Abstract
Dynamic contrast material-enhanced magnetic resonance imaging (DCE-MRI) of breasts is an important imaging modality in breast cancer diagnosis with higher sensitivity but relatively lower specificity. The objective of this study is to investigate a new approach to help improve diagnostic performance of DCE-MRI examinations based on the automated detection and analysis of bilateral asymmetry of characteristic kinetic features between the left and right breast. An image dataset involving 130 DCE-MRI examinations was assembled and used in which 80 were biopsy-proved malignant and 50 were benign. A computer-aided diagnosis (CAD) scheme was developed to segment breast areas depicted on each MR image, register images acquired from the sequential MR image scan series, compute average contrast enhancement of all pixels in one breast, and a set of kinetic features related to the difference of contrast enhancement between the left and right breast, and then use a multi-feature based Bayesian belief network to classify between malignant and benign cases. A leave-one-case-out validation method was applied to test CAD performance. The computed area under a receiver operating characteristic (ROC) curve is 0.78 ± 0.04. The positive and negative predictive values are 0.77 and 0.64, respectively. The study indicates that bilateral asymmetry of kinetic features between the left and right breasts is a potentially useful image biomarker to enhance the detection of angiogenesis associated with malignancy. It also demonstrates the feasibility of applying a simple CAD approach to classify between malignant and benign DCE-MRI examinations based on this new image biomarker.
Keywords: Breast diseases, Computer-aided diagnosis (CAD), MR mammography, Contrast enhancement, Kinetic feature analysis, Asymmetry
Introduction
Breast cancer is the most prevalent cancer with high mortality rate in women over the age of 40 worldwide [1], and scientific evidences have repeatedly shown that the effective detection and treatment of earlier cancer significantly reduced patients’ mortality and morbidity rates [2–4]. Although conventional X-ray mammography is the most widely used cancer screening and detection tool to date, it has a number of limitations; for example, cancer detection sensitivity of screening mammography is substantially reduced from the range of 98 to 100 % in fatty breasts (BIRADS 1) to 30 to 48 % in dense breasts (BIRADS 3 or 4) [5–7].
To increase cancer detection sensitivity in particular among the groups of premenopausal women with dense breasts or carrying higher risk genes, several other imaging modalities have been investigated and evaluated. Among them, the dynamic contrast-enhancement magnetic resonance imaging (DCE-MRI) of breast [8] has demonstrated significantly higher sensitivity in detecting early cancers depicted in dense breasts (e.g., increasing sensitivity from 33 to 59 % using mammography to 71 to 94 % when using DCE-MRI in a number of studies worldwide [9–11]). Despite the higher sensitivity, MRI actually has comparable or lower specificity than mammography [12, 13]. The higher false-positive rates do not only cause woman’s anxiety and also substantially increase health care costs by prompting a large number of unnecessary recalls for additional imaging workup and/or biopsies of the benign lesions [14]. As a result, in one large study involving 1,215 high-risk women, 512 (42.1 %) refused the offered breast DCE-MRI screening examinations because of a variety of reasons (including claustrophobia, refusing intravenous injection, and seriously concerning about additional biopsies or other procedures that may follow subsequently) [15]. Therefore, developing more accurate and reliable method to extract image features with higher discriminatory power in classifying between cases depicting malignant and benign breast lesions could be important to increase efficacy of applying DCE-MRI examinations to detection and diagnosis of breast cancer in the clinical practice.
To help improve diagnostic accuracy of DCE-MRI examinations, a number of research groups have developed and tested several computer-aided detection and/or diagnosis (CAD) approaches. The CAD schemes first extract and compute dynamic contrast enhancement features from the characteristic kinetic curves generated from a set of the selected pixels located inside the identified and segmented breast lesions, and then apply different machine learning classifiers to distinguish between the malignant and benign lesions [16–20]; for example, one study reported that using a leave-one-case-out testing method, a CAD scheme using a set of selected DCE-MRI features and a Bayesian artificial neural network yielded an area under a receiver operating characteristic (ROC) curve (AUC = 0.78 ± 0.04) when applying to a dataset involving 168 malignant and 45 benign breast lesions [21]. Although the previous studies focused on detection and analysis of characteristic kinetic curves computed from the pixels inside the segmented lesions, a recent study also showed that similar to a widely used image-based risk factor, namely, the mammographic density, the background parenchymal enhancement (BPE) evaluated or computed from the entire breast areas depicting on DCE-MRI images also carried useful or higher discriminatory information associated with cancer risk assessment [22] as well as the performance of cancer detection and staging [23].
These studies indicated that because of the difference of angiogenesis associated between malignant and benign lesions, adding BPE features computed from the whole breast could also increase discriminatory power or classification performance of the CAD schemes than using the dynamic contrast enhancement features computed only from the pixels inside the lesions. However, such assumption has not been tested in CAD development to date. Meanwhile, researchers also recognized that similar to the assessment of mammographic density [24], BPE values computed from breast DCE-MRI also depend on menstrual cycle at the examination date and other different life style factors of the individual woman. Hence, without knowing or considering women’s life style factors, using BPE values computed from DCE-MRI images to assess cancer risk or the likelihood of cancer detection could be biased or unreliable [25]. To overcome this limitation, we investigated and tested a different approach in this study.
