Abstract
Amplification of a cDNA product by quantitative PCR (qPCR) is monitored by a fluorescent signal proportional to the amount of produced amplicon. The qPCR amplification curve usually displays an exponential phase followed by a non-exponential phase, ending with a plateau. Contrary to prevalent interpretation, we demonstrate that under standard qPCR conditions, the plateau can be explained by depletion of the probe through Taq polymerase- catalysed hydrolysis. Knowing the probe concentration and the fluorescence measured at the plateau, a specific fluorescence can thus be calculated. As far as probe hydrolysis quantitatively reflects amplicon synthesis, this, in turn, makes it possible to convert measured fluorescence levels in the exponential phase into concentrations of produced amplicon. It follows that the absolute target cDNA concentration initially engaged in the qPCR can be directly estimated from the fluorescence data, with no need to refer to any calibration with known concentrations of target DNA.
INTRODUCTION
Quantitative real-time PCR (qPCR) is widely used for absolute or relative quantification of gene expression, and is a critical tool for basic research, biotechnology and genetic diagnostics (1–3). This methodology is particularly suited because of its conceptual simplicity and practical ease (4). The interpretation of amplification curves rests on the existence of an exponentially growing phase in which the PCR efficiency E (here defined as the factor by which the amplicon concentration is multiplied at each cycle) should be theoretically constant and equal to 2. For some undemonstrated reasons, the efficiency is, in most systems, significantly lower than 2. Current methods for relative and absolute quantification in qPCR have been developed to take this deviation into account (5–7). On the other hand, the non-exponential phase and the plateau of the amplification curve are either not considered in these methods, or are described by a phenomenological model (8) which does not explain the non-exponential shape of the curve. A progressive decrease of efficiency necessarily reflects a decline of DNA polymerase activity. It may result either from a true molecular inhibition (enzyme instability, inhibition by end product, etc.), or from a lack of factors required by the PCR to proceed (depletion of primers or of fluorescent probe, template unavailability due to incomplete denaturation or to product reannealing, etc.). Apart from early experiments which were unable to explain the plateau by a single factor (9), it has been more recently proposed that the main factor contributing to the plateau phase consists of the binding of DNA polymerase to its amplification products (10). The aim of our work was to further investigate the plateau effect. The unexpected outcome of our findings is that the interpretation of the plateau value may serve to calibrate the signal fluorescence. The useful consequence of this internal calibration is that the analysis of an amplification curve may lead to the absolute quantification of gene expression without the need for any calibration curve constructed with DNA plasmid standard samples.
MATERIALS AND METHODS
Real-time PCR
Using the Primer Express software program (Applied Biosystems), we designed PCR primers and probes (Eurogentec) for the amplification of DNA derived from three different transcripts. (i) Insulin-like growth factor (IGF1): accession number, EMBL X04480; forward primer, GCTCCGGAAGCAACACTCA; reverse primer, GCTATGGCTCCAGCATTCG; probe, CCACAATGCC TGTCTGAGGTGCCCT. (ii) Caveolin1 (CAV1): accession number, EMBL AK057982; forward primer, AGCTGA GGCAGAAGCAAGTGT; reverse primer, TGTTTAGG TCGCGGTTGAC; probe, ACGCGCACACCAAGGA GATCGAC. (iii) RAP1-GTPase-activating protein 1 (RAP1GA1): accession number, EMBL BC054490; forward primer, GCCCAAGTCGGAGAACTCATC; reverse primer, TGCGGTCTCCGCTCTGTT; probe, CCCCAGAGATG CCCACGACCA.
Taqman probes carried a 5′ FAM reporter label and a 3′ DABCYL quencher group. The enzyme was activated by heating for 10 min at 95°C. A two-step PCR procedure was used, 60 s at 60°C and 15 s at 95°C for 50 cycles in a PCR mix containing 2 µl of cDNA template, 1× qPCR Mastermix (RT-QP2X-03, Eurogentec), 300 nM of each primer and 100 nM probe in a total volume of 30 µl (except where otherwise stated). The fluorescence intensity of the reporter label was normalized to the rhodamine derivative ROX as a passive reference label present in the buffer solution. The system generated a kinetic amplification plot based upon the normalized fluorescence. All reactions were performed in an Abi-Prism 7700 (Applied Biosystems).
