Abstract
Gibberellin (GA) 2-oxidases play an important role in the GA catabolic pathway through 2β-hydroxylation. There are two classes of GA2oxs, i.e., a larger class of C19-GA2oxs and a smaller class of C20-GA2oxs. In this study, the gene encoding a GA 2-oxidase of rice, Oryza sativa GA 2-oxidase 5 (OsGA2ox5), was cloned and characterized. BLASTP analysis showed that OsGA2ox5 belongs to the C20-GA2oxs subfamily, a subfamily of GA2oxs acting on C20-GAs (GA12, GA53). Subcellular localization of OsGA2ox5-YFP in transiently transformed onion epidermal cells revealed the presence of this protein in both of the nucleus and cytoplasm. Real-time PCR analysis, along with GUS staining, revealed that OsGA2ox5 is expressed in the roots, culms, leaves, sheaths and panicles of rice. Rice plants overexpressing OsGA2ox5 exhibited dominant dwarf and GA-deficient phenotypes, with shorter stems and later development of reproductive organs than the wild type. The dwarfism phenotype was partially rescued by the application of exogenous GA3 at a concentration of 10 µM. Ectopic expression of OsGA2ox5 cDNA in Arabidopsis resulted in a similar phenotype. Real-time PCR assays revealed that both GA synthesis-related genes and GA signaling genes were expressed at higher levels in transgenic rice plants than in wild-type rice; OsGA3ox1, which encodes a key enzyme in the last step of the bioactive GAs synthesis pathway, was highly expressed in transgenic rice. The roots of OsGA2ox5-ox plants exhibited increased starch granule accumulation and gravity responses, revealing a role for GA in root starch granule development and gravity responses. Furthermore, rice and Arabidopsis plants overexpressing OsGA2ox5 were more resistant to high-salinity stress than wild-type plants. These results suggest that OsGA2ox5 plays important roles in GAs homeostasis, development, gravity responses and stress tolerance in rice.
Introduction
Gibberellins (GA) are plant hormones that govern many aspects of plant biology, including seed germination, stem elongation, leaf expansion, flowering transition, seed development and apical dominance [1]–[7]. There are more than 100 different GAs, but most of these are precursors and degradation products [8]. Bioactive GAs in higher plants include GA1, GA3, GA4 and GA7 [8]. Plants exhibiting the typical GA-deficiency phenotype are dwarfed, with small, dark green leaves, retarded growth and late flowering [9]–[12].
The GA biosynthesis pathway has long been a subject of study, and the genes encoding the main enzymes in each step of the GA biosynthesis and catabolism pathways have been identified in Arabidopsis thaliana and rice (Oryza sativa) [8], [13], [14]. In rice, several GA-related mutants have been studied in detail [10], [15]–[17]. GAs are synthesized from trans-geranylgeranyl diphosphate (GGDP) [8], [18] in three steps. In the first step, GGDP is transformed into the tetracyclic hydrocarbon ent-kaurene via ent-copalyl diphosphate (CDP) through two types of diterpene cyclases in plastids, copalyl diphosphate synthase (CPS) and ent-kaurene synthase (KS). Next, two membrane-associated P450 monooxygenases, ent-kaurene oxidase (KO) and ent-kaurenoic acid oxidase (KAO), help convert ent-kaurene into GA12 in the endoplasmic reticulum. The last step of GA synthesis involves soluble 2-oxoglutarate-dependent dioxygenase (2ODDs). In this step, C20 is oxidized and removed, leading to the formation of C19-GAs such as GA9 and GA20; the formation of these C19 GAs is catalyzed by GA 3-oxidase in the cytosol [8], [18], [19]. The main degradation pathway for GAs is catalyzed by GA 2-oxidase (GA2ox), a 2β-hydroxylation enzyme that hydroxylates C-2 of active GAs. These GA2oxs are encoded by a small gene family that has been identified in Arabidopsis, spinach and rice [8], [10], [20], [21]. These GA2oxs are classified into two subgroups based on the substrates that they act on, i.e., C19GA2oxs and C20GA2oxs. C19GA2oxs can hydroxylate the C-2 of active C19-GAs (GA1 and GA4), as well as C19-GA precursors such as GA9 and GA20, to produce the inactive forms of GAs, i.e., GA8, GA34, GA29 and GA51. The subgroup C20GA2oxs only acts on C20-GA precursors, such as GA12 and GA53, to form GA110 and GA97 [21], [22] , but not C19-GAs. These C20GA2oxs contain three unique, conserved amino acid motifs that are not present in the C19GA2oxs subgroup [21]. The C20GA2oxs includes two Arabidopsis thaliana proteins (AtGA2ox7 and AtGA2ox8), one soybean (Glycine max [L.] Merr) protein (GmGA2ox4), one spinach (Spinacia oleracea) protein (SoGA2ox3) and three rice (Oryza sativa) proteins (OsGA2ox5, OsGA2ox6 and OsGA2ox9) [21]–[24]. The physiological functions of these C20GA2oxs have been studied in some plant species. The overexpression of AtGA2ox7 and AtGA2ox8 produce a dwarf phenotype with reduced GA levels, while ectopic expression of AtGA2ox7 and AtGA2ox8 in transgenic tobacco (Nicotiana tabacum) leads to a dwarf phenotype [22]. A similar phenotype was also observed in rice overexpressing OsGA2ox6 [25]. These results suggest that C20GA2oxs reduce the level of bioactive GAs in plants. Another mechanism of GA degradation has recently been reported. The rice ELONGATED UPPERMOST INTERNODE gene (OsEUI) encodes a cytochrome P450 monooxygenase that catalyzes epoxidation of the C16, 17 double bond, which results in decreased levels of bioactive GAs [26], [27].
In this study, we report the functional characterization of the OsGA2ox5 gene, which encodes a C20GA2ox enzyme in rice. Overexpression of OsGA2ox5 in rice and Arabidopsis plants produced a dwarf phenotype with retarded growth; the application of exogenous GA3 rescued the GA-deficient phenotype. GA biosynthesis and GA signaling pathway genes were up-regulated in transgenic rice plants, especially OsGA3ox1, the last enzyme in the synthesis of bioactive GAs. We also found out that OsGA2ox5 functions in salinity resistance and gravity responses.
Materials and Methods
Plant Materials and Growing Conditions
The rice cultivar Zhonghua 11 (Oryza sativa L. subsp. japonica) was used for rice transformation. Rice plants were grown in a greenhouse at 28°C. Arabidopsis thaliana ecotype Col-0 was used as the wild type. Plants were grown on soil or on plates containing MS medium under LD (16 h light/8 h dark) condition at 22°C.
Rice seeds were surface sterilized for 5 min with ethanol (75% v/v) and 30 min with commercially diluted (1∶3 v/v) NaOCl, followed by several rinses with sterile water. Germination was carried out for 72 h on sterile MS medium in the dark at 28°C. The plants were then grown at 28°C-day/25°C-night, under a 12-h-light/12-h-dark cycle and at a relative humidity of 50%.
