Abstract
The relationship between chromatin modification and lymphocyte development is still poorly understood. Here we show a correlation between methylation of lysine 4 on histone 3 (H3-K4) and activation of several loci required for the pre-B cell to immature B-cell developmental transition. A critical step in this transition is the induction of V(D)J recombination at the Igκ locus. Upon activation of Igκ recombination, a >10-fold enrichment of both di- and trimethylated H3-K4 is observed at Jκ targeting signals, but not at an analogous targeting signal in the T-cell receptor α locus or, surprisingly, at several Vκ signals. However, H3-K4 methylation is restricted to the actively recombining fraction of Jκ recombination targeting signals, consistent with a direct relationship between H3-K4 methylation and signal activity. Correlations between increased H3-K4 methylation and induction of transcription are also observed at some, but not all, loci where transcription is induced. H3-K4 methylation may therefore be a widely used but not universal means for controlling chromatin activity in this developmental transition.
INTRODUCTION
Lymphocyte development uniquely requires the programmed activation of different loci for targeted chromosomal rearrangement as well as transcription (1). Rearrangement is required for assembly of mature immunoglobulin (Ig) and T-cell receptor (TCR) genes, and is regulated at many levels. Rearrangement of TCR loci is restricted to T cells, just as rearrangement of Ig loci is largely restricted to B cells. Within B-cell development, Ig heavy chain rearrangement occurs first, and is required for the pro-B to pre-B transition, while transition of pre-B cells to immature B cells requires subsequent rearrangement of one of either light chain locus (Igκ and Igλ) (reviewed in 2,3). Within light chain loci, only one allele is activated for recombination, while the other is kept in an inactive state, which helps ensure that only one light chain is expressed in a given cell (allelic exclusion) (4).
Accurate control of where and when V(D)J recombination occurs is therefore critical for normal B-cell development, and is largely dependent on the accessibility of DNA to the RAG1/2 nuclease complex within its cellular context, chromatin (reviewed in 5,6). Chromatin accessibility is mediated, in part, by nucleosome remodeling activities [e.g., SWI/SNF complex (7)]. Remodeling in turn is controlled through covalent modification of histones with a wide variety of functional groups, but most typically through acetylation or methylation of key lysine residues on histone tails. Recent work suggests an important role for histone modifications in lymphocyte development. Loci active in V(D)J recombination have nucleosomes with hyperacetylated histones, and enhancement of histone acetylation levels increases V(D)J recombination activity (reviewed in 8).
Histone methylation is another potentially important regulator of both V(D)J recombination and transcription in lymphocyte development. Methylation of H3-K79 is exclusive to loci that are active in V(D)J recombination in a given cell type (9): high levels of methylated H3-K79 are detected at the IgH locus but not TCR loci in pro-B cells, and at the TCR loci but not IgH locus in pro-T cells. Conversely, methylation of H3-K9 correlates with the loci that are inactive for V(D)J recombination in a given cell type, and thus the modification is likely to be inhibitory (10). A direct functional link between histone methylation and V(D)J recombination comes from an experiment where the H3-K27 methyltransferase, Ezh2, was deleted in developing B cells. Ezh2 deletion results in an altered pattern of V(D)J recombination, such that recombination to the distal portion of the IgH locus, containing members of the VhJ558 gene family, is not observed (11). The regulatory role of the cytokine IL-7 in V(D)J recombination may be mediated in part through the methylation of H3-K27 (12).
Methylation of H3-K4, a well-studied marker of euchromatin, is also a strong candidate for establishing or maintaining chromatin accessibility for V(D)J recombination (reviewed in 13). H3-K4 can be mono-, di- or trimethylated (di-Me-H3-K4 or tri-Me-H3-K4), but the functional impact of having different degrees of H3-K4 methylation in chromatin is still poorly understood. The presence of di-Me-H3-K4 can correlate with loci active or potentially active in transcription (14,15). A similar correlation between di-Me-H3-K4 and V(D)J recombination can be seen, as peaks of di-Me-H3-K4 are found flanking IgH and TCR loci at developmental stages preceding active recombination of these loci (10).
Tri-Me-H3-K4 is restricted to actively transcribing genes, but unlike di-Me-H3-K4, is typically not detected at a locus prior to transcriptional activation (16). In yeast, tri-Me-H3-K4 may be dependent on an initial round of transcription, but promotes subsequent rounds of transcription by facilitating recruitment of chromatin remodeling complexes (17,18). Loci may thus progress from a potentially active or ‘poised’ state, marked by isolated peaks of di-Me-H3-K4, to one maintained in an active state and marked by tri-Me-H3-K4.
