The Tannat grape berry is used to produce high-quality wines with an intense purple color and remarkable antioxidant properties. Through reference-guided assembly of the genome combined with de novo assembly of the transcriptome, we found that the variety-specific genes that might contribute substantially to the unique characteristics of the Tannat berry are not present in the reference genome.
Abstract
The grapevine (Vitis vinifera) cultivar Tannat is cultivated mainly in Uruguay for the production of high-quality red wines. Tannat berries have unusually high levels of polyphenolic compounds, producing wines with an intense purple color and remarkable antioxidant properties. We investigated the genetic basis of these important characteristics by sequencing the genome of the Uruguayan Tannat clone UY11 using Illumina technology, followed by a mixture of de novo assembly and iterative mapping onto the PN40024 reference genome. RNA sequencing data for genome reannotation were processed using a combination of reference-guided annotation and de novo transcript assembly, allowing 5901 previously unannotated or unassembled genes to be defined and resulting in the discovery of 1873 genes that were not shared with PN40024. Expression analysis showed that these cultivar-specific genes contributed substantially (up to 81.24%) to the overall expression of enzymes involved in the synthesis of phenolic and polyphenolic compounds that contribute to the unique characteristics of the Tannat berries. The characterization of the Tannat genome therefore indicated that the grapevine reference genome lacks many genes that appear to be relevant for the varietal phenotype.
INTRODUCTION
Grapevine (Vitis vinifera) cv Tannat is a grapevine cultivar originally from southwestern France that is now mainly cultivated in Uruguay. The first Tannat vines were introduced into Uruguay in the 1870s by European immigrants, but since the 1970s, many of the plants have been replaced with new French Tannat commercial clones, allowing Uruguay to produce high-quality red wines (Carrau, 1997).
Tannat is the main grape berry produced for Gers wines and is the V. vinifera cultivar richest in tannins (Boido et al., 2011). Tannat seeds contain high levels of tannins, and Tannat skins at maturity contain high levels of anthocyanins. The total flavan-3-ol content of Tannat seeds (1946 mg/kg; Boido et al., 2011) is 6 times higher than the content reported for Pinot Noir (317 mg/kg; Mattivi et al., 2009) under similar experimental conditions. Tannat grapes therefore produce a colorful wine, with high acidity and a high tannin content that is suitable for long-term aging (Alcalde-Eon et al., 2006; Boido et al., 2006, 2011). Tannat berries have other unusual characteristics relating to the high content of phenolic and polyphenolic compounds (e.g., the presence of compounds such as malvidin, delphinidin, and petunidin monoglucosides), which confer more intense pigments to wine (Alcalde-Eon et al., 2006), and much higher levels of resveratrol compared with cultivars such as Pinot Noir, Merlot, and Cabernet (Gu et al., 1999; Carrau et al., 2011).
Polyphenols have many useful biological functions in plants (Kutchan, 2005), including defense against biotic stress (Bhattacharya et al., 2010), reflecting their antimicrobial, antifungal, and antiherbivore properties (Dixon et al., 2002; Chong et al., 2009). Polyphenols acquired from the moderate consumption of red wine also provide pharmaceutical and nutritional benefits to humans (Lin and Weng, 2006), such as helping to prevent cancer and reducing the inflammation associated with coronary artery disease (Khan et al., 2010). Proanthocyanidins are the principal vasoactive polyphenols in red wine; they induce the endothelium-dependent dilatation of blood vessels and suppress the synthesis of the vasoconstrictive peptide endothelin-1 (Fitzpatrick et al., 1993; Hashimoto et al., 2001). The proanthocyanidin content of local red wines correlates strongly with increased longevity in areas such as Gers in France and Nuoro in Italy (Corder et al., 2006).
State-of-the-art sequencing technology combined with the availability of a grapevine reference genome now allows the characterization of genetic variations that affect the properties of wine (Myles et al., 2010; Zhang et al., 2012). However, recent studies have shown that reliance on a single reference genome may underestimate the variability among different genotypes (Springer et al., 2009; Swanson-Wagner et al., 2010; Gan et al., 2011). Plant genomes contain core sequences that are common to all individuals, as well as dispensable sequences comprising partially shared and nonshared genes that contribute to intraspecific variation (Morgante et al., 2007). The dispensable genome is more difficult to characterize because one of the main drivers is transposon activity, which influences genome size (Ammiraju et al., 2007; Ågren and Wright, 2011) and the expression and regulation of genes (Kobayashi et al., 2004). However, it is now suspected that such variability is not restricted to the number and location of transposable elements but may also involve functional sequences as previously shown in prokaryotes (Medini et al., 2005). Similarly in humans, Asian and African individuals possess ∼5 Mb of population-specific DNA encoding 141 human RefSeq sequences that cannot be mapped against the current reference genome, suggesting that the human pan-genome would include an additional 19 to 40 Mb of novel information (Li et al., 2010). In Arabidopsis thaliana, 221 genes that are not present in the reference Columbia-0 genome have been identified in other ecotypes (Gan et al., 2011; Schneeberger et al., 2011). More recently, the presence in specific cultivars of hundreds of genes not shared with the reference genome has been demonstrated through the analysis of transcripts assembled de novo from RNA sequencing (RNA-Seq) data in both maize (Zea mays; Hansey et al., 2012) and grapevine (Venturini et al., 2013).
We investigated the genetic basis of the unique phenotypic characteristics of Tannat berries, which are most notable for their high content of polyphenolic compounds, by sequencing the Uruguayan Tannat clone UY11 from the vines introduced into Uruguay in the 1870s (Gonzalez Techera et al., 2004). UY11 belongs to a predominantly homozygous group of clones including the French commercial clone 399, one of the most widely grown clones in Uruguay (Gonzalez Techera et al., 2004). We also produced an exhaustive annotation of Tannat genes by RNA-Seq analysis and identified many genes that are not shared with the reference genome and that are involved in the synthesis of phenolic and polyphenolic compounds.
RESULTS
Genome Sequencing, Assembly, and Analysis of Variants
Genomic DNA from the Uruguayan Tannat clone UY11 was used to generate 322,786,617 Illumina reads (2 × 100) representing 134-fold base pair coverage and ∼260-fold physical coverage of the grapevine genome (see Supplemental Table 1 online). The genome was assembled using a hybrid approach based on iterative realignment to the reference genome and integration of de novo–assembled contigs with a reference genome (Gan et al., 2011) with the Pinot Noir PN40024 genome sequence as a reference (Jaillon et al., 2007). The reconstruction of 482 Mb of the Tannat genome (see Supplemental Table 1 online) required 11 cycles of iterative read mapping combined with de novo assembly (see Supplemental Figure 1 online). The reads were aligned with the final assemblies, identifying 8.6 Mb of polymorphic regions lacking read coverage that probably reflect complex polymorphisms as previously reported (Gan et al., 2011). The N50 length (the contig length at which 50% of the entire assembly is represented by contigs equal to or longer than this value) of contiguous read coverage between polymorphic regions was 97.2 kb.
To report complex alleles consistently, we defined all variants against the 12x grapevine reference genome assembly (Grimplet et al., 2012). There were 2,087,275 single-base differences between Tannat and PN40024, as well as 240,355 (11.5%) ambiguous calls. When the same procedure was applied to the Corvina cultivar, we found 426,415 ambiguous calls among 2,048,530 single-base differences (∼20.8%), which is twice the number of heterozygous single-nucleotide polymorphisms (SNPs) found in the Tannat cultivar despite the similar total number of SNPs. These results concur with microsatellite data suggesting that Tannat has a lower degree of heterozygosity than other grapevine cultivars (Gonzalez Techera et al., 2004).