RNA Extraction and Real-time PCR Assays
Total RNA was extracted from root, stem, leaf, sheath, and panicles using the TRIzol reagent (Invitrogen) for analysis of OsGA2ox5 mRNA expression. To analyze the transcription levels of gibberellin metabolism and signal pathway genes, 3-week-old WT and OsGA2ox5-ox rice seedlings were harvested and subjected to RNA extraction using the TRIzol reagent (Invitrogen). The RNA was reverse-transcribed using an oligo (dT) 18 primer and AMV reverse transcriptase (Toyobo) according to the manufacture’s protocol. Real-Time PCR was performed using CFX96 (Bio-Rad, USA) and SYBR Green I (CWBIO); the Real-time PCR assays were performed in triplicate for each cDNA sample. The data were normalized using the rice marker gene OsActin. All primers used in this study are listed in supplemental Table S1.
Construction of POsGA2ox5:GUS Vector and Staining
The promoter region of OsGA2ox5, 3,500-bp upstream of ATG (POsGA2ox5), was amplified from the rice genome by PCR using KOD polymerase (Toyobo) and inserted upstream of the GUS gene at the Xba I-Sma I sites of the p1300GN-GUS vector. The primers used are OsGA2ox5 gusF and OsGA2ox5 gusR (sangon) (the specific primers are listed in supplemental Table S1). The POsGA2ox5 :GUS construct was transfected into A. tumefaciens EHA105 by heat shock, followed by transformation of rice embryonic calli, as described previously [28]. GUS staining was used to investigate the level of OsGA2ox5 expression in the T1 generation of POsGA2ox5 :GUS transgenic rice. Transgenic plant samples were incubated in GUS staining solution (100 mmol/L NaH2PO4 buffer pH 7.0, 0.5% Triton X-100, 0.5 mg/ml X-Gluc and 20% methanol) overnight at 37°C. After staining, the tissues were rinsed and photographed.
Overexpression of OsGA2ox5 in Rice and Arabidopsis
The full-length CDS of OsGA2ox5 was amplified using primers OsGA2ox5F and OsGA2ox5R (sangon) and cloned in the vector pMD-18T (TaKaRa); the sequence was confirmed by DNA sequencing. The OsGA2ox5 CDS from the sequenced clone was removed by digestion and cloned into modified binary vector pHB [29]. The binary vector pHB-OsGA2ox5 was transformed into Agrobacterium strain EHA105 and transfected into rice embryonic calli as described previously [28]; this vector was used to transform Arabidopsis thaliana ecotype Columbia-0 using previously described methods [30]. The transgenic plants were selected using hygromycin. The T1 plants were confirmed by PCR using the following specific primers for the hygromycin phosphotransferase (HPT) gene: 5-TGTCCTGCGGGTAAATAGC-3 and 5-TGCTCCATACAAGCCAACC-3 (AY836546). To analyze of OsGA2ox5 gene expression level in transgenic plants, 3-week-old WT and OsGA2ox5-ox rice seedlings were harvested and subjected to RNA extraction using the TRIzol reagent (Invitrogen). RT-PCR was performed with oligos OsGA2ox5RTF and OsGA2ox5RTR (Table S1) using Taq DNA polymerase (TaKaRa).
Southern Blot
Southern blot was used to analyze the transgenic plants. 20 μg of total genomic DNA from leaf tissue of transgenic plants and wild type plants was digested with appropriate restriction endonuclease Hind III (only one recognition site in T-DNA sequence). DNA fragments were separated by electrophoresis on a 1% (w/v) agarose gel and then transferred to a nylon membrane (Amersham Bioscience) according to standard protocols. Dig-high DNA labeling kit I (Roche) was used to label the Hygromycin DNA probes.
Subcellular Localization of OsGA2ox5
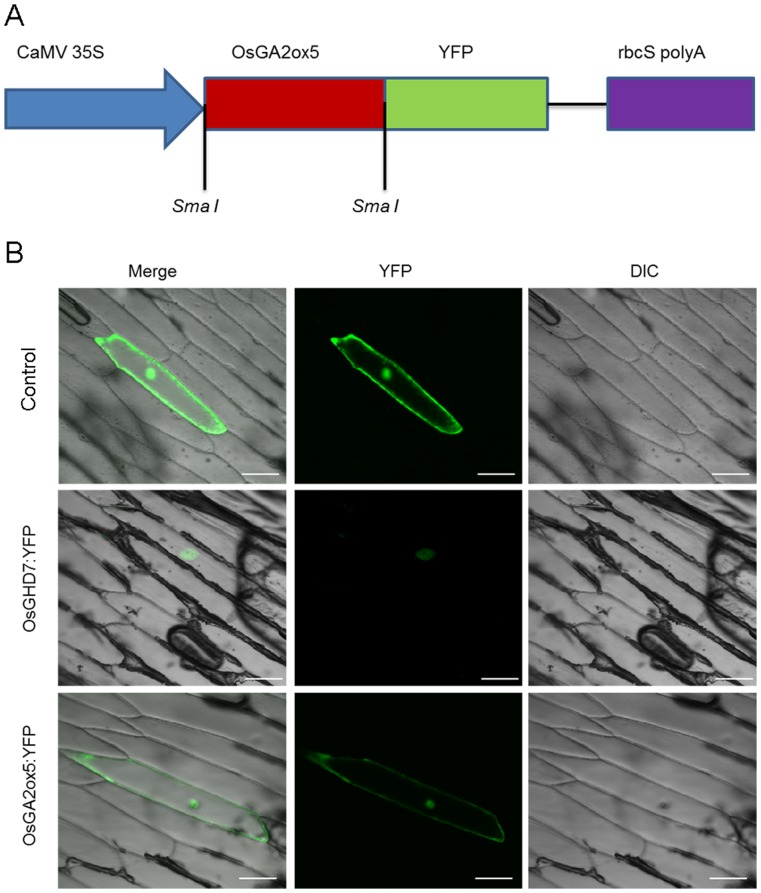
The coding region of OsGA2ox5 was amplified using the primer pair OsGA2ox5-pA7YFPF and OsGA2ox5-pA7YFPR (sangon) (the specific primers are listed in supplemental Table S1) and cloned into pA7-YFP [31], generating the OsGA2ox5-YFP fusion under the control of the CaMV 35S promoter. A previously study demonstrated that OsGHD7 [32] is a nuclei protein and localized in nuclei only, so we used OsGHD7 as a positive control. The OsGHD7 coding sequence was fused in frame to the N-terminus of YFP under the control of the CaMV 35S promoter. Then, OsGA2ox5-YFP, OsGHD7-YFP fused construct and pA7-YFP vectors were used to transiently transform onion epidermal cells by particle bombardment[33] using a particle gun system (PDS-1000/He; Bio-Rad). After 24 h, the epidermal cells were examined for YFP fluorescence under a scanning confocal microscope (Zeiss LSM510; Carl Zeiss Micro-Imaging GmbH, Jena, Germany).
Longitudinal Section Microscopic Analysis
The second leaf sheaths from 5-day-old WT and OX rice plants which were cut into 1mm width were rinsed in 75% ethanol at room temperature overnight. Then the samples were cleared for 24 h in a chloralhydrate solution (chloralhydrate-H2O-glycerol, 8:2:1, w:v:v) and detected in microscopic (Leica MZ95).