Our approach is developed based on the following underlying assumptions. First, since humans naturally show bilateral symmetry in paired morphological traits including two (left and right) breasts, breast tissue pattern asymmetry is an important radiographic image phenotype that is highly related to the abnormal biological processes leading to cancer development [26]. Second, since many potential biases or dependent factors in assessing breast density using mammograms and/or PBE using DCE-MRI images should equally affect on image features computed from the left and right breast, subtraction of characteristic kinetic features computed from all enhanced breast tissue pixels depicted on two bilateral breasts should help reduce or minimize many individualized biases to achieve more robust results because we compare the feature difference of two bilateral breasts of the same women not among the different women at different ages or having different genetic factors and life cycles or styles. As a result, the primary hypothesis of this study is that developing a new CAD scheme that enables to extract and compute bilateral asymmetry (or difference) of dynamic contrast enhancement related features on the left and right breast regions segmented from DCE-MRI images may provide a new feature (or a classification index) with more reliable and higher discriminatory power to help distinguish between the cases depicting malignant and benign lesions. The purpose of this study is to test this hypothesis. If successful, this new feature can provide useful supplementary information to be applied to the existing CAD schemes and help improve classification performance.
Materials and Methods
Image Dataset
The image dataset assembled and used in this study was retrospectively collected. It includes 130 DCE-MRI examinations acquired from 130 women who previously underwent breast cancer diagnosis and treatment at Zhejiang Cancer Hospital in the City of Hangzhou, Zhejiang Province, China. This study was approved by a human subject administration office (IRB). All the patient identifiers were removed in the research. Although many of these cancer patients had multiple DCE-MRI examinations before and after therapeutic treatment, all images selected in this dataset were acquired before the therapeutic treatment.
We divided these cases into two groups. Group one includes 80 cases depicting cancer (with biopsy and pathology-verified malignant lesions) and group two involves 50 cases that were highly suspicious in imaging examinations but later proved by biopsy as benign lesions. In each malignant case of this dataset, only one breast depicts malignant lesions and another breast is negative or benign. The majority of women whose DCE-MRI examination images are selected in this dataset are relatively younger. The mean and standard deviations of these women’s age are 48.7 ± 8.8 and 43.0 ± 9.5 for groups one and two, respectively.
During each breast DCE-MRI examination, the woman was scanned in prone position using a Siemens MRI machine (MAGNETOM Espree-Pink 1.5T) equipped with a standard double-breast MRI coil. The DCE-MRI examination protocol adopted in the hospital first acquires the pre-contrast (baseline) series of T1- and T2-weighted 3D image scans. After injection of Gd-DTPA contrast agent with a dose of 0.2 mmol/kg body weight and a saline flush of 20 ml intravenously at the same flow rate of 2 ml/s, three post-contrast series of 3D MR image scans and data acquisitions were then performed. The first two post-contrast series (namely T1 and T2) use the same image scanning process and thus generate images with the same spatial resolution as the pre-contrast (termed T0) images with a data matrix of 512 × 512 pixels. All three (T0 to T2) image scan series also have the same number of 88 image slices representing the same in-depth resolution. The third post-contrast series (T3) generates high in-depth resolution images involving 160 image slices. In this study, we only used three image scan series (T0, T1, and T2) in kinetic feature computation and data analysis process.
A CAD Scheme
We developed and tested a new CAD scheme to automatically detect and classify between the malignant and benign DCE-MRI examinations using a new image biomarker or classification index based on the bilateral asymmetry (difference) of contrast enhancement features computed from all pixels depicted on the left and right breast areas segmented from each DCE-MRI image slice. Specifically, the scheme includes following four basic image processing and data analysis steps namely (1) segmenting left and right breast areas, (2) registering images acquired from the sequential MR image scan series, (3) computing the bilateral asymmetry (or difference) value of the dynamic contrast enhancement features from the kinetic curves of all pixels between the left and right breasts, and (4) applying a multi-feature based Bayesian belief network (BBN) to produce a likelihood score for classifying each testing case into malignant or benign case group.
Breast Area Segmentation
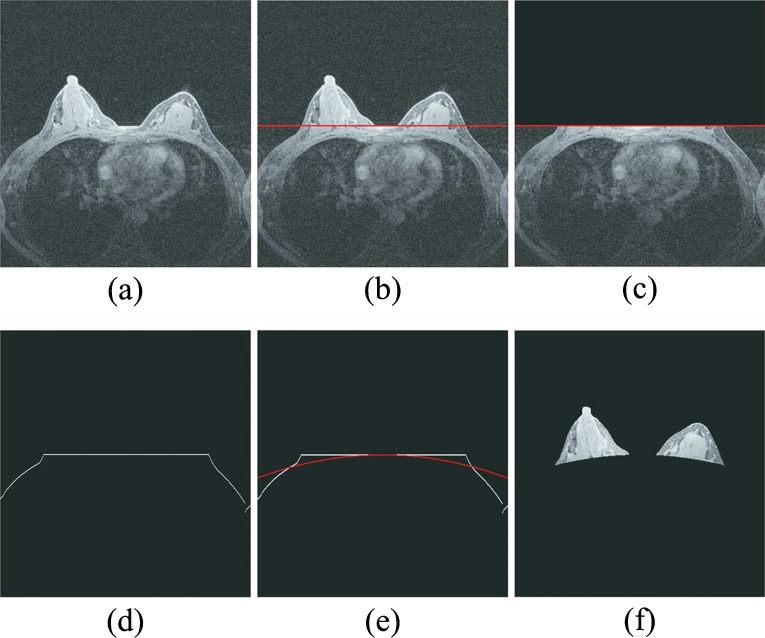
To identify the characteristic kinetic curves and compute average contrast enhancement (or wash-in and wash-out) ratios of all enhanced pixels on the left and right breasts, CAD scheme automatically segments breast areas depicted on each breast MR image slice. Hence, the contrast enhancement of pixels located behind the chest wall will not affect the final case classification result. For this purpose, we designed the following image processing procedures to segment breast area depicted on each MR image slice automatically (as shown in Fig. 1).