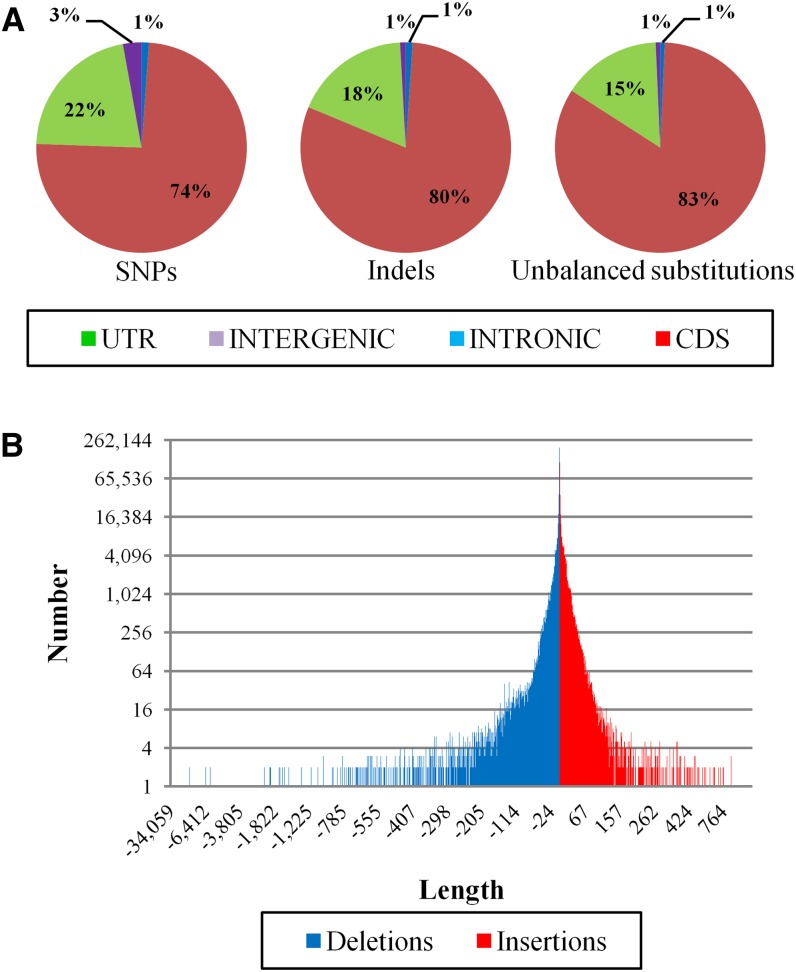
As expected, most of the identified SNPs were located outside known genes or within introns, with few located in coding sequences (CDSs) or untranslated regions (Figure 1A). The frequency of SNPs within genes (0.2572%), CDSs (0.2408%), and untranslated regions (0.3336%) was lower than the whole genome frequency (0.4292%). The low frequency of SNPs in CDSs was attributed to the conservation of protein functions. We identified 20,843 genes containing SNPs, and these were enriched for major functional categories such as protein Tyr kinase activity, regulation of development, protein binding, cellular component organization or biogenesis, and transferase activity (see Supplemental Table 2 online). We also identified 644,772 insertion/deletion polymorphisms (indels) and 42,471 unbalanced substitutions representing the replacement of the PN40024 reference sequence with a different sequence. Most of these variants were located in intergenic regions, and the portion that mapped within known CDSs was lower than for SNPs (Figures 1A). This concurs with the greater potential impact of these types of variants on protein function. Although 57.5% of indels and unbalanced substitutions were shorter than 6 bp in length, 1.9% exceeded 100 bp (Figure 1B). Overall, the Tannat genome was 4.5 Mb (1%) shorter than the PN40024 genome, probably reflecting the limitations of current technologies for the detection of long insertions (Gan et al., 2011). Although most of the deletions were located in intergenic regions, 7.29% of the deleted bases were considered as parts of genes in PN40024, and 99 genes were notable for the deletion of >50% of the PN40024 sequence length.
Figure 1.
Variation in the Tannat Genome.
(A) Classification of SNPs, indels, and unbalanced substitutions based on the V. vinifera PN40024 genome and annotation. UTR, untranslated region.
(B) Distribution of lengths of deletions and insertion.
Gene Annotation
A naive projection of the coordinates of the 29,971 nuclear protein-coding genes from PN40024 (V1 annotation; http://plants.ensembl.org/Vitis_vinifera/Info/Index) onto the Tannat genome predicted that 46.78% of the proteins contained sequence differences. Most of the changes were predicted not to affect protein functionality, and, from a total of 29,971 known genes, 22,983 were confirmed to lack deleterious mutations or structural alterations and were therefore transferred to the Tannat genome annotation. The remaining 6988 genes (23.3%) were predicted to encode proteins affected by deletions, truncations, or other disruptions (see Supplemental Figure 2 online). For these genes, it was not possible to transfer the reference annotation in a reliable manner.
To reannotate the Tannat genome, we performed an RNA-Seq analysis using a panel of four tissues/developmental stages (whole berry, skin at two developmental stages, and seeds), thus generating 395,863,776 2 × 100 reads (see Supplemental Table 3 online). We focused on berry tissues at early developmental stages because Tannat berries are rich in tannins, which are produced mostly prior to veraison (when berries begin to change color and enlarge) (Downey et al., 2003a; Conde et al., 2007). The expression data provided experimental support for 16,169 of the 22,983 genes reliably transferred from PN40024 to Tannat (see Supplemental Data Set 1 online). Reference-based assembly of the RNA-Seq data (Trapnell et al., 2012) allowed us to identify 81,759 putative transcripts and to reannotate 5796 genes corresponding to loci transferred from the V1 annotation that appeared to be disrupted in the Tannat genome, as well as 2866 nonannotated protein-coding genes with homology to sequences in the National Center for Biotechnology Information (NCBI) nonredundant (NR) and/or Vitis vinifera Gene Index (VvGI) databases. We ultimately characterized 31,645 loci including 13,932 genes (44%) encoding multiple protein isoforms (file available in General Transfer Format at http://ddlab.sci.univr.it/files/Tannat/annotation.gtf). The features of newly annotated (or reannotated) genes, such as the average mRNA length (1662 nucleotides) and the average number of exons per gene (6.77), were similar to those of PN40024 and other plant species (see Supplemental Table 4 online).
The gene annotations were validated further using the nonredundant core eukaryotic genes (CEGs) from the CEG mapping approach (CEGMA) pipeline (Parra et al., 2009). The presence of 232 of 248 CEGs (93.5%) in the Tannat gene set confirmed the quality of the annotation (see Supplemental Figure 3 online).
Transcriptome Assembly and Identification of Tannat-Specific Genes
We did not initially identify protein-coding genes that were present in the Tannat genome but missing from the PN40024 reference genome, probably because the assembly method used made it difficult to incorporate long insertions into the Tannat genome sequence. We therefore performed a de novo assembly of the RNA-Seq reads to generate a NR set of 114,786 transcripts (see Supplemental Table 5 online) with an average size of 1491 nucleotides, which is similar to the corresponding value for the PN40024 annotation (1331 nucleotides) and the Tannat annotation (1554 nucleotides).
We validated the assembled transcriptome using the NR core gene set (Parra et al., 2009). The high percentage of CEGs represented in the data set (95%) confirmed that the Tannat gene set based on the assembled transcriptome was almost complete. Furthermore, the highest fraction of CEGs detected compared with the Tannat annotation (93.5%) suggested that the assembled transcriptome contained more genes than the annotated Tannat genome. Among the 114,786 transcripts, a large fraction (88%) mapped with high confidence to the reconstructed Tannat genome. BLAST queries of the 13,707 transcripts that could not be mapped to the reconstructed genome against the NCBI NR protein database (BLAST Expect value [E-value] ≤1*10−5; http://www.ncbi.nlm.nih.gov/BLAST/) identified 731 sequences as contaminants (see Supplemental Table 6 online). We found that 9302 of the remaining 12,976 putative transcripts matched expressed grapevine sequences represented in the VvGI database v8.0 or other plant proteins, and these were considered as novel grapevine transcripts potentially restricted to V. vinifera cv Tannat. Out of the 9302 putative transcripts, 5052 had a high coding potential and a full open reading frame with defined start and stop codons (Kong et al., 2007).
Among the 5052 high-confidence putative protein-coding transcripts that could not be mapped against the Tannat genome, 4501 were validated by comparison with raw Tannat genomic reads. After clustering and manual inspection, these transcripts were grouped into 3035 genes, which were compared with raw PN40024 genomic reads in order to identify genes that were still hidden in the unassembled portion of the PN40024 genome. This comparison revealed that the reference genome assembly lacks 1162 genes and that Tannat possesses a set of 1873 genes that are not shared with PN40024 (Table 1). Therefore, the Tannat genome appears to comprise 28,779 genes that are annotated on the reference genome (referred to as known genes), 4028 genes previously unannotated or not assembled in the reference genome (referred to as novel genes), and 1873 genes that appear to be unique to Tannat (varietal genes).
Table 1. Summary of Genes in V. vinifera cv Tannat.
| Classification | No. of Genes |
|---|---|
| Known (V1 annotation) | 28,779 |
| Novel on assembly | 2,866 |
| Novel outside assembly | 1,162 |
| Varietal | 1,873 |
| Total | 34,680 |
All 34,680 genes mapping to the reconstructed genome or obtained from de novo transcriptome assembly were functionally annotated using the NCBI NR protein database, VvGI 8.0, Gene Ontology, and the Kyoto Encyclopedia of Genes and Genomes (see Supplemental Data Set 1 online). For genes transferred from the V1 annotation, we integrated the VitisNet annotations (Grimplet et al., 2012). Definitions could be assigned by homology searches to 20,170 of the genes (∼58%). We also assigned Gene Ontology terms to 21,442 genes (61.8%), and 8543 genes encoding enzymes were classified by Enzyme Commission number. The gene categories are summarized in Supplemental Figure 4 online, revealing a higher proportion (>50%) of genes related to metabolic and cellular processes as well as binding and catalytic molecular functions.
Expansion of Gene Families Related to Polyphenol Biosynthesis
The Tannat cultivar is characterized by the accumulation of high levels of polyphenols in the berry skin and seed. We developed a hypothesis that the unusual metabolic profile of Tannat berries may reflect the amplification of genes encoding enzymes representing the phenylpropanoid pathway and its derivatives. Therefore, we checked the Tannat gene set for the presence of new family members in pathways related to phenol and polyphenol biosynthesis.