Exogenous GA3 Treatment
14-day-old WT (ZH11) and OsGA2ox5-ox plants were incubated in 1/2 MS medium containing 1 μM GA3 (sangon). The seedling height (from the base to the leaf cap) was measured at day 7 after GA3 treatment. Plants grown in a greenhouse were sprayed with 10 µM GA3 three times a week.
Root Gravitropism Analysis
The seeds were surface-sterilized and sown on half-strength MS medium containing 0.45% phytagel. Four-day-old seedlings with radicals approximately 6 cm in length were subjected to gravitropism analysis. Light-grown wild-type and OsGA2ox5-ox seedlings were displaced by 90° and monitored for the orientation of the primary root caps. The vertical position is represented by 90°, and the horizontal position is represented by 0°. The seedlings were reoriented by 90°, and images of the roots were captured at 0 h, 0.5 h, 1 h, 2 h, 3 h, 4 h and 5 h. The degrees of curvature were measured from the digital images using Image J software (http://rsbweb.nih.gov/ij/).
Staining of Starch Granules
Starch granules in the root cap were visualized with 1% I2-KI solution in 4-day-old seedlings grown on 1/2 MS. Roots were stained for 1 minute, rinsed with water, cleared with 50% chloral hydrate for 45 seconds and photographed with Leica MZ95.
For the resin section, the 3 mm-length of root caps were obtained from plants grown on MS culture medium for 4 days. They were vacuum infiltrated for 1 h in 2.5% glutaral-dehyde, in 0.05 M phosphate buffer, pH 7.4 at room temperature and then in 4 °C overnight. Samples were then subsequently dehydrated in a graded acetone series at room temperature and embedded in 812 resin. Blocks were polymerized at 70°C for 24 hours, and cut into 1 μm sections on a RM2265 microtome (Leica, Heidelberg, Germany). For amyloplast staining, slides with sections attached were immersed in 0.5% periodic acid solution for 10 min, rinsed with distilled water, and then immersed in Schiff’s reagent (0.5% aniline red, 0.01M HCl, 1% sodium metabisulfite) for 15 min. then slides were rinsed, dried mounted by neutral balsam and observed microscopically.
Starch Extraction and Quantification
Four-day-old seedlings with radicals approximately 6 cm in length were subjected to starch analysis. Roots cap segments were excised about 1 cm and pooled into samples from 10 plants each. Starch was extracted with 0.7 M perchloric acid and the insoluble fraction was cleared with 80% (v/v) ethanol three times then resuspended in water as described [34]. Samples were boiled for 10 min then starch was measured using the Starch (GO/P) Assay Kit(sigma)according to the manufacturer’s instructions.
Salt Stress Treatment
WT and OsGA2ox5-ox rice seeds were incubated in water for two days, followed by incubation in water supplemented with 100 mM or 140 mM NaCl for 1 week. For Arabidopsis, Col-0 and OsGA2ox5-ox transgenic seeds were planted in Petri dishes containing solidified 1/2 MS medium and grown for two weeks. The seedlings were then transferred to 1/2 MS medium containing 170 mM NaCl; the seedlings were photographed three weeks later.
Results
OsGA2ox5 is Widely Expressed in Various Rice Tissues
To determine the expression pattern of OsGA2ox5 in rice, we analyzed OsGA2ox5 expression in rice plants by real-time PCR using OsGA2ox5-specific primers. Rice OsGA2ox5 was detected in the root, leaf, culm, sheath and young panicles of rice seedlings (Fig. 1A). To confirm the expression pattern of OsGA2ox5, an expression vector containing GUS (ß-glucuronidase) driven by the OsGA2ox5 promoter was constructed and transformed into Zhonghua 11 (Oryza sativa L. subsp. japonica). Consistent with the results of real-time PCR assays, the transgenic plants showed GUS staining in the roots, culms, leaves, sheaths and panicles (Fig. 1B). These tissues always get staining at positions of a cut and suggest that this is mechanical.
Figure 1. Expression pattern of OsGA2ox5 in vivo.
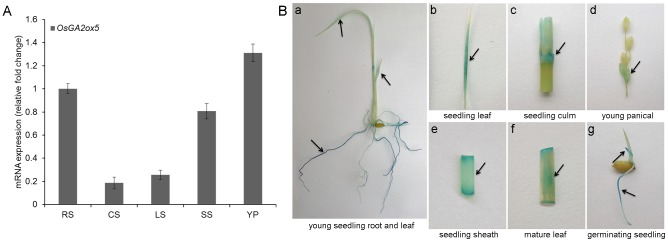
(A) Real-time PCR analysis of OsGA2ox5 in various organs of wild-type plants. RS, seedling root; CS, seedling culm; LS, seedling leaf; SS, seeding sheath; YP, young panical. The expression is relative to that of OsActin. Values are expressed as the average ± SD of three technical replicates, and the amount of OsGA2ox5 in roots was set at 1.0; (B) Histochemical analysis of POsGA2ox5:GUS gene activities in different tissues and organs of rice. The promoter region of OsGA2ox5, 3,500-bp upstream of ATG (POsGA2ox5) was inserted upstream of the GUS gene at the Xba I-Sma I sites of the p1300GN-GUS vector. Arrows indicated the expression tissues of OsGA2ox5.
OsGA2ox5 is Localized to the Cytoplasm and Nucleus
To determine the subcellular localization of the OsGA2ox5 protein, The OsGA2ox5 coding sequence was fused in frame to the N-terminus of YFP (Fig. 2A). The OsGHD7 coding sequence was also fused in frame to the N-terminus of YFP under the control of the CaMV 35S promoter. The subcellular localization of the OsGA2ox5-YFP was examined through a transient expression of OsGA2ox5-YFP in onion epidermal cells. An examination of yellow florescence by confocal laser-scanning microscopy showed that YFP alone localized at the nucleus and cytosol of onion epidermal cells and the yellow fluorescent signal of OsGHD7 was detected exclusively in the nucleus of the onion epidermal cells, while OsGA2ox5-YFP was localized to the same region as YFP alone, i.e., the cytoplasm and nucleus (Fig. 2B). More than 30 YFP positive cells were detected. OsGA2ox5-YFP exhibited cytoplasm and nucleus localization in all these cells. These results demonstrated that OsGA2ox5 is a cytoplasm- and nuclear-localized protein.
Figure 2. Localization of OsGA2ox5-YFP protein.
(A) Diagram of the inserted region of the vector pA7:: OsGA2ox5::YFP; (B) Subcellular localization of OsGA2ox5. OsGA2ox5 was detected both in the cytoplasm and nucleus, the nucleus marker protein OsGHD7 was detected exclusively in the nucleus of onion epidermal cells and the control YFP showing signal both in cytoplasm and nucleus. DIC (Differential Interference Contrast), referring to bright field images of the cells.