In order to demonstrate the aetiology of the plateau, a slight modification of the protocol was applied. After 35 cycles, the reaction was stopped and a further addition of 1 µl containing either water, 1.55 µM probe (50 nM final) or 27.9 µM primers (900 nM final) was done before restarting the reaction for 15 more cycles.
Plasmid preparation
IGF1 cDNA was obtained from mouse thyroid RNA extract. Plasmids containing IGF1 cDNA (pCR 2.1), CAV1 cDNA (pIRES CAV1 puro, kindly provided by Dr S. Costagliola) and RAP1GA1 cDNA (pMT2-HA-RG glu, kindly provided by Dr F. Zwartkruis) were linearized by restriction with XbaI, EcoRI and SalI, respectively. The copy number was estimated by optical density according to the exact molar mass derived from the plasmidic and the amplicon sequences. Different dilutions were made to obtain 102, 103, 104, 105 and 106 copies in 2 µl.
Dilution curve construction
Serially diluted samples (dilution factors of 1, 10, 100, 1000 and 10 000) were submitted to a qPCR run. The Ct values were estimated at a threshold value of 0.1, which was at least twice as high as the detection limit corresponding to 10 times the value of the standard deviation of the fluorescences in cycles 1–10.
RESULTS
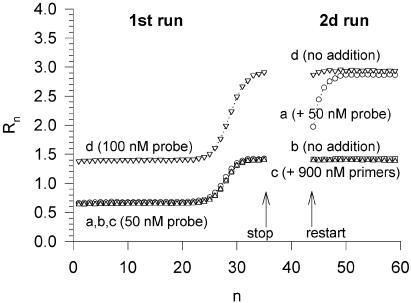
In an attempt to question the proposal (10) that the plateau results from a complete molecular inhibition of the polymerase, an experiment was designed to test whether a further addition of a limiting factor at the time when the plateau was reached was capable of reinitiating the amplification process. When a cDNA sample was amplified by qPCR using 900 nM of primers and 50 nM of fluorescent Taqman probe which hybridized to the IGF1 sequence, the amplification curve reached a plateau after about 30 cycles (curves a–c in Fig. 1). After 35 cycles, the qPCR run was interrupted to allow a further addition of a final probe concentration of 50 nM. As shown in Figure 1, the probe supplement led to an additional amplification which converged on the plateau obtained with an initial probe concentration of 100 nM (compare curves a and d in Fig. 1). These results demonstrate that, under these commonly used qPCR conditions, the plateau of the amplification curve is due to the depletion of a limiting factor (here the fluorescent probe) and not to any intrinsic loss of Taq polymerase activity.
Figure 1.
Two-step qPCR allowing two successive additions of a limiting factor. Each sample initially contained primer concentrations of 900 nM and probe concentrations of 50 nM (samples a–c) or 100 nM (sample d) in a volume of 30 µl. Baseline fluorescence was not subtracted from the measured Rn fluorescence. The first qPCR run was stopped after 35 cycles. A further 1 µl volume of water was added to each sample. It contained a final concentration of 50 nM probe in sample a, and a final concentration of 900 nM primers in sample c. A second qPCR run of 15 cycles was then started. Only sample a, in which the probe concentration was increased, showed significantly rising fluorescence in the second run.