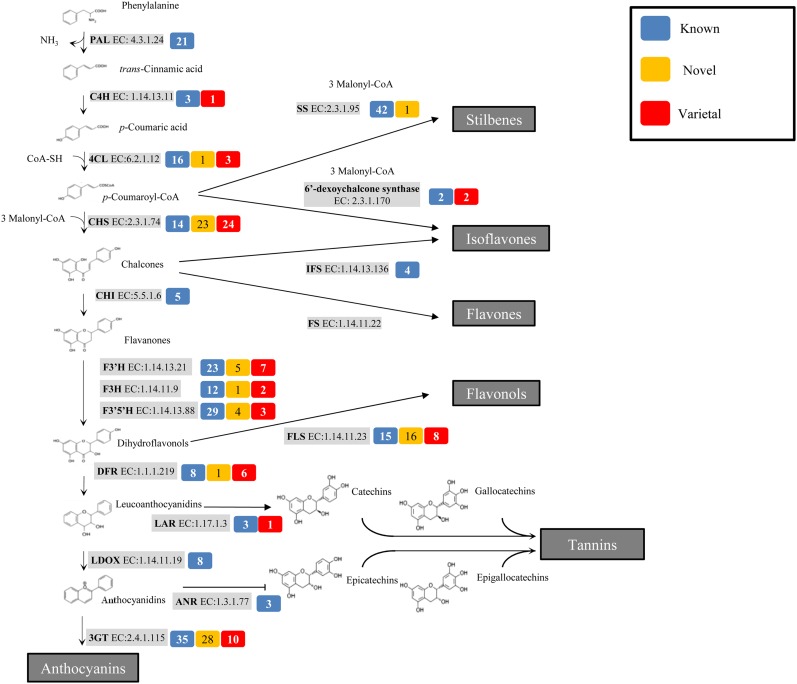
This approach identified 148 novel genes and 141 varietal genes corresponding to 23 different enzymes (see Supplemental Data Set 2 online). These included genes encoding some of the key enzymes in the phenylpropanoid pathway, such as cinnamate-4-hydroxylase, 4-coumarate:CoA ligase (4CL), and chalcone synthase (CHS). The CHS gene family in particular showed remarkable overrepresentation, with 47 new genes compared with 14 in the current V1 annotation (Figure 2; see Supplemental Data Set 2 online). Gene families encoding key enzymes in the flavonoid pathway were also expanded: Flavonoid 3′ hydroxylase (F3′H) was overrepresented, with 12 new genes compared with 23 genes in current annotation; three new flavonone 3-hydroxylase (F3H) genes were added to the 12 known genes; both the dihydroflavonol reductase (DFR) and flavonol synthase (FLS) families were expanded extensively, with the former adding seven new genes to the eight already present in the current annotation and the latter adding 24 members to the current 15 (Figure 2; see Supplemental Data Set 2 online). Finally, we identified 38 and 12 new genes similar to anthocyanidin 3-O-glucosyltransferases and 2'-hydroxyisoflavone reductases, respectively, compared with the 35 and 15 genes, respectively, in the current V1 annotation (Figure 2; see Supplemental Data Set 2 online).
Figure 2.
Schematic Diagram of Metabolic Pathways Involved in Polyphenol Biosynthesis.
For each enzyme, the number of known genes (blue box), novel genes (yellow box), and varietal genes in Tannat (red box) is reported. FS, flavone synthase; IFS, 2-hydroxyisoflavanone synthase; LDOX, leucocyanidin oxygenase; ANR, anthocyanidin reductase.
Expression of Genes Related to Polyphenol Biosynthesis
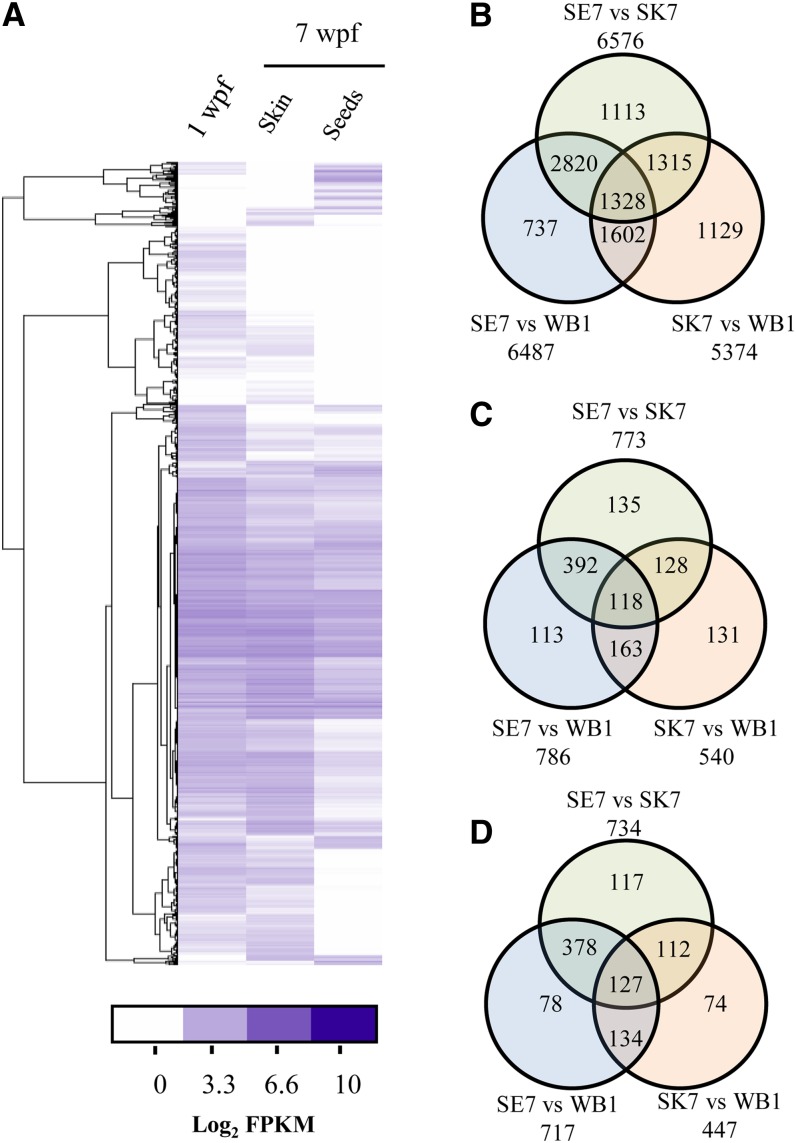
We investigated the expression of each of the 34,680 genes in the whole berry at 1 week post flowering (wpf) and in skins and seeds at 7 wpf (Figure 3A; see Supplemental Data Set 1 online). Most of the genes were expressed in at least one of the samples (29,982; fragments per kilobase of transcript per million fragments mapped [FPKM] > 0.01), whereas ∼65% (22,764) were expressed in all samples. Approximately 35% of the genes (12,244) were differentially expressed (false discovery rate < 5%, absolute of the logarithmic fold change [|Log2FC|] > 1) between the two developmental stages (1 and 7 wpf) or between the two tissues (seeds and skin) at 7 wpf (Figures 3B to 3D). The large proportion of modulated genes matches the plethora of biochemical and physiological changes that occur in different tissues during fruit development (Giovannoni, 2001). A greater proportion of varietal genes were differentially expressed compared with the whole data set (54% versus 35%).
Figure 3.
Analysis of Gene Expression in Tannat Berries.
(A) Hierarchical clustering of RNA-Seq read counts (Log2 FPKM) in three tissues of Tannat berries.
(B) Venn diagram showing numbers of commonly and uniquely differentially expressed known genes in pairwise comparisons of whole berry at 1 wpf (WB1), seeds at 7 wpf (SE7), and skin at 7 wpf (SK7).
(C) Venn diagram showing numbers of commonly and uniquely differentially expressed novel genes in pairwise comparisons among WB1, SE7, and SK7 samples.
(D) Venn diagram showing numbers of commonly and uniquely differentially expressed varietal genes in pairwise comparisons among WB1, SE7, and SK7 samples.
