Abstract
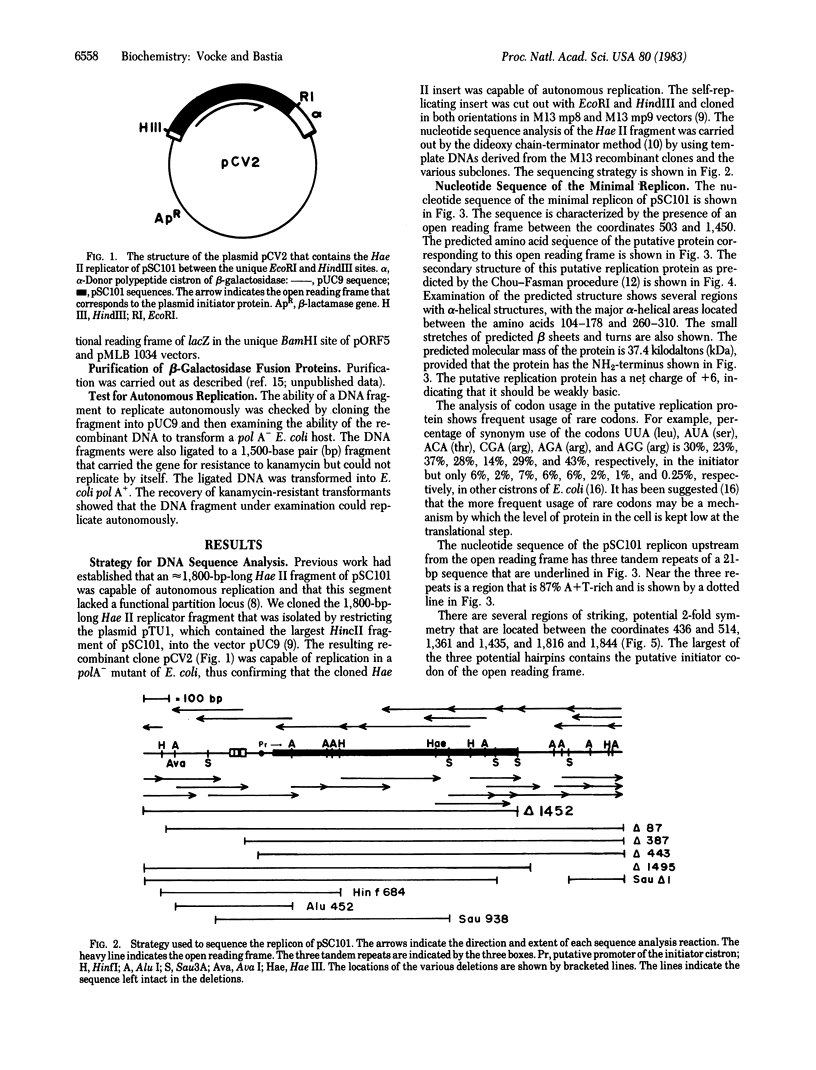
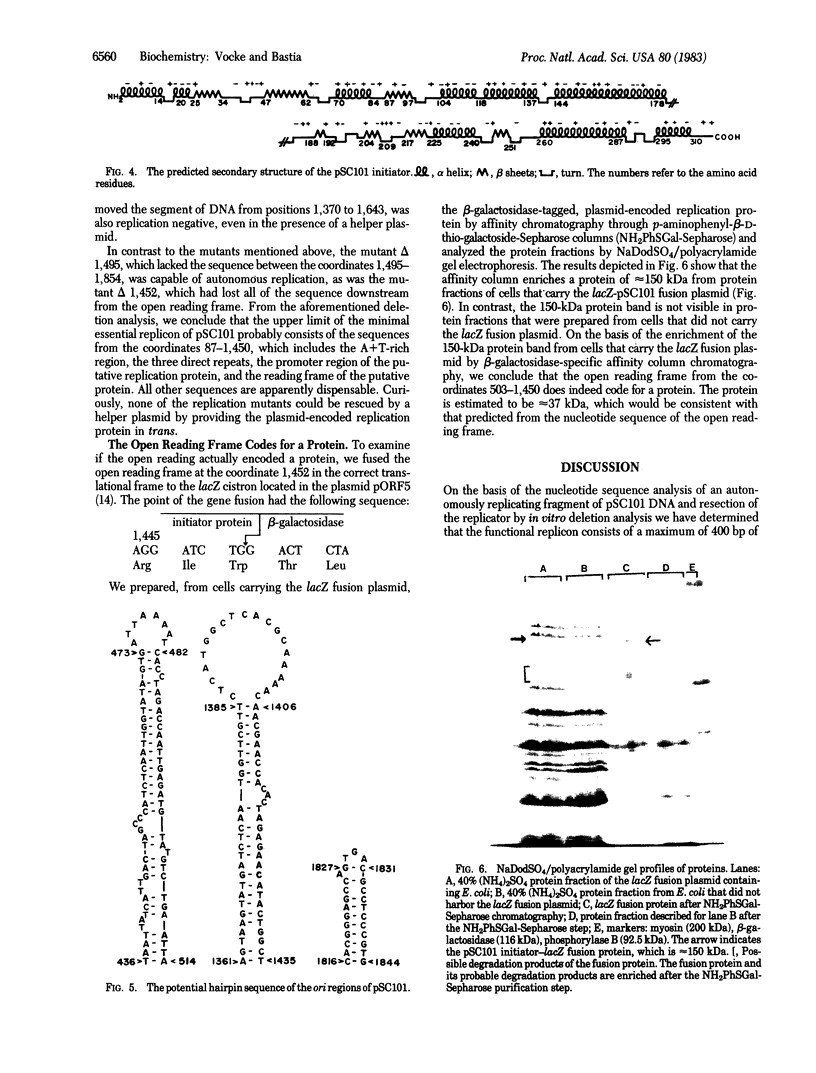
The replicon of the low copy number plasmid pSC101 has an obligatory requirement for the dnaA initiator protein of Escherichia coli as well as a plasmid-encoded initiator protein. We have identified the cistron of the plasmid-encoded initiator by DNA sequence analysis. Fusion of the initiator cistron with the lacZ gene of E. coli yielded a fusion protein of approximately equal to 150 kilodaltons, thus confirming that the open reading frame detected by DNA sequence analysis actually encoded a 37.5-kilodalton protein. Deletion of 26 amino acid residues from the COOH terminus of the plasmid initiator abolished autonomous replication from pSC101 origin. By in vitro deletion analysis we have shown that, although sequences downstream from the initiator cistron are dispensable, a maximum of 400 base pairs immediately upstream from the NH2-terminal region of the initiator is necessary for plasmid replication. These upstream sequences contain an A + T-rich region and three tandem repeats of a 21-base pair sequence; these features are characteristics of other replication origins.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cabello F., Timmis K., Cohen S. N. Replication control in a composite plasmid constructed by in vitro linkage of two distinct replicons. Nature. 1976 Jan 29;259(5541):285–290. doi: 10.1038/259285a0. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C. Recircularization and autonomous replication of a sheared R-factor DNA segment in Escherichia coli transformants. Proc Natl Acad Sci U S A. 1973 May;70(5):1293–1297. doi: 10.1073/pnas.70.5.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C. Revised interpretation of the origin of the pSC101 plasmid. J Bacteriol. 1977 Nov;132(2):734–737. doi: 10.1128/jb.132.2.734-737.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germino J., Bastia D. Interaction of the plasmid R6K-encoded replication initiator protein with its binding sites on DNA. Cell. 1983 Aug;34(1):125–134. doi: 10.1016/0092-8674(83)90142-3. [DOI] [PubMed] [Google Scholar]