Apply a Sobel filter to enhance the boundary pixels between breast skin and the air background recorded on each image slice.
Apply a morphological closing and smooth filter to remove isolated pixels (e.g., artificial noise) and generate a smoothed protruding curvature to segment between imaged skin (both breast and chest) and air background pixels depicted on each image slice.
Detect and fit a line passing through the segment curvature detected in step 2 in the central chest region between the left and right breasts (Fig. 1b) based on the fact that the chest boundary between two breasts is almost horizontal.
Temporally remove the breast areas located above the fitted line in step 3 from each image slice (Fig. 1c).
Detect and fit two chest skin surface curves beyond breast areas in both left and right side of breast regions (Fig. 1d).
Apply a parabolic model based curve fitting method to generate a complete segmentation curve to separate between breast and chest wall regions (Fig. 1e) in which the parameters of the parabolic model are defined based on the least square criterion.
Connect the first curve separating between breast skin and air background and the second fitted curve separating between breast tissue and chest wall or skin to generate two segmented regions that represent the left and right breast areas depicted on one DCE-MRI image slice (Fig. 1f).
Fig. 1.
Illustration of image processing steps to automatically segmenting bilateral breast areas depicted on one MR image slice which includes a showing a raw MR image slice, b detecting an initial line passing through the central section of chest wall, c removing breast areas above the initial line, d delineating the surface curve of the remaining area, e fitting a parabolic curve as a segmentation boundary curve, and f showing the left and right breast areas finally segmented from the MR image slice
Image Registration
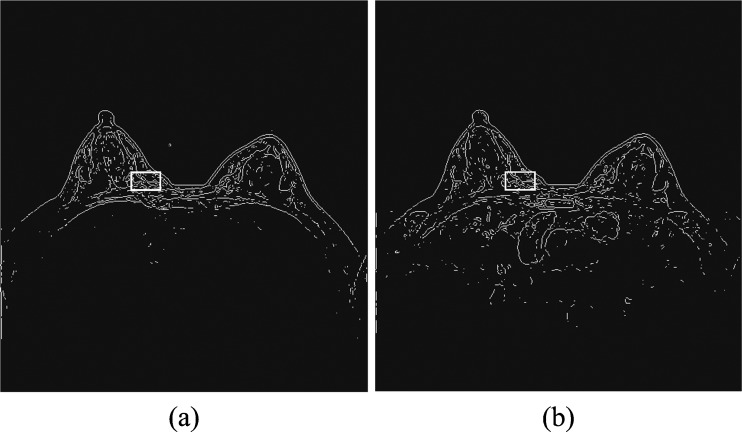
To help overcome the impact of potential body motion of a woman during the sequential imaging scanning procedure of a DCE-MRI examination on the reliability in computing kinetic (contrast enhancement) features and the accuracy of lesion classification [17, 27], we developed and implemented a simple and easy to performed image registration process in our CAD scheme. Since in our DCE-MRI examination protocol, three image sequences (T0, T1, and T2) have the same number of image slices; for the simplicity, we ignored the possible motion along the vertical (z) direction in this study. Thus, our image registration process was only applied on three matched MR image slices acquired from three (T0, T1, and T2) image scanning sequences. Using each pre-contrast image slice (T0) as the reference, our scheme applies a rigid image registration method to shift image slices acquired from T1 and T2 scans accordingly to align with the corresponding T0 image slice. For this purpose, we tested several image matching and registration methods (including mutual information and Pearson correlation based template matching methods [28, 29]) and visually examined/compared the performance of these methods being directly applied to the original MR images. Given the image noise and gray level variation among the sequentially scanned MR images, we found that the registration results were often not reliable. Hence, we tested and implemented a new approach in this study. Taking Fig. 2 as an illustration example, our image registration algorithm is explained as follows.
Apply a Sobel filter to process each image slice to enhance the edge curves of the fibro-glandular tissue structure within breast area and use an adaptive threshold embedded in the Sobel filter program to generate a binary image (Fig. 2). The image registration was then conducted based on the converted binary images instead of the original gray level images.
Open a rectangular window and apply it to scan over the generated binary image in baseline (T0) examination. During the scan, the scheme detects and counts the number of nonzero pixels within the covered window. The scheme then searches for and identifies one location of the image where the window includes the maximum number of nonzero pixels. This window is considered depicting the maximum information and is named as a reference window in the T0 image slice with the center coordinate of (xc and yc). Based on our experiment and data analysis results, size of the reference window was experimentally determined with 40 × 25 pixels (Fig. 2a).
Apply a new floating (or scanning) window with the same size as the reference window selected in T0 image slice to the matched image slice acquired from either T1 or T2 imaging scan sequence. By assuming that only small image shift is required because of the restricted slight motion of the woman during the whole DCE-MRI scanning process, the floating window is first mapped to be centered at the same location of (xc and yc) as the baseline (T0) image slice and then scanned within a range of (xc ± Δx and yc ± Δy) in which Δx = 5 and Δy = 5 in T1 or T2 imaging slice. By computing correlation coefficient between two regions defined by the scanning window in T1 or T2 image and the reference window in T0 image at different locations, the scheme determines the final center location of this scanning window (Fig. 2b) on the post-contrast image slice (T1 or T2). At this location, the correlation coefficient of the two (reference and scanning) windows reaches the maximum level.
Register one pre-contrast (T0) and one matched post-contrast (T1 or T2) image slices by linearly shifting T1 or T2 image slice to align with T0 image. The registration target is to make the reference window identified in step 2 and the scanned window determined in step 3 being exactly matched (or overlapped) on the sequential MR images.
Fig. 2.