Overexpression of OsGA2ox5 Produces a Severe Dwarf Phenotype
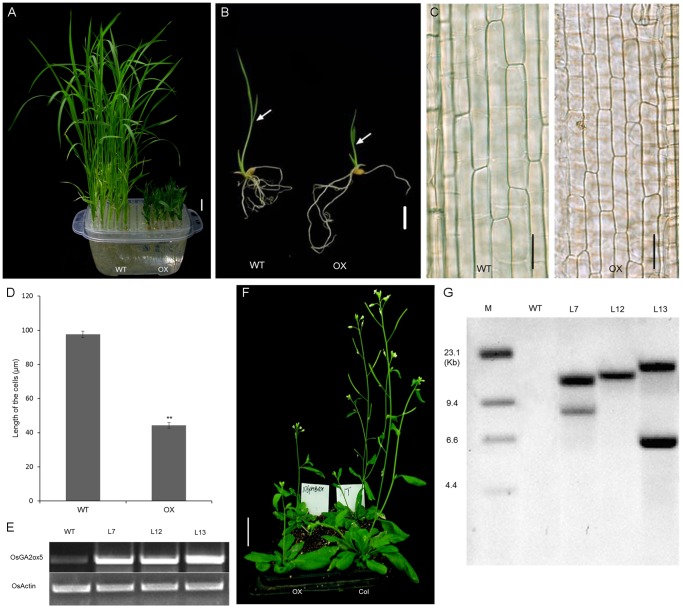
To analyze the roles of OsGA2ox5 in plants, OsGA2ox5 was inserted into the pHB vector and overexpressed in Arabidopsis and rice under the control of the double CaMV 35S promoter. Transformants were selected based on hygromycin resistance. Stable inherited homozygous T3 plants of three independent transgenic lines (L7, L12, and L13) were examined by Southern blot analysis (Fig. 3E). Southern blotting produced one band in L12, while two bands were observed in L7 and L13 and no band was observed in the WT. These results suggested L12 was single integration in the genome and L13, L17 were double integration in the genome of transgenic plants. But all of those transgenic lines showing similar phenotype which means that the transgenic plants phenotypes were caused by OsGA2ox5. Moreover, RT-PCR analysis revealed that OsGA2ox5 was overexpressed in both transgenic rice and transgenic Arabidopsis plants (Fig. 3D). The OsGA2ox5-ox rice plants exhibited a severe dwarf phenotype (Fig. 3A), as previously reported [23]. There was no obvious difference in root length between two-week-old WT and OsGA2ox5-ox plants, but the height of the transgenic plants was 75% lower than that of the WT. Longitudinal section analysis of the second leaf sheaths revealed that the cells of the OsGA2ox5-ox plants were markedly shorter and smaller than those of the WT (Fig. 3B). The flowering and heading stages were delayed by approximately 20 days in the OsGA2ox5-ox plants, and the spike length was shorter, compared with those of the WT. Also, the seeds of OsGA2ox5-ox were small and irregularly shaped, light green and not well filled. Similar results also were observed in transgenic Arabidopsis, such as slow growth and late flowering compared with the WT (Fig. 3C).
Figure 3. Phenotype of OsGA2ox5-overexpressing plants.
(A) Phenotype of WT (left) and dwarf OsGA2ox5-ox plants (right). 2-week-old water cultured seedlings were used for photograph; (B) Arrows indicate the boundary between the second leaf sheath and the blade of 5-day-old water cultured seedlings. Bar = 1 cm; (C) Longitudinal sections of the elongated regions of the second leaf sheath of WT (left) and OsGA2ox5-ox plants (right). Bar = 25 µm; (D) Quantitative measurement of the cell length of second leaf sheath in WT and OX (n = 20). Error bars show standard errors (SE). Asterisks indicated significant differences at P <0.01 compared with the wild type by Student’s t test; (E) Ectopic expression of OsGA2ox5 in Arabidopsis. Left is OsGA2ox5 transgenic plants and wide type Arabidopsis (Col) is on the right. Plants photographed are 4-weeks-old. Bar = 2.5 cm; (F) Expression level of OsOsGA2ox5 in transgenic rice; WT was used as a control; (G) Southern blotting analysis of transgenic plants. Restriction endonuclease Hind III was used to digest the genomic DNA from the leaf tissue. M, molecular marker; WT, wild type; L7, L12, L13, three transgenic lines.
The Dwarf Phenotype of Transgenic Plants is Rescued by the Application of Exogenous GA3
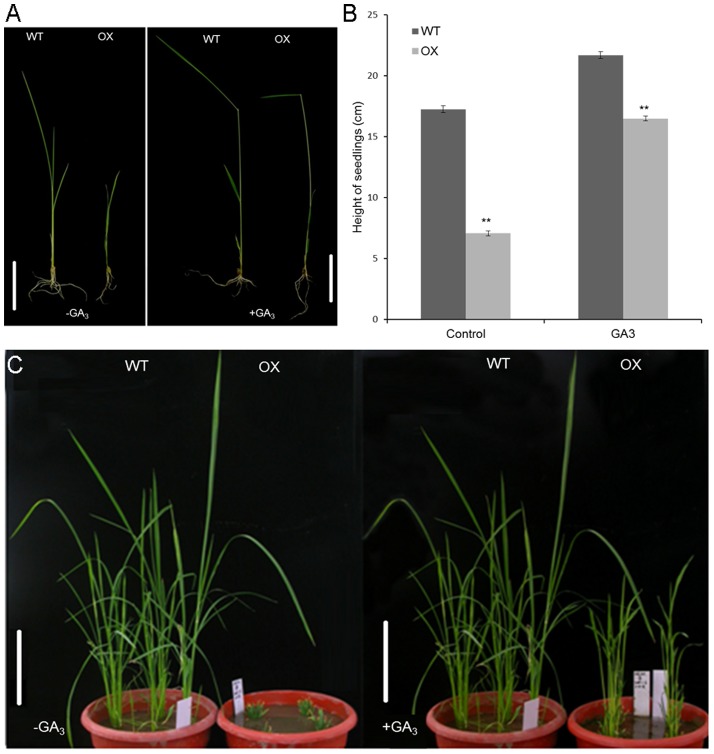
According to homology analysis result (Fig. S1) and previous data, we deduced that OsGA2ox5 can degrade C20-GA precursors. Overexpression of OsGA2ox5 produced a severe dwarf phenotype, dark-green leaves and late flowering, which are all typical of GA-deficiency mutants. To investigate the responsiveness of OsGA2ox5-ox and WT plants to exogenous bioactive gibberellin, GA3, we cultivated WT and OsGA2ox5-ox plants in MS liquid medium containing 1 µM GA3 for 7 days. GA3 partially restored the height of OsGA2ox5-ox plants (Fig. 4A and B). Plants grown in a greenhouse and sprayed with exogenous GA3 exhibited a similar phenotype (Fig. 4C); the spike length, grain number and 1,000-grain weight were higher in plants sprayed with exogenous GA3 than in the control (Figure S2).
Figure 4. Exogenous GA3 effectively reverses the GA-deficiency phenotype.
(A) Response to the application of GA3 in the plants. Two-week-old plants cultivated in MS liquid medium containing 1 µM GA3 or no GA3 for 1 week. Bar = 2 cm; (B) Plant elongation of OsGA2ox5-ox and WT seedlings treated with GA3. Plant height was measured at day 7 after GA3 treatment. Results represent three independent experiments with similar results. Error bars show standard errors (SE). Asterisks indicate significant difference at P <0.01 compared with the wild type by Student’s t test; (C) 1-month-old OsGA2ox5-ox and WT plants grown in a greenhouse and sprayed with exogenous GA3. Bar = 10 cm.