Here we address dynamic changes in H3-K4 di- and trimethylation during V(D)J recombination by employing a cell line in which initiation of V(D)J recombination can be manipulated. This cell line was transformed with the temperature-sensitive Abelson Murine leukemia virus (ts-Ab-MuLV), is already rearranged at the heavy chain locus, and undergoes only low levels of recombination at light chain loci when grown at the permissive temperature. However, a shift to the non-permissive temperature mimics several aspects of the pre-B to immature B-cell transition (19,20). This transition includes high levels of recombination at light chain loci, as well as the transcriptional activation of several factors required for the pre-B to immature B-cell transition in bone marrow pre-B cells (21).
We show a strong association between initiation of recombination and H3-K4 methylation. We also observe increased H3-K4 methylation at some sites where transcription is activated, suggesting a role for this modification in general re-programming of chromatin during B-cell development.
MATERIALS AND METHODS
Antibodies
Two rabbit polyclonals recognizing trimethyl H3-K4 were used for chromatin immunoprecipitation (ChIP) assays [Abcam (see Fig. 4) and Upstate (all other figures)]. The anti-dimethyl H3-K4 rabbit polyclonal antisera was a gift from Yi Zhang, UNC-Chapel Hill. Specificity of both di- and trimethyl H3-K4 antibodies in ChIP assays was confirmed by the ability of appropriate peptides to block immunoprecipitation. Purified mouse IgG1 (Pharmingen) was used as control for levels of non-specific DNA association in all ChIP experiments.
Figure 4.
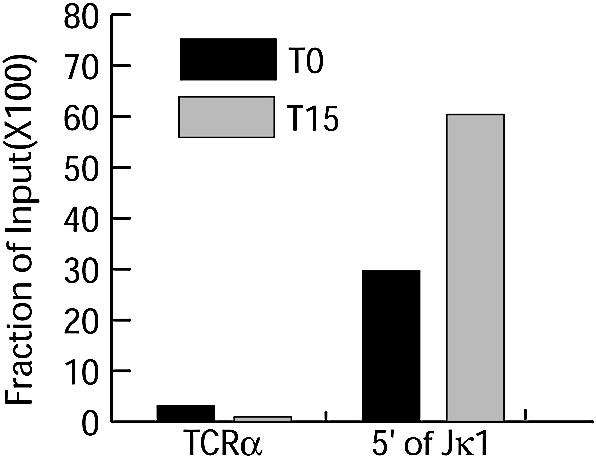
Levels of tri-Me-H3-K4 in primary pre-B cells. T0, immediately before IL-7 withdrawal; T15, 15 h after IL-7 withdrawal.
Cells and ChIP analysis
SP9 ts-Ab-MuLV cell line was established, maintained and induced as described previously (20). Pro-B lymphocytes were expanded in Opti-MEM containing 10% FBS (Gibco), 5 µM 2-mercaptoethanol, 1 × penicillin/streptomycin/glutamine (Gibco), IL-7 (1:100 dilution from J558-IL-7 supernatant, gift of F. Melchers) and c-kit ligand (KL; 1:500 dilution from MGF-CHO supernatant, gift of the Genetics Institute). Cultures were initiated with 1 × 106 B220+ bone marrow (BM) cells/ml isolated from C57Bl/6 BM by selection with anti-B220–magnetic beads on a magnetic column (Miltenyi Biotech., Inc) as described previously (22). To enrich for pre-B and immature B cells the pro-B lymphocytes were washed four times with PBS and replated at 1 × 106 cell/ml in media lacking IL-7 and KL. Cells were further processed for ChIP analysis as described previously (23). ChIP analysis was performed on extracts equivalent to 3.5 × 106 cells for ts-Ab-MuLV cells or 2 × 106 for primary cells, using 2 µg of purified antibody.
PCR analysis
For each reaction, input DNA template (equivalent to 1/5 of the amount used for immunoprecipitation) was serially diluted and amplified to ensure each PCR was in the linear range. Template equivalent to 1/6 of the material recovered from each immunoprecipitation was then compared with input DNA to establish a relative measure of recovered material, thus making comparison between each different PCR target site valid. PCR products were detected and quantified by inclusion of [33P]α-dCTP in reactions. When comparing different extracts, we corrected for recovered DNA by PCR of κ intron enhancer region (κE) from ChIP input samples.
RNA was prepared from 1 × 107 cells using a Qiagen RNeasy mini kit, according to the manufacturer’s instructions. RNA samples were DNase-treated and equalized for concentration according to optical density. RT–PCR was carried out with ∼100 ng total RNA per reaction as template, using the Qiagen OneStep RT–PCR kit (see primers, below). As with other PCR reactions, [33P]α-dCTP was included for purposes of detection and quantification.
