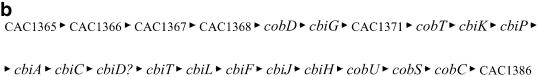Abstract
Prokaryotic genes are frequently organized in multicistronic operons (or transcriptional units, TUs), and usually the regulatory motifs for the whole TU are located upstream of the first TU gene. Although the number of sequenced genomes has increased dramatically, experimental information on TU organization is extremely limited. Even for organisms as extensively studied as Escherichia coli and Bacillus subtilis, TU annotation is far from complete. It therefore becomes imperative to rely on computational approaches to complement experimental information. Here we present a TU map for the obligate anaerobe Clostridium acetobutylicum ATCC 824. This map is largely based on the distance between pairs of consecutive genes but enhanced and refined by predictions of several types of promoters (σA, σE and σF/G) and rho-independent terminator structures. Based on the set of known C.acetobutylicum TUs, the presented TU map offers an 88% prediction accuracy.
INTRODUCTION
Genomic annotation for the ever-growing number of genomes is mostly limited to the level of single genes. Although this is an essential part of the annotation process, there is a need for more complete annotation, which, in prokaryotes, includes the discovery of the fundamental transcription units (TUs). Although the discovery of all single genes of an organism allows the determination of the complete set of intergenic regions (IRs) of a genome, IR prediction alone does not provide any information about TU organization unless specific knowledge about the IRs (length and binding motifs) is used, and this is the aim of this paper. As stated by Gelfand et al. (1) ‘An adequate understanding of cell functioning is impossible without a knowledge of the transcription regulatory circuits that exist in all prokaryotes… The first step in this direction is the identification of transcription regulatory sites (operators), which also helps in deciphering the operon organization of genes in poorly characterized genomes’. Despite the vital importance of TU annotation, it has mainly been confined to Escherichia coli (2–5), Bacillus subtilis (6) and partially for some other organisms (7).
There are three main approaches for TU prediction: distance-based models (8), signal discovery models (2) and order conservation among organisms (7). It is also possible to combine several of these approaches (4–6) or use microarray expression data (4) to predict and/or refine the TU map of an organism. Distance-based methods (3) rely on the different intergenic distances between consecutive genes depending on whether they belong to the same TU or not. In general, very short distances cannot accommodate all the signals and spacers required by a promoter structure, whereas longer intergenic DNA sequences are more likely to include a promoter structure.
Signal discovery methods (2) are based on the discovery of a promoter and/or terminator structure in the intergenic regions using a model. Such models range from simple consensus sequences to RNA structure models. Theoretically, such methods may identify the precise location of the promoter and/or terminator structures. However, lack of accurate predictors (models) for promoters other than for σA make these methods rather inaccurate and, thus, regulation of several important specialized programs (e.g. sporulation) may be difficult to elucidate. The same can be said of the discovery of terminator sequences whereby we are only aware of the existence of predictors for intrinsic termination (9,10) but not for rho-dependent termination.
Order conservation methods (7) are based on the discovery of gene clusters where gene order and orientation are conserved in two or more genomes. The main strength of such methods is that they do not rely on any kind of promoter and/or terminator prediction but only on the presence of homologous proteins between organisms, which makes it a good tool for discovering horizontal transfer events. The quality of the TU map obtained directly relates to the number of genomes used and the evolutionary distance between them. The main drawback with this approach is that implementations are computationally very demanding and can leave unannotated zones if no matches are found.
As stated above, the available literature on the prediction of the complete TU map of an organism other than E.coli is very limited. Furthermore, predictions of TU maps using more than one σ factor have not been reported in the literature. The objective of this paper is to present a complete TU map for Clostridium acetobutylicum ATCC 824. Clostridium acetobutylicum is a Gram-positive, spore-forming, obligate anaerobe with a high A–T base content (72%) that is able to ferment a wide variety of carbohydrates to acids and solvents. Its genome has been sequenced and computer annotated (11), and its physiology extensively studied. DNA array-based transcriptional analysis of this organism has been recently developed and is providing a large-scale understanding of important cellular programs (12–15), such as chemotaxis, motility, sporulation and stress response, for the first time among all clostridia. None of the other closely or distantly related clostridia is understood genetically (let alone genomically) any better than C.acetobutylicum, and thus one has to rely on B.subtilis as a prototypical organism for such studies. However, B.subtilis is not a strict anaerobe and is only distantly related to C.acetobutylicum. For example, although their sporulation/differentiation programs appear to be similar, many physiological and genetic differences exist (11,16). Thus, this organism has the potential to become the model organism for the analysis of not only other solventogenic clostridia, but also for clostridial pathogens such as C.botulinum and C.perfringens. In order to construct a TU map of C.acetobutylicum, we combined a distance-based method with several predictors of promoter and rho- independent termination sequences. The full TU map and additional data are available at NAR Online and at our website www.papoutsakisresearch.northwestern.edu as plain text gbk files that can be read using the last development version of Artemis (17) (http://www.sanger.ac.uk/Software/Artemis/).
METHODS, SEQUENCES AND ALGORITHMS
Computational resources
All scripts and programs were run on a dual booting (Windows XP/Linux Red Hat 9.0) Toshiba Satellite 2435-S255 laptop with a Pentium 4 CPU running at 2.40 GHz with 512 Mbytes of RAM.
Software
Matlab (version 5.3) scripts were created to extract the upstream IRs between pairs of genes and find motifs given a consensus sequence.
The RNAMotif program (18) was downloaded from http://www.scripps.edu/case. The RNAFold program is part of the Vienna RNA package version 1.4 (19). This package was downloaded from http://www.tbi.univie.ac.at/RNA/.
