Abstract
PARP-1 is a nuclear protein that has important roles in maintenance of genomic integrity. During genotoxic stress, PARP-1 recruits to sites of DNA damage where PARP-1 domain architecture initiates catalytic activation and subsequent poly(ADP-ribose)-dependent DNA repair. PARP-1 inhibition is a promising new way to selectively target cancers harboring DNA repair deficiencies. However, current inhibitors target other PARPs raising important questions concerning long-term off-target effects. Here we propose a new strategy that targets PARP-1 allosteric regulation as a selective way of inhibiting PARP-1. We found that disruption of PARP-1 domain-domain contacts through mutagenesis held no cellular consequences on recruitment to DNA damage or a model system of transcriptional regulation, but prevented DNA-damage dependent catalytic activation. Further, PARP-1 mutant overexpression in a pancreatic cancer cell line (MIA PaCa-2) increased sensitivity to platinum-based anti-cancer agents. These results not only highlight the potential of a synergistic drug combination of allosteric PARP inhibitors with DNA damaging agents in genomically unstable cancer cells (regardless of homologous recombination status), but also signify important applications of selective PARP-1 inhibition. Lastly, the development of a high-throughput (HT) PARP-1 assay is described as a tool to promote discovery of novel PARP-1 selective inhibitors.
Keywords: PARP, pancreatic cancer, allosteric, cisplatin, assay design
Introduction
Poly(ADP-ribose) polymerase-1 (PARP-1) is a multi-domain protein related to 16 other members of a family possessing ADP-ribosyl transferase catalytic domains with similar enzymatic and structural features (1). PARP-1 has multiple cellular functions including transcriptional regulation, cell death signaling, and DNA damage repair. PARP-1 recently has garnered attention as a therapeutic target in such lethal malignancies as pancreatic cancer (2).
PARP-1 has six domains (Figure 1). The N-terminus contains three zinc-binding domains: Zn1, Zn2, and Zn3. Zn1 and Zn2 bind to altered DNA structures (3), while Zn3 contains a structurally unique “zinc ribbon” fold that contributes to PARP-1 DNA damage-dependent activation through domain-domain contacts (4, 5). The central automodification domain contains a BRCT fold, and serves as a major site of PAR-modification. Toward the C-terminus, the WGR domain makes DNA and protein contacts that are important for catalytic activation (6, 7). The C-terminal catalytic region contains the active site where NAD+ is consumed to synthesize poly(ADP-ribose), or PAR, covalently modified onto nuclear targets such as PARP-1 itself (8). Under genotoxic stress, PARP-1 domains assemble on DNA damage and form a network of domain-domain contacts that imposes a structural distortion on the catalytic domain (7), leading to a dramatic increase in catalytic activity (9). This hyper-activation accounts for the majority of PAR accumulation at sites of DNA damage, leading to recruitment of repair proteins to facilitate DNA repair (10). PAR formation DNA repair under low to moderate levels of genomic stress, but can trigger cell death mechanisms upon excessive damage. These important roles of PARP-1 ensure maintenance of genomic integrity and provide protection from carcinogenesis of badly damaged cells.
