplimer of (2) (WT-arm), plus (3) (spacer) (781 bp), (6) final fusion amplimer of (4) (MT-arm+spacer) plus (5) (WT-arm+spacer) (1083 bp). B. Agarose gel electrophoresis of all the PCR products and the final product, the JAK2 MT/WT 1::1 cDNA construct. M indicates 100-bp ladder molecular marker. (1) amplimer of JAK2 cDNA V617F MT-arm (371 bp), (2) amplimer of JAK2 cDNA V617 WT-arm (371 bp), (3) DNA-spacer (473 bp, F8 gene part of IVS22), (4) fusion amplimer of (1) (MT-arm) plus (3) (spacer) (752 bp) and (5) final fusion amplimer of (4) (MT-arm+spacer) plus (spacer+WT-arm) (1023 bp). The final cDNA and gDNA constructs (i.e., A. [6] and B. [5]) were cloned and DNA sequenced. C and D show the relevant DNA sequences of the WT-arm (upper panel) and MT-arm (lower panel) of the gDNA and cDNA recombinant plasmids, respectively. E. Agarose gel electrophoresis showing the BsaXI restriction analysis of both constructs: (1) undigested gDNA, (2) BsaXI-digested gDNA, (3) undigested cDNA and (4) BsaXI-digested cDNA.
(PPT)
Experiments to check the structural specificity of the gDNA (A) and cDNA (B) reference plasmids. The experimental results are shown by the agarose gel electrophoresis analysis of the PCR products. The annealing temperatures used in the PCR amplification and the combined primer pairs are indicated at the top and bottom of each gel image, respectively.
(PPT)
JAK2 V617F and JAK2 WT DNA standard curves. A. cDNA. The upper curves show the PCR amplification cycle versus the fluorescence (530 nm) from triplicates of serial dilutions (i.e., 10−5, 10−6, 10−7 and 10−8) of the JAK2 cDNA MT:WT 1∶1 plasmid. The lower graphs show the corresponding log-transformed standard curves of the cDNA-plasmid concentration (arbitrary units, AUc associated with a specific dilution of the same plasmid) versus the crossing points for the JAK2 V617F mutation (left) and JAK2 WT (right), as indicated. Eff. indicates the efficiency of the real-time PCR amplification. Note that standard curves share the same cDNA-plasmid concentration units (AUc); therefore, these units may be added or canceled in relative quantification equations. B. gDNA. The upper curves show the PCR amplification cycle versus the fluorescence (530 nm) from triplicates of serial dilutions (i.e., 10−3, 10−4, 10−5, 10−6 and 10−7) of the JAK2 gDNA MT:WT 1∶1 plasmid. The lower graphs show the corresponding log-transformed standard curves of the gDNA-plasmid concentration (arbitrary units, AUg associated with a specific dilution of the same plasmid) versus the crossing point for the JAK2 V617F mutation (left) and JAK2WT (right), as indicated. Eff. indicates the efficiency of the real-time PCR amplification. Again, the standard curves share the same plasmid concentration units (AUg); therefore, these may be added or canceled in relative quantification equations.
(PPT)
Correlation analysis between qPCR results using gDNA and cDNA as substrates. There was a significant correlation between the allelic burden and expression levels of the JAK2V617F mutation (Spearman P<0.0002). Four cases with increased JAK2 V617F RNA expression levels (outliers) are indicated.
(PPT)
(DOC)