Abstract
Hypoxia-inducible factor 1 (HIF-1) is a major mediator of tumor physiology, and its activation is correlated with tumor progression, metastasis, and therapeutic resistance. HIF-1 is activated in a broad range of solid tumors due to intratumoral hypoxia or genetic alterations that enhance its expression or inhibit its degradation. As a result, decreasing HIF-1α expression represents an attractive strategy to sensitize hypoxic tumors to anticancer therapies. Here, we show that cyclin-dependent kinase 1 (CDK1) regulates the expression of HIF-1α, independent of its known regulators. Overexpression of CDK1 and/or cyclin B1 is sufficient to stabilize HIF-1α under normoxic conditions, whereas inhibition of CDK1 enhances the proteasomal degradation of HIF-1α, reducing its half-life and steady-state levels. In vitro kinase assays reveal that CDK1 directly phosphorylates HIF-1α at a previously unidentified regulatory site, Ser668. HIF-1α is stabilized under normoxic conditions during G2/M phase via CDK1-mediated phosphorylation of Ser668. A phospho-mimetic construct of HIF-1α at Ser668 (S668E) is significantly more stable under both normoxic and hypoxic conditions, resulting in enhanced transcription of HIF-1 target genes and increased tumor cell invasion and migration. Importantly, HIF-1α (S668E) displays increased tumor angiogenesis, proliferation, and tumor growth in vivo compared with wild-type HIF-1α. Thus, we have identified a novel link between CDK1 and HIF-1α that provides a potential molecular explanation for the elevated HIF-1 activity observed in primary and metastatic tumors, independent of hypoxia, and offers a molecular rationale for the clinical translation of CDK inhibitors for use in tumors with constitutively active HIF-1.
Keywords: CDK1, HIF-1α, angiogenesis, cell cycle, hypoxia
Introduction
Angiogenesis, cell survival, invasion, and enhanced glucose metabolism are hallmarks of tumorigenesis that involve the upregulation of genes that are frequently activated by hypoxia.1,2 Hypoxia, involving a reduction in the normal level of tissue oxygen, is frequently observed in cancer, and exposure to prolonged or severe hypoxic conditions can lead to cell death or promote selection for death resistance.3,4 While hypoxia can be toxic to both normal and cancer cells, cancer cells take particular advantage of their ability to initiate a transcriptional program that allows them to adapt to these harsh conditions, promoting survival and proliferation in a hypoxic environment.5 Hypoxia-inducible factor 1 (HIF-1) is a basic helix-loop-helix-PAS domain transcription factor that is a critical mediator of the cellular response to oxygen deprivation.6 HIF-1 is a heterodimer that consists of a constitutively expressed subunit, HIF-1β, and HIF-1α, a subunit whose expression is tightly regulated in an oxygen-dependent manner. In the presence of oxygen, cytoplasmic HIF-1α is rapidly hydroxylated on proline 402 (Pro402) and 564 (Pro564) by prolyl hydroxylase-domain protein 1–3 (PHDs).7,8 Following hydroxylation, HIF-1α is recognized by the von Hippel-Lindau (VHL) tumor suppressor protein, which recruits an E3 ubiquitin-protein ligase complex that leads to the ubiquitination and subsequent degradation of HIF-1α by the 26S proteasome.9,10 Under hypoxic conditions, HIF-1α is no longer rapidly degraded, allowing the protein to accumulate and enter the nucleus, where it dimerizes with HIF-1β. The HIF-1 heterodimer then binds to regulatory DNA sequences known as hypoxia response elements (HRE), and it recruits transcriptional co-activators to enhance the transcription of a plethora of target genes that promote angiogenesis, tumor progression, therapeutic resistance, and metastasis.11,12
In addition to intratumoral hypoxia, HIF-1α is frequently overexpressed in human cancers due to genetic alterations. Most notably, loss of VHL results in impaired ubiquitination and degradation of HIF-1α, leading to its constitutive expression under normoxic conditions, which results in the constitutive upregulation of hypoxia-inducible genes that drive angiogenesis and tumor growth.13 Alternatively, gain of function mutations in oncogenic signaling pathways, such as PI3K or RAS, enhance the transcription of HIF-1α and increase its steady-state levels.14,15 Post-translational modification of HIF-1α has also been shown to regulate its expression and function. For example, direct phosphorylation by ERK blocks the nuclear export of HIF-1α and ensures its efficient nuclear accumulation and full activation.16 It has also been reported that phosphorylation by glycogen synthase kinase 3 (GSK3) and Polo-like kinase 3 (PLK3) promote HIF-1α degradation, while binding to the cellular chaperone, HSP90, stabilizes HIF-1α.17-19 Thus, HIF-1α expression and the transcriptional activation of HIF-1 are controlled by both oxygen-dependent and oxygen-independent mechanisms in response to extracellular stimuli and the conditions of the cellular microenvironment. The identification of novel mechanisms that regulate HIF-1α expression is extremely valuable in the effort to understand tumor progression as well as to target HIF-1 as an anti-cancer therapeutic strategy.
In this study, we identify a novel mechanism controlling the stability of HIF-1α, independent of its known regulators and oxygen concentration. Specifically, we found that CDK1 directly phosophorylates HIF-1α at Ser668, increasing its steady-state levels under both normoxic and hypoxic conditions to promote the transcription of HIF-1 target genes. Expression of a phospho-mimetic construct of HIF-1α at Ser668 (S668E) enhanced tumor angiogenesis, proliferation, and tumor growth in vivo. Thus, CDK1-mediated phosphorylation of HIF-1α is important for regulating the steady-state levels of HIF-1α and provides a novel molecular explanation for the elevation of HIF-1 in primary and metastatic tumors, independent of hypoxia.
Results
Inhibition of CDK1/4 decreases HIF-1α expression at the post-translational level, independent of its known regulators
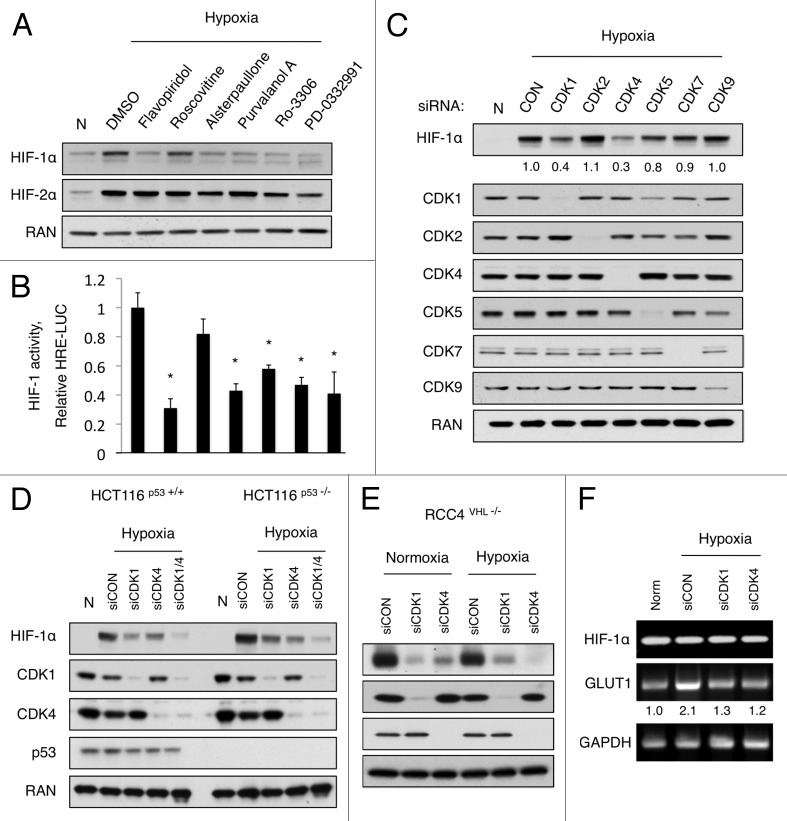
Due to the importance of HIF-1 activation for tumor progression and lethal metastases, the regulation of HIF-1α stability and the mechanisms responsible for its degradation warrant further investigation. A recent report from our lab revealed that SLM3, a novel compound that sensitizes cancer cells to TRAIL-induced cell death under hypoxic conditions, does so in part by inhibiting CDK1, which subsequently reduced HIF-1α levels.20 Thus, we tested whether other CDK inhibitors, which are known to block CDK1 activity and have shown promising results in preclinical and clinical trials, could also reduce HIF-1α expression. To this end, we assembled the following panel of relevant CDK inhibitors: flavopiridol, roscovitine, alsterpaullone, purvalanol A, Ro-3306 (CDK1-specific), and PD-0332991 (CDK4/6-specific).21,22 HCT116 colon carcinoma cells were exposed to hypoxia (0.2% O2) and treated with vehicle or the indicated inhibitor for 16 h, and western blotting was used to monitor HIF-1α protein levels. All of the CDK inhibitors tested significantly reduced HIF-1α protein levels, with the exception of Roscovitine, which showed only a modest effect (Fig. 1A). The protein levels of HIF-2α were not significantly changed by any of the CDK inhibitors, although modest inhibition was observed following treatment with PD-0332991. To assess the transcriptional activation of HIF-1, a parallel experiment was performed on HCT116 cells stably expressing a previously described HIF-1 reporter that drives luciferase expression (HRE-Luc) (Fig. 1B).20 To account for cell death or growth inhibition in response to treatment with these compounds, HCT116 cells constitutively expressing luciferase (CMV-Luc) were treated simultaneously, and the HRE-Luc signal was normalized to CMV-Luc levels. These results verified that HIF-1 activity correlated with HIF-1α levels, and was significantly reduced by multiple CDK inhibitors (Fig. 1B).