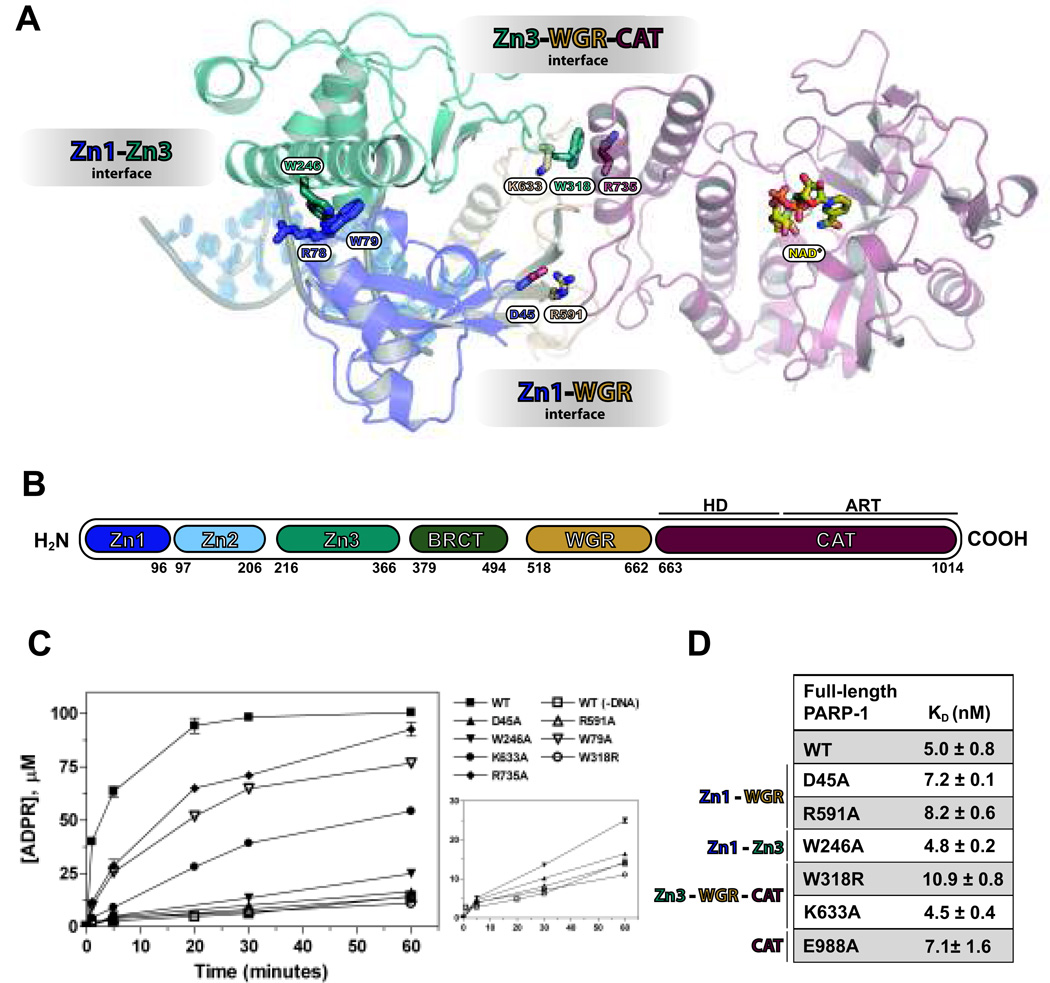
Figure 1.
PARP-1 domain-domain contacts are necessary for DNA-dependent activity, but not high-affinity DNA binding. (A) Structural representation of activated PARP-1 highlighting the interdomain interfaces. (B) Schematic of full-length PARP-1. (C) DNA-dependent PARP-1 catalytic activity. (D) DNA binding affinities of full-length PARP-1 mutants.
Therapeutic strategies targeting PARP-1 have promising applications for the treatment of some cancers, drawing considerable attention in recent years (11). PARP-1 inhibitors have been optimized to low nanomolar potency and show clinical utility when used as a single agent or in combination therapy to treat cancers with DNA repair deficiencies (12). While the clinical potential of PARP-1 inhibitors looks promising, therapeutic benefit versus adverse effects stemming from targeting other PARPs is not well understood. For instance, a study profiled the binding of 185 current PARP inhibitors to the active sites of 13 PARP family members, and observed varying degrees of cross-selectivity (13). With such a high degree of conservation in the catalytic active sites of PARPs, it is not surprising that cross-selectivity exists among small molecules that target the substrate-binding site.
Toward achieving selective PARP-1 inhibition, we focused on PARP-1 domain unique to DNA damage-dependent activation. Zn1, Zn3, and WGR domains are essential for catalytic activation. A recent crystal structure of PARP-1 revealed that these domains assemble into a complex in the presence of damaged DNA (7). This structure has opened possibilities of selectively targeting damage-dependent activation through disruption of PARP-1 domain-domain contacts.
Here, we mutated key residues located at the interfaces between essential PARP-1 domains and observed no major changes in binding to DNA damage, but severe deficiencies in catalytic activation. A pancreatic cancer cell line carrying these deficiencies in PARP-1 showed an increase in sensitivity to platinum-based anticancer agents, suggesting a dominant negative effect that interferes with DNA damage processing. With prospective therapeutic applications of allosteric PARP-1 inhibition, we present a PARP-1 interdomain communication assay designed to identify new classes of selective PARP-1 inhibitors.
Materials and Methods
Gene Cloning and Mutagenesis
Full-length WT PARP-1 (1–1014) and ΔZn2 PARP-1 (97–206 deleted) were expressed with an N-terminal hexahistidine tag (pET 28). Zn1-Zn3 (1–366 with 97–206 deleted) and WGR-CAT (518–1014) were expressed with a C-terminal hexahistidine tag (pET24). QuickChange mutations (Stratagene) were verified by automated sequencing. PARP-1 pcDNA constructs were cloned as described (3). EGFP-PARP-1 was generated by sub-cloning the Nhe1/Xho1 fragment from pcDNA-PARP-1 into pEGFP-N1.
Protein Production
PARP-1 proteins were expressed in Escherichia coli and purified as described (14).
Transient Transfection and Immunofluorescent staining of MEFs
PARP-1−/− MEFs were treated as described (3). To induce DNA damage, H2O2 was added to cells for 10 minutes prior to fixation. HeLa cells were grown under the same conditions as the MEFs. Cells were transfected 24 hours later with 1µg DNA and 3µl Fugene® (Promega) in Serum-Free media following the recommended protocol.