Illustration of DCE-MRI sequential image registration based on alignment of two maximum information windows detected on the T0 (a) and T1 or T2 (b) image slices
Computing the Bilateral Difference of Characteristic Kinetic Features
After breast area segmentation on each image slice and registration of matched images in three MRI scanning sequences, CAD scheme counts all qualified nonzero (e.g., fibro-glandular tissue) pixels inside one entirely segmented (either left or right) breast area and computes two average characteristic kinetic (contrast enhancement) values. The first one is computed from the relative average difference of all registered pixel values between T0 and T1 images and the second one is computed from the difference between T0 and T2 images namely, 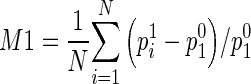 and
and 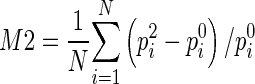 , where p0i, p1i, p2i are the pixel value of matched pixels (i) depicted on T0, T1, and T2 image slices, respectively, and N is the total number of nonzero pixels detected inside one 3D breast volume (segmented in all 2D MR image slices) in T0 MRI scanning sequence. CAD scheme then computes three image features related to bilateral asymmetry (difference) of characteristic kinetic features F1 = |M1LB − M1RB| in T1, F2 = |M2LB − M2RB| in T2 image scanning sequences, respectively, and their change, F3 = F3 − F1, between the left (LB) and right (RB) breasts.
, where p0i, p1i, p2i are the pixel value of matched pixels (i) depicted on T0, T1, and T2 image slices, respectively, and N is the total number of nonzero pixels detected inside one 3D breast volume (segmented in all 2D MR image slices) in T0 MRI scanning sequence. CAD scheme then computes three image features related to bilateral asymmetry (difference) of characteristic kinetic features F1 = |M1LB − M1RB| in T1, F2 = |M2LB − M2RB| in T2 image scanning sequences, respectively, and their change, F3 = F3 − F1, between the left (LB) and right (RB) breasts.
A Bayesian Belief Network
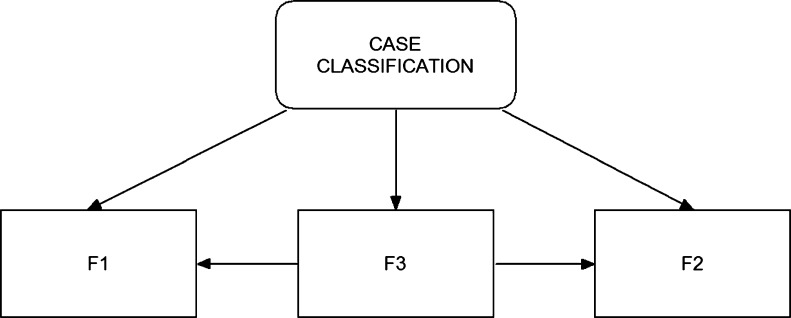
To optimally utilize these three image features (F1, F2, and F3) in classifying between the malignant and benign cases, we also designed and implemented a BBN in the final step of our CAD scheme (as shown in Fig. 3). BBN is a popular statistical learning method that has been investigated and applied in a number of CAD schemes for detecting breast cancer [30–32]. One unique advantage of the BBN approach is that the topology of the BBN represents the joint probability distribution of a problem domain by exploiting the dependencies between variables and capturing the knowledge of a given problem in a natural and efficient way [33]. Unlike many other types of machine learning methods or networks (i.e., artificial neural network and support vector machine) in which the iterative training or learning processes are required to minimize the errors to the specific targets, a BBN uses all available samples (data) once to compute a joint probability table installed in the network when the node topology of the network is determined. This reduces the risk of over-fitting and increases the robustness of the network. To compute these joint probabilities, each node (feature) must be represented by a relatively small number of discrete states.
Fig. 3.
An example of building a Bayesian belief network using all three bilateral asymmetric kinetic features to classify between malignant and benign cases
In our study, one feature computed from all training samples was divided into five discrete states with equal sample distribution [31]. Since the decision node (case classification in Fig. 3) has three potentially dependent nodes represented by features (F1, F2, and F3), a set of joint probabilities can be computed to generate the final joint-conditional probability table. When assuming all three features are used in the BBN, a set of the conditional probability values are computed as follows.
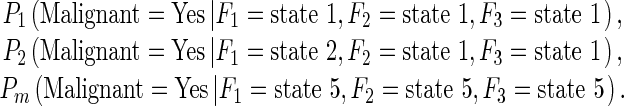 |
In this study, we built and tested several BBNs using the different number and combinations of these three features to identify an optimal network. Specifically, once the features (i.e., two or three in this study) were selected to build the BBN, the complete conditional probability table applied to build the network was automatically generated using a publicly available BBN optimization tool (the BN power constructor and predictor [34]). Using a pre-established conditional probability table, the BBN was then applied to classify testing cases based on a BBN-generated probability or likelihood score of the case being malignant (e.g., from 0 to 100 %).
Performance Assessment
We took the following steps to conduct the tasks of assessing classification performance of our CAD scheme. First, we plotted box-plot diagrams, took statistical t test to analyze and compare the discriminatory power of each image feature. Second, we computed correlation coefficients among different combinations of feature pairs (e.g., F1 and F2, F1 and F3, as well as F2 and F3). Third, after building and implement a BBN, we tested classification performance using a leave-one-case-out method by applied CAD scheme to our entire DCE-MRI testing dataset. A set of BBN or CAD-generated classification scores was recorded. The feature and classification scoring data were analyzed using a maximum likelihood algorithm based ROC program (ROCKIT, 0.9B beta version, http://www.radiology.uchicago.edu/krl/, University of Chicago, 1998 [35]). The areas under the ROC curves (AUC) and the corresponding 95 % confidence intervals (CI) were computed as performance assessment index. In addition, the positive predictive value (PPV) and the negative predictive value (NPV) were also computed and compared between using a single feature and the BBN.