GA Biosynthesis and Signaling is Regulated by OsGA2ox5 Expression
OsGA2ox5 acts on C20-GA precursors, resulting in reduced levels of bioactive GA synthesis in vivo. In rice, the overexpression of OsEUI and OsGA2ox6 altered the expression of GA signaling genes, and mutations in AtGA2ox7 and AtGA2ox8 resulted in the down regulated expression of GA5 [25], [26], [35]. To investigate whether overexpression of OsGA2ox5 also regulates the expression of GA biosynthesis and GA signaling genes, we used real-time PCR assays to detect the expression of genes encoding GA20oxidase, GA3oxidase and GA2oxidase [36], [37] as well as OsSLR [16] and OsGIDs [17], [38]–[40]. The expression of all of these GA biosynthesis genes was up-regulated in OsGA2ox5-ox plants, especially the OsGA3ox1 gene, which encodes the enzyme that catalyzes the last step of GA synthesis (Fig. 5). Interestingly, the GA catalysis gene OsGA2ox1, GA signaling genes, the receptor gene OsGID1, the F-box gene OsGID2 and the gene encoding rice DELLA protein OsSLR, a negative GA regulator, were all up-regulated in OsGA2ox5-ox plants compared with the WT.
Figure 5. Gene expression analyses.
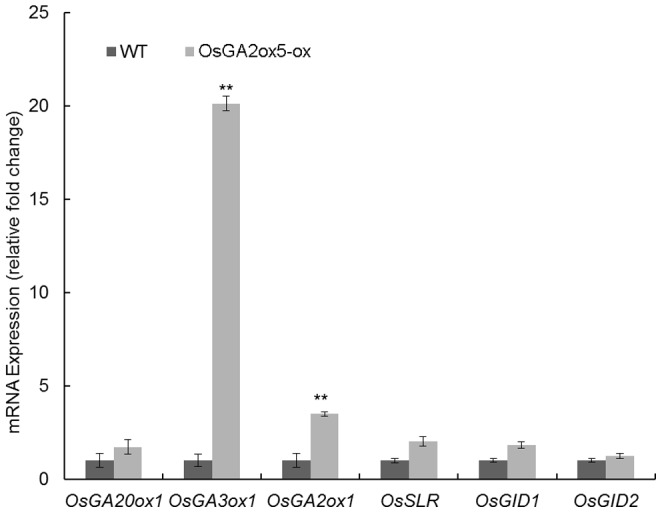
The transcription levels of gibberellin metabolism and signal pathway genes expression levels analyzed by Real-time PCR. The rice OsActin gene was used as an internal control. Data are the mean ±SE of three independent measurements with three repeats. Asterisks indicate significant difference at P <0.01 compared with the wild type by Student’s t test. The expression of all of these GA biosynthesis and GA signaling genes was up-regulated in OX plants, especially the OsGA3ox1 gene nearly 10-fold up-regulated. 3-week-old rice plants cultivated in water were used for experiments. OsGA20ox1 and OsGA3ox1 encode GA biosynthesis enzymes; OsGA2ox1 encodes an enzyme that functions in GA degradation; OsSLR encodes a negative GA regulator in GA signaling and OsGID1 and OsGID2 encode GA receptors in rice.
OsGA2ox5 Affects Root Starch Granule Development and Gravitropism
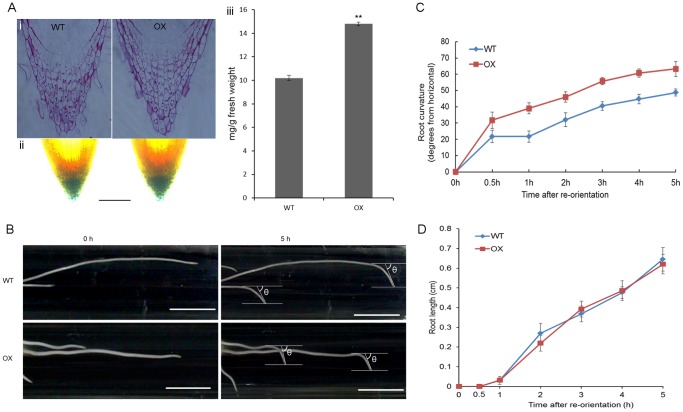
A previous study revealed that GA plus kinetin causes the complete destarching of amyloplasts [41]. In addition, the GA-degrading enzyme OsEUI also alters rice root granule development and gravity responses [42]. We want to know whether OsGA2ox5 had a similar effect on root starch granule development and gravitropism in rice. By extracting and quantifying the root cap starch, we found OsGA2ox5-ox plants generated more starch granules (14.9 mg/g) than the WT plants (10.2 g/mg). Then we using two methods to staining the root caps starch granule and depend on more than 20 staining results we found out that those increased starch granules in OsGA2ox5-ox plants were caused by increased cell layers in the enlarged root caps (Fig. 6A). Then, we analyzed the gravitropic response of WT and OsGA2ox5-ox roots in light-grown seedlings. OsGA2ox5-ox roots bent more quickly than the WT in response to gravity. The roots of OsGA2ox5-ox seedlings exhibited an accelerated gravity response; most OsGA2ox5-ox root caps were nearly vertical 5 h after rotation (Fig. 6B and C). And the roots growth rates showed no significant difference between WT and OX after reorientation (Fig. 6D), suggesting the quick bending of OsGA2ox5-ox roots than the WT in response to gravity was not due to the different root growth rates between them.
Figure 6. OsGA2ox5 affects root starch granule development and gravitropism.
(A) Starch staining and detection of wild-type and OsGA2ox5-ox plants root caps. i, Resin section of WT and OX plants root caps and ii, I2-KI staining of the WT and OX plants root caps. Figures showing that OX plants have increased cell layers in the enlarged root caps compared to WT; iii, starch content in root caps of WT and OX plants. Roots cap segments were excised about 1 cm and pooled into samples from 10 plants each for experiments. Results represent three independent experiments with similar results. Bar = 100µm. Asterisks indicate significant difference at P <0.01 compared with the wild type by Student’s t test; (B) Gravity response of light-grown 4-day-old WT and OX seedling roots. After reorientation the OX seedling roots bent faster than WT. θ indicated the roots bending angle of the WT and OX plants respectively after reorientation at 5h. Experiments were performed three times with similar results. Bar = 1 cm; (C) Time course of root gravitropical curvature (after reorientation). Light-grown wild-type and OsGA2ox5-ox seedlings were displaced by 90° and monitored for the orientation of the primary root caps. The vertical position is represented by 90°, and the horizontal position is represented by 0°. Data shown are the means ± SE of 30 seedlings; (D) Time course of root length (after reorientation). Data shown are the means ± SE of 20 seedlings.