Live cell microscopy and laser irradiation
Cells were sensitized post-transfection with 1µM BrdU in warm phenol-red free media (Ham’s F-12 with 25mM Hepes pH 8.0, 10% FBS) for 24 hours at 37°C, 5% CO2 before addition of Hoechst stain (10µg/mL). Experiments were performed using a Zeiss LSM-510 Meta Confocal laser scanning microscope equipped with a 405nm diode laser (set to 100% power) focused through 63×/1.4 NA oil immersion lens to locally irradiate nuclear sites for 1 second. Images were recorded by excitation with a 488nm argon laser (set to 10% power).
Drug Sensitivity Assays
Stably transfected MIA PaCa-2 cells (Figure S3) were seeded at low confluency and incubated at 37°C overnight. Cells were treated with drug and then grown to confluence (5–6 days). Cell viability was assessed by quantification of double-stranded DNA using Quant-iT PicoGreen (Invitrogen). Gemcitabine was purchased from Lilly. All other drugs were purchased from Sigma Aldrich.
Fluorescent Polarization DNA Binding Assay
Reactions were carried out as previously described using an 18-bp DNA duplex (5). For the PARP-1 release experiment, WT and mutant proteins (200 nM) were first incubated with the DNA duplex (100nM total DNA, 5% fluorescein labeled) for 30 min before addition of NAD+ (5mM) or H2O. Polarization was measured over time on a plate reader (Perkin Elmer).
Colorimetric PARP-1 Automodification Assay
This assay measures incorporation of biotinylated-NAD+ into PAR (14).
Androgen receptor (AR) reporter assay
AR ligand-induced transcriptional activity was measured by relative luciferase activity (RLU) as described (15).
Results and Discussion
Disruption of PARP-1 domain-domain contacts impairs catalytic activation without affecting high-affinity interaction with DNA damage
The essential Zn1, Zn3, and WGR domains each have low binding affinity for DNA damage, but in combination their collective affinity increases nearly 100-fold (Figure S1). The activated PARP-1 structure indicated that each of these domains forms contacts with DNA that are mutually compatible, consistent with their high collective DNA-binding affinity. We tested whether the contacts at the interfaces between the domains contributed to the collective assembly on DNA, thus forming a high-affinity interaction with DNA. Key residues located at domain interfaces were mutated (Figure 1A). Although the mutations had a severe impact on DNA damage-dependent catalytic activity (Figure 1C), they did not affect overall PARP-1 DNA-binding affinity (Figure 1D). The Zn2 domain is not essential for DNA-dependent PARP-1 activity (7); however Zn2 has high binding affinity that could potentially mask DNA-binding deficiencies of interdomain mutants. Thus, several mutants were tested in a PARP-1 construct with Zn2 deleted (ΔZn2). All mutants tested retained a high DNA-binding affinity (Figure S2), indicating that high-affinity binding to DNA mediated by the assembly of Zn1, Zn3, and WGR is independent from the allosteric regulation that triggers activation.