- Germino J., Bastia D. Primary structure of the replication initiation protein of plasmid R6K. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5475–5479. doi: 10.1073/pnas.79.18.5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germino J., Bastia D. The replication initiator protein of plasmid R6K tagged with beta-galactosidase shows sequence-specific DNA-binding. Cell. 1983 Jan;32(1):131–140. doi: 10.1016/0092-8674(83)90503-2. [DOI] [PubMed] [Google Scholar]
- Hashimoto T., Sekiguchi M. Isolation of temperature-sensitive mutants of R plasmid by in vitro mutagenesis with hydroxylamine. J Bacteriol. 1976 Sep;127(3):1561–1563. doi: 10.1128/jb.127.3.1561-1563.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasunuma K., Sekiguchi M. Replication of plasmid pSC101 in Escherichia coli K12: requirement for dnaA function. Mol Gen Genet. 1977 Sep 9;154(3):225–230. doi: 10.1007/BF00571277. [DOI] [PubMed] [Google Scholar]
- Hobom G., Grosschedl R., Lusky M., Scherer G., Schwarz E., Kössel H. Functional analysis of the replicator structure of lambdoid bacteriophage DNAs. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):165–178. doi: 10.1101/sqb.1979.043.01.023. [DOI] [PubMed] [Google Scholar]
- Kingsbury D. T., Helinski D. R. DNA polymerase as a requirement for the maintenance of the bacterial plasmid colicinogenic factor E1. Biochem Biophys Res Commun. 1970 Dec 24;41(6):1538–1544. doi: 10.1016/0006-291x(70)90562-0. [DOI] [PubMed] [Google Scholar]
- Konigsberg W., Godson G. N. Evidence for use of rare codons in the dnaG gene and other regulatory genes of Escherichia coli. Proc Natl Acad Sci U S A. 1983 Feb;80(3):687–691. doi: 10.1073/pnas.80.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacock P. A., Cohen S. N. Partitioning of bacterial plasmids during cell division: a cis-acting locus that accomplishes stable plasmid inheritance. Cell. 1980 Jun;20(2):529–542. doi: 10.1016/0092-8674(80)90639-x. [DOI] [PubMed] [Google Scholar]
- Messer W., Meijer M., Bergmans H. E., Hansen F. G., von Meyenburg K., Beck E., Schaller H. Origin of replication, oriC, of the Escherichia coli K12 chromosome: nucleotide sequence. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):139–145. doi: 10.1101/sqb.1979.043.01.020. [DOI] [PubMed] [Google Scholar]
- Miller C. A., Tucker W. T., Meacock P. A., Gustafsson P., Cohen S. N. Nucleotide sequence of the partition locus of Escherichia coli plasmid pSC101. Gene. 1983 Oct;24(2-3):309–315. doi: 10.1016/0378-1119(83)90091-4. [DOI] [PubMed] [Google Scholar]