Figure 1. CDK inhibitors decrease HIF-1α expression and HIF-1 activity independent of p53, VHL, or oxygen concentration. (A) HCT116 cells and (B) HCT116 cells stably expressing an HRE-Luc reporter were treated with the indicated compounds and exposed to hypoxia for 16 h. (A) HIF-1α expression levels were monitored by western blotting, and (B) HIF-1 activity was measured by luciferase activity. The bar graph represents HIF-1 activity after normalization to CMV-LUC. (C) HCT116 cells were transfected with the indicated siRNA for 48 h prior to exposure to hypoxia for 4 h. (D) HCT116 (WT and p53 −/−) and (E) RCC4 (VHL−/−) cells were treated with the indicated siRNA for 48 h and then exposed to hypoxia for 4 h. Protein was collected for western blotting, and (F) RNA was collected for RT-PCR analysis. (n = 3 for all experiments).
A majority of the CDK inhibitors that have been developed inhibit multiple CDKs with similar IC50.23 Therefore, we sought to identify which individual CDK(s) alter HIF-1α expression. HCT116 cells were transfected with control siRNA or siRNA against CDK1, CDK2, CDK4, CDK5, CDK7, and CDK9. After 48 h, the cells were incubated in hypoxia for 4 h, and western blotting was used to measure HIF-1α expression levels. Only knockdown of CDK1 or CDK4 significantly reduced HIF-1α levels (Fig. 1C), confirming that specific inhibition of CDK1 and/or CDK4 effectively blunts hypoxia-induced HIF-1α expression and HIF-1 transcriptional activity.
Next, we focused on identifying the mechanism by which CDK1 and CDK4 regulate HIF-1α expression. First, we asked whether CDK1/4-mediated downregulation of HIF-1α is controlled by p53 and/or VHL, which are known regulators of HIF-1α stability.10,24 HCT116 p53+/+ and HCT116p53 −/− colon cancer cell lines were transfected with scrambled siRNA (siCON) or siRNA targeting CDK1, CDK4, or both. Forty-eight hours after transfection, the cells were exposed to hypoxia for 4 h. Hypoxia-induced HIF-1α expression was decreased to a similar extent by knockdown of both CDK1 and CDK4, regardless of p53 status (Fig. 1D); it is noteworthy that the combination of CDK1 and CDK4 knockdown reduced HIF-1α to a greater extent than either siRNA alone. To determine whether VHL was required for CDK-mediated downregulation of HIF-1α, RCC4 cells, which constitutively express HIF-1α in normoxic conditions due to the loss of VHL function, were transfected with siCON, siCDK1, or siCDK4. Knockdown of CDK1 or CDK4 dramatically reduced HIF-1α levels in the absence of VHL under both normoxic and hypoxic conditions (Fig. 1E). Therefore, CDK1/4-mediated regulation of HIF-1α expression is independent of p53, VHL, and oxygen concentration.
Because protein levels are dictated by the rate of biosynthesis and degradation, we sought to determine whether CDK1/4 inhibition altered the rate of HIF-1α transcription. To this end, mRNA was collected from HCT116 p53+/+ cells in parallel with protein lysates. While knockdown of CDK1 and CDK4 dramatically reduced HIF-1α protein levels (Fig. 1D), RT-PCR analysis revealed no effect on HIF-1α mRNA levels (Fig. 1F). Furthermore, GLUT1 mRNA levels increased in response to hypoxia, indicative of HIF-1 activation, and knockdown of CDK1/4 reduced GLUT1 expression to near normoxic levels, verifying that the observed reduction in HIF-1α protein levels following CDK1 or CDK4 knockdown correlated with reduced HIF-1 activity (Fig. 1F). These data indicate that CDK1/4 inhibition reduces HIF-1α expression at the post-transcriptional level.
CDK1 inhibition increases the proteasomal degradation of HIF-1α
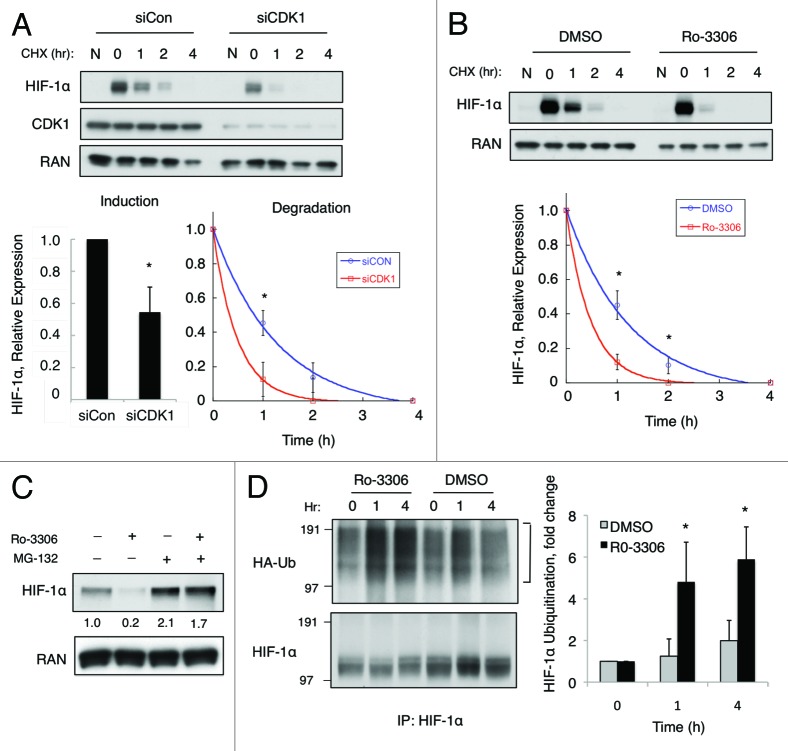
We next focused our attention on investigating the mechanism by which CDK1 regulates HIF-1α stability. First, we questioned whether CDK1 inhibition alters the rate of HIF-1α degradation. HCT116 cells were transfected with siCON or siCDK1 for 48 h, and the cells were placed in hypoxia for 4 h prior to the addition of cycloheximide (CHX), a global inhibitor of protein translation. Lysates were collected over a 4-h time course, and HIF-1α expression levels were monitored over time. Silencing of CDK1 not only blunted the induction of HIF-1α in response to hypoxia by approximately 50% (Fig. 2A, bar graph), but also significantly reduced the half-life of the cellular HIF-1α protein (siCON, t1/2 = 1.0 ± 0.1 h vs. siCDK1, t1/2 = 0.7 ± 0.1 h) (Fig. 2A). Because long-term knockdown of CDK1 affects various cellular processes, such as the cell cycle, we sought to assess whether acute inhibition of CDK1 kinase activity had similar effects on HIF-1α expression. HCT116 cells were exposed to hypoxia for 4 h and then pretreated with DMSO or Ro-3306 for 30 min prior to treatment with CHX. Following the addition of CHX, lysates were collected over a 4-h time course, and HIF-1α expression was monitored over time. Acute inhibition of CDK1 by Ro-3306 did not significantly alter the cell cycle profile (4 h), whereas long-term treatment (20 h) resulted in complete cell cycle arrest in G2/M-phase; in vitro kinase assays demonstrated that Ro-3306 effectively blocked CDK1 activity (Fig. S1A and 1B). In the presence of Ro-3306, HIF-1α was degraded at a significantly faster rate (t1/2 = 0.6 ± 0.1 h) compared with the DMSO-treated control (t1/2 = 1.0 ± 0.1 h) prior to significant cell cycle arrest (Fig. 2B). These data indicate that inhibition of CDK1 kinase activity rapidly decreases the stability of HIF-1α protein, independent of its effects on the cell cycle.
Figure 2. CDK1 inhibition increases the rate of HIF-1α ubiquitination and proteasomal degradation. (A) HCT116 cells were transfected with control or CDK1 siRNA for 48 h and exposed to hypoxia for 4 h prior to the addition of cycloheximide (CHX, 12.5 μg/mL). (B) HCT116 cells were exposed to hypoxia for 4 h and pretreated with DMSO or Ro-3306 (5 μM) for 30 min prior to the addition of CHX. Lysates were harvested at the indicated time points, HIF-1α expression was monitored by western blotting, and densitometry was used to determine the rate of decay. (C) HCT116 cells were treated with MG-132 (1 μM), Ro-3306, or the combination of both for 6 h in hypoxia, and HIF-1α expression was monitored by western blotting and densitometry. (D) HCT116 cells were transfected with HA-ubiquitin, pretreated for 30 min with MG-132 prior to the addition of Ro-3306 or DMSO, and lysates were collected at the indicated time points. HIF-1α was immunoprecipitated and ubiquitination was detected using the anti-HA antibody. The blot was stripped and reprobed using the HIF-1α antibody to determine the total amount of HIF-1α immunoprecipitated in each sample, and densitometry was used to assess the relative amount of ubiquitination at each time point (n = 3 for all experiments).