Allosteric mutant W318R localizes to sites of DNA damage, but is defective in damage-induced PAR synthesis and release from DNA
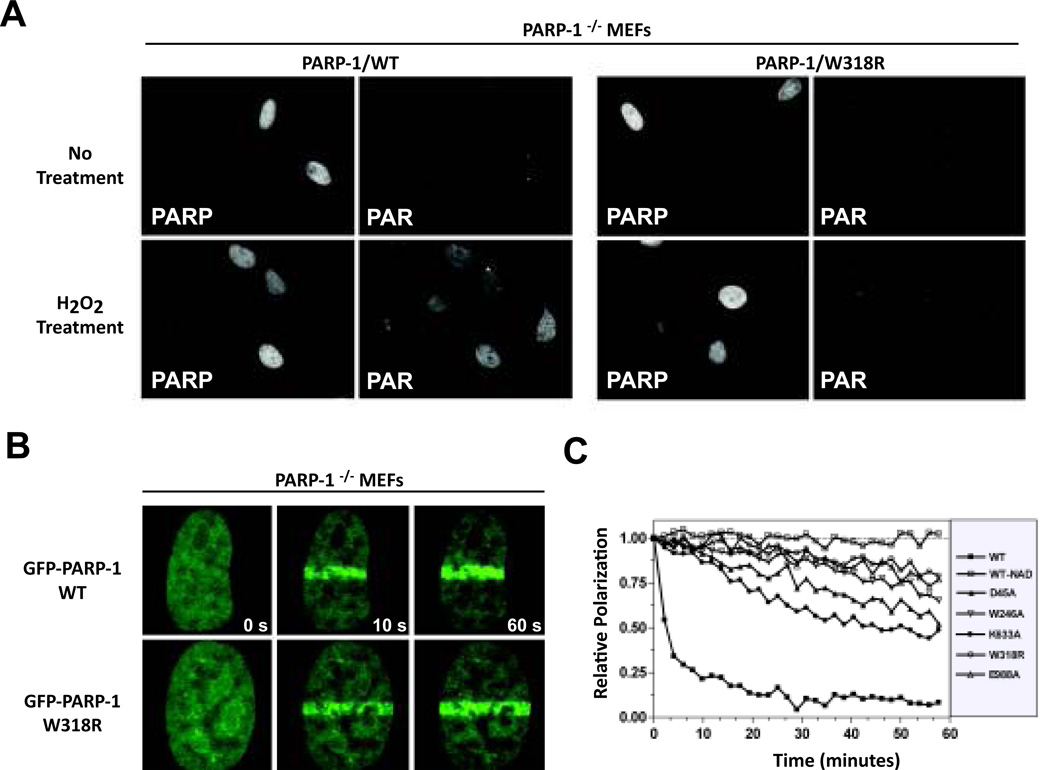
Cellular tests of PARP-1 function assessed the effect of disrupting allosteric activation. PARP-1/W318R was compared to PARP-1/WT, since it showed major deficiencies in DNA-dependent activation biochemically (Figure 1C) (5). In the absence of DNA damage, both localized to the nucleus of PARP-1−/− MEFs and did not produce a notable PAR signal using indirect immunohistochemistry (Figure 2A). Upon treatment with H2O2, PAR production was significantly increased in cells positive for the presence of PARP-1/WT but not PARP-1/W318R, indicating that the cellular deficiency of PARP-1/W318R resembles the biochemical observation. We next tested the ability of PARP-1/W318R to localize to damage sites. GFP-PARP-1 was transfected into HeLa cells. GFP-PARP-1/WT and GFP-PARP-1/W318R both recruited to DNA damage sites in a similar manner (Figure 2B), further supporting that recognition of damaged DNA is not affected by disruptions in PARP-1 allosteric communication.
Figure 2.
Cellular localization and activity of PARP-1 mutants. (A) Immunofluorescent detection of PARP localization and PAR formation in response to H2O2 treatment. (B) Localization of GFP-PARP-1 to sites of laser-induced damage in HeLa cells. (C) NAD+ dependent release of PARP-1 mutants from DNA in the presence of NAD+ (5mM).
DNA-activated PARP-1 consumes NAD+ in an automodification reaction that results in covalently attached PAR, which ultimately releases PARP-1 from DNA. PARP-1 inhibitors cause deficiencies in PARP-1 release from DNA (16). Prolonged residency at DNA damage sites is proposed to contribute to the genotoxic effect of PARP-1 inhibition, and to the synergistic effect of combining PARP-1 inhibitors with DNA-damaging agents. We tested whether interdomain mutants showed deficiencies in the mechanism of release from DNA damage. Upon addition of NAD+, the W318R mutant exhibited a drastic delay in release kinetics compared to WT (Figure 2C), similar to that of catalytic mutant E988A. Other interdomain mutants exhibited delayed release kinetics, consistent with their slower rates of PAR formation (Figure 1D). Collectively, cell-based and biochemical analyses indicate that disruption of domain-domain contacts critical for activation do not influence PARP-1 binding to DNA damage, and that the deficiency in DNA damage-dependent activation causes PARP-1 to remain engaged with DNA in a manner that is not effectively reversed by automodification.