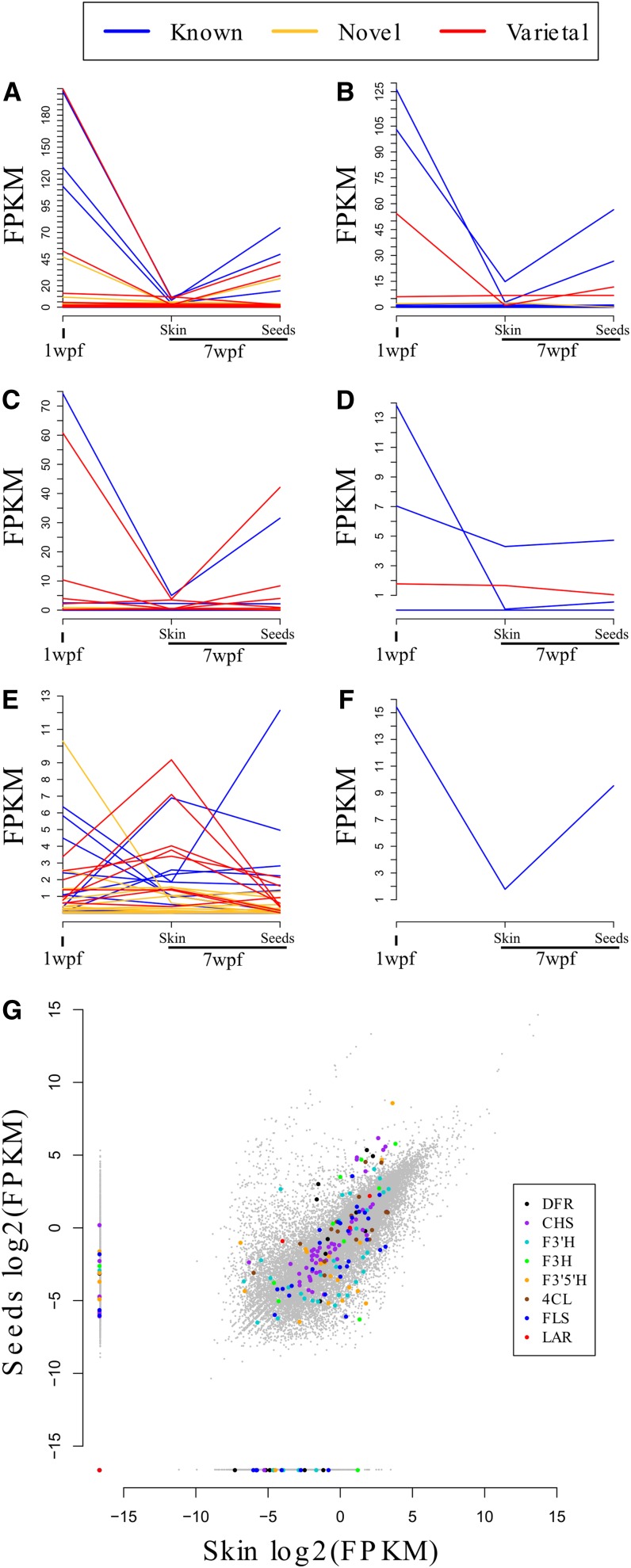
Most of the key genes representing the phenylpropanoid pathway and downstream pathways leading to flavonoids were strongly expressed, including CHS, F3H, and DFR (Figures 4A to 4F). The expression profiles were consistent with the biosynthesis of tannins in seeds during the first few weeks after fruit set (Bogs et al., 2005) and were highly correlated to the expression profile of MYBPA1, a gene encoding a transcription factor known to regulate berry tannin synthesis in grapevine (Bogs et al., 2007). A similar pattern was also observed for two annotated shikimate dehydrogenase genes and for two Ser carboxypeptidase-like genes, which are highly expressed at 1 wpf and strongly induced in seeds at 7 wpf compared with skin at the same time point. Genes encoding enzymes involved in the production of other flavonoids, such as FLS, which catalyzes the first committed step in flavonol biosynthesis, showed more diverse expression profiles. Some were expressed only in the berry skin and others only in the seeds (Figure 4G).
Figure 4.
Comparison of Gene Expression in Different Tannat Berry Tissues.
(A) to (F) Expression profiles of CHS (A), F3H (B), DFR (C), LAR (D), FLS (E), and MybPA1 (F) in whole berries at 1 wpf, skin at 7 wpf, and seeds at 7 wpf.
(G) Scatterplot of RNA-Seq expression values in seeds and skin at 7 wpf. Selected genes involved in polyphenol biosynthesis are shown in color as indicated.
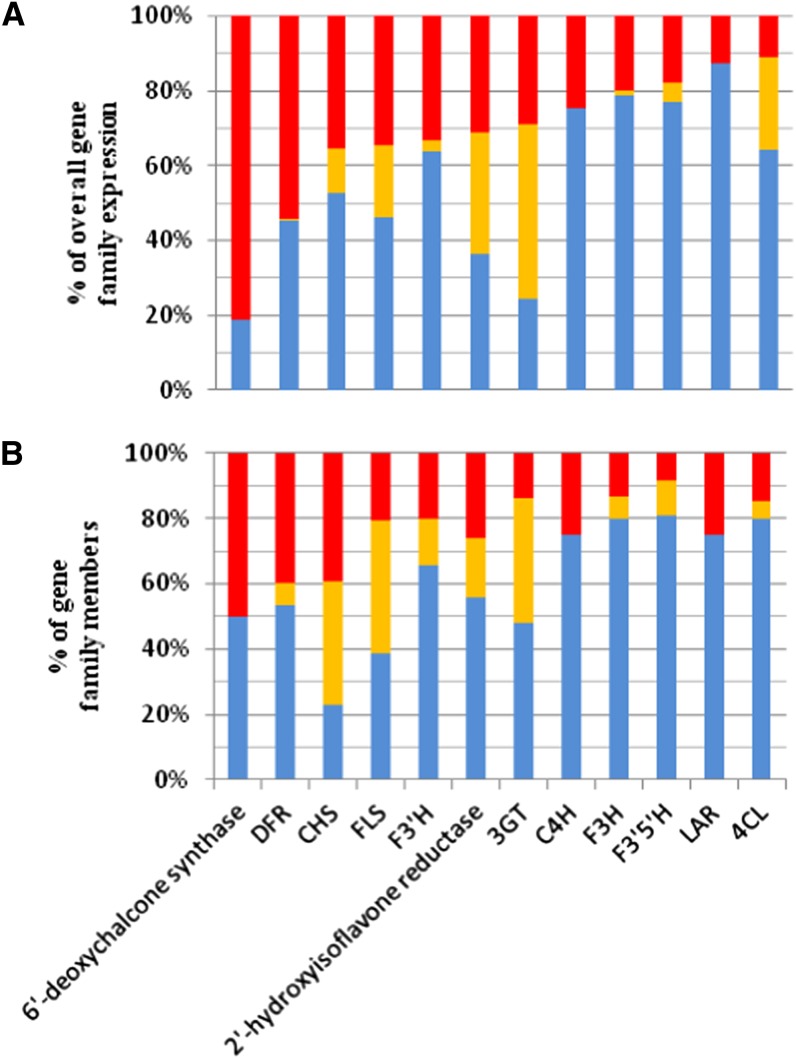
To evaluate the potential contribution of varietal genes to the extensive production and accumulation of tannins in Tannat, we determined the expression level of known, novel, and varietal genes relative to the overall expression level of each enzyme. Among the enzymes contributing to tannin biosynthesis, 14 were represented by the expression of varietal genes, with a contribution generally ranging from 11.16 to 81.24% (Figure 5A), but that in the case of 6'-deoxychalcone synthase reached 94% in the berry skin at 7 wpf (see Supplemental Data Set 2 online). The overall contribution to the expression of each enzyme was not strictly proportional to the number of gene copies among the known, novel, and varietal genes (Figure 5B), suggesting the significant differential regulation of genes encoding the same enzyme.
Figure 5.
Family Expansion and Expression Contribution of Genes Related to Polyphenol Biosynthesis.
(A) Percentage contribution of known, novel, and varietal genes to the overall expression of gene families encoding enzymes in the phenylpropanoid and flavonoid biosynthesis pathways.
(B) Percentage of known, novel, and varietal genes in gene families encoding enzymes in the phenilpropanoid and flavonoid biosynthesis pathways.
Regulation of Polyphenol Biosynthesis Pathway Genes
To investigate the regulation of polyphenol biosynthesis in Tannat berries, we analyzed four transcription factor superfamilies (basic helix-loop-helix [bHLH], R2R3 MYB, WD40, and WRKY) known to be involved in the regulation of this pathway (Lepiniec et al., 2006). The V1 annotation includes 106 bHLH, 131 R2R3 MYB, 56 WD40, and 61 WRKY genes. By contrast, with the observed expansion of enzyme gene families, we identified only two varietal genes representing the bHLH and R2R3 MYB superfamilies and four varietal genes and three novel genes representing the WD40 superfamily (see Supplemental Data Set 2 online). The contribution to overall expression levels by new genes (i.e., not present in the V1 annotation) in these four transcription factor superfamilies was low (1.4, 2.3, and 7% for the R2R3 MYB, bHLH, and WD40 superfamilies, respectively).
Origin of the Varietal Genes
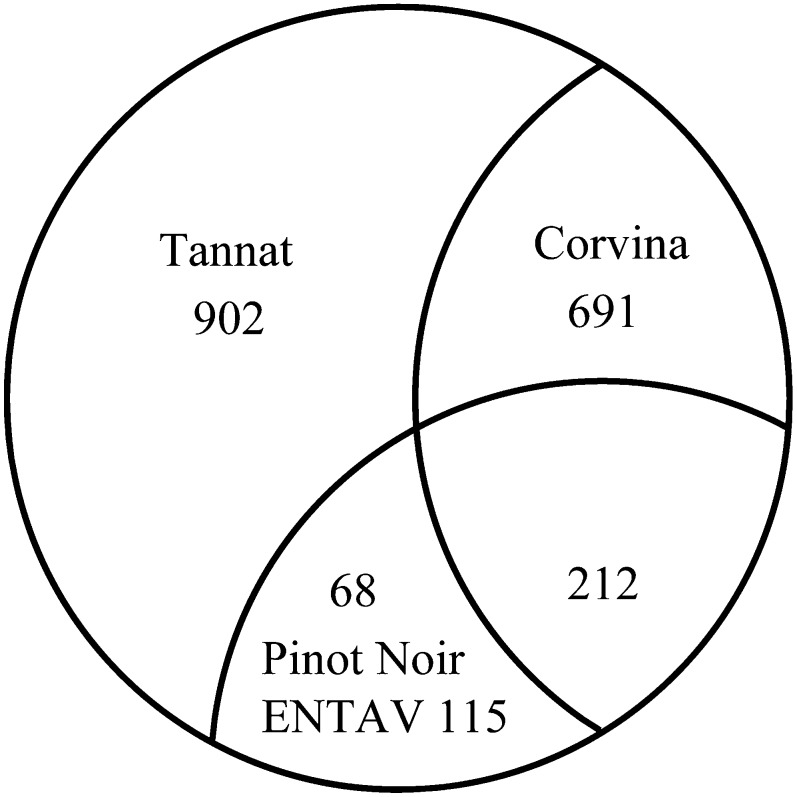
To gain more insight into the origins of the 1873 Tannat varietal genes, we compared them with genomic sequences available for Pinot Noir clone ENTAV 115 (Velasco et al., 2007) and with internally produced Illumina genomic reads from the Corvina cultivar (Figure 6). This revealed 280 Tannat varietal genes shared with the Pinot Noir clone ENTAV 115 that were probably lost by the PN40024 clone during the self-breeding process. Of the remaining genes, 691 were shared with Corvina and 902 were Tannat varietal genes. The presence of only partially overlapping differential sets of dispensable genes in the three cultivars concurs with the recent large-scale analysis of simple sequence repeat data showing that Corvina, Tannat, and Pinot Noir cultivars belong to different phylogenetic clades related by common ancestors (Cipriani et al., 2010).