Usually, an amplification curve is described by the normalized fluorescence increment ΔRn associated with the free fluorophore release during probe hydrolysis. The subtracted background level, which is due to the partially quenched fluorescence of the probe-bound fluorophore, is thus dependent on the sample probe concentration. The fact that different probe concentrations were used in our experimental design required that the results of Figure 1 were described by the crude normalized fluorescence Rn (no background subtraction). A quantitative look at curves a and d confirms that both the initial and the plateau values of the fluorescence, hence their difference ΔRn,plateau, are proportional to the total probe concentration. Knowing the probe concentration, an apparent specific fluorescence Δφ of the incremental signal ΔRn can be estimated:
Δφ = ΔRn,plateau/[probe]total 1
In fact, Δφ represents the difference between the specific fluorescence of the free fluorophore and the specific fluorescence of the probe-bound fluorophore. The measured incremental signal ΔRn can be converted into the corresponding concentration of hydrolysed probe, at any cycle:
[probe]hydrolysed = ΔRn/Δφ 2
As far as the synthesis of one amplicon molecule is accompanied by the hydrolysis of one probe molecule, equation 2 can be rewritten:
[amplicon]synthesized = ΔRn/Δφ 3
This strict parallelism between amplicon synthesis and probe hydrolysis happens if template elongation starts after the hybridization of probe to template. This should be the case at least in the exponential phase of the amplification where most of the probe molecules are still available, and for a probe for which a high Tm value ensures an efficient hybridization. If this is the case, the absolute initial target cDNA concentration in the sample can be directly estimated by the ratio between the chemical concentration of amplicon exponentially produced after n cycles (equation 3), and the amplification factor which is, in the exponential phase, equal to En:
[cDNA]initial = (ΔRn/Δφ)/En 4
Alternatively, one may prefer to characterize the target cDNA content in the sample by the initial copy number. Equation 4 and the explicit formulation of Δφ lead to
(copy number)initial = [(ΔRn.[probe]total)/(ΔRn,plateau.En)].V.N0 5
where V and N0 are the sample volume and the Avogadro’s number, respectively. Equation 5, which is valid for any detectable ΔRn value belonging to the exponential phase, shows that the calculation of the initial copy number just requires a single amplification curve obtained with a known and limiting probe concentration, and the efficiency value. Theoretically, the efficiency can be estimated by analysing the exponential phase of the same single amplification curve (6,7,11). However, when applying such an approach to our systems, we previously experienced very inaccurate efficiency estimates, simply because the exponential phase of most of our amplification curves did not contain more than three or four points above the background level. We have thus preferred to estimate the efficiency by analysing the slope of a dilution curve constructed in a preliminary experiment in which Ct values were determined from serially diluted samples (12). It must be stressed that this latter strategy does not require samples with a known amount of target DNA, since the dilution curve is only referring to sample dilution factors. Thus, the use of samples for which the target cDNA content will be determined should ensure a relevant estimate of PCR efficiency.
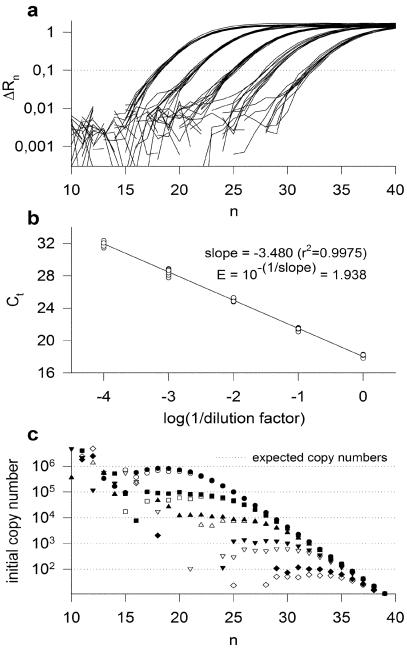
The robustness of the proposed method based on equation 5 has been tested by analysing qPCR curves obtained with samples containing known concentrations of cloned IGF1 DNA. However, these samples have been treated and analysed by our method as if their respective DNA contents were unknown. A first preliminary experiment was performed for estimating the PCR efficiency: five serially diluted samples (in decuple) were submitted to a qPCR run (Fig. 2a) and the dilution curve, constructed on the basis of the Ct values estimated at a threshold of 0.1, was analysed by linear regression, which led to an efficiency estimate of 1.938 (Fig. 2b) with the 95% confidence interval (CI) being 1.920–1.957. Each qPCR curve was then separately analysed to estimate the initial target DNA content of the corresponding sample: equation 5 was applied to the data point displaying the lowest detectable fluorescence signals (the detection limit being set to 10 times the standard deviation of the baseline fluorescence), and the resulting estimate of copy number was compared with the expected value (which was derived from the OD measurement of the plasmid preparation). In order to visualize the reliability of the results, a graphical representation was elaborated: equation 5 was repeatedly applied to all the data points of an amplification curve (as if they all were detectable and situated in the exponential phase characterized by the previously estimated efficiency of 1.938), and the result was plotted against the cycle number (Fig. 2c). The plot showed scattered values as long as the fluorescence level was below the detection limit, followed by a plateau value obtained with the detectable points of the exponential phase. At higher cycle number values, the points progressively deviated from the plateau since they were obtained assuming a constant efficiency of 1.938, which led to an overestimated amplification factor. The plateau value that such a plot unveils represents the copy number estimate of the sample. Figure 2c shows, for each dilution condition, the two curves leading to the lowest and the highest estimates of the copy number, respectively.
Figure 2.