Results
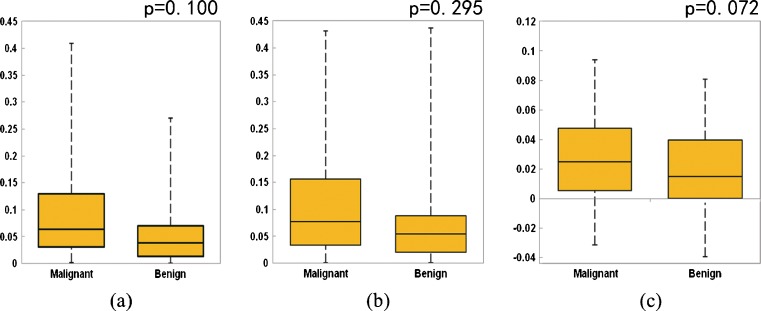
Figure 4 shows three sets of box-plots that illustrate the distributions of three kinetic features (F1, F2, and F3) in two groups of 80 malignant and 50 benign cases. The first two sets of box-plots (Fig. 4a, b) indicate that malignant cases in general have higher average bilateral contrast enhancement difference (asymmetry) between the two bilateral (left and right) breasts (F1 and F2). The third set of box-plots (Fig. 4c) shows that the asymmetrical level is also likely to increase faster in the malignant cases from T1 scan to T2 scan than the benign cases. The results indicate that in both T1 and T2 image scanning sequences, the malignant breasts typically have higher average kinetic (contrast) enhancement than the negative or benign breasts. However, the t test for each individual feature indicates that the differences as described above are not statistically significant (p > 0.05).
Fig. 4.
Box-plots showing distributions of three features (a F 1, b F 2, and c F 3) in two groups of malignant and benign cases with statistical t test p values
Using ROC data analysis method, Table 1 summarizes the computed AUC values and the corresponding 95 % CIs when one of three features was independently applied in classifying between two groups of malignant and benign cases. Comparing the classification results evaluated using AUC values, the results show no statistically significant differences between the AUC values generated using these three features. The computed two-tailed p values are 0.329, 0.055, and 0.082 between using these three feature pairs of F1 and F2, F1 and F3, and F2 and F3, respectively. Meanwhile, the correlation coefficients of the computed AUC values are 0.873, −0.238, and −0.634 when comparing AUC values generated using pairs of F1 and F2, F1 and F3, F2 and F3, respectively. Among these three features, using F1 yielded the highest classification performance (AUC = 0.657 ± 0.049). As a result, when using F1 to predict diagnostic outcome of 80 malignant and 50 benign cases verified in our dataset, this feature index classified that 103 cases were malignant and 27 cases were benign. The overall classification accuracy is 65.4 %, while the PPV is 0.67 (69/103) and the NPV is 0.59 (16/27).
Table 1.
Summary of AUC values and their 95 % confidence intervals (CI) using three individual features (F 1, F 2, and F 3)
| Feature | F 1 | F 2 | F 3 |
|---|---|---|---|
| AUC | 0.657 | 0.628 | 0.523 |
| 95 % CI | 0.557, 0.746 | 0.528, 0.718 | 0.423, 0.622 |
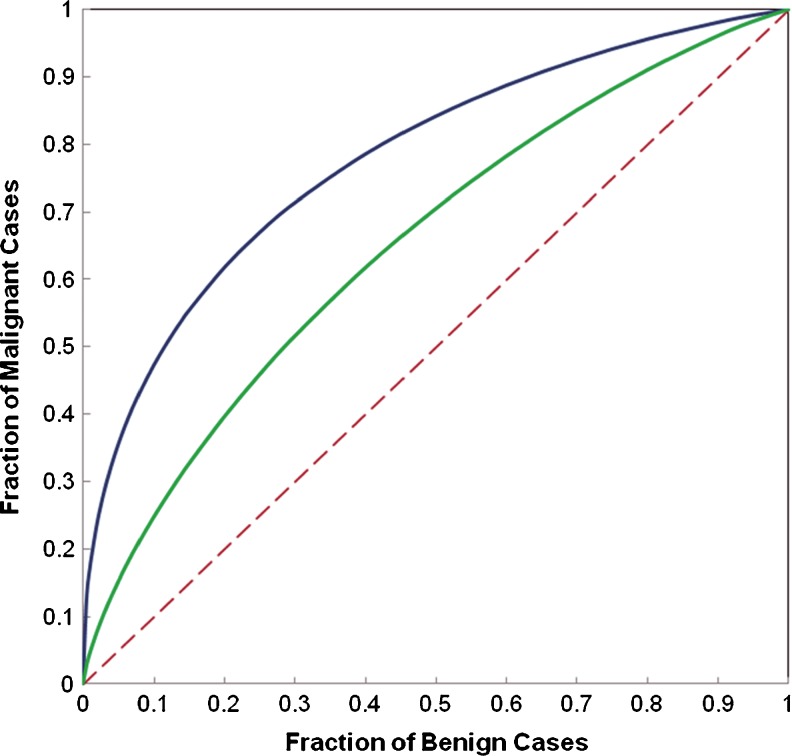
Table 2 summarizes and compares several performance levels of classification indices (including absolute classification accuracy, areas under ROC curve and 95 % confidence intervals, positive predictive and negative predictive values) when using three BBNs that involve either two or three computed DCE-MRI features. Given the high correlation between features F1 and F2, although using a BBN that combined these two features yielded greater AUC value (increasing from 0.657 ± 0.049 when using F1 only to 0.705 ± 0.046 when using a BBN that combines features of F1 and F2), the classification performance improvement is not statistically significant (with two-tailed p value = 0.092). However, when using the second BBN that combined two substantially low-correlated features (F1 and F3), the computed AUC value was significantly increased to 0.776 ± 0.041 (p < 0.01). Figure 5 plots and compares two ROC curves generated using the single feature (F1) and the BBN. The results also show that the BBN using all three features has little performance improvement (AUC = 0.779 ± 0.040) compared with the BBN that involves only two features of F1 and F3 (p = 0.907) indicating that F2 is a redundant feature.