Figure 6.
Categorization of the 1873 Genes Not Shared with PN40024.
Number of genes found in common among Tannat, Corvina, and Pinot Noir (ENTAV 115).
DISCUSSION
Because currently available sequencing technologies still produce reads that are too short for the accurate assembly of complex genomes (Schatz et al., 2012), we reconstructed the Tannat genome using a hybrid approach (Gan et al., 2011) that relied on both the de novo assembly of Illumina genomic reads and iterative mapping against the PN40024 reference genome (Jaillon et al., 2007). The highly contiguous genome assembly is comparable in length to the reference genome (98.9%), but divergence is demonstrated by the presence of more than two million SNPs and almost 700,000 indels or unbalanced substitutions. A striking feature of the Tannat genome is the low level of heterozygosity compared with common cultivated grapevine cultivars. The total number of SNPs is similar in Tannat and another representative cultivar (Corvina), but the Tannat genome contains only half as many ambiguous, potentially heterozygous bases, in agreement with previous reports (Gonzalez Techera et al., 2004). The relatively high level of homozygosity could reflect the geographic isolation of Tannat in southwestern France after its branching from Manseng Noir (Lacombe et al., 2013) and the resulting predominance of self-fertilization, but it may also indicate that Tannat is more tolerant to inbreeding (Lassalle, 1993) than Pinot varieties, where the viability of selfed progenies declines after a few generations (Bronner and Oliveira, 1991). Furthermore, the distribution of variants among Tannat genes indicates the fixation of mutations in genes related to specific biological processes such as development, the transfer of glycosyl groups, and protein Tyr kinase activity (see Supplemental Table 2 online). The enrichment of variation within particular functional classes of genes may reflect the environment in which Tannat was cultivated, and this is particularly interesting because genome-wide structural and gene content variations may drive the phenotypic variations that characterize different genotypes within a species (McHale et al., 2012).
The de novo assembly of the transcriptome from berry tissues sampled between flowering and veraison identified 1873 genes that we considered cultivar-specific because they are not shared with the reference genome. Although genes that are not conserved in a certain species are often assumed to be dispensable or redundant for development or survival, there is now evidence to indicate that such genes may have a strong impact on phenotype (Chen et al., 2013); therefore, we named them varietal genes.
Tannat berries are notable for the production and accumulation of polyphenols, particularly anthocyanidins, in berry skins at maturity and tannins in both seeds and to a lesser extent in berry skins. The biosynthesis of these substances involves a large set of enzymes that convert Phe into diverse phenylpropanoids and flavonoids (Dixon et al., 2002) and whose expression is tightly regulated during berry development (Boss et al., 1996; Bogs et al., 2005). The production and accumulation of tannins in the seeds and skins occurs mainly during the early phases of berry development (Conde et al., 2007; Boido et al., 2011). We identified 141 varietal genes encoding 19 enzymes involved in the production of polyphenols (see Supplemental Data Set 2 online). Some of them act in the early steps of the pathway and provide precursors for all the derivative branches, such as cinnamate-4-hydroxylase and 4CL (Dixon et al., 2002). These precursors are directed toward the flavonoid and isoflavonoid pathways by the enzymes CHS and chalcone isomerase, respectively. CHS catalyzes the condensation of 4-coumaroyl-CoA with three C2 units from malonyl-CoA to produce the C15 skeleton naringenin chalcone, which can be converted into flavones, isoflavones, flavonols, anthocyanidins, and tannins. We identified 24 varietal CHS genes in the Tannat genome, and this expansion compared with the reference genome may explain the higher polyphenol content of Tannat berries, since gene amplification is a well-documented mechanism to increase expression levels (Pollack et al., 1999; Wang et al., 2012). Interestingly, varietal CHS genes account for more than 30% of total CHS gene expression, including VVV_00520, which is one of the two most strongly expressed CHS genes in our data set, on par with the known gene CHS2.
Most of the gene families representing key enzymes in the flavonoid pathway showed some degree of expansion, including FLS, F3H, F3′H, and flavonoid 3′5′-hydroxylase (F3′5′H). The biosynthesis of flavonols is induced in the grapevine flower but decreases postanthesis and is then restored in berry skins after veraison (Downey et al., 2003b). The most common flavonols in Tannat berry skins are different glucosylated forms of quercetin (Boido et al., 2011), which together with myricetin have cytoprotective activities (Echeverry et al., 2004).
FLS catalyzes the first committed step in flavonol biosynthesis. We inspected the expression of two previously characterized FLSs (FLS1 and FLS2), and we found that, in agreement with previous reports (Downey et al., 2003b), the first was expressed at minimal levels at 1 wpf, while the second was not expressed at all in the analyzed stages. However, we identified two varietal FLS genes expressed specifically in the berry skin at 7 wpf (VVV_00613 and VVV_00659), suggesting that they are involved in the biosynthesis of specific flavonols or similar compounds in the skin during the early stages of Tannat berry development. Three flavonoid hydroxylases (F3H, F3′H, and F3′5'H) catalyze common steps in the flavonoid and anthocyanin biosynthesis pathways (Jeong et al., 2006). F3′H is also responsible for the conversion of kaempferol to quercetin, a natural antioxidant present in particular in red grape berries and wines. RNA-Seq data revealed that F3′5′H genes were expressed at double the level of F3′H genes in the Tannat berries, agreeing with the reported sevenfold increase in the ratio of 3′,5′-dihydroxy/3′-hydroxy anthocyanin derivatives compared with Pinot Noir and Corvina, perhaps reflecting differences in the activities of these pathways among different cultivars (Mattivi et al., 2006). Interestingly, three of the five most strongly expressed F3′5′H genes were varietal, suggesting that expansion within this family might explain the high 3′,5′-dihydroxy/3′-hydroxy anthocyanin ratio observed in Tannat berries.
Whereas FLS catalyzes the first step in the isoflavonoid pathway, DFR catalyzes a key step in the flavonoid pathway common to anthocyanin and tannin biosynthesis and is completely devoted to tannin biosynthesis before veraison. DFR was represented by six varietal genes that, in addition to the eight genes present in the reference genome, accounted for more than 50% of overall DFR expression with a peak of 62% in the seeds at 7 wpf. A varietal DFR (VVV_00044) was among the two most strongly expressed DFR genes in our data set and showed an expression pattern consistent with tannin biosynthesis during the early phases of berry development. This varietal gene was slightly more specific for the seed at 7 wpf compared with the known DFR gene (Boss et al., 1996).
Taken together, these data indicate that the higher production of tannins and polyphenols in Tannat berries may be associated with the expansion of gene families encoding relevant enzymes in the varietal component of the Tannat genome. It is also worth noting that the gene family expansion we observed was not directly proportional to the existing gene family size; for example, some families with numerous members such as 4CL (16 members in the current annotation) showed a modest expansion (one novel and three varietal genes), whereas some small families like 6’-deoxychalcone synthase (two members in the current annotation) doubled in size (see Supplemental Data Set 2 online). It is worth noting that gene expression levels do not always correlate with the related protein biosynthesis and/or activity levels or with the abundance of their target metabolites. However, previous results strongly suggest a high correlation of expression levels of polyphenol-related genes with the concentration of important polyphenol metabolites, including tannins, in grapevine (Dal Santo et al., 2013).
In sharp contrast with the expansion of several gene families encoding enzymes participating in polyphenol biosynthesis, transcription factor families involved in the regulation of these pathways showed little or no expansion of gene members, and the observed contribution of varietal genes to the overall expression of those families was very limited. The expression patterns of regulators known to be involved in the regulation of polyphenol biosynthesis were nevertheless consistent with current knowledge concerning the regulation of the pathway (see Supplemental Data Set 2 online). For example, the WD40 transcription factor WDR1 and the bHLH transcription factor MycA1, which are known to coregulate the biosynthesis of polyphenols in the early and late stages of berry development, were expressed at high levels at 1 wpf and repressed at 7 wpf, in agreement with previous reports (Matus et al., 2010). We also detected the expression of Myc1, which can induce the expression of key structural genes representing the flavonoid pathway (Hichri et al., 2010), with a pattern similar to that of MycA1 but at markedly lower FPKM values. Two of the most important regulators for the biosynthesis of grapevine proanthocyanidins, the R2R3 MYB factors MybPA1 and MybPA2, also showed expression profiles that agree with previous reports and match the temporal and spatial regulation of tannin biosynthesis (Bogs et al., 2007; Terrier et al., 2009). MybPA1 was the most strongly expressed R2R3 MYB factor both at 1 wpf and in the seeds at 7 wpf, whereas MybPA2 was moderately expressed at 1 wpf and almost completely silent at 7 wpf in both tissues.