Primer sequences and PCR reaction conditions for 5′ of Jκ1, RAG1 ORF and 5′ of Jα50 have been previously described (24). The following primer pairs all required a 55°C annealing step: IgκIVS, EJP58 (5′-CCCGACTTCAGTTCAATGGT-3′) and EJP59 (5′-ACCTGCACCTTACGTTGACC-3′); κE, EJP60 (5′-GACTTTCCGAGAGCCATCTG-3′) and EJP61 (5′-GCCTTAAGCCAGGGTCTGTA-3′); Jκ4, EJP62 (5′-CGGGGACAAAGTTGGAAATA-3′) and EJP63 (5′-CGC TCAGCTTTCACACTGAC-3′); RAG1 promoter, EJP64 (5′-AACAACCCTGAATGTTTCTGC-3′) and EJP65 (5′-TATCAGCTCTCACGCCCTTC-3′); GAPDH internal RT–PCR control, gapdown (5′-CAAAGTTGTCATGGATGACC-3′) and gapup (5′-CCATGGAGAAGGCTGGG-3′); 4 kb 5′ of Jκ1 (4 kb), EJP76 (5′-GGAACCCACAACAGCCAATA-3′) and EJP77 (5′-CCCACTTGTCCCTGAAGAAA-3′); Spi-B promoter, EJP84 (5′-CCTAGCCTGCTCTGAACCAC-3′) and EJP85 (5′-AGGAAACGCTGGACAGAAGA-3′); Spi-B ORF, EJP90 (5′-GCCTGGCCTCCAGGTGTA-3′) and EJP91 (5′-AAGAGGGCCTCGTCTGACTC-3′); IRF-4 promoter, EJP86 (5′-AGAAATGGCGACAGAGGAGA-3′) and EJP87 (5′-CCTTGACTTAGGCGGTTTCA-3′), also EJP96 (5′-GGAAGAAGAGGGGAAAATGG-3′) and EJP97(5′-CAA GCACAGTCCCCAAAGTT-3′); IRF-4 ORF, EJP88 (5′- CAGATGGGCTTTATGCCAAA-3′) and EJP89 (5′-TCT TTCTGGCCCTCTGCTAA-3′); Vκ4, EJP68 (5′-ACTGCA GCCAGTGGTTTGAT-3′) and EJP69 (5′-ATGAATATCC TGAAGCAAAACC-3′); Vκ21A, EJP78 (5′-CCTGCATC TCAACACAGAGC-3′) and EJP79 (5′-AAGGAGGATGC TGAGAGTGG-3′). Coding junctions were amplified with an annealing step of 58°C using DAR14 (5′-GGCTGCA GCTTCAGTGGCAGTGGATCAGGAAC-3′) and DAR24 (5′-CTTTGCCTTGGAGAGTGCCAGAATC-3′).
For ligation-mediated PCR (LM-PCR) analysis of signal ends, a linker was prepared from FM11 (5′-CACTTCA GATC-3′) and FM25 (5′-GCGGTGACTCGGGAGAT CTGAAGTG-3′), ligated to twice the amount of template DNA as was used for standard PCR analysis, and linker-ligated DNA recovered as previously described. In the first round of amplification, 1/4th of the ligated material was amplified for 20 cycles using FM25, DAR11 (24) and a 60°C annealing step for Jκ1 DSBs. 1/25th of the first round PCR was further amplified for 25 cycles using the same annealing temperatures and FM25 and DAR289 (5′-GTTAAGC TTTCGCCTACCCACTGCTC-3′) for Jκ1 DSBs. [33P]α-dCTP was added to the second round of PCR amplification for detection purposes.
RESULTS
H3-K4 di- and trimethylation patterns at the κ locus upon V(D)J recombination
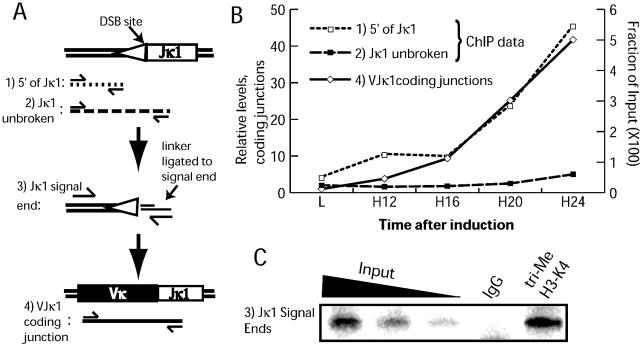
Initially, our ChIP analysis focused on histone modifications observed in chromatin immediately 5′ of the Jκ1 recombination targeting signal. We tested a number of histone modification-specific antibodies and confirmed earlier reports showing hyperacetylation on both H3 and H4, as well as a slight enrichment of methylated H3-K79 after recombination was induced. However, the modification with the most striking change after induction of recombination was methylation of H3-K4 (Fig. 1A, compare L to H). L represents extracts from ts-Ab-MuLV cells grown at permissive temperature, H represents extracts from cells grown at the non-permissive temperature for 24 h. Both di- and tri-Me-H3-K4 were increased >10-fold over the induction period (Fig. 1B and C) whereas other modifications only increased 2–5-fold (data not shown).