Table 2.
Summary of classification performance levels using three BBNs involving the combination of three features (F 1, F 2, and F 3)
| Feature | F 1 + F 2 | F 1 + F 3 | F 1 + F 2 + F 3 |
|---|---|---|---|
| Classification accuracy | 66.9 % | 70.8 % | 71.5 % |
| Areas under ROC curves | 0.705 | 0.776 | 0.779 |
| 95 % confidence interval | 0.611, 0.788 | 0.689, 0.847 | 0.692, 849 |
| Positive predictive value | 0.69 | 0.75 | 0.77 |
| Negative predictive value | 0.61 | 0.63 | 0.64 |
Fig. 5.
Comparison of ROC curves generated using a single feature (F 1) and a BBN that involves two features (F 1 and F 3). The areas under the reference (dashed) line and two ROC curves are 0.5, 0.657, and 0.776, respectively
In summary, using three simple characteristic kinetic features computed from the entirely segmented breast areas and a BBN classifier, our CAD scheme enabled to classify 93 out of 130 cases correctly in our testing dataset yielding an absolute classification accuracy of 71.5 %. Specifically, the scheme classified 81 cases as malignant cases in which 62 were correctly classified and 19 were misclassified, which results in PPV = 0.765. The scheme also predicted 49 as benign cases in which 31 were correct and 18 were incorrect making NPV = 0.633. Based on ROC analysis, AUC = 0.779 ± 0.040 and thus the scheme could yield 47 % classification sensitivity level at 90 % specificity or 62 % sensitivity level at 80 % specificity. As a result, adding a BBN, our scheme enables to achieve significantly higher classification performance than using only a single image feature (e.g., F1).
Discussion
DCE-MRI has been considered an important and valuable imaging modality used in breast cancer diagnosis and patient management (including treatment planning and efficacy assessment). However, the lower diagnostic specificity reduces the efficacy of applying DCE-MRI in the clinical practice. As a result, exploring and extracting more effective and non-redundant image features or biomarkers from DCE-MRI images to help improve performance in classifying between malignant and benign cases remains an attractive research topic in the biomedical imaging and CAD fields.
In this study, we investigated a new DCE-MRI image feature or biomarker that is related to the bilateral dynamic contrast enhancement asymmetry computed from the entire breast areas and tested its discriminatory power of classifying between malignant and benign DCE-MRI examinations. The experimental results showed that using this new feature, our CAD scheme enabled to yield a classification performance level (e.g., AUC≈0.78) that is quite comparable to the existing CAD schemes using a set of optimally selected DCE-MRI kinetic features that are computed from pixels located inside the segmented breast lesions [21]. Although the performance of our CAD scheme remains lower if it is used as a standalone tool, this study provides a new image biomarker or classification index that has never been used in any previously developed CAD schemes for DCE-MRI images. Hence, our approach and CAD scheme does not directly compete with the existing CAD schemes due to the use of totally different image features. If there is low correlation between the classification scores generated by our CAD scheme and the other existing CAD schemes (which needs to be verified in future studies), one could relatively easily add this new feature or optimally fuse the classification score of our CAD scheme into the existing CAD schemes without scheme retraining. This will have potential to significantly improve the overall CAD performance levels in classifying between the malignant and benign DCE-MRI examinations (cases). The similar approach has been investigated and used in our previous work to improve performance of CAD schemes for mammography by fusing with a bilateral mammographic density asymmetry-based classification index [36].
Although previous studies have shown that BPE computed from DCE-MRI examinations was a useful cancer risk prediction and/or a cancer diagnostic factor [22, 23], this study is different. We investigated and tested a new feature index that is generated based on the bilateral asymmetry of BPE feature values computed from the left and right breasts of one woman. Since similar to mammographic density, BPE could vary substantially in different examination date because of the change of woman’s menstrual cycle and also be influenced by many other woman’s life style factors (e.g., using hormonal agent, smoking, and drinking alcohol) [24, 25]. However, these potential biases or influences on BPE feature assessment should equally impact on the features computed from two bilateral breasts of the same woman, subtraction of two feature values computed from the left and right breasts should help cancel or reduce many of these assessment biases. As a result, this new image feature could be more robust. The similar approach has been previously investigated in assessing the association between mammographic density and breast cancer risk, which demonstrated that using bilateral mammographic density symmetry enabled a computerized prediction model to yield more robust and significantly higher discriminatory power than those using the average mammographic density assessed from two breasts in breast cancer risk prediction [37].