Sequences
The complete genome, coding sequences and annotation files for C.acetobutylicum ATCC 824 (11) were downloaded from the Genome Therapeutics Corporation website (http://www.genomecorp.com/programs/sequence_data_clost.shtml). The complete genome, coding sequences and annotation files for E.coli K12 (20) and B.subtilis (21) were downloaded from two NCBI ftp sites (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/Escherichia_coli_K12/ and ftp://ftp.ncbi.nih.gov/genomes/Bacteria/Bacillus_subtilis/), respectively.
A list of 100 experimentally known B.subtilis transcriptional units (22) was downloaded from http://www.cib.nig.ac.jp/dda/taitoh/bsub.operon.html. A list of 359 known E.coli transcriptional units was obtained from Sabatti et al. (4). The combined set of 459 TUs will be referred as our distance training set.
Data preparation
The location of the IR between pairs of consecutive C.acetobutylicum ATCC 824 genes was recorded and tabulated. Figure 1 shows how the IRs are defined for all possible combinations of coding strands between consecutive genes. Using a Matlab program, only IRs with a length greater than zero were extracted from the sequenced C.acetobutylicum genome.
Figure 1.
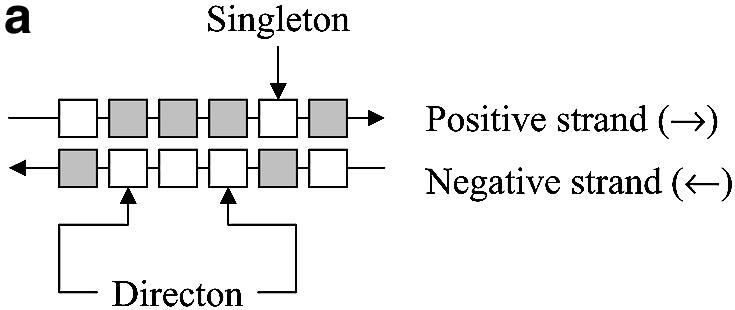
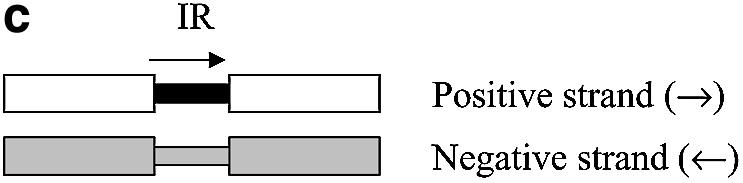
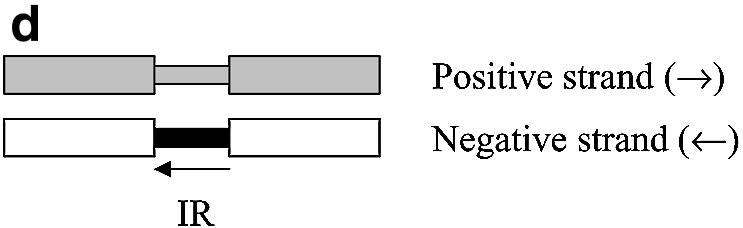
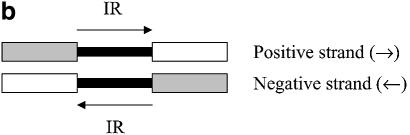
IR examples. Open boxes represent the coding region of a gene, and gray boxes represent non-coding regions. Small black boxes represent the IR region of the gene of interest. (a) A directon is a collection of adjacent genes on the same DNA strand with no intervening genes on the complementary strand. A singleton is a directon made up of only one gene. Definition of an IR: (b) at the start of a directon, both genes share the same IR but coded by different strands; (c) when the positive strand codes two consecutive genes; and (d) when two consecutive genes are coded by the negative strand.
Hidden Markov model (HMM) for σA, σE and σF/σG
Three HMMs were used to locate the position of the σA, σE and σF/σG promoters in the C.acetobutylicum genome. All three HMMs were generated using the DOS/Cygwin port of HMMER (23) v 2.2g (http://hmmer.wustl.edu/). The E.coli promoter regions were obtained from Hershberg et al. (24) (http://bioinfo.md.huji.ac.il/marg/promec), whereas the σA promoter regions of B.subtilis came from DBTBS (25). It should be noted that the E.coli promoter set also contains a small number of other promoters different from σA. The σE promoter regions were obtained from Eichenberger et al. (26). The σF promoter regions were obtained from DBTBS (25) (http://dbtbs.hgc.jp/) for B.subtilis and from Park et al. (27) for E.coli. Although all the promoters chosen were of the σF type, there exists some experimental evidence (28) that some σF promoters can also be recognized by σG, and vice versa. In what follows, we will refer to this HMM as the σF/σG HMM so as to address both possibilities.