Because the proteasomal degradation pathway is known to regulate HIF-1α protein stability, we tested whether CDK1 inhibition promotes HIF-1α degradation through the proteasome. HCT116 cells were treated with Ro-3306 alone or in combination with MG-132, a proteasome inhibitor, for 6 h under hypoxic conditions. As expected, treatment with Ro-3306 alone led to a significant decrease in the levels of HIF-1α (Fig. 2C, compare lanes 1 and 2). However, in the presence of a proteasome inhibitor (MG-132), Ro-3306 was unable to reduce HIF-1α levels, indicating that CDK1 inhibition reduces HIF-1α stability in a proteasome-dependent manner (Fig. 2C, compare lanes 2 and 4). To further investigate the role of the ubiquitin–proteasome pathway in regulating CDK1-mediated HIF-1α stability, we designed an assay to determine whether CDK1 inhibition increases the amount ubiquitin linked to HIF-1α. HCT116 cells were transfected HA-ubiquitin for 24 h and incubated with MG-132 for 30 min prior to treatment with either DMSO or Ro-3306 for the indicated times. HIF-1α was immunoprecipitated, and the amount of ubiquitin linked to HIF-1α was detected by immunoblotting with an anti-HA antibody. The western blot in Figure 2D revealed a robust increase in the amount of HA-ubiquitin conjugated to HIF-1α immunoprecipitated from cells treated with the CDK1 inhibitor compared with DMSO, within 1 h. Each blot was stripped and reprobed to determine the total amount of HIF-1α immunoprecipitated in each sample, and densitometry was used to calculate the relative amount of ubiquitin at each time point (Fig. 2D). These data reveal that HIF-1α ubiquitination significantly increases when CDK1 kinase activity is blocked, further suggesting that CDK1 stabilizes HIF-1α by interfering with its regulation by the ubiquitin–proteasome pathway.
CDK1 directly phosphorylates HIF-1α at Serine 668 in vitro
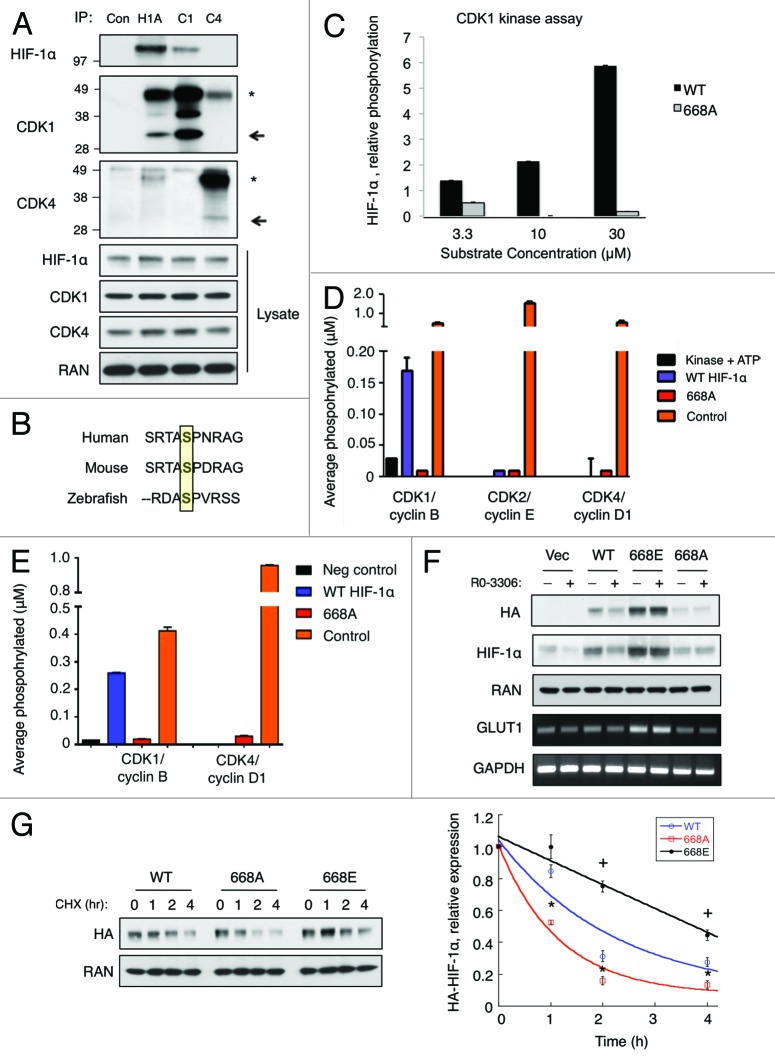
To gain further insight into how CDK1 may be modifying HIF-1α to affect its stability, we tested whether CDK1 or CDK4 and HIF-1α physically interact in cells. HCT116 cells were treated with MG-132 to block HIF-1α degradation and incubated in hypoxic conditions for 4 h prior to harvest. Endogenous HIF-1α, CDK1, or CDK4 were then immunoprecipitated, and western blotting was used to detect their interaction. We found that immunoprecipitated HIF-1α pulled down CDK1 under hypoxic conditions, and similar results were observed for the reciprocal immunoprecipitation experiment (Fig. 3A). However, we could not detect an interaction between HIF-1α and CDK4 under these conditions. Thus, CDK1 and HIF-1α physically interact in cells.
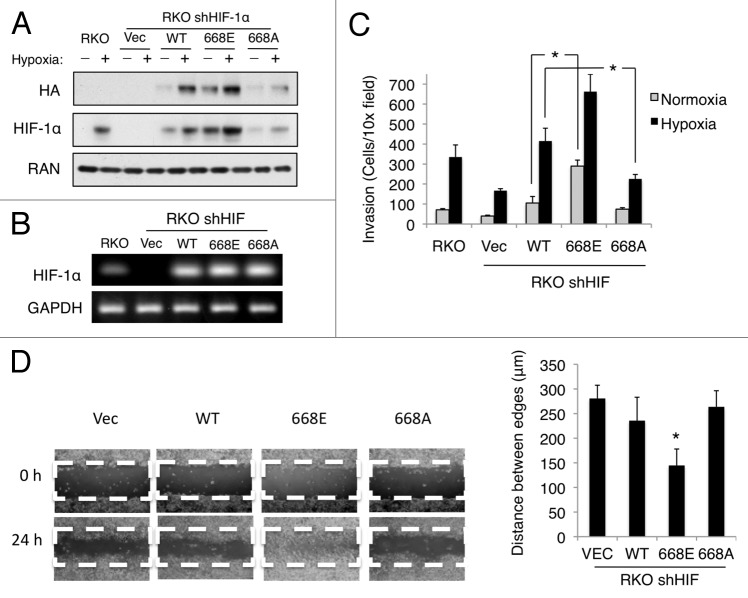
Figure 3. CDK1 regulates HIF-1α stability via direct phosphorylation of Ser668. (A) HCT116 cultured in hypoxia for 6 h in the presence of MG-132. Endogenous HIF-1α, CDK1, and CDK4 were immunoprecipitated, and western blotting was used to detect their reciprocal interaction. (B) Sequence alignment of Ser668 of HIF-1α; yellow box indicates the Ser668 residue in human HIF-1α. (C) Purified CDK1/cyclin B1 was incubated with increasing concentrations of WT or 668A peptides, and phosphorylation was plotted relative to control reactions lacking a substrate. The indicated purified CDK/cyclin complexes were incubated with no substrate, a positive control, (D) 30 μM of the WT or 668A peptides, or (E) purified full-length His-tagged WT or 668A proteins and average phosphorylation was plotted. (F) HCT116 cells were transfected with the vector or the indicated constructs of HIF-1α (WT, 668E, or 668A) for 24 h. Cells were then exposed to hypoxia and treated with DMSO or Ro-3306 (5 μM) for 6 h prior to harvest. (G) HCT116 cells were transfected with the indicated HA-tagged constructs of HIF-1α (WT, 668E, or 668A) for 24 h. Cells were exposed to hypoxia for 4 h prior to the addition of CHX, and lysates were collected at the indicated time points. HIF-1α expression was monitored by western blotting, and densitometry was used to determine the rate of decay. *P < 0.05 vs. (668A vs. WT); + P < 0.05 (668E vs. WT). (n = 3 for all experiments).