Transgenic Plants Overexpressing OsGA2ox5 Showed Increased Tolerance to High Salinity Stress
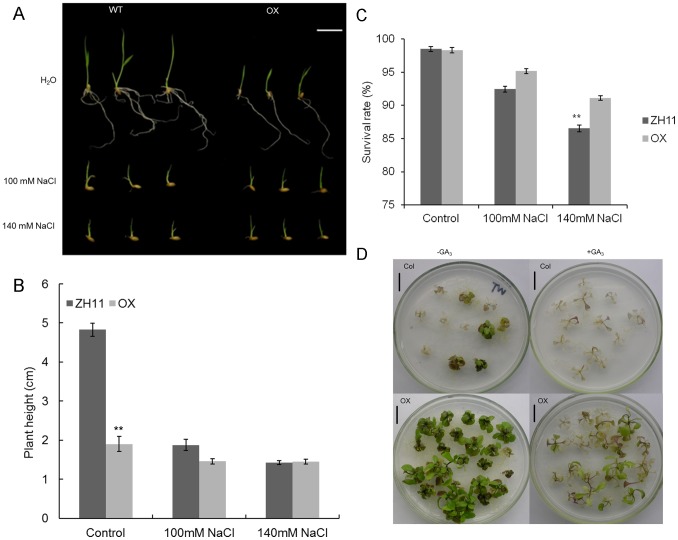
A previous study showed that AtGA2ox7 was directly activated by a transcription factor of the DREB1/CBF subfamily DDF1, and showed resistance to high salinity [43]. OsGA2ox5 shares high homology with AtGA2ox7 and may therefore also be responsive to salt stress. To test this, OsGA2ox5-ox transgenic rice plants that were geminated in water were transferred to water containing 100 mM or 140 mM sodium chloride (Fig. 7A). As shown, on day 7 after transfer, high salinity restricted plant growth, compared with plants grown in water. The height of the WT plants under 100 mM and 140 mM sodium chloride conditions were reduced by 53% and 60%, respectively, compared with water-grown plants, while the OsGA2ox5-ox plants exhibited only a 25% reduction in either 100 mM or 140 mM sodium chloride compared with the control (Fig. 7B). Consistently, more than 95% and 91% of the OsGA2ox5-ox plants survived under 100 mM and 140 mM sodium chloride conditions, whereas 92% and 86% of the Zhonghua 11 plants survived (Fig.7C). To clarify the role of OsGA2ox5 during high salinity stress, we also examined the growth of transgenic OsGA2ox5 Arabidopsis under high-salt conditions (Fig. 7D). Transgenic plants grown on 1/2 MS medium for 2 weeks were transferred to 1/2 MS medium containing 170 mM NaCl. After 21 days, most WT plants died, however, the survival rate of OsGA2ox5-ox plants was very high. When 1/2 MS medium containing 170 mM NaCl was supplied with 10 µM GA3, both WT and OsGA2ox5-ox plants showed reduced survival rates. These results demonstrate that GA reduces salinity tolerance, and OsGA2ox5 is related to salt-stress tolerance.
Figure 7. Effects of salinity stress on plant growth.
(A) Phenotype of WT and OX rice seedlings under salt treatment. Photographs were taken after 5 d of growth in water (control) and 100 mM or 140 mM NaCl. For each treatment, 20 seedlings were measured. Bar = 1 cm; (B) Statistics analysis of plants height under salt treatment. WT and OX plants stem height at 5 days of growth in various concentrations of NaCl. Bar = 1 cm. Asterisks indicate significant difference at P <0.01 compared with the wild type by Student’s t test; (C) Quantitative analysis of survival rates under salt treatment. The results are averages of three independent experiments with 30 plants per experiment. Asterisks indicate significant difference at P <0.01 compared with the wild type by Student’s t test; (D) Phenotype of wide type Arabidopsis (Col) and transgenic Arabidopsis with and without GA3 under salt treatment. Physiological changes in WT and OsGA2ox5-ox transgenic Arabidopsis plants transferred to plates containing 170 mM NaCl. Photographs were taken 3 weeks after transfer. +GA represents treatment with 10 µM GA3.
Discussion
The plant hormone GA is very important; GA homeostasis is essential for normal plant growth and development as well as environmental adaptation. In plants, the level of bioactive GAs is accurately maintained by the regulation of GA biosynthesis and catabolism. To date, two types of enzymes are known to regulate GA biosynthesis and catabolism. GA2oxs and OsEUI [21], [23], [27] act on bioactive GAs and their precursors to reduce the level of bioactive GAs, thereby maintaining GA homeostasis. GA2oxs catalyze the catabolism of bioactive GAs and GA precursors into inactive GAs by hydroxylating the C2 of C19-GAs and C20-GAs [21]. EUI P450 converts bioactive GA4 and its precursor GA9 into inactive 16, 17 epoxy-GAs by 16, 17-epoxidation [26], [27]. In this study, we cloned OsGA2ox5 from Oryza sativa. Real-Time PCR assays and GUS staining revealed that this gene is expressed in roots, culms, leaves, sheaths and panicles (Fig. 1A and B). The high level of OsGA2ox5-GUS expression in culm suggests that OsGA2ox5 functions in plant elongation. OsGA2ox5 is localized to both the nucleus and cytoplasm (Fig. 2), which is similar to the localization of OsGA2ox6 [25]. Previous studies have suggested that OsGA2ox5 only hydroxylates C20-GA substrates in rice, and OsGA2ox5 belongs to the subgroup C20GA2ox, which also contains AtGA2ox7, AtGA2ox8, SoGA2ox3 OsGA2ox6 and OsGA2ox9 [21], [23], [35]. Amino acid sequence alignment showed that OsGA2ox5 is similar to AtGA2ox8 and SoGA2ox3 and has the three unique conserved motifs (Fig. S1) [21]. Therefore, we deduced that OsGA2ox5 can act on C20-GA substrates to produce inactive GAs.
Overexpression OsGA2ox5 in rice and Arabidopsis produced plants that were dwarfed and dark green with retarded growth (Fig. 3A and C). The dwarf phenotype of OsGA2ox5–overexpressing rice plants was restored by exogenous GA3 treatment (Fig. 4A and C). This suggests that the overexpression of OsGA2ox5 decreases the level of bioactive GAs in rice, and the dwarf phenotype is caused by a shortage of bioactive Gas; these plants exhibit the typical GA-deficiency phenotype. Similar to the overexpression of AtGA2ox7 and AtGA2ox8 in Arabidopsis, and OsGA2ox6 in rice, OsGA2ox5-ox plants exhibited the dwarf phenotype [25], [35].