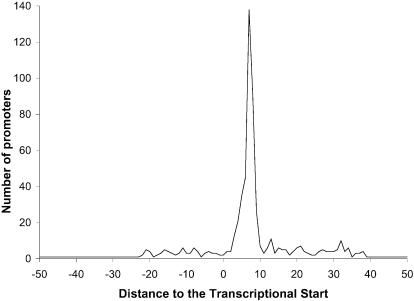
The σA HMM was created through an iterative process, where the properly aligned promoter regions of the i-th iteration were used as the HMM training set for the (i + 1)-st iteration. The determination of whether a promoter region was properly aligned was made by measuring the distance from the last nucleotide of the –10 part of the promoter to the transcriptional start site (TSS) for the highest scoring alignment of each sequence. If this distance was between 3 and 10 nt, the sequence was considered to be properly aligned. A total of 608 promoter regions was used, but 17 of them were discarded because the TSS location has not been experimentally determined, yielding only 591 (608 – 17) useful promoter regions. The information available for some of the TSSs indicates that they cover more than one nucleotide. For instance, the TSS for the first promoter of the E.coli gene pyrG spans seven consecutive nucleotides. In such cases, only the position of the first nucleotide was used in subsequent calculations. When the 591 useful promoter regions were aligned using the σA HMM, only 375 of them were properly aligned (63.5% of the 591 useful promoter regions). The distribution of the distances to the TSS for the whole set of 591 useful promoter regions is shown in Figure 2. With each alignment, the HMM returns a negative log-likelihood score (29), which represents how different the alignment of that sequence is from the alignment of a random sequence. This random sequence is generated from a previously specified null model. In our case, and given that we combine sequences from two microorganisms and that the resulting HMMs will be applied to another organism, we used a null model that assigns a 25% probability of the occurrence at each position to each one of the four nucleotides. Positive scores indicate that the identified promoter is clearly different from the null model, whereas negative scores imply the opposite.
Figure 2.
Distribution of distances from the last nucleotide of the –10 region to the transcriptional start site for the σA HMM predictions. Computations were based on the training set of 591 experimentally known E.coli and B.subtilis promoters discussed in the text.
A total of 59 genes with experimentally verified σE promoters regions from B.subtilis (26) were used to generate the σE HMM. Ten of these genes have two σE promoter regions, thus increasing the number of promoter regions used to generate the σE HMM to 69. We will refer to these 69 sequences as the σE HMM training set. Given the small number of sequences comprising the σE HMM training set, the IRs for all 59 B.subtilis genes in the training set were aligned using the σE HMM, in order to assess the degree to which the σE HMM captures the σE concept. For 49 of these 59 genes (83.1%), the best alignment matches the experimentally determined σE promoter region, yet only 22 out of the 49 successful alignments resulted in a positive score.
A total of 13 genes with experimentally verified σF/σG promoter regions from B.subtilis (25) and 12 from E.coli (27) were used to generate the σF/σG HMM. We will refer to these 25 sequences as the σF/σG HMM training set. As before, and given the small number of sequences in the σF/σG HMM training set, the IRs for all 25 genes in the training set were aligned using the σF/σG HMM, in order to assess the degree to which the σF/σG HMM captures the σF/σG promoter. In 21 cases (87.5 %), the best alignment matches the experimentally determined σF/σG promoter region, yet only 14 out of the 25 successful alignments show a positive score.
Finally, we estimated the degree of cross-talk among the designed HMMs by using each HMM’s training set as the input for the other two HMMs. In the case of the σA promoters, they were only seven cross-recognitions (three σA promoters were also recognized as σE promoters and four as σF promoters), whereas two promoters in the σE training set were also recognized as σF promoters. There were no cross-recognitions when the σF/σG training set was used as the test input.
Inter-TU and intra-TU distances
The IR length of each sequence in the distance training set was classified as intra-TU (when the IR separates two genes belonging to the same TU), inter-TU (when the IR separates genes in different TUs but both share the same coding strand) or inter-directon (when the IR separates genes coded by different strands) (Fig. 1). Usually the IR of the first gene of a directon signals the start of a TU, although there are some TUs with genes in more than one directon. In such cases, the directon between both parts of the TU is usually a singleton and it greatly overlaps one or more of the genes of the TU. Such events are uncommon; the ECOCYC database (30) contains only one relevant case, the metT-leuW-glnUW-metU-glnVX operon that overlaps with three open reading frames in the opposite direction. Thus, without loss of generality, we can assume that the IR belonging to the first gene of a directon contains at least one functional promoter and that its length is longer than the usual single promoter IR. This atypical length is due to the presence of the promoter of the neighbor directon as shown in Figure 1b, where the IR of the first gene of a directon is shared between two genes. Each gene has its own promoter (understood as the region where all the transcriptional signals are located), but each one is coded in a different strand. As it is not possible to clearly define where the IR of each gene ends and given the possibility that these zones could be overlapping to some extent, inter-directon distances were not used to calculate the inter-TU and intra-TU distance distributions.
We applied the above IR definitions to the distance training set, and we found 887 intra-TU distances (577 from E.coli and 310 from B.subtilis) and 502 inter-TU distances (372 from E.coli and 130 from B.subtilis). The number of inter-TU distances is greater than the number of TUs in the distance training set because each TU is flanked by two inter-TU distances, one separating the first gene of the TU from the previous one and another separating the last gene of the TU from the next one. As stated before, only inter-TU distances separating genes from the same directon were used.
Experimentally known C.acetobutylicum TUs
A list of the experimentally known TUs for C.acetobutylicum was compiled based on Medline referenced literature and it is available at NAR Online and at our website http://www.papoutsakisresearch.northwestern.edu.
Intrinsic termination prediction
Using a procedure similar to the one described previously, the position of the intergenic region downstream of each gene was tabulated, and those with a length greater than zero were extracted using a Matlab program. Potential intrinsic terminator structures were modeled after Lesnik et al. (10), and the RNAMotif (18) program was used to scan the previously extracted downstream intergenic regions for suitable candidates. The structure and ΔG° of putative terminators were assessed using RNAfold (19). ΔG° for the DNA–RNA hybrid was calculated as in Sugimoto et al. (31).