Disruption of PARP-1 allosteric regulation sensitizes pancreatic cancer cells to platinum-based agents
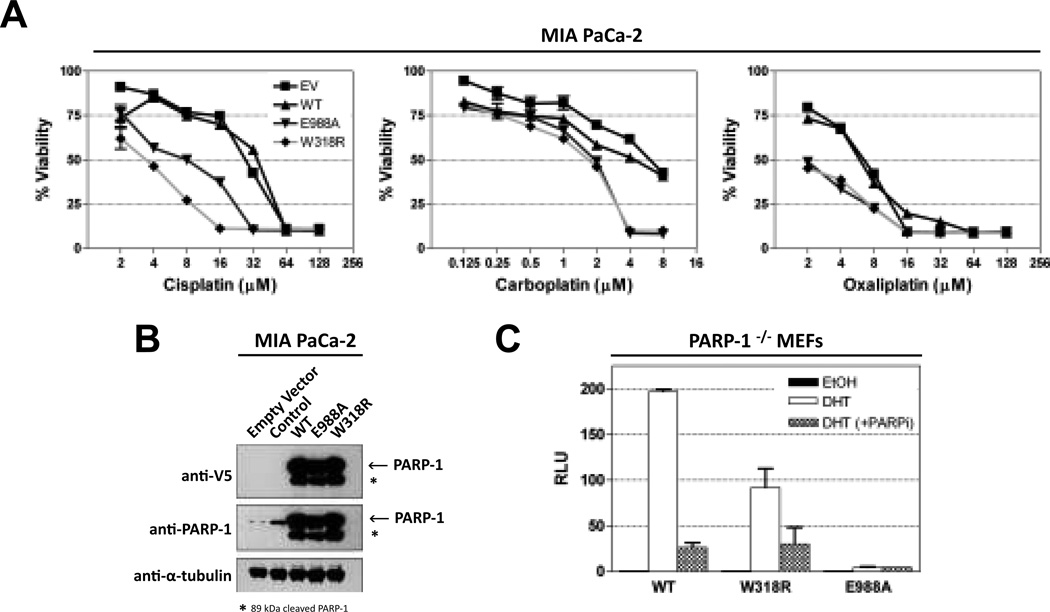
To determine whether allosteric disruption of PARP-1 catalytic activity could deliver comparable therapeutic potential as current PARP inhibitors, isogenic MIA PaCa-2 cells overexpressing PARP-1 mutants were treated with a variety of chemotherapeutic agents following two criteria: 1) they currently or previously had been part of a chemotherapeutic regimen for pancreatic cancer and 2) they are classified as DNA damaging agents. PARP-1/WT and PARP-1/W318R transfected cell lines were analyzed, as well as the cell line bearing the E988A mutation that targets a catalytic active site residue and should therefore mimic drugs that target the catalytic domain. Overexpression of PARP-1/WT had no effect in combination with chemotherapy (Figure 3A). However, overexpression of PARP-1/W318R or PARP-1/E988A rendered cells 4–8 times more sensitive to platinum-based anticancer agents (Figure 3A). Increased sensitivity was not seen in combination with other chemotherapeutic agents (Figure S3), indicating a preferential synergistic potential between PARP-1 inhibition and platinum-based therapies, in line with current clinical trial outcomes (17). In addition, allosteric disruption of catalytic activity appears to be equally effective at sensitizing cells as direct disruption at the catalytic domain. The molecular mechanisms responsible for sensitization with platinum agents are not completely understood. However, sensitization from this combination has been observed in MIA PaCa-2 cells (18) as well as other cell-based models (19–21) and is likely cell-type specific (22). One possible mechanism of action could relate to the ability of mutant PARP-1 to recognize platinum-DNA damage (23) and form trapped complexes that shield the toxic platinum-modified lesions from repair and promote accumulation of unrepaired damage.
Figure 3.
Functional consequences of inactivating PARP-1 interdomain communication. (A) Pancreatic cancer cells stably transfected with inactive PARP-1 mutants are sensitive to platinum-based agents compared to WT. (B) Western blot analysis of cells in panel (A) using anti-PARP (Trevigen) and anti-V5 (Invitrogen) antibodies. (C) Ligand-induced and control AR reporter activity were measured in transfected PARP-1−/− MEFs by relative luciferase activity as result of 3 independent replicates ±SE. Results are normalized to the control set to “1”. PARP inhibitor ABT-888 used at 2.5 µM.
Allosteric disruption of PARP-1 catalytic activity does not prevent AR transcriptional regulatory functions
In addition to the DNA damage response, PARP-1 is involved in a number of transcriptional regulation events (24). For example, PARP-1 catalytic activity is necessary for androgen receptor (AR) ligand-dependent transcription (15). To determine if allosteric regulation is specific to the DNA damage response, AR transcriptional function was assessed in PARP-1−/− MEFs transiently transfected with PARP-1/WT, PARP-1/W318R, and PARP-1/E988A. The catalytic mutant PARP-1/E988A is deficient in AR transcription compared to WT (Figure 3C) (15) whereas the interdomain mutant PARP-1/W318R exhibited substantial ligand-induced AR reporter activity. Furthermore, the ligand-induced AR reporter activity of PARP-1/W318R was decreased in the presence of PARP inhibitor ABT-888, similar to PARP-1/WT (Figure 3C). Thus, disruption of PARP-1 allosteric regulation shuts down DNA damage-dependent activity without altering the catalytic domain per se, thus allowing PARP-1 to maintain other important cellular functions outside of the DNA damage response.