We then examined the expression of genes encoding GA biosynthesis and metabolism enzymes, as well as GA signaling pathway genes (Fig. 5). The GA biosynthesis genes OsGA20ox1 and OsGA3ox1 were up-regulated in OsGA2ox5-ox plants compared with WT. The expression of OsGA20ox1 was slightly increased in OsGA2ox5-ox plants, while the expression of OsGA3ox1, which encodes the last key enzyme in the synthesis of bioactive GAs, was increased sharply (nearly 10-fold) in OsGA2ox5-ox plants. This may represent a type of feedback regulation required for plants to maintain a stable endogenous GA level, as the overexpression of OsGA2ox5 decreased the endogenous GA level, and plants must synthesize increasing amounts of bioactive GAs to maintain GA homeostasis. Therefore, GA synthesis genes were up-regulated in the OsGA2ox5-ox plants, especially OsGA3ox1. Consistent with this notion, the upregulation of GA20ox and GA3ox has been reported in GA-deficient and GA-insensitive mutants [44], [45]. We also examined the expression of OsGA2ox1, which is involved in GA catabolism. The expression of OsGA2ox1 was also increased in OsGA2ox5-ox plants, which was perhaps influenced by the increased expression of OsGA3ox1. Interestingly, the expression of GA signaling genes that were examined was up-regulated in the OsGA2ox5-ox plants. The expression of GID1 and GID2, which encode receptors of GA and the rice DELLA protein SLR, was also increased in OsGA2ox5-ox plants. Bioactive GAs interact with SLR by binding to its receptors (GID1, GID2), thereby decreasing the activity of SLR protein in GA signal transduction [17], [40]. Perhaps a similar mechanism occurs during the feedback regulation of GA biosynthesis, which is also influenced by altered levels of bioactive GAs. Decreased levels of bioactive GAs increase the expression level of GID1 and GID1, and this feedback stimulates the expression of SLR. This phenomenon was also identified in EUI-ox plants [42].
In addition, the root caps of OsGA2ox5-ox rice plants exhibited a stronger gravitropic response than WT plants (Fig. 6B and C), and not caused due to the growth speed (Fig. 6D), along with more starch granules (Fig. 6A). EUI-ox plants also have more starch granules than WT [42]. GA induces amylase activity, which can degrade starch [46]. Perhaps the increased gravitropism in OsGA2ox5-ox plants is due to altered levels of bioactive GAs.
We also found that the OsGA2ox5-ox plants showed increased stress tolerance. OsGA2ox5-ox seedlings grown in water containing 100 or 140 mM NaCl showed little decrease in growth (vs. water-grown seedlings) compared with WT rice plants grown under high salinity conditions (Fig. 7A). Arabidopsis plants ectopically expressing OsGA2ox5 also exhibited salinity resistance (Fig. 7C), which is consistent with the phenotype of AtGA2ox7-ox Arabidopsis plants [43]. Overexpression of the DWARF AND DELAYED FLOWERING 1 (DDF1) gene, which encodes an AP2 transcription factor of the DREB1/CBF subfamily, causes dwarfism and salinity tolerance. The AtGA2ox7 gene is strongly up-regulated in DDF1-overexpressing transgenic Arabidopsis, and its promoter has a DDF1 binding motif. This suggests that AtGA2ox7 is a direct target of the DDF1 transcriptional activator [47]. Transgenic AtGA2ox7-ox Arabidopsis also exhibits the salinity tolerance phenotype [43]. According to sequence analysis, some DREB1/CBF binding motifs are present in the promoter of OsGA2ox5 [48]. Therefore, perhaps the salinity tolerance of OsGA2ox5-ox transgenic plants is due, at least in part, to the influence of DREB1/CBF.
In summary, we demonstrated that the overexpression of OsGA2ox5 in plants produces a dwarf phenotype, and this phenotype is restored by exogenous GA3 treatment. The overexpression of OsGA2ox5 altered the expression levels of GA biosynthesis and GA signaling genes, leading to a series of responses, including increased stress resistance and increased gravitropism.
Supporting Information
Comparison of the deduced amino acid sequences of OsGA2ox5 with other GA2-oxidases. (A) Amino acid sequence alignment of rice GA2oxs (OsGA2ox1, OsGA2ox5, OsGA2ox6 and OsGA2ox9), Arabidopsis GA2oxs (AtGA2ox1, AtGA2ox7 and AtGA2ox8) and spinach GA2ox (SoGA2ox3) using the DNAMAN software. C20 GA2oxs (OsGA2ox5, OsGA2ox6, OsGA2ox9, AtGA2ox7, AtGA2ox8, and SoGA2ox3) contain three highly conserved sequence motifs (underlined with Roman numerals) that are absent in all C19 GA2oxs (OsGA2ox1 and OsGA2ox3 as examples for comparison). (B) Phylogenetic analysis of these GA2-oxidase proteins.
(TIF)
1000-grain weight of main panicle (A), grains number of each spike (B), Height of stem (C), Length of spike (D) of transgenic lines overexpressing OsGA2ox5 and wild type Zhonghua 11 under normal and GA3 condition. Twelve samples were measured for plant height, spike length and grains number of each line. 1,000-seed weight was measured in triplicate.
(TIF)
Primers.
(XLS)
Acknowledgments
We thank Tongtong Guo, Ling Chen, Xin Dong, Anrui Lu and Ke Zhang for technical assistance and Xianying Dou for critical reading of the manuscript.
Funding Statement
This work was supported by the National Basic Research Program of China (2011CB710902), National Natural Science Foundation of China (31070237), the National Scientific Program (2012AA101103-04), Strategic Priority Research Program of the Chinese Academy of Sciences (XDA04020202-15) and the China Manned Space Flight Technology Project. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Harberd NP, King KE, Carol P, Cowling RJ, Peng J, et al. (1998) Gibberellin: inhibitor of an inhibitor of...? Bioessays 20: 1001–1008. [DOI] [PubMed] [Google Scholar]
- 2. Silverstone AL, Chang C, Krol E, Sun TP (1997) Developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana. Plant J 12: 9–19. [DOI] [PubMed] [Google Scholar]
- 3. Yamaguchi S, Smith MW, Brown RG, Kamiya Y, Sun T (1998) Phytochrome regulation and differential expression of gibberellin 3beta-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10: 2115–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Itoh H (2002) The Gibberellin Signaling Pathway Is Regulated by the Appearance and Disappearance of SLENDER RICE1 in Nuclei. The Plant Cell Online 14: 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rebers M, Kaneta T, Kawaide H, Yamaguchi S, Yang YY, et al. (1999) Regulation of gibberellin biosynthesis genes during flower and early fruit development of tomato. Plant J 17: 241–250. [DOI] [PubMed] [Google Scholar]
- 6. Ogas J, Kaufmann S, Henderson J, Somerville C (1999) PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci U S A 96: 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yamaguchi S, Kamiya Y (2000) Gibberellin biosynthesis: its regulation by endogenous and environmental signals. Plant Cell Physiol 41: 251–257. [DOI] [PubMed] [Google Scholar]
- 8. Hedden P, Phillips AL (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5: 523–530. [DOI] [PubMed] [Google Scholar]
- 9. King RW, Evans LT (2003) Gibberellins and flowering of grasses and cereals: prizing open the lid of the "florigen" black box. Annu Rev Plant Biol 54: 307–328. [DOI] [PubMed] [Google Scholar]
- 10. Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, et al. (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134: 1642–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fleet CM, Sun TP (2005) A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr Opin Plant Biol 8: 77–85. [DOI] [PubMed] [Google Scholar]
- 12. Wang Y, Li J (2005) The plant architecture of rice (Oryza sativa). Plant Mol Biol 59: 75–84. [DOI] [PubMed] [Google Scholar]