RESULTS AND DISCUSSION
Distance-based probability function generation
The 887 intra-TU distances and 502 inter-TU distances from the distance training set were separately ranked by value and combined in several bins. Each bin was created by adding consecutive intra-TU and inter-TU distances until the number of distances of each set is represented at least a certain number of times, which we call the number of occurrences. Then the probability of occurrence of intra-TU distances was calculated and it was assumed that it was representative of the probability of finding an intra-TU distance in the whole interval of IR distances spanned by that bin. Assuming that there must be a progressive transition from IR lengths mainly separating genes that belong to the same TU (short distances) to IR lengths that mainly separate genes belonging to different TUs (long distances), the calculated probabilities were fitted using a standard sigmoidal function (equation 1) in order to smooth the computational results and lower the dependence of the outcome on a particular training set.

where p(d) represents the probability that two consecutive genes coded by the same strand belong to the same TU, d is the intergenic distance between two consecutive genes and k1, k2 and k3 are parameters.
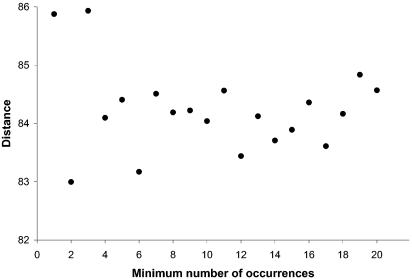
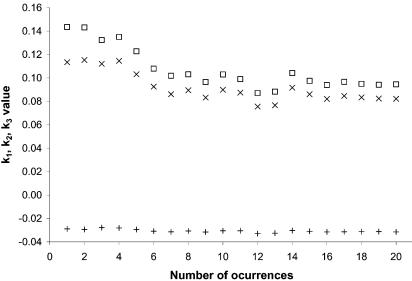
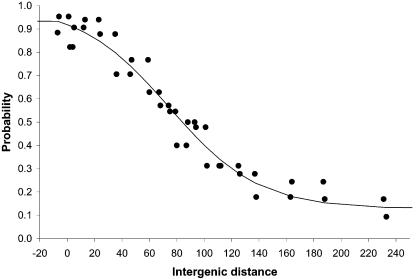
Previous reports (3) have used point-by-point plots instead of fitting the results to an analytical expression, thus making the process very sensitive to the particular set of IR distances used. As can be seen from Figure 3, the IR length required for a 50% probability that two consecutive genes coded by the same strand belong to the same TU is ∼84 bp. At the same time, the values of k1, k2 and k3 (Fig. 4) remain fairly constant for a number of occurrences higher than 8–10, thus suggesting that the number of occurrences is not a significant parameter in the fitting process.
Figure 3.
Distance between two genes that have a 50% probability of belonging to the same TU as a function of the minimum number of occurrences (see text for an explanation). Calculations are based on the training set of 887 intra-TU distances and 502 inter-TU distances from E.coli and B.subtilis as described in the text.
Figure 4.
Variation of the fitting parameters (equation 1) for the distance-based map as a function of the minimum number of occurrences. Crosses indicate parameter k1 values, open boxes indicate parameter k2 values and plus signs indicate parameter k3 values. This graph is based on the training set of 887 intra-TU distances and 502 inter-TU distances from E.coli and B.subtilis as described in the text.
Distance-based TU map creation
Based on Figure 4, the set of values chosen to estimate the probabilities for a TU start for the full set of C.acetobutylicum ATCC 824 IRs was k1 = 0.09, k2 = 0.10, k3 = –0.03. The distance-based probability function is shown in Figure 5 where the points were generated using a number of occurrences of 10. As stated earlier, it was assumed that TUs cannot have genes in different directons, and also that two consecutive genes coded by the same strand belong to the same TU if the intergenic distance between them yields a value of 0.5 or greater (≥50% probability) when used in equation 1. Given the flexibility exhibited by biological systems, this cut-off of 50% should be used with some latitude. We suggest that any IR length that yields a probability in the 40–60% range be catalogued as inconclusive, unless additional independent data exist to validate its classification.
Figure 5.
Probability that two genes belong to the same transcriptional unit given the intergenic distance between them. This graph is based on the training set of 887 intra-TU distances and 502 inter-TU distances from E.coli and B.subtilis as described in the text, with the number of occurrences set to 10.
Accuracy of prediction
The whole set of IR distances from the training set was scored using the distance-based probability function. Using a cut-off of 50%, 88% of the intra-TU distances and 74% of the inter-TU distances from the distance training set were classified properly. Only 8% of the distance training set shows a probability value in the range of 40–60%. Overall and applying the strict 50% criterion, 83.4% of the predictions based on the IR length were correct whereas 9.3% were false positives (the distance-based method returns a probability ≥50% that two consecutive genes belong to the same TU, but this contradicts the experimental evidence) and 7.3% were false negatives (the distance-based method predicts that the IR length is long enough to accommodate a promoter, but there is no experimental evidence to support it).
The probability of belonging to the same TU was calculated for each one of the 72 genes in the set of experimentally known C.acetobutylicum TUs; 87.5% of their IR lengths were predicted correctly, in close agreement with reported results (3) for E.coli.
Clostridium acetobutylicum TU map based on IR length
When applied to the C.acetobutylicum genome, a total of 2191 TUs were assigned, with 916 of them corresponding to either singletons or the start of a directon. In only 22 cases was a directon start or singleton not properly detected by the distance-based method as the start of a new TU.
σA, σE and σF/σG location using HMM
The HMMs described in the Methods were used to discover potential promoters in the C.acetobutylicum IRs, and any hit with a score equal or below zero was discarded. A total of 1537 potential σA, 696 potential σE and 621 potential σF/σG HMM promoters were found.