The fact that CDK1 and HIF-1α interact in vivo led us to question whether CDK1 modulates HIF-1α stability through direct phosphorylation. CDK1 is a proline residue-directed kinase that readily phosphorylates Ser/Thr-Pro sites in a number of substrates. Thus, to identify potential Ser/Thr residues that were likely to be modified by CDK1, we used in silico methods to analyze the amino acid sequence of HIF-1α for putative CDK1 phosphorylation consensus motifs (pS/T-P-x-R). Two potential CDK1 phosphorylation motifs were identified in the sequence of HIF-1α: Ser657 (ATSSPYR) and Ser668 (RTASPNR). The Ser657 site was previously identified as a target of PLK3, and mutation of this residue to an Ala enhances the stability of HIF-1α.19 Therefore, we focused on the other candidate site, Ser668. Sequence alignment revealed that the Ser668 residue is highly conserved in lower species, indicating that it may be of functional importance to HIF-1α (Fig. 3B). Importantly, in vivo phosphorylation of HIF-1α Ser668 was previously reported by mass spectrometry in a human gastric cancer cell line, MKN-45.25 To determine whether CDK1 can directly phosphorylate Ser668, we performed in vitro kinase assays using 15 aa peptides of the sequence surrounding the Ser668 residue: WT HIF (DTQSRTASPNRAGKGV) and, as a negative control, HIF-1α (S668A) (DTQSRTAAPNRAGKGV). Increasing concentrations (3.3 μM, 10 μM, and 30 μM) of these peptides were incubated with purified CDK1/Cyclin B and radiolabeled with ATP to determine whether HIF-1α Ser668 is a direct substrate of CDK1. CDK1 efficiently phosphorylated the WT HIF-1α peptide in a substrate concentration-dependent manner. However, the mutant HIF-1α (S668A) peptide was not phosphorylated by CDK1, verifying that CDK1 can phosphorylate a HIF-1α peptide specifically at the Ser668 residue (Fig. 3C). Furthermore, CDK2 and CDK4 were unable to phosphorylate the WT HIF peptide in vitro (Fig. 3D). Importantly, the results of our in vitro kinase assays were confirmed using full-length recombinant WT HIF-1α and HIF-1α (S668A); CDK1/cyclin B1 readily phosphorylated the WT protein, but not the 668A mutant, whereas CDK4/cyclin D1 was unable to phosphorylate either protein (Fig. 3E). Taken together, these data suggest that CDK1 directly and specifically phosphorylates HIF-1α at Ser668 in vitro.
CDK1-mediated regulation of HIF-1α expression is dependent on Ser668 phosphorylation
To test whether Ser668 phosphorylation is necessary for CDK1-mediated regulation of HIF-1α stability in vivo, HCT116 cells were transfected with vector control or HA-tagged constructs of WT HIF-1α, 668E, or 668A. After 24 h, the cells were treated with DMSO or Ro-3306, exposed to hypoxia for 6 h, and exogenous HIF-1α levels were monitored using an anti-HA antibody. Inhibition of CDK1 significantly reduced the levels of both endogenous and WT HIF-1α (Fig. 3F). In contrast, the protein levels of both 668E and 668A were refractory to CDK1 inhibition. Thus, the ability to modify the phosphorylation state of the Ser668 residue is required for CDK1-mediated regulation of HIF-1α expression. Next, we questioned whether the phosphorylation state of Ser668 alters the basal rate of HIF-1α degradation. HCT116 cells were transfected with each of the indicated HIF-1α constructs and exposed to hypoxia for 4 h prior to the addition of CHX. As expected, the 668E mutant protein (t1/2 = 3.5 ± 0.2 h) was significantly more stable than WT HIF-1α (t1/2 = 1.8 ± 0.2 h), while the 668A mutant protein (t1/2 = 0.9 ± 0.1 h) was significantly less stable (Fig. 3G). Thus, the phosphorylation state of HIF-1α at Ser668 plays a critical role in controlling both the rate of HIF-1α turnover and its steady-state level.
HIF-1α is stabilized by CDK1/cyclin B1 overexpression and in G2/M phase via phosphorylation of Ser668
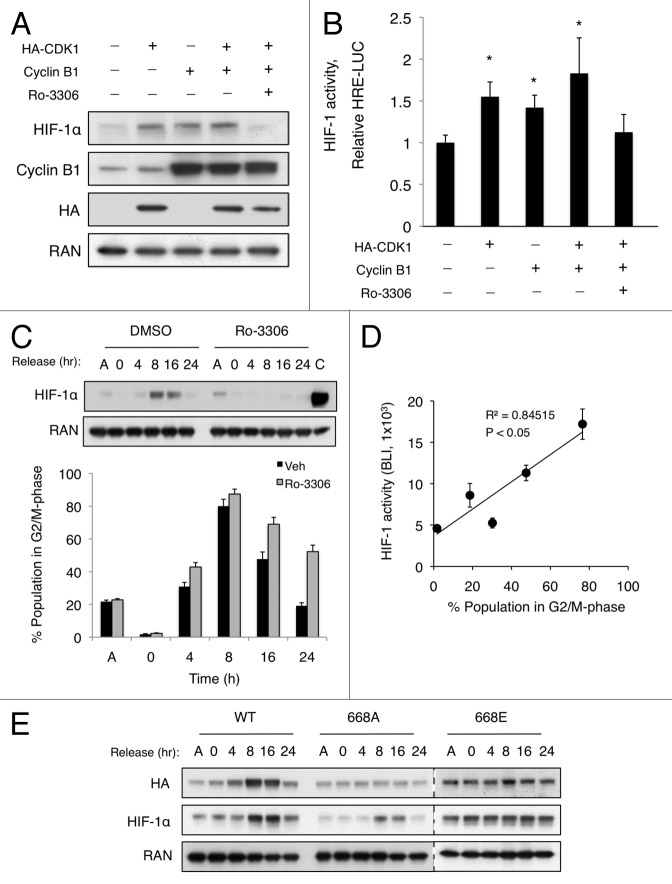
Next, we sought to determine whether overexpression of CDK1 and/or cyclin B1 was sufficient to stabilize HIF-1α in normoxic conditions. HCT116 cells were transfected with HA-CDK1, cyclin B1, or both for 24 h, and western blotting was used to monitor HIF-1α expression. Overexpression of CDK1 or cyclin B1 alone was sufficient to stabilize HIF-1α in normoxic conditions. Importantly, this effect was abolished when cells were treated with Ro-3306 for 4 h prior to harvest, indicating that CDK1 kinase activity was required to increase HIF-1α levels (Fig. 4A). Furthermore, we measured HIF-1 transcriptional activity in a parallel experiment using the HCT116 HRE-Luc cell line. As expected, in the presence of exogenous CDK1 or cyclin B1, HIF-1 activity was significantly increased, and inhibition of CDK1 blocked this activation (Fig. 4B). Taken together, these data reveal that overexpression of CDK1 and/or cyclin B1 stabilize HIF-1α and promote HIF-1 transcriptional activity in a CDK1-dependent manner.
Figure 4.
Overexpression of CDK1/cyclin B1 or accumulation of the G2/M-phase population is sufficient to stabilize HIF-1α. (A) HCT116 and (B) HCT116 HRE-Luc cells were transfected with HA-CDK1, cyclin B1 or both for 24 h, and DMSO or Ro-3306 (5 μM) were added for 6 h prior to harvest. (C) HCT116 cells were synchronized in S phase (double-thymidine block), and HIF-1a expression, phosphorylation (top), and cell cycle profile (bar graph, bottom) were monitored over time after release from S phase by western blotting and PI staining, respectively. A, asynchronous population; C, lysate from cells incubated in hypoxic conditions for 6 h. (D) HCT116 HRE-Luc cells were synchronized in S phase, released, and bioluminescence (BLI) was recorded at the indicated time points. Scatter plot represents HIF-1 activity as a function of the percentage of cells in G2/M phase (P < 0.05, as determined by linear regression analysis). (C) WT, 668A, and 668E constructs were transfected into HCT116 cells. After S-phase synchronization and release, as described previously, expression of exogenous and endogenous HIF-1α was determined by western blotting. (n = 3 for all experiments).
CDK1 is activated during late G2 phase to initiate mitosis, and it is necessary for proper cell cycle progression. Thus, we reasoned that HIF-1α expression and HIF-1 activity were likely to be regulated concomitant with CDK1 activation during progression through the cell cycle. To test this hypothesis, we synchronized HCT116 cells in S phase using a double-thymidine block. Then, the cells were released in fresh medium containing 10% serum, and cell lysates were collected at the indicated time points to assess HIF-1α expression. In parallel, cells were collected for propidium iodide (PI) staining and subsequent FACS analysis to determine the cell cycle profile at each time point. After 8 h release, the time at which a majority of the cell population was in G2/M phase (approximately 80%; Fig. 4A, bar graph; Fig. S2) and CDK1 highly active, HIF-1α was markedly stabilized under normoxic conditions. HIF-1α stability was sustained through 16 h and returned to basal levels by 24 h (Fig. 4C). To determine whether CDK1 activity was required for the cell cycle-dependent stabilization of HIF-1α, we performed a similar experiment in the presence of Ro-3306. CDK1 inhibition completely abolished the cell cycle-dependent increase in HIF-1α expression (Fig. 4C, bar graph). To assess the transcriptional activation of HIF-1, a parallel synchronization experiment was performed in HCT116 cells stably expressing HRE-Luc. As expected, as HIF-1α levels increased in G2/M phase, HIF-1 was activated. When the bioluminescence at each time point was plotted relative to the percentage of cells in G2/M phase, a statistically significant linear correlation was observed, suggesting that HIF-1 becomes active under normoxic conditions during G2/M phase (Fig. 4D). To determine whether phosphorylation of Ser668 was required for the stabilization of HIF-1α in G2/M phase, HA-tagged WT, 668A, or 668E constructs were overexpressed in HCT116 cells, and these cells were synchronized as described previously. Similar to endogenous HIF-1α, the expression of WT HIF-1α was markedly increased at 8 and 16 h, In contrast, the expression the 668A and 668E mutants remained unchanged, as assessed by relative levels of the HA-tagged protein (Fig. 4E). Thus, HIF-1α is stabilized and HIF-1 is activated via CDK1-mediated phosphorylation of Ser668 in G2/M phase.