Development of a high-throughput method to detect allosteric regulation
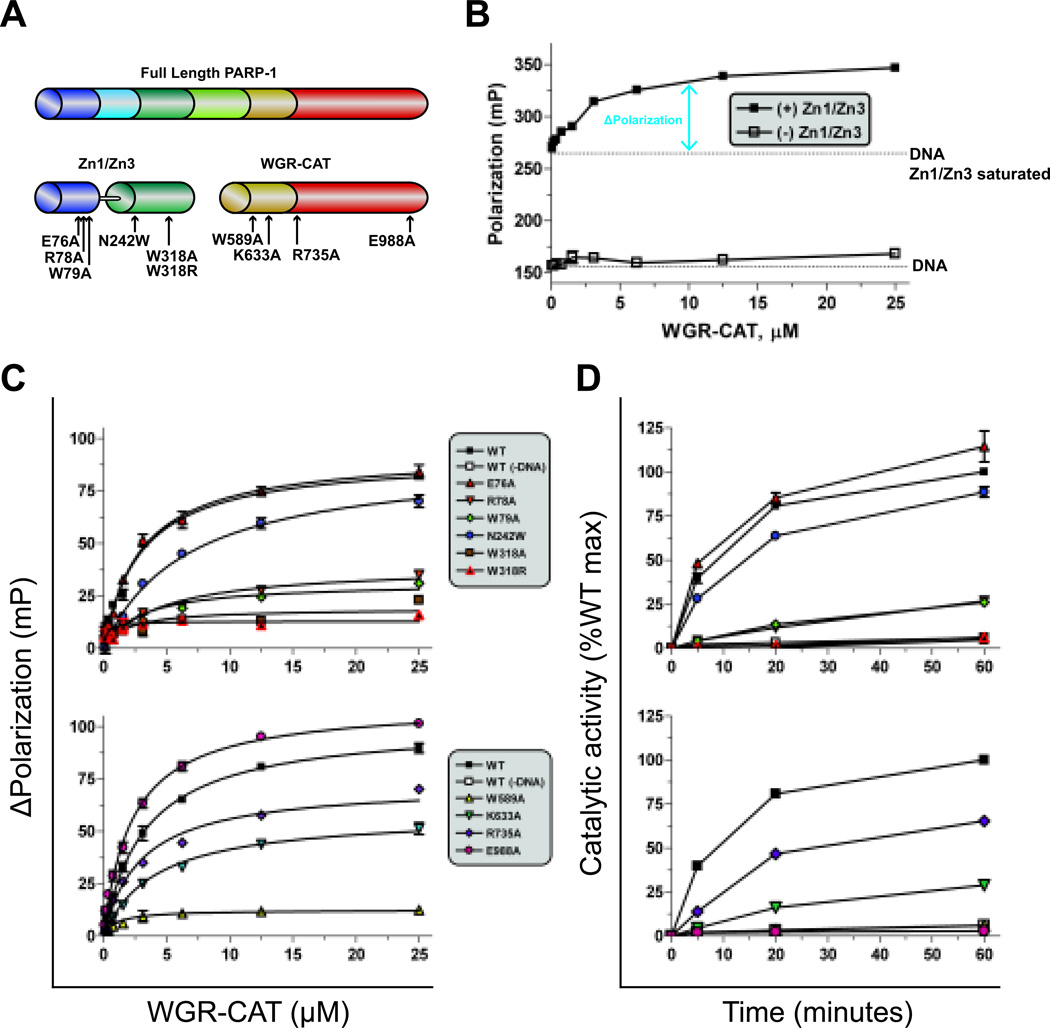
As PARP-1 inhibitors continue to hold tremendous promise as targeted anticancer agents, there is a growing interest in isoform-specific PARP-1 inhibitors. The unique structural aspects of the PARP-1 complex of essential domains is therefore of particular interest as a therapeutic target. Using the core components of this complex as our target model (Figure 4A), we developed a high-throughput assay that is capable of detecting complex formation in the presence of DNA. The assay is based on fluorescent polarization techniques, and detects the binding of WGR-CAT to a Zn1-Zn3 saturated DNA complex as an increase in fluorescence polarization (FP) (Figure 4B). To validate that the readout accurately represented allosteric activation, we tested several mutants at the domain interfaces. Four mutations were made along the Zn1-Zn3 interface: E76A, R78A, W79A, and N242W; two mutations were made along the Zn3-WGR-HD interface. Mutations in the Zn1-Zn3 construct did not affect their DNA binding affinity (Figure S4). When WGR-CAT was added to the Zn1-Zn3 variants, the maximum change in polarization (ΔPolarization) was notably lower for R78A, W79A, W318A, and W318R, although the estimated binding affinity was comparable to WT (Figure 4C). The deficiency in ΔPolarization correlated well with the observed deficiencies in DNA-dependent catalytic activation (Figure 4D). The N242W mutant was designed to mimic the steric clash of a potential small allosteric inhibitor bound at the Zn1-Zn3 interface. In this instance, the binding affinity of WGR-CAT dropped 2-fold, and consistently, the rate of PAR synthesis was reduced. Mutations made in WGR-CAT further validated the assay. When WGR residues K633 and R735 were mutated to alanine, decreases in maximum polarization were seen in the high-throughput assay that corresponded to the level of DNA-dependent PAR synthesis (Figures 4C and 4D). The mutant W589A is known to affect DNA binding of WGR, and showed no increase in polarization, which correlated well with its deficient DNA-dependent activation. Mutant E988A was used to show that direct disruption of the catalytic domain does not influence the assay readout, and therefore catalytic site inhibitors would not be detected by this assay.