Intrinsic termination prediction
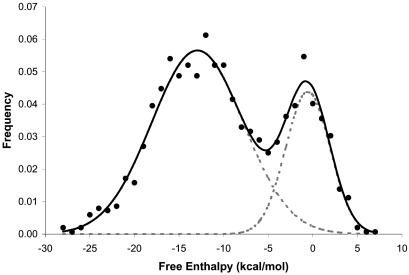
The free enthalpy distribution of the resulting intrinsic terminators is shown in Figure 6, which suggests the presence of two distinct overlapping populations, one centered around –0.6 kcal/mol and another around –12.9 kcal/mol, showing a point of intersection around –4.3 kcal/mol. Lesnick et al. (10) used a set of 109 intrinsic terminators with a free enthalpy of –4.0 kcal/mol or lower as a training set. The striking similarity between this –4.0 kcal/mol cut-off and the point of intersection of Figure 6 could be due to the use of the same model. However, given the clear difference between the two frequency distributions, it seems more likely that –4.0 kcal/mol could be the border between naturally functional intrinsic terminators and random occurrences due to the nucleotide distribution of the genome.
Figure 6.
Free enthalphy distribution for the computationally predicted potential intrinsic terminator structures in the C.acetobutylicum genome. For illustrative purposes, a potential overall frequency distribution (continuous line) and two partial frequency distributions (discontinuous gray lines) are shown.
Overall, there are 996 different genes that account for 1095 potential intrinsic terminators with a free enthalpy equal to or lower than –4.0 kcal/mol. Of these, 904 genes (82.6%) had only one potential intrinsic terminator, 85 genes (7.8%) had two potential intrinsic terminator, and seven genes (0.6%) had three potential intrinsic terminators. Among all of them, there are 66 pairs (12.1%) of potentially bi-directional terminator structures.
Combination of methods
In order to obtain the most accurate possible TU map for C.acetobutylicum ATCC 824, we combined the methods discussed so far as follows. (i) We located all singletons and first genes in each directon in the transcription sense. A TU starts in each of these genes. At this point, the TU map is composed of 916 different TUs. (ii) Using the distance-based method, we located pairs of genes with a probability lower than 50% of belonging to the same TU. A TU starts in each of these genes. After this step, the number of TUs increases to 2191. (iii) Those genes with a distance-based probability between 50 and 60% and either a promoter in their upstream region or an intrinsic terminator in front of their promoter were also chosen as the start of a TU. This third step increases the final number of TUs to 2268 (2140 on the chromosome and 128 in the pSOL1 megaplasmid).
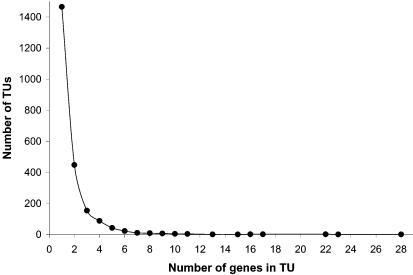
Figure 7 shows the distribution of genes per TU in the proposed TU map. Of the predicted TUs, 64.6% contain only one gene, and 91.2% of the TUs are composed of three or fewer genes. At the other extreme, there are 19 TUs containing 10 or more genes. The longest TU predicted contains 28 genes starting at CAC3135 and ending at CAC3108, and among them, 23 encode ribosomal proteins. Another long TU contains 22 genes (from CAC1365 to CAC1386) encoding enzymes for cobalt metabolism.
Figure 7.
Distribution of genes per TU in the proposed C.acetobutylicum TU map.
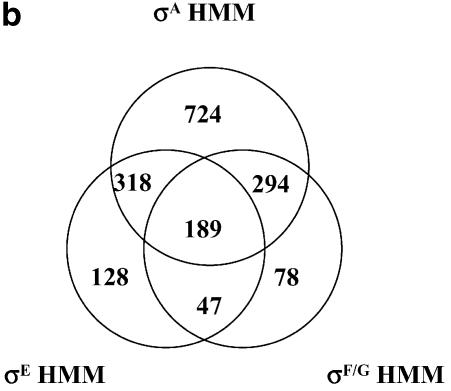
Figure 8a shows a Venn diagram with the contribution of each of the methods to the total number of TUs. There are 308 TUs only supported by the distance-based method (probability that two consecutive genes belong to the same TU of ≥50%). A total of 152 TUs are supported by the distance-based method and a terminator structure in front of its first gene. Thirty TUs are only supported by the existence of a terminator structure in front of its first gene and a distance-based probability between 50 and 60% that its first gene and the previous one belong to the same TU. Thirty-five TUs are only supported by the finding of an HMM-based promoter and distance-based probability between 50 and 60% that its first gene and the previous one belong to the same TU. A total of 1097 TUs are supported by the distance-based method and an HMM promoter. Twelve TUs are supported by an HMM promoter, a terminator and distance-based probability between 50 and 60%. There are 634 cases which are supported by the distance-based method (P = 50%), a terminator and an HMM promoter. Figure 8b show the details of the 1778 TUs with a known promoter (35 + 1097 + 12 + 634 = 1778) in the final TU map. A total of 724 genes have a unique σA promoter, 128 a unique σE promoter and 78 a unique σF/G promoter. We have been able to find two or more potential promoters in a total of 848 TUs. It is very likely that some of these TUs have more than one promoter either of the same kind or of a different kind. However, it should be emphasized that the promoters proposed here indicate a high probability of being factual rather than a fact. Experimental data and/or more sophisticated computational methods are needed to confirm or reject such predictions. A more detailed table covering the results is available as Supplementary Material and at our website http://www.papoutsakisresearch.northwestern.edu.
Figure 8.
(a) Contribution of the different methods to the final TU map. (b) Venn diagram of the final distribution of potential promoters in the proposed C.acetobutylicum TU map.