Phosphorylation of HIF-1α at Ser668 enhances tumor cell migration and invasion
Altering HIF-1α levels has been shown to increase tumor growth, angiogenesis, and invasion in colon carcinoma cells.26 Therefore, we assessed whether the phosphorylation state of Ser668 could influence the invasive potential of colon carcinoma cells, with the hypothesis that a more stable construct of HIF-1α, 668E, would augment tumor proliferation, migration, and invasion. To assay the effects of hypoxia and HIF-1α overexpression on these processes, we transfected RKO colorectal cancer cells in which HIF-1α was stably knocked down (shHIF) with shRNA-resistant WT HIF-1α, 668E, and 668A mutants. Despite having similar mRNA levels, 668E protein levels were greater than WT HIF-1α, particularly under normoxic conditions, whereas 668A expression was reduced compared with WT (Fig. 5A and B). Using this system, we assessed the effect of Ser668 phosphorylation on tumor cell invasion. The indicated cell lines were seeded onto a filter that was coated with Matrigel, exposed to normoxia or hypoxia for 24 h, and the number of cells that migrated through the 8-μm pores in the filter was counted. The invasiveness of the vector control cells significantly increased under hypoxic conditions compared with normoxia (Fig. 5C). Verifying previous reports, knockdown of HIF-1α greatly reduced tumor cell invasion under hypoxic conditions (Fig. 5C, Vec vs. shHIF in hypoxia). Expression of WT HIF-1α in the shHIF cell line restored hypoxia-induced invasion and led to a modest increase in invasiveness under normoxic conditions, (Fig. 5C, RKO vs. WT). Importantly, the 668E mutant cell line displayed significantly increased tumor cell invasion compared with WT under normoxic conditions (Fig. 5C, RKO or WT vs. 668E in normoxia). Under hypoxic conditions, 668E also increased invasiveness compared with WT, but this difference did not reach statistical significance. In contrast, the 668A cell line displayed significantly reduced invasion under hypoxic conditions (Fig. 5C, WT vs. 668A in hypoxia). Next, we assessed whether the phosphorylation state of Ser668 could effect tumor cell migration using a scratch assay. At 24 post-scratch, the 668E mutant cells migrated significantly further than the vector control or WT cell lines, whereas no significant difference was observed between the Vec, WT, and 668A (Fig. 5D). Taken together, these data indicate that the phosphorylation state of Ser668 significantly increases HIF-1α-mediated tumor cell migration and invasion.
Figure 5. Phosphorylation at Ser668 enhances tumor cell migration and invasion. (A) Wild-type RKO cells or RKO cells stably expressing HIF-1α shRNA (shHIF) were transfected with the indicated HIF-1α constructs (WT, 668A, or 668E), cultured in normoxia or hypoxia for 6 h, and endogenous and exogenous HIF-1α expression was monitored by western blotting and (B) RT-PCR. (C) The indicated cell lines were seeded into the top chamber of a Matrigel-coated filter in DMEM lacking serum, and DMEM containing 10% FBS was placed in the bottom chamber as a chemoattractant. The cells were then cultured in normoxia or hypoxia for 24 h, and the number of cells that migrated through the filter was counted by microscopy. (D) The indicated cell lines were plated in 6-well plates and allowed to grow to confluence. Each well was scratched with a 200 μl pipette tip, the cells were washed twice with PBS, and complete medium containing 10% serum was added back to the wells. Images were acquired immediately and 24 h after the scratch, and the distance between each edge was measured. *P < 0.05. (n = 3 for all experiments).
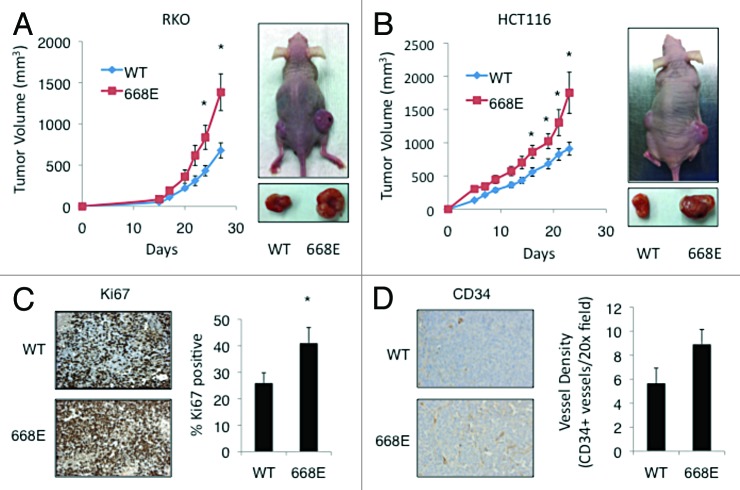
Phosphorylation at Ser668 enhances proliferation, angiogenesis, and tumor growth in vivo
Due to the importance of HIF-1 in the tumor microenvironment and on physiological processes that are critical for tumorigenesis, we sought to determine whether a phospho-mimetic of HIF-1α at Ser668 was sufficient to promote tumor growth in vivo. To this end, WT HIF-1α and 668E mutant constructs were stably expressed in both RKO and HCT116 colorectal cancer cells. HIF-1α mRNA levels, protein expression levels, and proliferation were assessed in both the RKO (Fig. S3A–C, respectively) and HCT116 (Fig. S4A–C, respectively) stable cell lines prior to inoculation. Interestingly, there was no difference in the proliferation rate of the cell lines stably expressing Vec, WT, and 668E in vitro. To assess in vivo tumor growth, 1 × 106 RKO or HCT116 cells stably expressing the indicated constructs of HIF-1α were injected subcutaneously into 4–6-wk-old female athymic nude mice. In order to minimize inter-mouse variability, mice were injected in the right or left flank with cells expressing WT or 668E, respectively, and tumor growth was measured with calipers 3 times a wk for approximately 4 wk. At the end of the study, the 668E tumors grew significantly faster and larger than the WT tumors in both cell lines, suggesting that a phospho-mimetic at the Ser668 site is sufficient to promote tumor growth in vivo (Fig. 6A and B). Surprisingly, in contrast to the results of our in vitro proliferation assays, immunohistochemical staining of Ki67 revealed that the 668E tumors had significantly higher proliferation compared with WT tumors (Fig. 6C). Furthermore, the 668E tumors exhibited enhanced angiogenesis, as determined by immunostaining for CD34 (Fig. 6D). Indicative of the increased pro-angiogenic potential of the 668E mutant compared WT, we observed elevated VEGF levels in the 668E vs. WT RKO cell line in normoxic and hypoxic conditions in vitro (Fig. S3D). Taken together, these findings suggest that the increased vascularity of the 668E tumors promoted a tumor microenvironment that was conducive to cell proliferation and growth.
Figure 6. Phosphorylation at Ser668 promotes angiogenesis, proliferation, and tumor growth in vivo. One million (A) RKO (WT, n = 7; 668E, n = 9) and (B) HCT116 (n = 10 for both groups) cells stably expressing WT or 668E were injected subcutaneously into the rear flanks of athymic nude mice. Tumor volumes were monitored over time by caliper measurements (graphs). At the end of the study, RKO tumors were harvested and immunostained with antibodies specific for (C) Ki67 and (D) CD34 to assess proliferation and angiogenesis, respectively. Percent Ki67 and vessel count were determined as described in the materials and methods section. *P < 0.05.
Discussion
The association between HIF-1 and therapeutic resistance, metastasis, and poor patient prognosis has been well documented, but is still not fully understood. In this report, we show that CDK1 increases the steady-state level of HIF-1α via direct phosphorylation of Ser668. As a result, inhibition of CDK1 significantly reduces HIF-1α protein levels and HIF-1 activation, independent of its known regulators. These findings provide new insights into pathways of HIF-1 regulation that are independent of hypoxia and support the development of CDK inhibitors as a novel therapeutic strategy for targeting HIF-1α-expressing tumors.