- Moore D. D., Denniston-Thompson K., Kruger K. E., Furth M. E., Williams B. G., Daniels D. L., Blattner F. R. Dissection and comparative anatomy of the origins of replication of lambdoid phages. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):155–163. doi: 10.1101/sqb.1979.043.01.022. [DOI] [PubMed] [Google Scholar]
- Murotsu T., Matsubara K., Sugisaki H., Takanami M. Nine unique repeating sequences in a region essential for replication and incompatibility of the mini-F plasmid. Gene. 1981 Nov;15(2-3):257–271. doi: 10.1016/0378-1119(81)90135-9. [DOI] [PubMed] [Google Scholar]
- Müller U. R., Wells R. D. Intercistronic regions in phi X174 DNA. I. Construction of mutants with altered intercistronic regions between genes J and F. J Mol Biol. 1980 Jul 25;141(1):1–24. doi: 10.1016/s0022-2836(80)80026-x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz J., Silhavy T. J., Berman M. L., Fiil N., Emr S. D. A previously unidentified gene in the spc operon of Escherichia coli K12 specifies a component of the protein export machinery. Cell. 1982 Nov;31(1):227–235. doi: 10.1016/0092-8674(82)90422-6. [DOI] [PubMed] [Google Scholar]
- Stalker D. M., Kolter R., Helinski D. R. Nucleotide sequence of the region of an origin of replication of the antibiotic resistance plasmid R6K. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1150–1154. doi: 10.1073/pnas.76.3.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalker D. M., Thomas C. M., Helinski D. R. Nucleotide sequence of the region of the origin of replication of the broad host range plasmid RK2. Mol Gen Genet. 1981;181(1):8–12. doi: 10.1007/BF00338997. [DOI] [PubMed] [Google Scholar]
- Steers E., Jr, Cuatrecasas P., Pollard H. B. The purification of beta-galactosidase from Escherichia coli by affinity chromatography. J Biol Chem. 1971 Jan 10;246(1):196–200. [PubMed] [Google Scholar]
- Tolun A., Helinski D. R. Direct repeats of the F plasmid incC region express F incompatibility. Cell. 1981 Jun;24(3):687–694. doi: 10.1016/0092-8674(81)90095-7. [DOI] [PubMed] [Google Scholar]
- Tsurimoto T., Matsubara K. Purified bacteriophage lambda O protein binds to four repeating sequences at the lambda replication origin. Nucleic Acids Res. 1981 Apr 24;9(8):1789–1799. doi: 10.1093/nar/9.8.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]