It should be noted that so far we have been unable to assign a promoter to 490 TUs. A total of 308 of these TUs were assigned in steps (i) and (ii), but the HMMs were unable to recognize any promoter in the presumed promoter region of these genes. The remaining 182 TUs were assigned in step (iii) and belong to genes with a distance-based probability between 50 and 60% with an intrinsic terminator just upstream the first gene in the TU. They could be TUs under the control of σ factors other than σA, σE and/or σF/G, or the promoter might have a longer separation between the –35 and –10 regions. For instance, promoters with active 0A boxes [the binding sites of the Spo0A protein, the master regulator of differentiation in bacilli, clostridia an other organisms (32)] often exhibit poor conservation and longer distances than their counterparts without 0A boxes (33). Also there are 12 predicted σA promoters, 14 predicted σE promoters, 13 predicted σF/G promoters and 168 predicted terminators located in the IR of a pair of genes that our distance-based method assigns as belonging to the same TU with >60% probability. These high numbers of potential terminator structures that remain unassigned could be either real structures, random occurrences due to the overlapping of both intrinsic terminator populations, or intrinsic terminators that only appear in the absence or presence of appropriate stimuli.
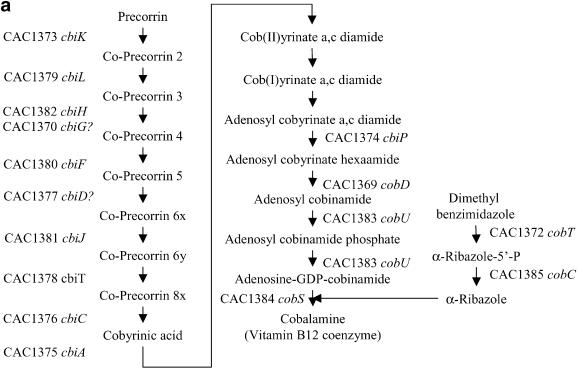
We predicted the TU structure of the C.acetobutylicum ATCC 824 genome by combining intergenic distance, promoter prediction and rho-independent terminator prediction. It is not possible to conduct a TU by TU explanation of the results mainly due to the lack of experimental data on most of the predicted TUs and also due to the large size of the data set. We chose to discuss just two interesting TU cases. The first one shows the B12 biosynthetic pathway that is almost exclusively encoded on a 22 gene-long TU. The second case demonstrates how a pathogenic clostridium can be used to gain insight into its non-pathogenic counterparts such as C.acetobutylicum.
Genes CAC1365–CAC1386 form a 22 gene-long TU related to cobalt metabolism that contains most of the anaerobic biosynthetic pathway of vitamin B12 coenzyme (cobalamine) including some genes encoding a cobalt transport system. Large operons related to B12 metabolism have been reported in the past (34), and recently a comparative genomics study among prokaryotes regarding the metabolism and regulation of vitamin B12 has been published (35). Figure 9 shows the cobalamine biosynthetic pathway [as presented in Rodionov et al. (35)] with the systematic name for each gene in the TU. We have been able to find two probable promoters upstream of the first gene of the TU, a σA-type and a σF-type promoter. The actual annotation of the C.acetobutylicum genome contains three genes of unknown function, CAC1366, CAC1371 and CAC1386, and a gene CAC1369 (hisC) apparently not related to cobalamin metabolism. It can be shown (35) that CAC1366, CAC1369 and CAC1371 are in fact related to cobalamine metabolism. Our results predict that CAC1386 also belongs to the same TU, although apparently it is not related to the anaerobic biosynthetic pathway of vitamin B12. The current annotation for CAC1386 is a ‘Zn-dependent hydrolase, glyoxylase family’. Glyoxalases usually have two zinc ions per molecule needed for catalytic activity (36).
Figure 9.
(a) Cobalamine biosynthetic pathway based on the KEGG database (39). The systematic names of the C.acetobutylicum genes of the cobalamine operon are shown. Conversion of Cob(II)yrinate a,c diamide into Cob(I)yrinate a,c diamide is carried out by CAC0018 (not shown). The gene or genes responsible for the conversion of Cob(I)yrinate a,c diamide into adenosyl cobyrinate a,c diamide are unknown. (b) Genome organization of the B12 TU in C.acetobutylicum. CAC1365, CAC1366, CAC1367 and CAC1368 encode the high-affinity-ATP-dependent cobalt transport system cbiMNQO, CAC1371 encodes an l-threonine kinase needed for the biosynthesis of l-threonine-3P which is a substrate for CobD. The function of CAC1386 is still unknown, but it might encode the conversion of Cob(I)yrinate a,c diamide into adenosyl cobyrinate a,c diamide.
Flagellar and chemotaxis genes in C.acetobutylicum are organized in several long TUs, CAC2225–CAC2215 (CAC2225-cheWDBRACYW-fliMY, with a potential σF promoter upstream of CAC2225), CAC2214–CAC2204 (flgm-CAC2213-flgKL-CAC2210-csrA-CAC2208-CAC2207-fliSD-CAC2204 with potential σF and σA promoters upstream of flgm), CAC2165–CAC2155 (flgBC-fliEFGHLJKD-CAC2155, with potential σF, σE and σA promoters upstream of CAC2155) and CAC2154–CAC2139 (flgE-flbD-fliLZPQR-flhAF-CAC2145-CAC2144-sigD-CAC2142-CAC2141-flgG-flgG, with a potential σE promoter upstream of CAC2154). Genes identified by their systematic name (CACXXXX) were not assigned to a known flagellar/chemotaxis protein when the C.acetobutylicum genome was annotated in 2001 (11). However, based on a BLAST (37) search against the C.tetani genome annotated in 2003 (38), we have been able to annotate some of these uncharacterized genes, which show a high degree of homology to flagellar proteins in C.tetani, as is shown in Table 1. The organization of the genes included in these four TUs is very similar between the two genomes regarding gene order and even at a TU level (a complete table regarding these four TUs and their arrangement can be found as Supplementary Material).