Despite encouraging experimental results, this is a preliminary study that has several limitations. First, the size of our image dataset is relatively small and it may not adequately cover the diversity of cases in the real clinical environment. Hence, the performance and reliability level of our CAD scheme needs to be further tested using larger and more diverse datasets in future studies. Second, the malignant case group in our dataset does not include the cases with bilateral malignancy. All malignant cases in this dataset have only one malignant breast and one negative (or benign) breast. Although the percentage of cancer cases with two malignant breasts is small in the clinical practice, how the inclusion of the cases with bilateral malignancy affects performance of our scheme has not been tested. Third, considering the special DCE-MRI examination protocol used in Zhejiang Cancer Hospital (in China), we only computed kinetic features (enhancement values) in two time post- contrast points (M1 and M2), which makes it unable to generate an entire kinetic curve. Although in this study we found that features computed from the first and the second post-contrast image scanning time points (F1 and F2) are highly correlated, whether involving more time points in the kinetic curves could help further improve the classification performance of this type of schemes need to be later investigated and verified with the diverse image datasets acquired from different DCE-MRI examination protocols. Fourth, we have not investigated the correlation between this new image feature and other image features extracted from the targeted lesions by other existing CAD schemes. Thus, whether fusion between the classification scores generated by the BBN of our scheme and those generated by the existing CAD schemes could help significantly improve CAD performance in diagnosis of DCE-MRI examinations is also an interesting research topic in the future studies.
In summary, we investigated in this study a new type of kinetic image features computed from breast DCE- MRI images and reported our preliminarily experimental results. The study demonstrated that a new image biomarker or classification index based on the bilateral asymmetry of characteristic kinetic features computed from the entire left and right breast areas segmented from breast DCE-MRI images provided a relatively higher discriminatory power to classify between the malignant and benign cases. Since this type of features or classification index has never been investigated and applied in previous CAD schemes for DCE-MRI images to date, it may provide useful supplementary information in future CAD development. However, to realize the ultimate goal of adding this new image feature or a classification index generated by our CAD scheme to improve diagnostic performance in DCE-MRI examinations using CAD approach, much work is needed including building large and diverse image databases, exploring and selecting optimal image features to more effectively detect bilateral asymmetry of characteristic kinetic image features, and evaluating the performance and reliability of CAD schemes after integrating or fusing with this or similar image features in future studies.
Acknowledgments
This work is supported in part by grants from the National Natural Science Foundation of China (61271063), 973 Program (2013CB329502), National Distinguished Young Research Scientist Award (60788101), and Grant CA160205 from the National Cancer Institute, National Institutes of Health, USA.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Cady B, Michaelson JS. The life-sparing potential of mammographic screening. Cancer. 2001;91:1699–1703. doi: 10.1002/1097-0142(20010501)91:9<1699::AID-CNCR1186>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 3.Tabar L, Vitak B, Chen HH, et al. Beyond randomized controlled trials: organized mammographic screening substantially reduces breast carcinoma mortality. Cancer. 2001;91:1724–1731. doi: 10.1002/1097-0142(20010501)91:9<1724::AID-CNCR1190>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 4.Smith RA, Cokkindes V, Brooks D, et al. Cancer screening in the United States, 2011. CA Cancer J Clin. 2011;61:8–30. doi: 10.3322/caac.20096. [DOI] [PubMed] [Google Scholar]
- 5.Mandelson MT, Oestreicher N, Porter PL, et al. Breast density as a predictor of mammographic detection: comparison of interval-and screen-detected cancers. J Natl Cancer Inst. 2000;92:1081–1087. doi: 10.1093/jnci/92.13.1081. [DOI] [PubMed] [Google Scholar]
- 6.Kolb TM, Lichy J, Newhouse JH. Comparison of the performance of screening mammography, physical examination, and breast US and evaluation of factors that influence them: an analysis of 27,825 patient evaluations. Radiology. 2002;225:165–175. doi: 10.1148/radiol.2251011667. [DOI] [PubMed] [Google Scholar]
- 7.Berg WA, Gutierrez L, NessAiver MS, et al. Diagnostic accuracy of mammography, clinical examination, US, and MR imaging in preoperative assessment of breast cancer. Radiology. 2004;233:830–849. doi: 10.1148/radiol.2333031484. [DOI] [PubMed] [Google Scholar]
- 8.Saslow D, Boetes C, Burke W, et al. American Cancer Society guidelines for breast screening with MRI as an adjunct to mammography. CA Cancer J Clin. 2007;57:75–89. doi: 10.3322/canjclin.57.2.75. [DOI] [PubMed] [Google Scholar]
- 9.Leach MO, Boggis CR, Dixon AK, et al. Screening with magnetic resonance imaging and mammography of a UK population at high familial risk of breast cancer: a prospective multicentre cohort study (MARIBS) Lancet. 2005;365:1769–1778. doi: 10.1016/S0140-6736(05)66646-9. [DOI] [PubMed] [Google Scholar]
- 10.Kuhl CK, Schrading S, Leutner CC, et al. Mammography, breast ultrasound, and magnetic resonance imaging for surveillance of women at high familial risk for breast cancer. J Clin Oncol. 2005;23:8469–8476. doi: 10.1200/JCO.2004.00.4960. [DOI] [PubMed] [Google Scholar]