Analysis of qPCR results according to the proposed experimental strategy. Fifty samples containing either 106, 105, 104, 103 or 102 copies of IGF1 DNA were submitted to qPCR using a probe concentration of 100 nM and primer concentrations of 300 nM. (a) Amplification curves for the 50 samples (five conditions in decuple). A threshold value of 0.1 was used to estimate the Ct values. (b) Dilution curve constructed on the basis of the sample dilution factors (1, 10, 100, 1000 and 10 000) and of the corresponding Ct values. The PCR efficiency was estimated through the linear regression of the dilution curve (r2 = 0.9975). Estimates of slope and efficiency were equal to –3.480 and 1.938, respectively. (c) Plot of the initial copy number estimates against the cycle number, as resulting from the application of equation 5 to all the points of the amplification curve. For each dilution factor, two curves were plotted, which gave the highest estimate (filled symbols) and the lowest estimate (open symbols) of the copy number, respectively. The dotted lines represent the five expected copy numbers.
The data obtained in Figure 2 were also analysed by the classical method which used a calibration curve constructed by serially diluting the sample containing the known OD measured concentration of IGF1 DNA (Table 1). Then, each individual sample was considered as unknown. Since the same samples were used as standard points and unknowns, the copy numbers estimated by the classical method had to converge in average on the expected value, as was in fact observed (compare columns 1 and 2 of Table 1). Both classical and proposed methods led to similarly dispersed copy number estimates with coefficients of variation lower than 30% (compare the standard deviations of columns 2 and 3 of Table 1). The estimate means obtained by the proposed method showed a systematic deviation of ∼15% from the expected values. However, this deviation could be explained by the uncertainty of the efficiency estimate: taking the 95% CI for the efficiency estimate into account, it appeared that the corresponding CI for the copy number estimates extended over the expected copy number, with an exception in the case of the highest copy number value (see last column of Table 1). The observation that the upper limit of the CI was about twice the lower limit conformed with a Ct uncertainty of about 1 unit.
Table 1. Comparison of the proposed method and the classical method based on the analysis of the 50 IGF1 amplification curves presented in Figure 2.
| Expected number (OD measurement) | Estimated number: mean ± SD (n = 10) | 95% CI | |
| |
Classical method |
Proposed method |
Proposed method |
| E = 1.938 | E = 1.957–1.920 | ||
| 102 | (1.00 ± 0.19) × 102 | (0.82 ± 0.16) × 102 | (0.60–1.10) × 102 |
| 103 | (1.04 ± 0.29) × 103 | (0.88 ± 0.26) × 103 | (0.64–1.09) × 103 |
| 104 | (1.00 ± 0.11) × 104 | (0.85 ± 0.10) × 104 | (0.67–1.07) × 104 |
| 105 | (1.09 ± 0.12) × 105 | (0.84 ± 0.10) × 105 | (0.69–1.03) × 105 |
| 106 | (0.95 ± 0.10) × 106 | (0.75 ± 0.05) × 106 | (0.63–0.89) × 106 |
The classical method converted the Ct value (obtained at a threshold value of 0.1) into the copy number (column 2) by using the calibration curve constructed with standard samples obtained by serial dilutions of a sample for which the DNA content was determined by OD measurement (column 1). The proposed method calculated the copy number by applying equation 5 to the points of the exponential phase for which the fluorescence level exceeded the detection limit of 0.05, i.e. 10 times the value of the standard deviation of the fluorescences in cycles 1–10. The copy number estimates (column 3) were calculated using an efficiency value of 1.938 (as obtained in Fig. 2b). The last column represents the 95% CI as obtained by using the corresponding interval for the efficiency estimate.