In conclusion, Tannat appears to rely on a standard palette of transcription factors to increase the level of polyphenols through the activation of an expanded set of genes encoding the enzymes involved in this pathway. As recent studies have shown that parts of the genome that are not shared among all genotypes of a species can contain functional genes, the observed amplification of genes encoding key enzymes in the polyphenol biosynthesis pathway in a cultivar characterized by very high levels of polyphenolic compounds suggests that the dispensable genome of grapevine contains many genes that can contribute to the establishment of intervarietal differences in phenotype.
METHODS
Sample Collection
Unexpanded young leaves were collected from the old Uruguayan Tannat clone UY11 of grapevine (Vitis vinifera) (Gonzalez Techera et al., 2004) in a vineyard in Canelones, southern Uruguay. The samples were frozen in liquid nitrogen in the field and stored at –80°C.
Ten berry clusters from the French Tannat commercial clone 717 (ENTAV nomenclature: ENTAV 1995) were collected 1, 5, and 7 wpf during the 2011 to 2012 growing season, from a vineyard of 2000 plants in Melilla, Montevideo, southern Uruguay. The clusters were sampled randomly from 10 different plants, excluding the borders of the vineyard. Ten berries were randomly selected from each cluster and pooled with berries from the other plants on the same vineyard site, resulting in three independent pools of 30 berries for each developmental stage. Skin and seeds sampled at each time point were frozen separately in liquid nitrogen, except at 1 wpf, where the whole berries were used because the seeds were not fully developed.
Nucleic Acid Extraction
Frozen leaves were ground to powder under liquid nitrogen. Total DNA was extracted using the Qiagen Plant DNeasy kit (Qiagen) according to the manufacturer’s instructions. The purity and quantity of the DNA were determined using a Nanodrop 1000 spectrophotometer (Thermo Scientific). Frozen berries were ground to powder using liquid nitrogen, and total RNA was extracted using the Spectrum Plant Total RNA kit (Sigma-Aldrich) according to the manufacturer’s instructions. The purity and quantity of the RNA were determined using a Nanodrop 1000 spectrophotometer (Thermo Scientific). RNA integrity was determined using a Bioanalyzer 2100 (Agilent) with RNA 6000 Nano Kit I (Agilent).
Library Preparation and Sequencing
Genomic DNA (6 µg) was mixed with 750 μL of nebulization buffer (Illumina) and fragmented for 6 min in a nebulizer (Life Technologies) using compressed nitrogen at 35 p.s.i. to produce fragments with a typical size range of 150 to 700 bp and a peak at 500 bp. The sheared DNA was purified using the QIAquick PCR purification kit (Qiagen) and eluted in 30 μL of water. The quality of the fragmented DNA was determined using the Agilent DNA 1000 kit on an Agilent 2100 bioanalyzer, and DNA libraries were prepared using TruSeq DNA sample preparation kits (Illumina). Illumina RNA-Seq libraries were prepared from 2.5 µg of total RNA per sample according to the manufacturer’s instructions.
The DNA-Seq and RNA-Seq libraries were size-selected at 350 to 550 bp using the Pippin Prep DNA size selection system (Sage Science). Library quality was determined using the Agilent High Sensitivity DNA kit on the Agilent 2100 bioanalyzer, and the quantity was determined by quantitative PCR using the KAPA Library Quantification kit (KapaBiosystems). Libraries were then pooled in equimolar concentrations and sequenced with the TruSeq Sequencing by Synthesis Kit v3-HS and TruSeq Paired End Cluster Kit v3-cBot-HS (Illumina) using an Illumina HiSequation 1000 sequencer according to the manufacturer’s instructions to generate 100-bp paired-end reads.
Genome Assembly, Identification, and Analysis of Polymorphic Regions
The genomic sequences were assembled using the IMR/DENOM v0.3.3 pipeline (Gan et al., 2011) with default parameters and the 12x PN40024 genome as a reference (Jaillon et al., 2007). Chloroplast and mitochondrial sequences were excluded from the final reconstruction.
To identify polymorphic regions, we aligned genomic reads to the reconstructed Tannat genome using Burrows-Wheeler Aligner (BWA) with default parameters (Li and Durbin, 2010). Alignments were processed with SAMtools v0.1.18 (Li et al., 2009) to produce a histogram of the sequence coverage at each position in the genome. We identified polymorphic regions in the final genome as regions with a contiguous read coverage lower than four (Gan et al., 2011). The effects of polymorphisms detected in the IMR/DENOM pipeline on the functionality of proteins encoded by the V1 grapevine annotation were predicted using Variant Effect Predictor with EnsEMBL database v64 (McLaren et al., 2010).
The gene space of the assembled genome was assessed by aligning CEGs (Parra et al., 2009) with the Tannat genome using BLAST with a 65% identity threshold (Altschul et al., 1990).
Gene Annotation
V1 annotation gene models were translated to the reconstructed Tannat genome by taking structural variations in the reference genome into account and adjusting the coordinates accordingly using bespoke software (available on request). The reference-guided reconstruction of transcripts was performed using TopHat v2.0.6 (Trapnell et al., 2009) and Cufflinks v2.0.2 (Trapnell et al., 2010) as described (Trapnell et al., 2012) and was compared and merged with the translated V1 annotation using the Cuffcompare withol in the Cufflinks suite. We clustered the loci reconstructed by Cufflinks based on a comparison with sequences in the NCBI NR protein database using BLASTX (E-value ≤ 10−5) and VvGI database v8.0 (E-value ≤ 10−5) and manual inspection of alignments.
De Novo Assembly of Tannat Transcripts and the Identification of New Genes
De novo assembly of transcripts from RNA-Seq data was performed using the Velvet/Oases assembler with a k-mer value of 49, a minimum contig length of 200, an insert length of 310, and a standard deviation of 100 (Zerbino and Birney, 2008; Schulz et al., 2012). These parameters were optimized for scaffold N50 and total base coverage, after running the assembler across a range of parameters. Redundancy among assembled sequences was first removed using cd-hit-est with a threshold of 90% sequence identity (Li and Godzik, 2006).
We then applied five different filters to identify de novo–assembled transcripts missing from the genome assembly. Transcripts were aligned against the Tannat genome using GMAP v2012-11-07 with default parameters (Wu and Watanabe, 2005), and those with at least 80% of coverage and identity were discarded. Contaminant sequences were identified by comparison with NCBI NR database sequences using BLASTX (E-value ≤ 1*10−5) and were discarded. Putative grapevine transcripts were identified by sequence identity with VvGI using BLASTX (E-value ≤ 1*10−5). The coding potential of transcripts was computed with CPC v0.9 (Kong et al., 2007), and those with a complete open reading frame including start and stop codons and more than 50 codons overall were retained as high-confidence putative protein-coding transcripts. The Tannat genomic reads were aligned against transcripts using BWA with default parameters and the alignments were processed with BedTools suite v2.17.0 (Quinlan and Hall, 2010) to produce a histogram of the sequence coverage at each position in the transcripts. Those with <80% sequence coverage were discarded.
De novo–assembled transcripts that passed the above filters were classified as missing from the genome assembly and compared with different sets of genomic sequences to ascertain their presence in other genotypes. For PN40024, we obtained both the set of Sanger sequences used for the current version of the genome (downloaded from ftp://ftp-private.ncbi.nlm.nih.gov/pub/TraceDB/vitis_vinifera/) and two different sets of Illumina genomic reads (downloaded from http://urgi.versailles.inra.fr/galaxy/u/nchoisne/h/pn40024-public-data). For Pinot Noir ENTAV115, we downloaded the final assembled sequences from GenBank (accession numbers AM423240 to AM489403; Velasco et al., 2007). PN40024 Sanger reads were aligned with BLAT v34x12 against the transcripts to find matches >100 bp with no gaps and sequence coverage ≥80% (Kent, 2002), whereas the other sets of sequences were aligned with BWA using default parameters. A histogram of the sequence coverage at each position was computed with genomeCoverageBed (Quinlan and Hall, 2010). Transcripts covered (≥80%) by PN40024 genomic reads were classified as novel (shared with Pinot Noir but not previously annotated), whereas the remainder were classified as varietal.
The novel and varietal transcripts were clustered based on similarity to sequences present in NR, VvGI, and the Tannat genome. A multiple alignment was performed for each cluster using mafft v6.864b (Katoh and Toh, 2008) and manually checked for consistency. Final clusters were considered to be genes. The gene space for genes missing from the assembled genome was assessed as described above.
Functional Annotation of Genes
Genes were functionally annotated by integrating the V1 manual annotation (Grimplet et al., 2012) and automatic annotations performed with Blast2GO (Conesa et al., 2005). Novel and varietal genes without assigned Enzyme Commission numbers were manually classified by BLAST using known polyphenol-related genes as reference sequences and discarding hits with an E-value >1*10−10.