Figure 1.
Levels of tri-Me-H3-K4 detected at selected sites. (A) ChIP analysis of extracts from ts-Ab-MuLV cells. L, extracts from ts-Ab-MuLV cells grown at permissive temperature; H, extracts from ts-Ab-MuLV cells grown at non-permissive temperature for 24 h. Additional 5- and 2-fold dilutions of H input DNA are shown to confirm PCR is in the linear range. Input DNA template (left of figure) was compared to DNA template immunoprecipitated with a non-specific control antibody (IgG) or using the tri-Me-H3-K4 antibody, and template detected using 5′ of Jκ1 or TCR primer pairs as noted. (B) Levels of tri-Me-H3-K4 at selected sites. Approximate locations of primer pairs noted on the locus diagram (details in Materials and Methods). The amount of immunoprecipitated DNA, averaged from two independent inductions, for a given primer pair is expressed as a fraction of input DNA for both L and H samples. Error bars represent standard deviation. (C) Levels of di-Me-H3-K4 at selected sites from one representative experiment of two. Primer pair locations as in (B).
In contrast, levels of both di- and tri-Me-H3-K4 at an analogous site in the recombinationally inactive TCR α locus, 5′ of TCR Jα50 (TCRα) remained low and unaffected by the induction of V(D)J recombination (Fig. 1A). Very high levels (>30% of input DNA) of tri-Me-H3-K4 at a site in the IgH locus are evident prior to induction of recombination, and levels only slightly increase (<2-fold) after shift to the non-permissive temperature (unpublished data).
Activation of recombination at the Igκ locus has been linked to increased chromatin accessibility, and to initiation of a germline transcript that starts ∼4 kb upstream of Jκ1 and continues to Cκ. Significantly increased enrichment of both di- and tri-Me-H3-K4 was restricted to a portion of the germline transcript, including a site near the germline transcript promoter (4 kb, Fig. 1B and C) and the majority of the Igκ J segments (Jκ1 and Jκ4, Fig. 1B and C). However, methylated H3-K4 does not extend for the entire length of the germline transcript, as levels of di- and tri-Me-H3-K4 are low and remain unchanged at the Cκ intron–exon boundary after induction of recombination (unpublished data). Levels of −tri-Me-H3-K4 also remain low at the κ intron enhancer (κE, ∼4 kb downstream of Jκ5, Fig. 1B). Levels of di-Me-H3-K4 at the κE are initially somewhat higher than other κ locus sites, but are reduced 2-fold after induction of recombination. Levels of H3-K4 methylation remain low at a site 9 kb upstream of Jκ1 (IgκIVS; midway between Jκ1 and the most proximal Vκ) as well as a number of sites near Vκ segments. Vκ sites tested included a site immediately 3′ of Vκ21A, the most J proximal Vκ (Fig. 1B and C), and a primer pair common to sites immediately 3′ of two members of the Vκ4 family (aa4 and kn4), located approximately in the center of the Vκ locus. Increased methylation of H3-K4 is thus restricted to a ∼7 kb region of the κ locus that roughly correlates with the 5′ portion of the germline transcript.
H3-K4 trimethylation is specific for actively recombining loci
We tracked how closely tri-Me-H3-K4 enrichment correlates with recombination over an interval during which recombination increases exponentially. Our results show increases in tri-Me-H3-K4 at the Jκ1 signal flank which very closely paralleled increases in recombination to this signal (Fig. 2B, compare 5′ of Jκ1 ChIP and VJκ1 coding junctions). As expected, H3-K4 methylation at the TCRα locus was almost undetectable throughout the time course (data not shown).
Figure 2.
Ordering of recombination and H3-K4 methylation near Jκ1. (A) Approximate location of PCR primers used in (B) and (C); numbers identify each PCR product used in subsequent panels. (B) Relative levels of coding junctions from input DNA are plotted against levels of tri-Me-H3-K4 detected at the indicated sites before (L) or after shift of cells to the non-permissive temperature (H) for 12, 16, 20 or 24 h. Initial levels of coding junctions at L were defined as 1. Data are from a representative experiment. (C) ChIP analysis of extracts from ts-Ab-MuLV cells grown at non-permissive temperature for 24 h. Additional 5- and 2-fold dilutions of input DNA are shown to confirm LM-PCR is in the linear range; see (A) for cartoon representation of LM-PCR. Input DNA template (left of panel) was compared to DNA template immunoprecipitated with a non-specific control antibody (IgG) or using the tri-Me-H3-K4 antibody.