Table 1. Clostridium acetobutylicum genes (with an unknown function) inside flagellar operons and their C.tetani counterparts.
| Gene name | C.acetobutylicum | C.tetani | E-value | C.tetani function |
|---|---|---|---|---|
| CAC2225 | CAC2225 | CTC01736 | 1 × 10–108 | Conserved protein |
| CAC2213 | CAC2213 | CTC01725 | 5 × 10–20 | Conserved protein |
| CAC2210 | CAC2210 | CTC01721 | 9 × 10–32 | Conserved protein |
| CAC2208 | CAC2208 | CTC01720 | 2 × 10–11 | Flagellin |
| CAC2207 | CAC2207 | No match | ||
| CAC2204 | CAC2204 | CTC01716 | 4 × 10–04 | Hypothetical protein |
| CAC2155 | CAC2155 | CTC01668 | 5 × 10–30 | Putative flagellar hook-associated protein |
| CAC2145 | CAC2145 | CTC01655 | 1 × 10–85 | Flagellar synthesis regulator fleN |
| CAC2144 | CAC2144 | CTC01654 | 4 × 10–26 | Flagellar protein |
| CAC2142 | CAC2142 | CTC01652 | 6 × 10–04 | Conserved protein |
| CAC2141 | CAC2141 | No match |
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants BES-9905669 and BES-0331402 from the National Science Foundation.
REFERENCES
- 1.Gelfand M.S., Koonin,E.V. and Mironov,A.A. (2000) Prediction of transcription regulatory sites in Archaea by a comparative genomic approach. Nucleic Acids Res., 28, 695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yada T., Nakao,M., Totoki,Y. and Nakai,K. (1999) Modeling and predicting transcriptional units of Escherichia coli genes using hidden Markov models. Bioinformatics, 15, 987–993. [DOI] [PubMed] [Google Scholar]
- 3.Salgado H., Moreno-Hagelsieb,G., Smith,T.F. and Collado-Vides,J. (2000) Operons in Escherichia coli: genomic analyses and predictions. Proc. Natl Acad. Sci. USA, 97, 6652–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabatti C., Rohlin,L., Oh,M.-K. and Liao,J.C. (2002) Co-expression pattern from DNA microarray experiments as a tool for operon prediction. Nucleic Acids Res., 30, 2886–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bockhorst J., Qiu,Y., Glasner,J.D., Liu,M., Blattner,F.R. and Craven,M. (2003) Predicting bacterial transcription units using sequence and expression data. Bioinformatics, 19, i34–i43. [DOI] [PubMed] [Google Scholar]
- 6.de Hoon M.J.L., Imoto,S., Kobayashi,K., Ogasawara,N. and Miyano,S. (2004) Predicting the operon structure of Bacillus subtilis using operon length, intergene distance and gene expression information. PSB Online Proceedings, 9, 276–287. [DOI] [PubMed] [Google Scholar]
- 7.Ermolaeva M.D., White,O. and Salzberg,S.L. (2001) Prediction of operons in microbial genomes. Nucleic Acids Res., 29, 1216–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreno-Hagelsieb G. and Collado-Vides,J. (2002) A powerful non-homology method for the prediction of operons in prokaryotes. Bioinformatics, 18, S329–S336. [DOI] [PubMed] [Google Scholar]
- 9.d'Aubenton-Carafa Y., Brody,E. and Thermes,C. (1990) Prediction of rho-independent Escherichia coli transcription terminators: a statistical analysis of their RNA stem–loop structures. J. Mol. Biol., 216, 835–858. [DOI] [PubMed] [Google Scholar]
- 10.Lesnik E.A., Sampath,R., Levene,H.B., Henderson,T.J., McNeil,J.A. and Ecker,D.J. (2001) Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res., 29, 3583–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nölling J., Breton,G., Omelchenko,M.V., Makarova,K.S., Zeng,Q., Gibson,R., Lee,H.M., Dubois,J., Qiu,D., Hitti,J. et al. (2001) Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol., 183, 4823–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomas C.A., Welker,N.E. and Papoutsakis,E.T. (2003) Overexpression of groESL in Clostridium acetobutylicum results in increased solvent production and tolerance, prolonged metabolism and changes in the cell’s transcriptional program. Appl. Environ. Microbiol., 69, 4951–4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomas C.A., Alsaker,K.V., Bonarius,H.P.J., Hendriksen,W.T., Yang,H., Beamish,J.A., Paredes,C.J. and Papoutsakis,E.T. (2003) DNA array-based transcriptional analysis of asporogenous nonsolventogenic Clostridium acetobutylicum strains SKO1 and M5. J. Bacteriol., 185, 4539–4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tummala S.B., Junne,S.G. and Papoutsakis,E.T. (2003) Antisense RNA downregulation of coenzyme A transferase combined with alcohol-aldehyde dehydrogenase overexpression leads to predominantly alcohologenic Clostridium acetobutylicum fermentations. J. Bacteriol., 185, 3644–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tummala S.B., Junne,S.G., Paredes,C.J. and Papoutsakis,E.T. (2003) Transcriptional analysis of product concentration driven changes in cellular programs of recombinant Clostridium acetobutylicum strains. Biotechnol. Bioeng., 84, 842–854. [DOI] [PubMed] [Google Scholar]