HIF-1α expression and the cell cycle
A growing body of literature has confirmed that HIF-1α is overexpressed in tumors compared with normal tissue.27 Our results demonstrate that CDK1 directly phosphorylates HIF-1α at the Ser668 residue, suggesting that the expression of HIF-1α could be controlled in a cell cycle-dependent manner during tumor growth, even prior to the evolution of significant hypoxia associated with bulky tumor growth in vivo. Several groups have reported that colon cancer cells undergo partial G1 arrest and that proliferation is altered in response to hypoxia, in which case, CDK1 activity would be reduced in these cells.28,29 While we observe that CDK1 activation and phosphorylation of Ser668 affect HIF-1 stability in both hypoxic and normoxic conditions, the physiological effects are much more evident in normoxic conditions (i.e., enhanced invasion; statistical significance is only observed between WT and 668E in normoxic conditions [Fig. 5C]). We believe that this finding is due to the fact that under hypoxic conditions, HIF-1α levels are above the threshold necessary to fully activate HIF-1 transcription, and the increase in protein expression observed with the 668E construct does not significantly enhance HIF-1 activity. In contrast, in normoxic conditions, wild-type HIF-1α is essentially absent, whereas the basal levels of the 668E mutant are sufficient to activate the transcription of HIF-1 target genes. Therefore, it is likely that CDK1-mediated phosphorylation and stabilization of HIF-1α plays a much more important role in promoting the oncogenic effects of HIF-1 in normoxic conditions where CDK1 is aberrantly activated.
The relevance of Ser668 phosphorylation in tumorigenesis
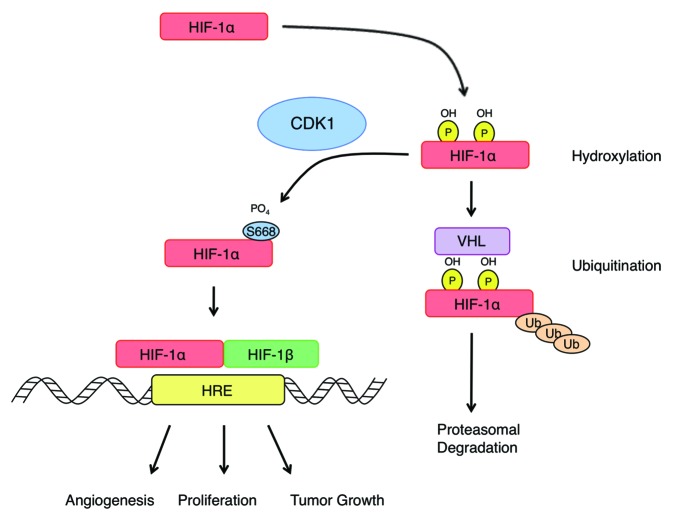
The constitutive activation of oncogenes is a common theme underlying the initiation and progression of human tumors. The frequent overexpression of HIF-1α in cancer is commonly the result of mutations that inactivate its negative regulators, such as VHL, or upregulation of oncogenic pathways, including PI3K or RAS.30 However, there are instances where HIF-1α is highly expressed in tumors, independent of hypoxia or its known regulators. The identification of Ser668 phosphorylation as an important regulator of HIF-1α provides a novel mechanism to explain the constitutive expression of HIF-1α in some tumors, most of which also have increased CDK activity. The results of our overexpression experiments indicate that mutation of Ser668 to a negative charge is sufficient to promote HIF-1α expression in normoxic conditions, activate the transcription of HIF-1 target genes, and promote tumor growth and invasion in vivo (Fig. 7). Therefore, mutation at this site could provide a selective advantage for tumor cells and may be retained in aggressive tumors and metastatic cells. To our knowledge, there have been no reported mutations of the Ser668 residue in human cancer, but due to the apparent importance of Ser668 in maintaining the proper steady-state levels of HIF-1α, a thorough sequencing effort in a broad range of tumor types is needed to determine whether this site is mutated in human cancers and whether mutations evolve in metastatic tumors or drug-treated tumors. Also, alterations in the expression level and activity of CDK1 have been reported in human cancers via several different biological mechanisms. For example, cyclin B1 overexpression is observed in many cancers, including non-small cell lung cancer, esophageal cancer, colorectal cancer, and breast cancer, among others, and it has been shown to be associated with high-grade tumors, advanced disease, and poor patient prognosis.31-34 CDK1 activity is also acutely regulated by CDK activating enzymes; deregulation or overexpression of Cdc25 isoforms has been shown to promote the assembly and activity of the CDK1-cyclin B1 complex,35 and it is commonly correlated with progressive disease and poor prognosis.36 Thus, CDK1 is frequently activated in human cancers through various mechanisms, which promotes the phosphorylation of CDK1 substrates, including HIF-1α. As a result, in addition to its canonical role in cell cycle progression, the enhanced expression or activation of CDK1 observed in tumors could promote an aggressive phenotype and poor patient prognosis through constitutive activation of HIF- 1.
Figure 7. CDK1 regulates HIF-1α protein stability and HIF-1 activity to promote tumor growth. Under normoxic conditions, CDK1 directly phosphorylates HIF-1α at Ser668, which inhibits the proteasomal degradation of HIF-1α. Phosphorylation at this site increases the steady-state levels of HIF-1α and the expression of HIF-1 target genes that promote physiological processes associated with invasion and tumor growth.
CDK inhibition as a strategy to oppose HIF-1 in cancer
It has been well established that HIF-1 inhibition sensitizes solid tumors to radiation, standard chemotherapies, and targeted agents.37,38 Therefore, the use of CDK inhibitors to reduce HIF-1 activity may be a useful strategy in combination with therapies that are currently used in the clinic. Here, we show that inhibition of CDK1 and CDK4 dramatically reduces HIF-1 activity in both normoxic and hypoxic environments. Thus, inhibition of CDK1/4 represents a novel approach for the treatment of cancer cells with constitutively active HIF-1, regardless of whether HIF-1α is overexpressed as a result of intratumoral hypoxia or the deregulation of genetic mechanisms. Due to their role in the cell cycle and association with apoptotic pathways, there has been great interest in the development of CDK inhibitors as cancer therapeutics. Our preliminary data indicate that CDK1 inhibition selectively downregulates HIF-1α expression. Sequence alignment revealed that the Ser668 regulatory residue identified in HIF-1α is not present in HIF-2α, a closely related α subunit protein of the HIF-1 family. The fact that this CDK1-controlled residue is not present in HIF-2α explains why HIF-2α protein levels remained largely unchanged in response to treatment with various CDK inhibitors, while HIF-1α levels were dramatically decreased (Fig. 1A). Therefore, CDK1 inhibitors are specific inhibitors of HIF-1α, which may limit their efficacy in some tumor types, such as renal cell carcinoma, which commonly express only HIF-2α.
It is noteworthy that inhibition of CDK4 reduced the expression of both HIF-1α and HIF-2α (Fig. 1A), which makes it a more promising approach for reducing HIF-1 activity clinically. This study provides rationale for the further development of CDK1 and/or CDK4 inhibitors as a novel approach to negate HIF-1 signaling and sensitize tumors to chemotherapy and targeted agents.
In summary, we identified a novel mechanism regulating the protein stability and steady-state levels of HIF-1α, independent of hypoxia or its known regulators (Fig. 7). Specifically, CDK1 directly phosphorylates HIF-1α at Ser668, which inhibits its proteasomal degradation, thus providing a potential molecular explanation for the elevation of HIF-1 in primary and metastatic tumors, independent of hypoxia. These data implicate CDK1 as driver of key hallmarks of cancer, including angiogenesis, cell survival and invasion, through the stabilization of HIF-1α, and provide a novel molecular rationale for the clinical translation of CDK inhibitors for use in tumors with constitutively active HIF-1.
Materials and Methods
Plasmids and siRNA
HA-HIF-1α,39 HA-CDK1,40 and cyclin B141 expression constructs were purchased from Addgene, and HA-Ubiquitin was a generous gift from Dr Alexandra C Newton (University of California).42 Point mutations were introduced into HA-HIF-1α using the Quikchange Lightning site-directed mutagenesis kit (Aligent Technologies) and verified by DNA sequencing. His-tagged WT and 668A constructs were generated by subcloning the indicated HIF-1α DNA sequence into N-terminal pQE-30 vector (Qiagen). shRNA-resistant constructs of HIF-1α were generated by introducing silent mutations via changing the 3′ nucleotide of 4 amino acids within the shRNA seed sequence. Control, CDK1, CDK2, CDK4, CDK7, and CDK9 targeting siRNAs were purchased from Santa Cruz Biotech. CDK2 and CDK5 siRNA were purchased from Cell Signaling Technology.
Materials and antibodies
Cycloheximide, MG-132, and thymidine were purchased from Sigma. Flavopiridol, roscovitine, and PD-0332991 were purchased from Selleck Chemicals. Alsterpaullone and purvalanol A were purchased from Sigma, and Ro-3306 was purchased from Santa Cruz Biotech. All of the listed inhibitors were dissolved in dimethyl sulfoxide (DMSO). The antibodies to HIF-1α, HIF-2α, and RAN were purchased from BD Transduction Laboratories. CDK2 and CDK5 antibodies were purchased from Cell Signaling Technology. An anti-HA monoclonal antibody was purchased from Covance. Antibodies to CDK1, CDK4, CDK7, CDK9, and p53 (DO-1) were purchased from Santa Cruz Biotech. The antibody against cyclin B1 was purchased from Leica Microsystems. Antibodies against CD34 and Ki67 were purchased from DAKO. All other materials and chemicals were reagent-grade.