The PCR primer pair used in all previous figures to assess chromatin near Jκ1 (5′ of Jκ1) amplifies DNA independent of whether recombination has yet initiated at Jκ1. In the ts-Ab-MuLV model, less than half of all Igκ alleles undergo recombination at this signal. To determine if tri-Me-H3-K4 enrichment occurs preferentially within the fraction of alleles active in recombination, we altered the location of the downstream primer of the 5′ of Jκ1 pair such that the PCR product now spans the Jκ1 break site (Fig. 2A). The latter PCR is thus directed towards the same region, but is specific for chromatin that is still unbroken at Jκ1. Parallel analysis of the same immunoprecipitated DNA with these two primer pairs gives strikingly different results. In contrast to the ∼10-fold enrichment of tri-Me-H3-K4 observed using the 5′ of Jκ1 primer pair, analysis using the Jκ1 unbroken primer pair shows consistently low levels of tri-Me-H3-K4 (Fig. 2B). By inference, H3-K4 trimethylation is thus restricted to alleles either immediately prior to or after cleavage of the chromosomes at recombination signals. Consistent with this correlation, tri-Me-H3-K4 can be readily detected at Jκ1 signal ends when immunoprecipitated DNA is analyzed using a LM-PCR assay specific for this species (Fig. 2C).
H3-K4 trimethylation at other loci important for B-cell development
Previous studies addressing histone modifications in the context of lymphocyte development focused primarily on links to V(D)J recombination, and thus loci active in V(D)J recombination. We applied ChIP analysis to three loci where transcription is activated after a shift to the non-permissive temperature to determine whether tri-Me-H3-K4 enrichment was also associated with activation of transcription in this cell line. Using RT–PCR we confirmed previous work (21) indicating that transfer of cells to the non-permissive temperature activates transcription of RAG1, Spi-B and IRF-4 genes (Fig. 3A). Tri-Me-H3-K4 is enriched >10-fold in a site within the Spi-B gene (Fig. 3B), and a site within the RAG1 gene undergoes a 4-fold increase in tri-Me-H3-K4 (Fig. 3B), correlating transcriptional activation and H3-K4 trimethylation in this model for these two genes. Surprisingly, while transcription of IRF-4 is induced, levels of tri-Me-H3-K4 at several sites within the IRF-4 gene, including both the promoter and transcribed DNA, are not (Fig. 3B). ChIPs for each gene were repeated at least three times.
Figure 3.
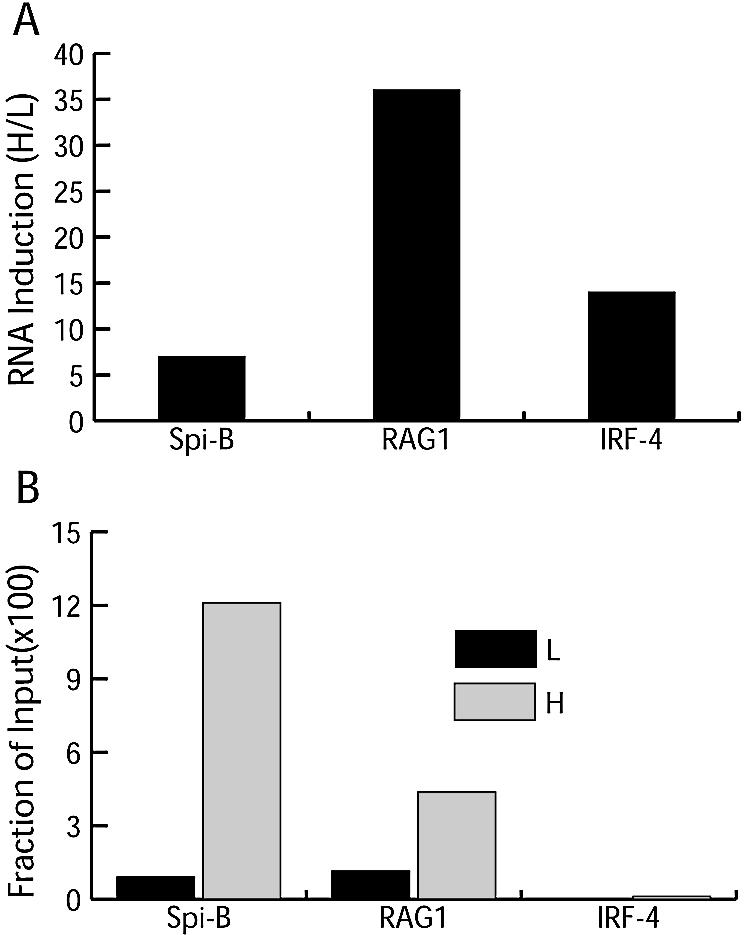
Tri-Me-H3-K4 and activation of transcription. (A) Relative levels of RNA before (L) and after (H) shifting to the non-permissive temperature using RT–PCR and ORF primer pairs for each gene, as indicated in the Materials and Methods. A GAPDH RT–PCR was used to correct for differences in RNA between samples. (B) ChIP data showing levels of tri-Me-H3-K4 in Spi-B, RAG1 and IRF-4 as assessed by ORF primer pairs for each gene.