- 13. Olszewski N, Sun TP, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14 Suppl: S61–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun TP, Gubler F (2004) Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol 55: 197–223. [DOI] [PubMed] [Google Scholar]
- 15. Ueguchi-Tanaka M, Fujisawa Y, Kobayashi M, Ashikari M, Iwasaki Y, et al. (2000) Rice dwarf mutant d1, which is defective in the alpha subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc Natl Acad Sci U S A 97: 11638–11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, et al. (2001) slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13: 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, et al. (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898. [DOI] [PubMed] [Google Scholar]
- 18. Hedden P, Kamiya Y (1997) GIBBERELLIN BIOSYNTHESIS: Enzymes, Genes and Their Regulation. Annu Rev Plant Physiol Plant Mol Biol 48: 431–460. [DOI] [PubMed] [Google Scholar]
- 19. Lange T (1998) Molecular biology of gibberellin synthesis. Planta 204: 409–419. [DOI] [PubMed] [Google Scholar]
- 20. Thomas SG, Phillips AL, Hedden P (1999) Molecular cloning and functional expression of gibberellin 2- oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci U S A 96: 4698–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee DJ, Zeevaart JA (2005) Molecular cloning of GA 2-oxidase3 from spinach and its ectopic expression in Nicotiana sylvestris. Plant Physiol 138: 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schomburg FM, Bizzell CM, Lee DJ, Zeevaart JA, Amasino RM (2003) Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 15: 151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lo SF, Yang SY, Chen KT, Hsing YI, Zeevaart JA, et al. (2008) A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20: 2603–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Suo H, Ma Q, Ye K, Yang C, Tang Y, et al. (2012) Overexpression of AtDREB1A causes a severe dwarf phenotype by decreasing endogenous gibberellin levels in soybean [Glycine max (L.) Merr]. PLoS One 7: e45568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang J, Tang D, Shen Y, Qin B, Hong L, et al. (2010) Activation of gibberellin 2-oxidase 6 decreases active gibberellin levels and creates a dominant semi-dwarf phenotype in rice (Oryza sativa L.). Journal of Genetics and Genomics 37: 23–36. [DOI] [PubMed] [Google Scholar]
- 26. Luo A, Qian Q, Yin H, Liu X, Yin C, et al. (2006) EUI1, encoding a putative cytochrome P450 monooxygenase, regulates internode elongation by modulating gibberellin responses in rice. Plant Cell Physiol 47: 181–191. [DOI] [PubMed] [Google Scholar]
- 27. Zhu Y, Nomura T, Xu Y, Zhang Y, Peng Y, et al. (2006) ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18: 442–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282. [DOI] [PubMed] [Google Scholar]
- 29. Mao J, Zhang YC, Sang Y, Li QH, Yang HQ (2005) From The Cover: A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc Natl Acad Sci U S A 102: 12270–12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 31. Zang A, Xu X, Neill S, Cai W (2010) Overexpression of OsRAN2 in rice and Arabidopsis renders transgenic plants hypersensitive to salinity and osmotic stress. J Exp Bot 61: 777–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xue W, Xing Y, Weng X, Zhao Y, Tang W, et al. (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40: 761–767. [DOI] [PubMed] [Google Scholar]
- 33.Scott A, Wyatt S, Tsou PL, Robertson D, Allen NS (1999) Model system for plant cell biology: GFP imaging in living onion epidermal cells. Biotechniques 26: 1125, 1128–1132. [DOI] [PubMed]
- 34. Delatte T, Trevisan M, Parker ML, Zeeman SC (2005) Arabidopsis mutants Atisa1 and Atisa2 have identical phenotypes and lack the same multimeric isoamylase, which influences the branch point distribution of amylopectin during starch synthesis. Plant J 41: 815–830. [DOI] [PubMed] [Google Scholar]
- 35. Schomburg FM (2002) Overexpression of a Novel Class of Gibberellin 2-Oxidases Decreases Gibberellin Levels and Creates Dwarf Plants. The Plant Cell Online 15: 151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, et al. (2002) Green revolution: a mutant gibberellin-synthesis gene in rice. Nature 416: 701–702. [DOI] [PubMed] [Google Scholar]
- 37. Spielmeyer W, Ellis MH, Chandler PM (2002) Semidwarf (sd-1), "green revolution" rice, contains a defective gibberellin 20-oxidase gene. Proc Natl Acad Sci U S A 99: 9043–9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, et al. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698. [DOI] [PubMed] [Google Scholar]
- 39. Nakajima M, Shimada A, Takashi Y, Kim YC, Park SH, et al. (2006) Identification and characterization of Arabidopsis gibberellin receptors. Plant J 46: 880–889. [DOI] [PubMed] [Google Scholar]
- 40. Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, et al. (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Busch MB, Sievers A (1990) Hormone treatment of roots causes not only a reversible loss of starch but also of structural polarity in statocytes. Planta 181: 358–364. [DOI] [PubMed] [Google Scholar]
- 42. Zhang Y, Zhu Y, Peng Y, Yan D, Li Q, et al. (2008) Gibberellin homeostasis and plant height control by EUI and a role for gibberellin in root gravity responses in rice. Cell Res 18: 412–421. [DOI] [PubMed] [Google Scholar]
- 43. Magome H, Yamaguchi S, Hanada A, Kamiya Y, Oda K (2008) The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J 56: 613–626. [DOI] [PubMed] [Google Scholar]
- 44. Cowling RJ, Kamiya Y, Seto H, Harberd NP (1998) Gibberellin dose-response regulation of GA4 gene transcript levels in Arabidopsis. Plant Physiol 117: 1195–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu YL, Li L, Wu K, Peeters AJ, Gage DA, et al. (1995) The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression. Proc Natl Acad Sci U S A 92: 6640–6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gubler F, Kalla R, Roberts JK, Jacobsen JV (1995) Gibberellin-regulated expression of a myb gene in barley aleurone cells: evidence for Myb transactivation of a high-pI alpha-amylase gene promoter. Plant Cell 7: 1879–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Magome H, Yamaguchi S, Hanada A, Kamiya Y, Oda K (2004) dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J 37: 720–729. [DOI] [PubMed] [Google Scholar]
- 48. Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, et al. (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290: 998–1009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of the deduced amino acid sequences of OsGA2ox5 with other GA2-oxidases. (A) Amino acid sequence alignment of rice GA2oxs (OsGA2ox1, OsGA2ox5, OsGA2ox6 and OsGA2ox9), Arabidopsis GA2oxs (AtGA2ox1, AtGA2ox7 and AtGA2ox8) and spinach GA2ox (SoGA2ox3) using the DNAMAN software. C20 GA2oxs (OsGA2ox5, OsGA2ox6, OsGA2ox9, AtGA2ox7, AtGA2ox8, and SoGA2ox3) contain three highly conserved sequence motifs (underlined with Roman numerals) that are absent in all C19 GA2oxs (OsGA2ox1 and OsGA2ox3 as examples for comparison). (B) Phylogenetic analysis of these GA2-oxidase proteins.
(TIF)
1000-grain weight of main panicle (A), grains number of each spike (B), Height of stem (C), Length of spike (D) of transgenic lines overexpressing OsGA2ox5 and wild type Zhonghua 11 under normal and GA3 condition. Twelve samples were measured for plant height, spike length and grains number of each line. 1,000-seed weight was measured in triplicate.
(TIF)
Primers.
(XLS)