Cell transfection and immunoblotting
HCT116 and HCT116 p53−/− cell lines were a gift from the laboratory of Dr Bert Vogelstein (Johns Hopkins University) and were maintained in McCoy 5A medium (Cellgro) containing 10% FBS (Hyclone) and 1% penicillin/streptomycin. RCC4 cells were a generous gift from Dr Celeste Simon (University of Pennsylvania),43 and the RKO shHIF-1α cell line was a kind gift from Dr Nicolas Denko (Stanford University).44 These cells were maintained in DMEM containing 10% FBS and 1% penicillin/streptomycin. RKO and HCT116 stable cell lines were transfected with the indicated HIF-1α constructs and selected for 2 wk in Geneticin (500 μg/ml, Gibco). Surviving cells were pooled and cultured in the presence of 100 μg/ml Geneticin for all subsequent experiments. All cell lines were maintained at 37 °C in 5% CO2. When indicated, cells were maintained in a hypoxia environment (0.2 or 0.5% O2) using a hypoxia chamber (In vivo2, Ruskinn). Transient transfection of DNA and siRNA was performed using Lipofectaimine 2000 and Lipofectamine RNAiMAX transfection reagents, respectively (Invitrogen), according to the manufacturer’s protocol. For immunoblotting, cultured cells were lysed in buffer A (50 mM Na2HPO4, 1 mM sodium pyrophosphate, 20 mM NaF, 2 mM EDTA, 2 mM EGTA, 1% Triton X-100, 1 mM DTT, 200 μM benzamidine, 40 μg ml−1 leupeptin, and 1 mM PMSF, pH 7.4), and protein yield was determined using a Bio-Rad Protein Assay kit. Lysates containing equal amounts of protein were analyzed by SDS-PAGE, and individual blots were probed using the indicated antibody. Densitometric analysis was performed with the NIH ImageJ analysis software (version 1.63). Where indicated, membranes were stripped using Restore stripping buffer (Invitrogen), according to the manufacturer’s protocol.
Immunoprecipitation
HCT116 cells were transiently transfected with the indicated constructs. Approximately 24–36 h post-transfection, cells were lysed in buffer B (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 5 mM EDTA, 1% Triton X-100, 10 mM sodium pyrophosphate, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium vanadate). Five percent of the total detergent-solubilized cell lysate was quenched in SDS sample buffer for further analysis, and the remaining detergent-solubilized cell lysate was incubated with appropriate antibody overnight at 4 °C, followed by a 2 h incubation with Ultra-link protein A/G-agarose (Pierce). The immunoprecipitates were washed 3 times in buffer B and proteins were separated using SDS-PAGE and analyzed by immunoblotting.
Cellular ubiquitination assays
HCT116 cells were transfected with HA-uiquitin (0.5 μg), and the cells were treated with MG-132 and either Ro-3306 or DMSO for the indicated times before harvest with buffer B containing 10 mM N-ethylmaleimide to preserve the ubiquitinated species. After centrifugation at 13 000 × g for 5 min, the supernatants were incubated with anti-HIF-1α antibody overnight at 4 °C, and then Ultralink protein A/G beads for 2 h. The immunocomplexes were washed 3 times with buffer B containing 10 mM N-ethylmaleimide. Each sample was resuspended in 1× lysis buffer, incubated at 95 °C for 5 min, and analyzed by immunoblotting with the indicated antibodies.
In vitro kinase assays
His-tagged full-length HIF-1α and His-HIF-1α (S668A) were purified using Ni-NTA resin (Thermo Scientific), according to the manufacturer’s protocol. Sixteen amino acid peptides surrounding the Ser668 residue were synthesized for WT-HIF-1α (DTQSRTASPNRAGKGV) and HIF-1α (S668A) (DTQSRTAAPNRAGKGV). In vitro kinase assays were performed by Reaction Biology Corporation, as previously described.45 Parallel reactions were performed with the appropriate positive (CDK1/2, Histone H1 [20 μM] and CDK4, Rb [3 μM]) and negative (no substrate) controls. Kinase activity was detected using the P81 filter binding method.
RT-PCR analysis
MRNA was isolated from cell lysates using the RNeasy mini kit (Qiagen), and RT-PCR reactions were performed on equal amounts of starting material (1 μg RNA) using the OneStep RT PCR kit (Qiagen), according to the manufacturer’s suggested parameters. The following primers were used for RT-PCR experiments: HIF-1α, Fwd – GGCGCGAACG ACAAGAAAAA, Rev – CCTTATCAAG ATGCGAACTC AC; GLUT1, Fwd – GCCAGAAGGA GTCAGGTTCA A, Rev – TCCTCGGAAA GGAGTTAGAT CC; VEGF, Fwd – AGGGCAGAAT CATCACGAAG T, Rev – AGGGTCTCGA TTGGATGGCA; GAPDH, Fwd – TGTGGGCATC AATGGATTTG G, Rev – ACACCATGTA TTCCGGGTCA AT.
Synchronization
Cells were synchronized in S phase by double-thymidine block. Briefly, HCT116 cells were plated at low confluence (approximately 50%) in 12-well plates and allowed to adhere overnight. The next day, the cells were incubated in 2 mM thymidine for 16 h, complete medium for 8 h, thymidine for 16 h, and then released by adding complete medium. When indicated, Ro-3306 was added at the time the cells were released from the second thymidine block, and WT, 668A, or 668E mutants were transfected into cells after the first thymidine block.
Invasion assays
Invasion assays were performed using the Biocoat Tumor Invasion System (BD Biosciences), according to the manufacturer’s instructions. Briefly, RKO cells expressing the indicated constructs plated in DMEM without FBS at 2.5 × 104 cells/well into the apical chambers. The chemo-attractant (DMEM containing 10% FBS) was added to the basal chamber, and the cells were incubated at normoxia or hypoxia for 24 h. Non-invading cells were removed by scrubbing the apical side of the membrane 2 times with a Q-tip, and the cells that migrated to the basal side of the membrane were stained using the Diff-Quik staining kit (Fisher Scientific). The membranes were then mounted on microscope slides, and the number of invading cells was manually counted. Cells were counted as number of cells/field at 10× magnification, and at least 3 fields were counted for each membrane. Data represent the mean ± s.e. of at least 3 independent experiments.
Migration assays
The indicated cell lines were plated in 6-well plates and allowed to grow to confluence. Then, each well was scratched with a 200-μl pipette tip, and the cells were washed twice with PBS. Complete medium containing 10% serum was added back to the wells, and the cells were incubated for an additional 24 h. Images were acquired immediately after the scratch and after 24 h, and the distance between each edge was measured.
In vivo studies
Hairless severe combined immunodeficient (SCID) mice were housed and maintained in accordance with the Institutional Animal Care and Use Committee and state and federal guidelines for the humane treatment and care of laboratory animals. RKO and HCT116 cells stably expressing either WT HIF-1α or the 668E mutant were injected subcutaneously into the rear flanks of mice at a density of 1 × 106 cells per injection in PBS/Matrigel (v:v) in 200 μL total volume. Tumor volumes were monitored over time by caliper measurements. At the end of the study, tumors were harvested, fixed, and embedded in paraffin, sectioned and stained with hematoxylin and eosin (H&E), or immunostained with antibodies specific for Ki67 and CD34. The percentage of Ki67-positive cells was calculated using a Prism and Reflector Imaging Spectroscopy System (PARISS) hyperspectral imaging system, as described previously.46 After CD34 staining, 2 independent investigators who were blinded to the identity of the samples scored vessel density as the number of CD34+ vessels present per 20× field (4 fields were counted on 3 separate tumors from the WT and 668E groups).