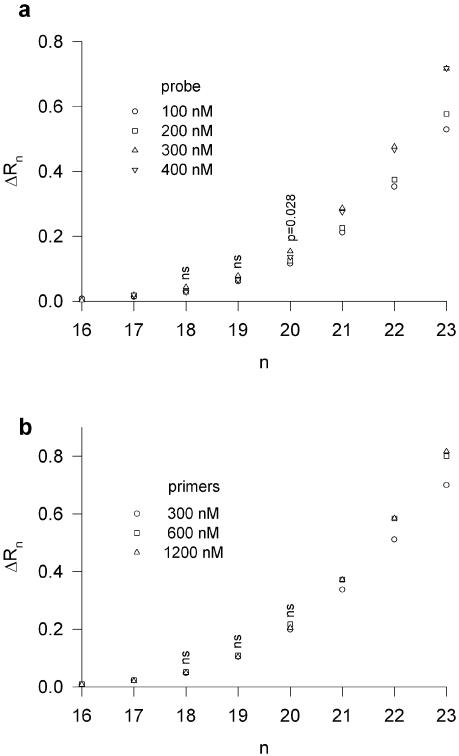
Several other systems consisting of different DNA targets were analysed according to the same strategy. In most cases, the proposed method significantly underestimated and never overestimated the target DNA content determined by OD measurement. Table 2 reports the results obtained with two particular systems: the representative CAV1 system which showed a moderate deviation of 30% from the expected values, and the RAP1GA1 system which more deeply underestimated the expected values by ∼75%. After checking that such deviations could not always be explained by errors of efficiency estimate, we considered the possibility that the experimental systems would not fulfil the two theoretical constraints involved in the proposed method, namely in the exponential phase of the amplification process, (i) the synthesis of one amplicon molecule must be accompanied by the hydrolysis of one probe molecule; and (ii) the primers should hybridize to the whole target content. To check the fulfilment of these constraints, we had to demonstrate that neither the probe nor the primers were limiting factors, at least during the exponential phase of the amplification. We thus verified that the fluorescence levels of the few measurable points in the exponential phase were not dependent on the concentrations of probe and primers. Figure 3 shows a typical result obtained with samples containing 106 copies of RAP1GA1 DNA for which the detectable exponential phase was achieved in cycles 18–20. For these cycles, the increase of probe and primer concentrations did not significantly enhance the fluorescence levels, with a minor exception at cycle 20 where a small increase was observed at the highest probe concentrations. These results confirmed that probe and primers were not limiting and that the experimental systems fulfilled the constraints of our methodology.
Table 2. Comparison of the proposed method and the classical method based on the analysis of 50 amplification curves obtained either with CAV1 DNA or RAP1GA1 DNA, according to the same protocol as that used with IGF1 DNA (see Table 1).
| Expected number (OD measurement) | Estimated number: mean ± SD (n = 10) | Target | |
| |
Classical method |
Proposed method |
|
| 102 | (1.02 ± 0.19) × 102 | (0.75 ± 0.17) × 102 | CAV1 |
| 103 | (0.89 ± 0.10) × 103 | (0.68 ± 0.08) × 103 | CAV1 |
| 104 | (1.02 ± 0.07) × 104 | (0.75 ± 0.11) × 104 | CAV1 |
| 105 | (1.04 ± 0.09) × 105 | (0.73 ± 0.09) × 105 | CAV1 |
| 106 | (0.97 ± 0.09) × 106 | (0.64 ± 0.03) × 106 | CAV1 |
| 102 | (0.98 ± 0.27) × 102 | (0.25 ± 0.07) × 102 | RAP1GA1 |
| 103 | (1.25 ± 0.27) × 103 | (0.32 ± 0.05) × 103 | RAP1GA1 |
| 104 | (0.87 ± 0.14) × 104 | (0.25 ± 0.04) × 104 | RAP1GA1 |
| 105 | (0.94 ± 0.14) × 105 | (0.22 ± 0.02) × 105 | RAP1GA1 |
| 106 | (1.09 ± 0.13) × 106 | (0.26 ± 0.03) × 106 | RAP1GA1 |
Figure 3.
Amplification curves (from cycle 16 to cycle 23) obtained with 106 copies of RAP1GA1 DNA and different concentrations of probe and primers. Under these conditions, the first point above the detection limit was obtained after 18 cycles and the copy number was estimated in the exponential phase between cycles 18 and 20. Each point represents the mean of a triplicate. (a) A constant primer concentration of 300 nM was used with four different probe concentrations, as indicated. The correlation between fluorescence and probe concentration was not significant at cycles 18 and 19 (P > 0.1, n = 12), and was significant at cycle 20 (P = 0.028, n = 12). (b) A constant probe concentration of 100 nM was used with three different primer concentrations, as indicated. No significant correlation between fluorescence and primer concentration was observed at cycles 18, 19 and 20 (P > 0.5, n = 9).