Figure 4.
A high-throughput (HT) fluorescent polarization (FP) assay to detect allosteric PARP-1 inhibitors. (A) Schematic of PARP-1 proteins used in this assay (Zn1-Zn3 and WGR-CAT), mapped with mutation sites. (B) Dose response of WGR-CAT binding to fluorescein-labeled DNA alone or to fluorescein-labeled DNA saturated with Zn1-Zn3. (C) WGR-CAT binding in the FP assay in the presence of Zn1-Zn3 (top) or WGR-CAT (bottom) mutations. (D) Catalytic activity of Zn1-Zn3 and WGR-CAT combinations using the colorimetric assay with mutations made in Zn1-Zn3 (top) or WGR-CAT (bottom).
Perspective
While the use of PARP-1 inhibitors has been widely deployed, the therapeutic benefit from pan-PARP inhibition (i.e., targeting multiple PARP family proteins) as opposed to specific PARP-1 inhibition is not clear. The functional roles among PARPs are diverse, and cross-inhibition creates the potential for deleterious off-target effects (13, 25). On the other hand, increased therapeutic effectiveness can often accompany the complexities of polypharmacology, as exemplified by promiscuous kinase inhibitors (26). Selective pharmacological targeting of PARP-1 (and other PARPs) will serve useful in distinguishing therapeutic effectiveness from toxicity in complex diseases.
In this study, disruption of allosteric regulation was used to model selective inhibition of PARP-1. Single-point mutations at domain interfaces were used to disrupt allosteric regulation. These mutants do not affect high affinity binding to DNA damage, although they do have a pronounced effect on DNA-dependent catalytic activation. In a cellular environment, these observations are paralleled as mutant PARP-1 recruits to sites of DNA damage, but fails to generate detectable PAR. Interestingly, the role of PARP-1 in AR-dependent transcription was not severely affected, revealing applications of allosteric inhibitors in reducing potential off-target effects. We also find that high-affinity DNA binding combined with catalytic deficiency stalls release from DNA in the presence of NAD+. The prolonged occupancy of mutant PARP-1 likely contributes to sensitization of DNA damaging agents by retaining unrepaired DNA damage.
We have identified a synergistic combination between allosterically disrupted PARP-1 and platinum-based agents in pancreatic cancer cells with a wild-type BRCA2-related DNA damage repair pathway. We believe allosteric inhibition will likely hold therapeutic potential in other types of cancers when used as combination therapy (in a BRCA2-wild type setting) or as monotherapy, especially in the context of BRCA1/2/Fanconi anemia-deficient tumors (27). We have developed a high-throughput assay to detect the allosteric activation status of PARP-1 to facilitate the identification of pharmacological inhibitors. This study initiates an innovative pipeline to identify an optimal compound that will disrupt PARP-1 function in cancer cells.
Supplementary Material
Acknowledgments
This work was supported by funds from the American Cancer Society (RSG0918301 to J.M.P and RSG1011901 to J.R.B), the NIH (R01087282 to J.M.P.), and the Prostate Cancer Foundation (Movember Challenge Award to K.E.K). J.D.S is supported by a Ruth L. Kirschstein National Research Service Award, and M.J.S is supported by a Prostate Cancer Foundation Young Investigator Award.