Enrichment of tri-Me-H3-K4 in primary cells
We also addressed changes in tri-Me-H3-K4 using a primary cell model for the pre-B to immature B-cell transition. Pro-B cells were selectively expanded from bone marrow for 4 days in the presence of IL-7 and KL (T0), after which immature B cells were enriched by withdrawal of IL-7 (25). Enrichment for pre-B cells was verified by detection of an ∼30-fold increase in levels of light chain rearrangement between 0 and 15 h after IL-7 withdrawal (data not shown). Limited amounts of material meant that only histone methylation at the 5′ of Jκ1 and 5′ of TCR Jα50 sites could be assessed by ChIP analysis. Initial levels of tri-Me-H3-K4 were already enriched at 5′ of Jκ1 relative to TCR Jα50, but an additional 2-fold increase in methylation was observed 15 h after IL-7 withdrawal (Fig. 4). Therefore, as observed in the ts-Ab-MuLV cell line, increased H3-K4 methylation correlates with increased recombination in primary cells. However, changes in H3-K4 methylation in primary cells are much less striking than that observed in the ts-Ab-MuLV cell line, probably due to greater heterogeneity in the initial primary cell population.
DISCUSSION
We link enrichment of both di- and tri-Me-H3-K4 with the activation of both recombination and transcription at several loci important for development of B cells using a transformed cell line. Experiments using primary cells indicate this enrichment is not unique to the transformed cell line model. These correlations provide important insight into the potential control mechanisms of V(D)J recombination and transcriptional activation during B-cell development.
Using the transformed cell line, we see a >10-fold enrichment of tri-Me-H3-K4 over a portion of the germline transcript that includes Jκ recombination signals. A progressive increase in tri-Me-H3-K4 detected near Jκ1 occurs in parallel with an increase in recombination to that signal. Remarkably, tri-Me-H3-K4 is also excluded from chromatin-containing Jκ1 signals not active or not yet active in recombination. Recombination and histone methylation near Jκs are thus very closely linked in time.
Previous work has linked induction of a germline transcript and increased chromatin accessibility with initiation of V(D)J recombination, but whether germline transcription is the cause or consequence of increased chromatin accessibility is unclear (26). We suggest that the germline transcript is in part the cause of increased accessibility: initiation of germline transcription may serve as a trigger for the marking of chromatin over Jκs with tri-Me-H3-K4. We show tri-Me-H3-K4 methylation and recombination are essentially coincident, arguing tri-Me-H3-K4’s ability to promote recruitment of remodeling complexes (17) may be the critical step in making Jκs accessible to RAG proteins.
However, we detect no significant increases in tri-Me-H3-K4 near several Vκ recombination signals. This observation is consistent with a lack of correlation between recombination activity of VH gene segments and whether the same VH segments have a germline transcript (27). Recent work suggests a RAG1/2 multimer that can cleave both signals assembles first on a single signal; the second signal is recruited into this complex and cleavage ensues (28,29). In the Igκ locus, tri-Me-H3-K4 and its ability to promote chromatin activity may thus be essential only for recognition of the first signal (in this case a Jκ). Subsequent recruitment of a Vκ recombination signal to this pre-formed complex may not be dependent on tri-Me-H3-K4.
A previous study identified peaks of di-Me-H3-K4 flanking other antigen receptor loci (IgH, TCRβ) poised for rearrangement (10). Possibly consistent with this observation, we detect before induction of recombination a modest enrichment of di-Me-H3-K4 at the κE relative to other sites in the Igκ locus. However, neither the prior study nor ours detected significant peaks of di-Me-H3-K4 in the more widely distributed VH and Vκ regions. Clearly, the techniques used here could not be practically employed to systematically cover the entire Vκ region (>3 Mbp), thus a peak of di-Me-H3-K4 in this region may have been missed.
Enrichment of tri-Me-H3-K4 after induction in this cell line is not restricted to loci active in recombination. As previously shown, transcription of RAG1, IRF-4 and Spi-B genes is also activated after induction of ts-Ab-MuLV cells (21). We demonstrate that activation of transcription is accompanied by enrichment of tri-Me-H3-K4 at these genes. Although our RT–PCR analysis confirms activation of IRF-4 transcription, we detect no enrichment for tri-Me-H3-K4 in this gene, after assessing the status of two sites covering the majority of this gene’s promoter as well as at a site approximately in the middle of the gene’s transcript. However, it is still possible that the pattern of trimethylation in the IRF-4 gene is more focused either than has been observed here for RAG1 and Spi-B genes (Fig. 3), or has been previously observed in other mammalian genes (30). Alternatively, IRF-4 may be an example of an RNA polymerase II-transcribed gene in which methylation of H3-K4 is not associated with activation of transcription.