Statistical analysis
All western blots shown are representative of at least 3 independent experiments. Exponential decay curves were used to determine the half-life of HIF-1α. Differences between groups were determined by the Student t test and linear regression analysis. The data are presented as the mean ± S.E., and a P value < 0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by NIH grants, CA135273 and CA123258, and start-up funds from the Penn State Hershey Cancer Institute to WSE-D. This work was also supported by an NIH/NCI sponsored Ruth L Kirstein Postdoctoral NRSA fellowship to NAW (award number: F32CA174138). WSE-D. is an American Cancer Society Research Professor.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/cc/article/26930
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/26930
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Semenza GL. HIF-1 and human disease: one highly involved factor. Genes Dev. 2000;14:1983–91. [PubMed] [Google Scholar]
- 3.Semenza GL. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci STKE. 2007;2007:cm8. doi: 10.1126/stke.4072007cm8. [DOI] [PubMed] [Google Scholar]
- 4.Harris AL. Hypoxia--a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 5.Semenza GL. Hypoxia-inducible factor 1 and cancer pathogenesis. IUBMB Life. 2008;60:591–7. doi: 10.1002/iub.93. [DOI] [PubMed] [Google Scholar]
- 6.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92:5510–4. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Semenza GL. Hydroxylation of HIF-1: oxygen sensing at the molecular level. Physiology (Bethesda) 2004;19:176–82. doi: 10.1152/physiol.00001.2004. [DOI] [PubMed] [Google Scholar]
- 8.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–40. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 9.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–8. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 10.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–5. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 11.Wenger RH, Stiehl DP, Camenisch G. Integration of oxygen signaling at the consensus HRE. Sci STKE. 2005;2005:re12. doi: 10.1126/stke.3062005re12. [DOI] [PubMed] [Google Scholar]
- 12.Rankin EB, Giaccia AJ. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 2008;15:678–85. doi: 10.1038/cdd.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baldewijns MM, van Vlodrop IJ, Vermeulen PB, Soetekouw PM, van Engeland M, de Bruïne AP. VHL and HIF signalling in renal cell carcinogenesis. J Pathol. 2010;221:125–38. doi: 10.1002/path.2689. [DOI] [PubMed] [Google Scholar]
- 14.Zhong H, Chiles K, Feldser D, Laughner E, Hanrahan C, Georgescu MM, Simons JW, Semenza GL. Modulation of hypoxia-inducible factor 1alpha expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: implications for tumor angiogenesis and therapeutics. Cancer Res. 2000;60:1541–5. [PubMed] [Google Scholar]
- 15.Richard DE, Berra E, Gothié E, Roux D, Pouysségur J. p42/p44 mitogen-activated protein kinases phosphorylate hypoxia-inducible factor 1alpha (HIF-1alpha) and enhance the transcriptional activity of HIF-1. J Biol Chem. 1999;274:32631–7. doi: 10.1074/jbc.274.46.32631. [DOI] [PubMed] [Google Scholar]
- 16.Mylonis I, Chachami G, Samiotaki M, Panayotou G, Paraskeva E, Kalousi A, Georgatsou E, Bonanou S, Simos G. Identification of MAPK phosphorylation sites and their role in the localization and activity of hypoxia-inducible factor-1alpha. J Biol Chem. 2006;281:33095–106. doi: 10.1074/jbc.M605058200. [DOI] [PubMed] [Google Scholar]
- 17.Flügel D, Görlach A, Michiels C, Kietzmann T. Glycogen synthase kinase 3 phosphorylates hypoxia-inducible factor 1alpha and mediates its destabilization in a VHL-independent manner. Mol Cell Biol. 2007;27:3253–65. doi: 10.1128/MCB.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isaacs JS, Jung YJ, Mimnaugh EG, Martinez A, Cuttitta F, Neckers LM. Hsp90 regulates a von Hippel Lindau-independent hypoxia-inducible factor-1 alpha-degradative pathway. J Biol Chem. 2002;277:29936–44. doi: 10.1074/jbc.M204733200. [DOI] [PubMed] [Google Scholar]
- 19.Xu D, Yao Y, Lu L, Costa M, Dai W. Plk3 functions as an essential component of the hypoxia regulatory pathway by direct phosphorylation of HIF-1alpha. J Biol Chem. 2010;285:38944–50. doi: 10.1074/jbc.M110.160325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin WG., Jr. Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell. 2002;1:237–46. doi: 10.1016/S1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 21.van den Heuvel S, Harlow E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science. 1993;262:2050–4. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- 22.Hagting A, Jackman M, Simpson K, Pines J. Translocation of cyclin B1 to the nucleus at prophase requires a phosphorylation-dependent nuclear import signal. Curr Biol. 1999;9:680–9. doi: 10.1016/S0960-9822(99)80308-X. [DOI] [PubMed] [Google Scholar]
- 23.Chen D, Gould C, Garza R, Gao T, Hampton RY, Newton AC. Amplitude control of protein kinase C by RINCK, a novel E3 ubiquitin ligase. J Biol Chem. 2007;282:33776–87. doi: 10.1074/jbc.M703320200. [DOI] [PubMed] [Google Scholar]
- 24.Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol Cell Biol. 2003;23:9361–74. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–97. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 26.Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1039–45. doi: 10.1038/nbt.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dolloff NG, Ma X, Dicker DT, Humphreys RC, Li LZ, El-Deiry WS. Spectral imaging-based methods for quantifying autophagy and apoptosis. Cancer Biol Ther. 2011;12:349–56. doi: 10.4161/cbt.12.4.17175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayes PA, Dolloff NG, Daniel CJ, Liu JJ, Hart LS, Kuribayashi K, Allen JE, Jee DI, Dorsey JF, Liu YY, et al. Overcoming hypoxia-induced apoptotic resistance through combinatorial inhibition of GSK-3β and CDK1. Cancer Res. 2011;71:5265–75. doi: 10.1158/0008-5472.CAN-11-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, Heimbrook DC, Chen L. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci U S A. 2006;103:10660–5. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fry DW, Harvey PJ, Keller PR, Elliott WL, Meade M, Trachet E, Albassam M, Zheng X, Leopold WR, Pryer NK, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther. 2004;3:1427–38. [PubMed] [Google Scholar]
- 31.Shapiro GI. Cyclin-dependent kinase pathways as targets for cancer treatment. J Clin Oncol. 2006;24:1770–83. doi: 10.1200/JCO.2005.03.7689. [DOI] [PubMed] [Google Scholar]
- 32.Ravi R, Mookerjee B, Bhujwalla ZM, Sutter CH, Artemov D, Zeng Q, Dillehay LE, Madan A, Semenza GL, Bedi A. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1alpha. Genes Dev. 2000;14:34–44. [PMC free article] [PubMed] [Google Scholar]
- 33.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40(Database issue):D261–70. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krishnamachary B, Berg-Dixon S, Kelly B, Agani F, Feldser D, Ferreira G, Iyer N, LaRusch J, Pak B, Taghavi P, et al. Regulation of colon carcinoma cell invasion by hypoxia-inducible factor 1. Cancer Res. 2003;63:1138–43. [PubMed] [Google Scholar]
- 35.Talks KL, Turley H, Gatter KC, Maxwell PH, Pugh CW, Ratcliffe PJ, Harris AL. The expression and distribution of the hypoxia-inducible factors HIF-1alpha and HIF-2alpha in normal human tissues, cancers, and tumor-associated macrophages. Am J Pathol. 2000;157:411–21. doi: 10.1016/S0002-9440(10)64554-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell. 2007;11:335–47. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goda N, Ryan HE, Khadivi B, McNulty W, Rickert RC, Johnson RS. Hypoxia-inducible factor 1alpha is essential for cell cycle arrest during hypoxia. Mol Cell Biol. 2003;23:359–69. doi: 10.1128/MCB.23.1.359-369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim WY, Kaelin WG. Role of VHL gene mutation in human cancer. J Clin Oncol. 2004;22:4991–5004. doi: 10.1200/JCO.2004.05.061. [DOI] [PubMed] [Google Scholar]
- 39.Yoshida T, Tanaka S, Mogi A, Shitara Y, Kuwano H. The clinical significance of Cyclin B1 and Wee1 expression in non-small-cell lung cancer. Ann Oncol. 2004;15:252–6. doi: 10.1093/annonc/mdh073. [DOI] [PubMed] [Google Scholar]
- 40.Nozoe T, Korenaga D, Kabashima A, Ohga T, Saeki H, Sugimachi K. Significance of cyclin B1 expression as an independent prognostic indicator of patients with squamous cell carcinoma of the esophagus. Clin Cancer Res. 2002;8:817–22. [PubMed] [Google Scholar]
- 41.Wang A, Yoshimi N, Ino N, Tanaka T, Mori H. WAF1 expression and p53 mutations in human colorectal cancers. J Cancer Res Clin Oncol. 1997;123:118–23. doi: 10.1007/BF01269890. [DOI] [PubMed] [Google Scholar]
- 42.Aaltonen K, Amini RM, Heikkilä P, Aittomäki K, Tamminen A, Nevanlinna H, Blomqvist C. High cyclin B1 expression is associated with poor survival in breast cancer. Br J Cancer. 2009;100:1055–60. doi: 10.1038/sj.bjc.6604874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Timofeev O, Cizmecioglu O, Settele F, Kempf T, Hoffmann I. Cdc25 phosphatases are required for timely assembly of CDK1-cyclin B at the G2/M transition. J Biol Chem. 2010;285:16978–90. doi: 10.1074/jbc.M109.096552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boutros R, Lobjois V, Ducommun B. CDC25 phosphatases in cancer cells: key players? Good targets? Nat Rev Cancer. 2007;7:495–507. doi: 10.1038/nrc2169. [DOI] [PubMed] [Google Scholar]
- 45.Moeller BJ, Dreher MR, Rabbani ZN, Schroeder T, Cao Y, Li CY, Dewhirst MW. Pleiotropic effects of HIF-1 blockade on tumor radiosensitivity. Cancer Cell. 2005;8:99–110. doi: 10.1016/j.ccr.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 46.Wouters A, Pauwels B, Lardon F, Vermorken JB. Review: implications of in vitro research on the effect of radiotherapy and chemotherapy under hypoxic conditions. Oncologist. 2007;12:690–712. doi: 10.1634/theoncologist.12-6-690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.