Footnotes
The authors declare no conflict of interest
References
- 1.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 2.Lowery MA, Kelsen DP, Stadler ZK, Yu KH, Janjigian YY, Ludwig E, et al. An emerging entity: Pancreatic adenocarcinoma associated with a known BRCA mutation: Clinical descriptors, treatment implications, and future directions. Oncologist. 2011;16(10):1397–1402. doi: 10.1634/theoncologist.2011-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langelier MF, Planck JL, Roy S, Pascal JM. Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: Structural and functional insights into DNA-dependent PARP-1 activity. J Biol Chem. 2011;286(12):10690–10701. doi: 10.1074/jbc.M110.202507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Langelier MF, Servent KM, Rogers EE, Pascal JM. A third zinc-binding domain of human poly(ADP-ribose) polymerase-1 coordinates DNA-dependent enzyme activation. J Biol Chem. 2008;283(7):4105–4114. doi: 10.1074/jbc.M708558200. [DOI] [PubMed] [Google Scholar]
- 5.Langelier MF, Ruhl DD, Planck JL, Kraus WL, Pascal JM. The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J Biol Chem. 2010;285(24):18877–18878. doi: 10.1074/jbc.M110.105668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37(11):3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langelier MF, Planck JL, Roy S, Pascal JM. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012;336(6082):728–732. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 9.Alvarez-Gonzalez R, Althaus FR. Poly(ADP-ribose) catabolism in mammalian cells exposed to DNA-damaging agents. Mutat Res. 1989;218(2):67–74. doi: 10.1016/0921-8777(89)90012-8. [DOI] [PubMed] [Google Scholar]
- 10.Malanga M, Althaus FR. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem Cell Biol. 2005;83(3):354–356. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- 11.Javle M, Curtin NJ. The role of PARP in DNA repair and its therapeutic exploitation. Br J Cancer. 2011;105(8):1114–1122. doi: 10.1038/bjc.2011.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferraris DV. Evolution of poly(ADP-ribose) polymerase-1 (PARP-1) inhibitors. from concept to clinic. J Med Chem. 2010;53(12):4561–4584. doi: 10.1021/jm100012m. [DOI] [PubMed] [Google Scholar]
- 13.Wahlberg E, Karlberg T, Kouznetsova E, Markova N, Macchiarulo A, Thorsell A, et al. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat Biotechnol. 2012;30(3):283–288. doi: 10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]
- 14.Langelier MF, Planck JL, Servent KM, Pascal JM. Purification of human PARP-1 and PARP-1 domains from escherichia coli for structural and biochemical analysis. Methods Mol Biol. 2011;780:209–226. doi: 10.1007/978-1-61779-270-0_13. [DOI] [PubMed] [Google Scholar]
- 15.Schiewer MJ, Goodwin JF, Han S, Brenner JC, Augello MA, Dean JL, et al. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2012;2(12):1134–1149. doi: 10.1158/2159-8290.CD-12-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 2012;72(21):5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pishvaian MJ, Wang H, Zhuang T, He AR, Hwang JJ, Hankin A, et al. A phase I/II study of ABT-888 in combination with 5-fluorouracil (5-FU) and oxaliplatin (ox) in patients with metastatic pancreatic cancer (MPC). | 2013 gastrointestinal cancers symposium | abstracts | meeting library. J.Clin.Oncol. 2012;30(suppl 34) abstr 147. [Google Scholar]
- 18.Jacob DA, Bahra M, Langrehr JM, Boas-Knoop S, Stefaniak R, Davis J, et al. Combination therapy of poly (ADP-ribose) polymerase inhibitor 3-aminobenzamide and gemcitabine shows strong antitumor activity in pancreatic cancer cells. J Gastroenterol Hepatol. 2007;22(5):738–748. doi: 10.1111/j.1440-1746.2006.04496.x. [DOI] [PubMed] [Google Scholar]
- 19.Donawho CK, Luo Y, Luo Y, Penning TD, Bauch JL, Bouska JJ, et al. ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clin Cancer Res. 2007;13(9):2728–2737. doi: 10.1158/1078-0432.CCR-06-3039. [DOI] [PubMed] [Google Scholar]
- 20.Rottenberg S, Jaspers JE, Kersbergen A, van der Burg E, Nygren AO, Zander SA, et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci U S A. 2008;105(44):17079–17084. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olaussen KA, Adam J, Vanhecke E, Vielh P, Pirker R, Friboulet L, et al. PARP1 impact on DNA repair of platinum adducts: Preclinical and clinical read-outs. Lung Cancer. 2013;80(2):216–222. doi: 10.1016/j.lungcan.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 22.Guggenheim ER, Ondrus AE, Movassaghi M, Lippard SJ. Poly(ADP-ribose) polymerase-1 activity facilitates the dissociation of nuclear proteins from platinum-modified DNA. Bioorg Med Chem. 2008;16(23):10121–10128. doi: 10.1016/j.bmc.2008.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu G, Chang P, Lippard SJ. Recognition of platinum-DNA damage by poly(ADP-ribose) polymerase-1. Biochemistry. 2010;49(29):6177–6183. doi: 10.1021/bi100775t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kraus WL. Transcriptional control by PARP-1: Chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol. 2008;20(3):294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welsby I, Hutin D, Leo O. Complex roles of members of the ADP-ribosyl transferase super family in immune defences: Looking beyond PARP1. Biochem Pharmacol. 2012;84(1):11–20. doi: 10.1016/j.bcp.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 26.Ghoreschi K, Laurence A, O'Shea JJ. Selectivity and therapeutic inhibition of kinases: To be or not to be? Nat Immunol. 2009;10(4):356–360. doi: 10.1038/ni.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Heijden MS, Brody JR, Dezentje DA, Gallmeier E, Cunningham SC, Swartz MJ, et al. In vivo therapeutic responses contingent on fanconi anemia/BRCA2 status of the tumor. Clin Cancer Res. 2005;11(20):7508–7515. doi: 10.1158/1078-0432.CCR-05-1048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.