- 16.Harris L.M., Welker,N.E. and Papoutsakis,E.T. (2002) Northern, morphological and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824. J. Bacteriol., 184, 3586–3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rutherford K., Parkhill,J., Crook,J., Horsnell,T., Rice,P., Rajandream,M.-A. and Barrell,B. (2000) Artemis: sequence visualisation and annotation. Bioinformatics, 16, 944–945. [DOI] [PubMed] [Google Scholar]
- 18.Macke T.J., Ecker,D.J., Gutell,R.R., Gautheret,D., Case,D.A. and Sampath,R. (2001) RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res., 29, 4724–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofacker I.L., Fontana,W., Stadler,P.F., Bonhoeffer,L.S., Tacker,M. and Schuster,P. (1994) Fast folding and comparison of RNA secondary structures. Monatsh. Chem., 125, 167–188. [Google Scholar]
- 20.Blattner F.R., Plunket III,G., Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1462. [DOI] [PubMed] [Google Scholar]
- 21.Kunst F., Ogasawara,N., Moszer,I., Albertini,A.,M., Alloni,G., Azevedo,V., Bertero,M.,G., Bessières,P., Bolotin,A., Borchert,S. et al. (1997) The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature, 390, 249–256. [DOI] [PubMed] [Google Scholar]
- 22.Itoh T., Takemoto,K., Mori,H. and Gojobori,T. (1999) Evolutionary instability of operon structures disclosed by sequence comparisons of complete microbial genomes. Mol. Biol. Evol., 16, 332–346. [DOI] [PubMed] [Google Scholar]
- 23.Eddy S.R. (1998) Profile hidden Markov models. Bioinformatics, 14, 755–763. [DOI] [PubMed] [Google Scholar]
- 24.Hershberg R., Bejerano,G., Santos-Zavaleta,A. and Margalit,H. (2001) PromEC: an updated database of Escherichia coli mRNA promoters with experimentally identified transcriptional start sites. Nucleic Acids Res., 29, 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishii T., Yoshida,K.-i., Terai,G., Fujita,Y. and Nakai,K. (2001) DBTBS: a database of Bacillus subtilis promoters and transcription factors. Nucleic Acids Res., 29, 278–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eichenberger P., Jensen,S.T., Conlon,E.M., van Ooij,C., Silvaggi,J., González-Pastor,J.E., Fujita,M., Ben-Yehuda,S., Stragier,P., Liu,J.S. et al. (2003) The σE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol., 327, 945–972. [DOI] [PubMed] [Google Scholar]
- 27.Park K., Choi,S., Ko,M. and Park,C. (2001) Novel σF-dependent genes of Escherichia coli found using a specified promoter consensus. FEMS Microbiol. Lett., 202, 243–250. [DOI] [PubMed] [Google Scholar]
- 28.Haldenwang W.G. (1995) The sigma factors of Bacillus subtilis. Microbiol. Rev., 59, 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrett C., Hughey,R. and Karplus,K. (1997) Scoring hidden Markov models. CABIOS, 13, 191–199. [DOI] [PubMed] [Google Scholar]
- 30.Karp P.D., Riley,M., Saier,M., Paulsen,I.T., Collado-Vides,J., Paley,S.M., Pellegrini-Toole,A., Bonavides,C. and Gama-Castro,S. (2002) The EcoCyc Database. Nucleic Acids Res., 30, 56–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugimoto N., Nakano,S.-i., Katoh,M., Matsumura,A., Nakamuta,H., Ohmichi,T., Yoneyama,M. and Sasaki,M. (1995) Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry, 34, 11211–11216. [DOI] [PubMed] [Google Scholar]
- 32.Molle V., Fujita,M., Jensen,S.T., Eichenberger,P., González-Pastor,J.E., Liu,J.S. and Losick,R. (2003) The Spo0A regulon of Bacillus subtilis. Mol. Microbiol., 50, 1683–1701. [DOI] [PubMed] [Google Scholar]
- 33.Lewis R.J., Krzywda,S., Brannigan,J.A., Turkenburg,J.P., Muchova,K., Dodson,E.J., Barak,I. and Wilkinson,A.J. (2000) The trans-activation domain of the sporulation response regulator Spo0A revealed by X-ray crystallography. Mol. Microbiol., 38, 198–212. [DOI] [PubMed] [Google Scholar]
- 34.Roth J.R., Jeffrey,G.L., Rubenfield,M., Kieffer-Higgings,S. and Church,G.M. (1993) Characterization of the cobalamin (vitamin B12) biosynthetic genes of Salmonella typhimurium. J. Bacteriol., 175, 3303–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodionov D.A., Vitreschak,A.G., Mironov,A.A. and Gelfand,M.S. (2003) Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem., 278, 41148–41159. [DOI] [PubMed] [Google Scholar]
- 36.Daiyasu H., Osaka,K., Ishino,Y. and Toh,H. (2001) Expansion of the zinc metallo-hydrolase family of the β-lactamase fold. FEBS Lett., 503, 1–6. [DOI] [PubMed] [Google Scholar]
- 37.Altschul S.F., Gish,W., Miller,W., Meyers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 38.Bruggemann H., Baumer,S., Fricke,W.F., Wiezer,A., Liesegang,H., Decker,I., Herzberg,C., Martinez-Arias,R., Merkl,R., Henne,A. et al. (2003) The genome sequence of Clostridium tetani, the causative agent of tetanus disease. Proc. Natl Acad. Sci. USA, 100, 1316–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanehisa M., Goto,S., Kawashima,S. and Nakaya,A. (2002) The KEGG databases at GenomeNet. Nucleic Acids Res., 30, 42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.