Thus far, links between H3-K4 methylation and B-cell development discussed here and elsewhere have largely been limited to correlations, possibly because the H3-K4 histone methyltransferases in mammals have pleiotropic effects. The mixed lineage leukemia (MLL) gene product, required for control of vertebrate HOX gene expression and thus many developmental programs (31), is one candidate for establishing the patterns of H3-K4 methylation we observe here. MLL has H3-K4 methyltransferase activity (32,33), but is unable to generate tri-Me-H3-K4 in vitro (33). However, MLL has links to B-cell development, as translocations in the MLL gene can cause malignancy in pre-B cells (reviewed in 34,35). Translocation deletes the MLL methyltransferase domain, suggesting pre-B-cell malignancy could be promoted by disruption of patterns of methylated H3-K4. Clearly, definitive assessment of the role of H3-K4 methylation in B-cell development awaits the tools to block or perturb this chromatin modification.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Yi Zhang and Brian Strahl (UNC) for advice and the gift of antibodies to Me-H3-K4, Dr Yung Chang (Arizona State University) for the gift of the ts-Ab-MuLV cell line, Stephanie Nick McElhinny for critical reading of the manuscript, and members of the Ramsden lab for helpful discussion. This work was supported by public health service grants CA84442 to D.A.R. and CA99978 to B.K.
REFERENCES
- 1.Mostoslavsky R., Alt,F.W. and Bassing,C.H. (2003) Chromatin dynamics and locus accessibility in the immune system. Nature Immunol., 4, 603–606. [DOI] [PubMed] [Google Scholar]
- 2.Bassing C.H., Swat,W. and Alt,F.W. (2002) The mechanism and regulation of chromosomal V(D)J recombination. Cell, 109 (Suppl.), S45–55. [DOI] [PubMed] [Google Scholar]
- 3.Gellert M. (2002) V(D)J recombination: RAG proteins, repair factors and regulation. Annu. Rev. Biochem., 71, 101–132. [DOI] [PubMed] [Google Scholar]
- 4.Schlissel M. (2002) Allelic exclusion of immunoglobulin gene rearrangement and expression: why and how? Semin. Immunol., 14, 207–226. [DOI] [PubMed] [Google Scholar]
- 5.Roth D.B. and Roth,S.Y. (2000) Unequal access: regulating V(D)J recombination through chromatin remodeling. Cell, 103, 699–702. [DOI] [PubMed] [Google Scholar]
- 6.Muegge K. (2003) Modifications of histone cores and tails in V(D)J recombination. Genome Biol., 4, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwon J., Morshead,K.B., Guyon,J.R., Kingston,R.E. and Oettinger,M.A. (2000) Histone acetylation and hSWI/SNF remodeling act in concert to stimulate V(D)J cleavage of nucleosomal DNA. Mol. Cell, 6, 1037–1048. [DOI] [PubMed] [Google Scholar]
- 8.Krangel M.S. (2003) Gene segment selection in V(D)J recombination: accessibility and beyond. Nature Immunol., 4, 624–630. [DOI] [PubMed] [Google Scholar]
- 9.Ng H.H., Ciccone,D.N., Morshead,K.B., Oettinger,M.A. and Struhl,K. (2003) Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc. Natl Acad. Sci. USA, 100, 1820–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morshead K.B., Ciccone,D.N., Taverna,S.D., Allis,C.D. and Oettinger,M.A. (2003) Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc. Natl Acad. Sci. USA, 100, 11577–11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su I.H., Basavaraj,A., Krutchinsky,A.N., Hobert,O., Ullrich,A., Chait,B.T. and Tarakhovsky,A. (2003) Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nature Immunol., 4, 124–131. [DOI] [PubMed] [Google Scholar]
- 12.Huang J. and Muegge,K. (2001) Control of chromatin accessibility for V(D)J recombination by interleukin-7. J. Leukoc. Biol., 69, 907–911. [PubMed] [Google Scholar]
- 13.Sims R.J. 3rd, Nishioka,K. and Reinberg,D. (2003) Histone lysine methylation: a signature for chromatin function. Trends Genet., 19, 629–639. [DOI] [PubMed] [Google Scholar]
- 14.Strahl B.D., Ohba,R., Cook,R.G. and Allis,C.D. (1999) Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl Acad. Sci. USA, 96, 14967–14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein B.E., Humphrey,E.L., Erlich,R.L., Schneider,R., Bouman,P., Liu,J.S., Kouzarides,T. and Schreiber,S.L. (2002) Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl Acad. Sci. USA, 99, 8695–8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santos-Rosa H., Schneider,R., Bannister,A.J., Sherriff,J., Bernstein,B.E., Emre,N.C., Schreiber,S.L., Mellor,J. and Kouzarides,T. (2002) Active genes are tri-methylated at K4 of histone H3. Nature, 419, 407–411. [DOI] [PubMed] [Google Scholar]