Expression of Polyphenol Pathway Genes
RNA-Seq reads were aligned against the Tannat genome and gene sequences with BWA using default parameters. We counted the mapping reads and calculated FPKM expression values for known, novel, and varietal genes using the R package DESeq with default parameters (Anders and Huber, 2010) to assess which genes were differentially expressed among the developmental stages and tissues under analysis (false discovery rate < 5%, |Log2FC| > 1).
Accession Numbers
Genomic sequence data from this article can be found in the NCBI Sequence Read Archive under accession numbers SRR863595 and SRR863618. Transcriptome sequence data from this article can be found in the NCBI Sequence Read Archive under accession numbers SRR866531, SRR866540, SRR866544, SRR866568, SRR866569, SRR866570, SRR866571, SRR866572, SRR866574, SRR866575, and SRR866576. Transcriptome shotgun assembly data from this article can be found in the NCBI Transcriptome Shotgun Assembly database under accession number GAKH00000000. Variation data are downloadable as IMR/DENOM sdi files from http://ddlab.sci.univr.it/files/Tannat/corvina.sdi and http://ddlab.sci.univr.it/files/Tannat/tannat.sdi. Annotations of known and novel genes can be downloaded from http://ddlab.sci.univr.it/files/Tannat/annotation.gtf. Mappings between varietal gene IDs and transcripts accession numbers can be downloaded from http://ddlab.sci.univr.it/files/Tannat/putative_transcripts2genes.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Number of Variants Identified at Each Iterative Mapping Step with IMR.
Supplemental Figure 2. Genes Affected by Potentially Disruptive Mutations.
Supplemental Figure 3. CEGMA Analysis of Tannat Genes.
Supplemental Figure 4. GO Gene Classification.
Supplemental Table 1. V. vinifera cv Tannat Genome Sequencing and Assembly Statistics.
Supplemental Table 2. Enrichment Analysis of Genes Containing SNPs.
Supplemental Table 3. V. vinifera cv Tannat Transcriptome Sequencing Statistics.
Supplemental Table 4. Comparison of Tannat Annotation with Other Plant Annotations.
Supplemental Table 5. V. vinifera cv Tannat Transcriptome Assembly Statistics.
Supplemental Table 6. Classification of Contaminants Present in de Novo–Assembled Transcripts Not Mapping on the Tannat Genome.
Supplemental Data Set 1. Definitions, GO and KEGG Annotations, Expression Values, and Differential Expression Tests for All 34,680 Tannat Genes.
Supplemental Data Set 2. Gene Number and Expression Levels for Gene Families Related to Polyphenol Biosynthesis; Expression Levels for Selected Genes Related to Polyphenol Biosynthesis.
Acknowledgments
We thank Didier Merdinoglu for providing us with DNA of PN40024 and the Institut National de la Recherche Agronomique for providing access to PN40024 raw Illumina genomic reads within the framework of GrapeReSeq, PLANT-KBBE2008. This work was supported by Fondazione Cariverona (Completamento e Attività del Centro di Genomica Funzionale Vegetale), a Joint Project 2012 between Biomolecular Research Genomics and the University of Verona, Ministero delle Politiche Agricole Alimentari e Forestali (Valorizzazione dei Principali Vitigni Autoctoni Italiani e dei loro Terroir-Vigneto), and Regione Veneto (Valorizzazione della Tipicità dei Vitigni Autoctoni e dei Vini Veneti-Valvive), and benefited from the networking activities within the European-funded COST ACTION FA1106. We also thank the support of Consejo Superior de Investigaciones Cientificas Group 656 of UdelaR and ANII of Uruguay for supporting the travels and PhD fellowship of C.D.S.
AUTHOR CONTRIBUTIONS
C.D.S. performed research, prepared samples, and analyzed data. G.Z. performed research, analyzed data, contributed new computational tools, and wrote the article. A.F. and L.V. analyzed data and wrote the article. A.M. and A.D.M. analyzed data and contributed new computational tools. G.B., P.T., C.A., and E.Z. prepared samples. E.B. and E.D. contributed new analytic tools. C.G. prepared samples and wrote and reviewed the article. M.P. reviewed the article. F.C. designed the research. M.D. designed the research and wrote the article.
Glossary
- RNA-Seq
RNA sequencing
- SNP
single-nucleotide polymorphism
- CDS
coding sequence
- NCBI
National Center for Biotechnology Information
- NR
nonredundant
- VvGI
Vitis vinifera Gene Index
- CEG
core eukaryotic gene
- 4CL
4-coumarate:CoA ligase
- CHS
chalcone synthase
- F3′H
flavonoid 3′ hydroxylase
- F3H
flavonone 3-hydroxylase
- FLS
flavonol synthase
- DFR
dihydroflavonol reductase
- wpf
week post flowering
- FPKM
fragments per kilobase of transcript per million fragments mapped
- bHLH
basic helix-loop-helix
- F3′5′H
flavonoid 3′5′-hydroxylase
References
- Ågren J.A., Wright S.I. (2011). Co-evolution between transposable elements and their hosts: A major factor in genome size evolution? Chromosome Res. 19: 777–786 [DOI] [PubMed] [Google Scholar]
- Alcalde-Eon C., Boido E., Carrau F., Dellacassa E., Rivas-Gonzalo J.C. (2006). Pigment profiles in monovarietal wines produced in Uruguay. Am. J. Enol. Vitic. 57: 449–459 [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Ammiraju J.S.S., et al. (2007). Evolutionary dynamics of an ancient retrotransposon family provides insights into evolution of genome size in the genus Oryza. Plant J. 52: 342–351 [DOI] [PubMed] [Google Scholar]
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A., Sood P., Citovsky V. (2010). The roles of plant phenolics in defence and communication during Agrobacterium and Rhizobium infection. Mol. Plant Pathol. 11: 705–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogs J., Downey M.O., Harvey J.S., Ashton A.R., Tanner G.J., Robinson S.P. (2005). Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves. Plant Physiol. 139: 652–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogs J., Jaffé F.W., Takos A.M., Walker A.R., Robinson S.P. (2007). The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol. 143: 1347–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boido E., Alcalde-Eon C., Carrau F., Dellacassa E., Rivas-Gonzalo J.C. (2006). Aging effect on the pigment composition and color of Vitis vinifera L. Cv. Tannat wines. Contribution of the main pigment families to wine color. J. Agric. Food Chem. 54: 6692–6704 [DOI] [PubMed] [Google Scholar]
- Boido E., García-Marino M., Dellacassa E., Carrau F., Rivas-Gonzalo J.C., Escribano-Bailón M.T. (2011). Characterisation and evolution of grape polyphenol profiles of Vitis vinifera L. cv. Tannat during ripening and vinification. Aust. J. Grape Wine Res. 17: 383–393 [Google Scholar]
- Boss P.K., Davies C., Robinson S.P. (1996). Expression of anthocyanin biosynthesis pathway genes in red and white grapes. Plant Mol. Biol. 32: 565–569 [DOI] [PubMed] [Google Scholar]
- Bronner A., Oliveira A. (1991). Création et étude de lignées chez le pinot noir (Vitis vinifera L.). J. Int. Sci. Vigne Vin. 3: 133–148 [Google Scholar]
- Carrau, F., Boido, E., Gaggero, C., Medina, K., Disegna, E., and Dellacassa, E. (2011). Vitis vinifera Tannat, chemical characterization and functional properties. Ten years of research. In Multidisciplinary Approaches on Food Science and Nutrition for the XXI Century, Rosana Filip, ed (Kerala, India: Transworld Research Network), pp. 53–71. [Google Scholar]
- Carrau F.M. (1997). The emergence of a new Uruguayan wine industry. J. Wine Res. 8: 179–185 [Google Scholar]
- Chen S., Krinsky B.H., Long M. (2013). New genes as drivers of phenotypic evolution. Nat. Rev. Genet. 14: 645–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J., Poutaraud A., Hugueney P. (2009). Metabolism and roles of stilbenes in plants. Plant Sci. 177: 143–155 [Google Scholar]
- Cipriani G., et al. (2010). The SSR-based molecular profile of 1005 grapevine (Vitis vinifera L.) accessions uncovers new synonymy and parentages, and reveals a large admixture amongst varieties of different geographic origin. Theor. Appl. Genet. 121: 1569–1585 [DOI] [PubMed] [Google Scholar]
- Conde C., Silva P., Fontes N., Dias A.C.P., Tavares R.M., Sousa M.J., Agasse A., Delrot S., Gerós H. (2007). Biochemical changes throughout grape berry development and fruit and wine quality. Food 1: 1–22 [Google Scholar]
- Conesa A., Götz S., García-Gómez J.M., Terol J., Talón M., Robles M. (2005). Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676 [DOI] [PubMed] [Google Scholar]
- Corder R., Mullen W., Khan N.Q., Marks S.C., Wood E.G., Carrier M.J., Crozier A. (2006). Oenology: Red wine procyanidins and vascular health. Nature 444: 566. [DOI] [PubMed] [Google Scholar]
- Dal Santo S., Tornielli G.B., Zenoni S., Fasoli M., Farina L., Anesi A., Guzzo F., Delledonne M., Pezzotti M. (2013). The plasticity of the grapevine berry transcriptome. Genome Biol. 14: r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R.A, Achnine L, Kota P, Liu C.J, Reddy M.S.S, Wang L. (2002). The phenylpropanoid pathway and plant defence: A genomics perspective. Mol. Plant Pathol. 3: 371–390 [DOI] [PubMed] [Google Scholar]
- Downey M.O., Harvey J.S., Robinson S.P. (2003a). Analysis of tannins in seeds and skins of Shiraz grapes throughout berry development. Aust. J. Grape Wine Res. 9: 15–27 [Google Scholar]