- 11.Sardanelli F, Podo F, D’Agnolo G, et al. Multicenter comparative multimodality surveillance of women at genetic-familial high risk for breast cancer (HIBCRIT study): interim results. Radiology. 2007;242:698–715. doi: 10.1148/radiol.2423051965. [DOI] [PubMed] [Google Scholar]
- 12.Kriege M, Brekelmans CT, Boetes C, et al. Efficacy of MRI and mammography for breast-cancer screening in women with a familial or genetic predisposition. N Engl J Med. 2004;351:427–437. doi: 10.1056/NEJMoa031759. [DOI] [PubMed] [Google Scholar]
- 13.Kriege M, Brekelmans CT, Boetes C, et al. Differences between first and subsequent rounds of the MRISC breast cancer screening program for women with a familial or genetic predisposition. Cancer. 2006;106:2318–2326. doi: 10.1002/cncr.21863. [DOI] [PubMed] [Google Scholar]
- 14.Warner E. Intensive radiologic surveillance: a focus on the psychological issues. Ann Oncol. 2004;15:143–147. doi: 10.1093/annonc/mdh657. [DOI] [PubMed] [Google Scholar]
- 15.Berg WA, Blume JD, Adams AM, et al. Reasons women at elevated risk of breast cancer refuse breast MR imaging screening: ACRIN 6666. Radiology. 2010;254:79–87. doi: 10.1148/radiol.2541090953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibbs P, Turnbull LW. Textural analysis of contrast-enhanced MR image of the breast. Magn Reson Med. 2003;50:92–98. doi: 10.1002/mrm.10496. [DOI] [PubMed] [Google Scholar]
- 17.Chen W, Giger ML, Bick U, Newstead GM. Automatic identification and classification of characteristic kinetic curves of breast lesions on DCE-MRI. Med Phys. 2006;33:2878–2887. doi: 10.1118/1.2210568. [DOI] [PubMed] [Google Scholar]
- 18.Meinel LA, Stolpen AH, Berbaun KS, Reinhardt JM. Breast MRI lesion classification: improved performance of human readers with a backpropagation neural network computer-aided diagnosis (CAD) system. J Magn Reson Imaging. 2007;25:89–95. doi: 10.1002/jmri.20794. [DOI] [PubMed] [Google Scholar]
- 19.Williams TC, DeMartini WB, Partridge SC, et al. Breast MR imaging: computer-aided evaluation program for discriminating benign from malignant lesions. Radiology. 2007;244:94–103. doi: 10.1148/radiol.2441060634. [DOI] [PubMed] [Google Scholar]
- 20.Bhooshan N, Giger ML, Jansen SA, Li H, Lan L, Newstead GM. Cancerous breast lesions on dynamic contrast-enhanced MR images: computerized characterization for image-based prognostic markers. Radiology. 2010;254:680–690. doi: 10.1148/radiol.09090838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan Y, Giger ML, Li H, Bhooshan N, Sennett CA. Multimodality computer-aided breast cancer diagnosis with FFDM and DCE-MRI. Acad Radiol. 2010;17:1158–1167. doi: 10.1016/j.acra.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King V, Brooks JD, Bernstein JL, Reiner AS, Pike MC, Morris EA. Background parenchymal enhancement at breast MR imaging and breast cancer risk. Radiology. 2011;260:50–60. doi: 10.1148/radiol.11102156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uematsu T, Kasami M, Watanabe J. Does the degree of background enhancement in breast MRI affect the detection and staging of breast cancer? Eur Radiol. 2011;21:2261–2267. doi: 10.1007/s00330-011-2175-6. [DOI] [PubMed] [Google Scholar]
- 24.Harvey J, Bovbjerg VE. Quantitative assessment of mammographic breast density: relationship with breast cancer risk. Radiology. 2004;230:29–41. doi: 10.1148/radiol.2301020870. [DOI] [PubMed] [Google Scholar]
- 25.Kopans DB. Basic physics and doubts about relationship between mammographically determined tissue density and breast cancer risk. Radiology. 2008;246:348–353. doi: 10.1148/radiol.2461070309. [DOI] [PubMed] [Google Scholar]
- 26.Scutt D, Lancaster GA, Manning JT. Breast asymmetry and predisposition to breast cancer. Breast Cancer Res. 2006;8:R14. doi: 10.1186/bcr1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim M, Wu G, Shen D. Hierarchical alignment of breast DCE-MR images by groupwise registration and robust feature matching. Med Phys. 2012;39:353–366. doi: 10.1118/1.3665705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Filev P, Hadjiiski L, Sahiner B, Chan HP, Helvie MA. Comparison of similarity measures for the task of template matching of masses on serial mammograms. Med Phys. 2005;32:515–529. doi: 10.1118/1.1851892. [DOI] [PubMed] [Google Scholar]
- 29.Wang XH, Park SC, Zheng B. Improving performance of content-based image retrieval schemes in searching for similar breast mass regions: an assessment. Phys Med Biol. 2009;54:949–961. doi: 10.1088/0031-9155/54/4/009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kahn CE, Robert LM, Wang K, et al. Construction of a Bayesian network for mammographic diagnosis of breast cancer. Comput. Biol. Med. 1997;27:1929–1940. doi: 10.1016/S0010-4825(96)00039-X. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Zheng B, Good WF, King JK, Chang Y. Computer-assisted diagnosis of breast cancer using a data-driven Bayesian belief network. Int J Med Informatics. 1999;54:115–126. doi: 10.1016/S1386-5056(98)00174-9. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Lederman D, Tan J, Zheng B. Computer-aided detection: the impact of machine learning classifier and image feature selection on scheme performance. Int J Intell Inf Process. 2010;1:30–40. [Google Scholar]
- 33.Mitchell TM. Machine learning. Boston, MA: McGraw-Hill; 1997. [Google Scholar]
- 34.Cheng J: BN Power constructor. University of Alberta, Edmonton, Alberta, Canada, 2001. Available from http://www.cs.ualberta.ca/~jcheng/bnsoft.htm
- 35.Metz CE: ROCKIT 0.9B beta version. University of Chicago, 1998. Available from http://www-radiology.uchicago.edu/krl/
- 36.Wang X, Li L, Xu W, Liu W, Lederman D, Zheng B. Improving performance of computer-aided detection of subtle breast masses using an adaptive cueing method. Phys Med Biol. 2012;57:561–575. doi: 10.1088/0031-9155/57/2/561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng B, Sumkin JH, Zuley ML, Wang X, Klym AH, Gur D. Bilateral mammographic density asymmetry and breast cancer risk: a preliminary assessment. Eur J Radiol. 2012;81:3222–3228. doi: 10.1016/j.ejrad.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]