- 17.Santos-Rosa H., Schneider,R., Bernstein,B.E., Karabetsou,N., Morillon,A., Weise,C., Schreiber,S.L., Mellor,J. and Kouzarides,T. (2003) Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol. Cell, 12, 1325–1332. [DOI] [PubMed] [Google Scholar]
- 18.Ng H.H., Robert,F., Young,R.A. and Struhl,K. (2003) Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell, 11, 709–719. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y.Y., Wang,L.C., Huang,M.S. and Rosenberg,N. (1994) An active v-abl protein tyrosine kinase blocks immunoglobulin light-chain gene rearrangement. Genes Dev., 8, 688–697. [DOI] [PubMed] [Google Scholar]
- 20.Chang Y. and Brown,M.L. (1999) Formation of coding joints in V(D)J recombination-inducible severe combined immune deficient pre-B cell lines. Proc. Natl Acad. Sci. USA, 96, 191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muljo S.A. and Schlissel,M.S. (2003) A small molecule Abl kinase inhibitor induces differentiation of Abelson virus-transformed pre-B cell lines. Nature Immunol., 4, 31–37. [DOI] [PubMed] [Google Scholar]
- 22.Kee B.L., Rivera,R.R. and Murre,C. (2001) Id3 inhibits B lymphocyte progenitor growth and survival in response to TGF-beta. Nature Immunol., 2, 242–247. [DOI] [PubMed] [Google Scholar]
- 23.Chen H., Lin,R.J., Xie,W., Wilpitz,D. and Evans,R.M. (1999) Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell, 98, 675–686. [DOI] [PubMed] [Google Scholar]
- 24.Perkins E.J., Nair,A., Cowley,D.O., Van Dyke,T., Chang,Y. and Ramsden,D.A. (2002) Sensing of intermediates in V(D)J recombination by ATM. Genes Dev., 16, 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rolink A., Kudo,A., Karasuyama,H., Kikuchi,Y. and Melchers,F. (1991) Long-term proliferating early pre B cell lines and clones with the potential to develop to surface Ig-positive, mitogen reactive B cells in vitro and in vivo. EMBO J., 10, 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stanhope-Baker P., Hudson,K.M., Shaffer,A.L., Constantinescu,A. and Schlissel,M.S. (1996) Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell, 85, 887–897. [DOI] [PubMed] [Google Scholar]
- 27.Angelin-Duclos C. and Calame,K. (1998) Evidence that immunoglobulin VH-DJ recombination does not require germ line transcription of the recombining variable gene segment. Mol. Cell. Biol., 18, 6253–6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones J.M. and Gellert,M. (2002) Ordered assembly of the V(D)J synaptic complex ensures accurate recombination. EMBO J., 21, 4162–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mundy C.L., Patenge,N., Matthews,A.G. and Oettinger,M.A. (2002) Assembly of the RAG1/RAG2 synaptic complex. Mol. Cell. Biol., 22, 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider R., Bannister,A.J., Myers,F.A., Thorne,A.W., Crane-Robinson,C. and Kouzarides,T. (2004) Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nature Cell Biol., 6, 73–77. [DOI] [PubMed] [Google Scholar]
- 31.Yu B.D., Hess,J.L., Horning,S.E., Brown,G.A. and Korsmeyer,S.J. (1995) Altered Hox expression and segmental identity in Mll-mutant mice. Nature, 378, 505–508. [DOI] [PubMed] [Google Scholar]
- 32.Milne T.A., Briggs,S.D., Brock,H.W., Martin,M.E., Gibbs,D., Allis,C.D. and Hess,J.L. (2002) MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell, 10, 1107–1117. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura T., Mori,T., Tada,S., Krajewski,W., Rozovskaia,T., Wassell,R., Dubois,G., Mazo,A., Croce,C.M. and Canaani,E. (2002) ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell, 10, 1119–1128. [DOI] [PubMed] [Google Scholar]
- 34.Ernst P., Wang,J. and Korsmeyer,S.J. (2002) The role of MLL in hematopoiesis and leukemia. Curr. Opin. Hematol., 9, 282–287. [DOI] [PubMed] [Google Scholar]
- 35.Collins E.C. and Rabbitts,T.H. (2002) The promiscuous MLL gene links chromosomal translocations to cellular differentiation and tumour tropism. Trends Mol. Med., 8, 436–442. [DOI] [PubMed] [Google Scholar]