DISCUSSION
Quantitative real-time PCR is a tool of choice to estimate the absolute or relative amount of a target cDNA. Up to now, an absolute quantification, when needed, required the construction of a calibration curve based on PCR results obtained with samples for which the amount of target DNA is known (13). For that purpose, DNA-containing plasmids should be acquired, with obvious practical and economical inconvenience. Moreover, the reliability of such an approach rests on the high purity of the standard samples, since the calibration of the method is based on the determination of the target DNA content by OD measurement, which can be biased by the presence of contaminating DNA. An alternative possibility to this approach would be to determine the specific fluorescence of the used fluorophore in order to convert the qPCR fluorescent signal into the produced amount of amplicon. Such a strategy was very recently proposed (12) but, again, standard samples containing known amounts of target DNA were required to determine the specific fluorescence. On the contrary, our approach proposes to infer the specific fluorescence from the sole amplification curve and in the appropriate experimental conditions of the qPCR run. This useful methodological improvement results from the finding that the plateau of the amplification curve is due to the depletion of the fluorescent TaqMan probe, and not to any amplicon accumulation-dependent inhibition of Taq polymerase (10). In this case, the calibration of the assay rests on the accurate determination of the probe concentration based on the OD measurement of the available pure probe sample. Thus, bias in the calibration procedure used in our approach can be easily avoided. The accuracy with which the DNA copy number can be determined is mainly dependent on the CI of the efficiency estimate. We can propose, as a rule of thumb, that the copy number estimate may be biased, at worst, by a factor of 2, and that the imprecision is characterized by a maximal variation of 30% between identical samples. Last but not least, the method presents the economical advantage of requiring a low concentration of probe to ensure that a plateau is reached within a minimum number of amplification cycles.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank G. Vassart and M. Parmentier for their helpful advice, and S. Giraud for technical assistance. A.d.K. and J.C.G. are Research Associate and Research Fellow of the Fonds National de la Recherche Scientifique, respectively. This work was supported by the Action de Recherche Concertée (CFB), the Belgium Programme of Interuniversity Poles of Attraction (initiated by the Belgium State, Prime Minister’s Office), the Association Recherche Biomédicale et Diagnostic, the Queen Elisabeth Medical Foundation (FMRE-Neurobiology 02-04), the Fund for Medical Scientific Research (FRSM-Belgium 3.4507.02) and the Alice et David Van Buuren Foundation.
REFERENCES
- 1.Bustin S.A. (2002) Quantification of mRNA using real-time reverse transcription PCR (RT–PCR): trends and problems. J. Mol. Endocrinol., 29, 23–39. [DOI] [PubMed] [Google Scholar]
- 2.Klein D. (2002) Quantification using real-time PCR technology: applications and limitations. Trends Mol. Med., 8, 257–260. [DOI] [PubMed] [Google Scholar]
- 3.Schmittgen T.D. (2001) Real-time quantitative PCR. Methods, 25, 383–385. [DOI] [PubMed] [Google Scholar]
- 4.Heid C.A., Stevens,J., Livak,K.J. and Williams,P.M. (1996) Real time quantitative PCR. Genome Res., 6, 986–994. [DOI] [PubMed] [Google Scholar]
- 5.Pfaffl M.W. (2001) A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res., 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramakers C., Ruijter,J.M., Deprez,R.H. and Moorman,A.F. (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett., 339, 62–66. [DOI] [PubMed] [Google Scholar]
- 7.Peirson S.N., Butler,J.N. and Foster,R.G. (2003) Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res., 31, e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu W. and Saint,D.A. (2002) Validation of a quantitative method for real time PCR kinetics. Biochem. Biophys. Res. Commun., 294, 347–353. [DOI] [PubMed] [Google Scholar]
- 9.Morrison C. and Gannon,F. (1994) The impact of the PCR plateau phase on quantitative PCR. Biochim. Biophys. Acta, 1219, 493–498. [DOI] [PubMed] [Google Scholar]
- 10.Kainz P. (2000) The PCR plateau phase—towards an understanding of its limitations. Biochim. Biophys. Acta, 1494, 23–27. [DOI] [PubMed] [Google Scholar]
- 11.Tichopad A., Dilger,M., Schwarz,G. and Pfaffl,M.W. (2003) Standardised determination of real-time PCR efficiency from a single reaction set-up. Nucleic Acids Res., 31, e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rutledge R.G. and Côté,C. (2003) Mathematics of quantitative kinetic PCR and the application of standard curves. Nucleic Acids Res., 31, e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfaffl M.W. and Hageleit,M. (2001) Validities of mRNA quantification using recombinant RNA and recombinant DNA external calibration curves in real-time RT–PCR. Biotech. Lett., 23, 275–282. [Google Scholar]