- Downey M.O., Harvey J.S., Robinson S.P. (2003b). Synthesis of flavonols and expression of flavonol synthase genes in the developing grape berries of Shiraz and Chardonnay (Vitis vinifera L.). Aust. J. Grape Wine Res. 9: 110–121 [Google Scholar]
- Echeverry C., Blasina F., Arredondo F., Ferreira M., Abin-Carriquiry J.A., Vasquez L., Aspillaga A.A., Diez M.S., Leighton F., Dajas F. (2004). Cytoprotection by neutral fraction of tannat red wine against oxidative stress-induced cell death. J. Agric. Food Chem. 52: 7395–7399 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick D.F., Hirschfield S.L., Coffey R.G. (1993). Endothelium-dependent vasorelaxing activity of wine and other grape products. Am. J. Physiol. 265: H774–H778 [DOI] [PubMed] [Google Scholar]
- Gan X., et al. (2011). Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477: 419–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni J. (2001). Molecular biology of fruit maturation and ripening. Annu. Rev. Plant Physiol. 52: 725–749 [DOI] [PubMed] [Google Scholar]
- Gonzalez Techera A., Jubany S., Ponce de Leon I., Boido E., Dellacassa E., Carrau F.M., Hinrichsen P., Gaggero C. (2004). Molecular diversity within clones of cv. Tannat (Vitis vinifera). Vitis 43: 179–185 [Google Scholar]
- Grimplet J., Van Hemert J., Carbonell-Bejerano P., Díaz-Riquelme J., Dickerson J., Fennell A., Pezzotti M., Martínez-Zapater J.M. (2012). Comparative analysis of grapevine whole-genome gene predictions, functional annotation, categorization and integration of the predicted gene sequences. BMC Res. Notes 5: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., Creasy L., Kester A., Zeece M. (1999). Capillary electrophoretic determination of resveratrol in wines. J. Agric. Food Chem. 47: 3223–3227 [DOI] [PubMed] [Google Scholar]
- Hansey C.N., Vaillancourt B., Sekhon R.S., de Leon N., Kaeppler S.M., Buell C.R. (2012). Maize (Zea mays L.) genome diversity as revealed by RNA-sequencing. PLoS ONE 7: e33071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M., Kim S., Eto M., Iijima K., Ako J., Yoshizumi M., Akishita M., Kondo K., Itakura H., Hosoda K., Toba K., Ouchi Y. (2001). Effect of acute intake of red wine on flow-mediated vasodilatation of the brachial artery. Am. J. Cardiol. 88: 1457–1460, A9. [DOI] [PubMed] [Google Scholar]
- Hichri I., Heppel S.C., Pillet J., Léon C., Czemmel S., Delrot S., Lauvergeat V., Bogs J. (2010). The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Mol. Plant 3: 509–523 [DOI] [PubMed] [Google Scholar]
- Jaillon O., et al. French-Italian Public Consortium for Grapevine Genome Characterization (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Jeong S.T., Goto-Yamamoto N., Hashizume K., Esaka M. (2006). Expression of the flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase genes and flavonoid composition in grape (Vitis vinifera). Plant Sci. 170: 61–69 [Google Scholar]
- Katoh K., Toh H. (2008). Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9: 286–298 [DOI] [PubMed] [Google Scholar]
- Kent W.J. (2002). BLAT—The BLAST-like alignment tool. Genome Res. 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N., Adhami V.M., Mukhtar H. (2010). Apoptosis by dietary agents for prevention and treatment of prostate cancer. Endocr. Relat. Cancer 17: R39–R52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S., Goto-Yamamoto N., Hirochika H. (2004). Retrotransposon-induced mutations in grape skin color. Science 304: 982. [DOI] [PubMed] [Google Scholar]
- Kong L., Zhang Y., Ye Z.Q., Liu X.Q., Zhao S.Q., Wei L., Gao G. (2007). CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 35 (Web Server issue): W345–W349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutchan T.M. (2005). A role for intra- and intercellular translocation in natural product biosynthesis. Curr. Opin. Plant Biol. 8: 292–300 [DOI] [PubMed] [Google Scholar]
- Lacombe T., Boursiquot J.M., Laucou V., Di Vecchi-Staraz M., Péros J.P., This P. (2013). Large-scale parentage analysis in an extended set of grapevine cultivars (Vitis vinifera L.). Theor. Appl. Genet. 126: 401–414 [DOI] [PubMed] [Google Scholar]
- Lassalle D. (1993). Nouvéaux cépages INRA pour le sud de la France. Progrès Agricole et Viticole 110: 511–517 [Google Scholar]
- Lepiniec L., Debeaujon I., Routaboul J.-M., Baudry A., Pourcel L., Nesi N., Caboche M. (2006). Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 57: 405–430 [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., et al. (2010). Building the sequence map of the human pan-genome. Nat. Biotechnol. 28: 57–63 [DOI] [PubMed] [Google Scholar]
- Li W., Godzik A. (2006). Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659 [DOI] [PubMed] [Google Scholar]
- Lin, J.K., and Weng, M.S. (2006). Flavonoids as nutraceuticals. In The Science of Flavonoids, E. Grotewold, ed (New York: Springer), pp. 213–238. [Google Scholar]
- Mattivi F., Guzzon R., Vrhovsek U., Stefanini M., Velasco R. (2006). Metabolite profiling of grape: Flavonols and anthocyanins. J. Agric. Food Chem. 54: 7692–7702 [DOI] [PubMed] [Google Scholar]
- Mattivi F., Vrhovsek U., Masuero D., Trainotti D. (2009). Differences in the amount and structure of extractable skin and seed tannins amongst red grape varieties. Aust. J. Grape Wine Res. 15: 27–35 [Google Scholar]
- Matus J.T., Poupin M.J., Cañón P., Bordeu E., Alcalde J.A., Arce-Johnson P. (2010). Isolation of WDR and bHLH genes related to flavonoid synthesis in grapevine (Vitis vinifera L.). Plant Mol. Biol. 72: 607–620 [DOI] [PubMed] [Google Scholar]
- McHale L.K., Haun W.J., Xu W.W., Bhaskar P.B., Anderson J.E., Hyten D.L., Gerhardt D.J., Jeddeloh J.A., Stupar R.M. (2012). Structural variants in the soybean genome localize to clusters of biotic stress-response genes. Plant Physiol. 159: 1295–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W., Pritchard B., Rios D., Chen Y., Flicek P., Cunningham F. (2010). Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 26: 2069–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medini D., Donati C., Tettelin H., Masignani V., Rappuoli R. (2005). The microbial pan-genome. Curr. Opin. Genet. Dev. 15: 589–594 [DOI] [PubMed] [Google Scholar]
- Morgante M., De Paoli E., Radovic S. (2007). Transposable elements and the plant pan-genomes. Curr. Opin. Plant Biol. 10: 149–155 [DOI] [PubMed] [Google Scholar]
- Myles S., Chia J.M., Hurwitz B., Simon C., Zhong G.Y., Buckler E., Ware D. (2010). Rapid genomic characterization of the genus vitis. PLoS ONE 5: e8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G., Bradnam K., Ning Z., Keane T., Korf I. (2009). Assessing the gene space in draft genomes. Nucleic Acids Res. 37: 289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack J.R., Perou C.M., Alizadeh A.A., Eisen M.B., Pergamenschikov A., Williams C.F., Jeffrey S.S., Botstein D., Brown P.O. (1999). Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat. Genet. 23: 41–46 [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. (2010). BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz M.C., Witkowski J., McCombie W.R. (2012). Current challenges in de novo plant genome sequencing and assembly. Genome Biol. 13: 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K., et al. (2011). Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proc. Natl. Acad. Sci. USA 108: 10249–10254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz M.H., Zerbino D.R., Vingron M., Birney E. (2012). Oases: Robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28: 1086–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer N.M., et al. (2009). Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 5: e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, Wagner R.A, Eichten S.R, Kumari S, Tiffin P, Stein J.C, Ware D, Springer N.M. (2010). Pervasive gene content variation and copy number variation in maize and its undomesticated progenitor. Genome Res. 20: 1689–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrier N., Torregrosa L., Ageorges A., Vialet S., Verriès C., Cheynier V., Romieu C. (2009). Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 149: 1028–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R., et al. (2007). A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE 2: e1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturini L., et al. (2013). De novo transcriptome characterization of Vitis vinifera cv. Corvina unveils varietal diversity. BMC Genomics 14: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang X., Paterson A.H. (2012). Genome and gene duplications and gene expression divergence: A view from plants. Ann. N. Y. Acad. Sci. 1256: 1–14 [DOI] [PubMed] [Google Scholar]
- Wu T.D., Watanabe C.K. (2005). GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21: 1859–1875 [DOI] [PubMed] [Google Scholar]
- Zerbino D.R., Birney E. (2008). Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18: 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Mao L., Wang H., Brocker C., Yin X., Vasiliou V., Fei Z., Wang X. (2012). Genome-wide identification and analysis of grape aldehyde dehydrogenase (ALDH) gene superfamily. PLoS ONE 7: e32153. [DOI] [PMC free article] [PubMed] [Google Scholar]