The Arabidopsis aba2, abi3, della pentuple, and som mutant seeds germinate even at high temperature. This work shows that ABI3, ABI5, and DELLA target to the SOM promoter and mediate high-temperature signaling to activate the expression of SOM in imbibed seeds.
Abstract
Seeds monitor the environment to germinate at the proper time, but different species respond differently to environmental conditions, particularly light and temperature. In Arabidopsis thaliana, light promotes germination but high temperature suppresses germination. We previously reported that light promotes germination by repressing SOMNUS (SOM). Here, we examined whether high temperature also regulates germination through SOM and found that high temperature activates SOM expression. Consistent with this, som mutants germinated more frequently than the wild type at high temperature. The induction of SOM mRNA at high temperature required abscisic acid (ABA) and gibberellic acid biosynthesis, and ABA-INSENSITIVE3 (ABI3), ABI5, and DELLAs positively regulated SOM expression. Chromatin immunoprecipitation assays indicated that ABI3, ABI5, and DELLAs all target the SOM promoter. At the protein level, ABI3, ABI5, and DELLAs all interact with each other, suggesting that they form a complex on the SOM promoter to activate SOM expression at high temperature. We found that high-temperature-inducible genes frequently have RY motifs and ABA-responsive elements in their promoters, some of which are targeted by ABI3, ABI5, and DELLAs in vivo. Taken together, our data indicate that ABI3, ABI5, and DELLAs mediate high-temperature signaling to activate the expression of SOM and other high-temperature-inducible genes, thereby inhibiting seed germination.
INTRODUCTION
Plant growth depends on ambient environmental conditions (e.g., light, moisture, nutrients, oxygen, and temperature); therefore, seeds need to monitor the environment to determine the proper timing of germination (Finch-Savage and Leubner-Metzger, 2006; Holdsworth et al., 2008; Weitbrecht et al., 2011). Light and temperature play critical roles in regulating seed germination, but different plant seeds show different responses to light. For example, Arabidopsis thaliana seeds germinate well in the light, whereas barley (Hordeum vulgare) seeds germinate well in the dark (Jacobsen et al., 2002; Gubler et al., 2008; Seo et al., 2009). The effective light spectra also differ between Arabidopsis and barley, with red light-promoting seed germination in Arabidopsis and blue light-inhibiting seed germination in barley (Gubler et al., 2008; Seo et al., 2009). Different plant species also have different optimal germination temperatures. For example, Arabidopsis and barley seeds germinate well at 10 to 20°C but show decreased germination at higher temperatures (Ali-Rachedi et al., 2004; Toorop et al., 2005; Leymarie et al., 2008; Mei and Song, 2010), whereas some tropical plant species and halophytes in the cold desert have higher optimal germination temperatures (Martinez et al., 1992; Khan and Gul, 2006).
Plants use several different photoreceptors to monitor different spectra of light. Phytochromes perceive red and far-red light; cryptochromes, phototropins, and zeitlupes perceive blue and UV-A light; and UV RESISTANCE LOCUS8 perceives UV-B light (Christie, 2007; Nagatani, 2010; Yu et al., 2010; Heijde and Ulm, 2012; Ito et al., 2012). Among these photoreceptors, the phytochromes are the major photoreceptors responsible for promoting seed germination in response to red, far-red, and blue light. Arabidopsis possesses five different phytochromes, three of which promote seed germination in response to different light conditions: phytochrome A (phyA) mediates the very-low-fluence response and the far-red high-irradiance response, phyB mediates the low-fluence response, and phyE mediates low-fluence response and far-red high-irradiance response to promote seed germination (Shinomura et al., 1994, 1996; Hennig et al., 2002; Oh et al., 2004). Immediately downstream of phytochromes, PHYTOCHROME-INTERACTING FACTOR1 (PIF1; also known as PHYTOCHROME-INTERACTING FACTOR3-LIKE5 [PIL5]) a basic helix-loop-helix transcription factor, inhibits seed germination in the absence of active phytochromes (Oh et al., 2004).
A previous genome-wide binding site analysis coupled with microarray analysis indicated that PIL5 binds to the promoters of 166 target genes and regulates their expression levels either positively or negatively in imbibed Arabidopsis seeds (Oh et al., 2009). The identified direct target genes included hormone signaling genes, such as GA-INSENSITIVE (GAI), REPRESSOR OF GA1-3 (RGA), HONSU (HON), ABSCISIC ACID-INSENSITIVE3 (ABI3), ABI5, AUXIN RESPONSE FACTOR18, CYTOKININ RESPONSE FACTOR1, and JASMONATE-ZIM-DOMAIN PROTEIN1 (JAZ1); cell wall–modifying enzyme-encoding genes, including EXPANSIN8 (EXP8), EXP10, and XYLOGLUCAN ENDOTRANSGLYCOSYLASE/HYDROLASE28; and other known germination-regulating genes, including SOMNUS (SOM), BOTRYTIS SUSCEPTIBLE1 INTERACTOR, INDETERMINATE DOMAIN1/ENHYDROUS, ARABIDOPSIS NAC DOMAIN CONTAINING PROTEIN2/ARABIDOPSIS THALIANA ACTIVATION FACTOR1, and PLANT U-BOX19 (Jensen et al., 2008; Kim et al., 2008, 2013; Oh et al., 2009; Bergler and Hoth, 2011; Feurtado et al., 2011; Liu et al., 2011; Park et al., 2011, 2013). Changes in the expression levels of these direct target genes can alter the expression levels of many indirect target genes of PIL5, including gibberellic acid (GA) and abscisic acid (ABA) biosynthetic genes, resulting in decreased GA levels and increased ABA levels in the absence of activated phytochromes (Oh et al., 2007; Kim et al., 2008). Thus, phytochromes appear to promote seed germination by coordinating hormone signals and loosening cell wall properties through PIL5.
The ability to monitor temperature is ecologically important for the seasonal timing of seed germination. The seeds of many winter annual plants (e.g., Arabidopsis) germinate in the cool temperatures of autumn, but not in the hot temperatures of summer (Baskin and Baskin, 1983), indicating that plant seeds have thermoreceptors that sense and transmit temperature signals for seed germination. However, we do not yet fully understand the mechanism through which plant species sense temperature to regulate seed germination.
Downstream of the thermoreceptors, the hormones ABA and GA, which critically regulate light-dependent seed germination, play key roles in mediating the high-temperature-induced signals that regulate seed germination (Yoshioka et al., 1998; Gonai et al., 2004; Tamura et al., 2006; Argyris et al., 2008; Toh et al., 2008, 2012; Chiu et al., 2012). High temperature increases ABA levels in imbibed Arabidopsis seeds both by activating the expression of ABA biosynthetic genes, such as ABA DEFICIENT1 (ABA1), NINE-CIS-EPOXYCAROTENOID DIOXYGENASE2 (NCED2), NCED5, and NCED9, and by repressing the expression of ABA catabolic genes, such as CYP707A1, CYP707A2, and CYP707A3. High temperature also decreases GA levels by repressing the expression of GA biosynthetic genes, such as GIBBERELLIN 20-OXIDASE1 (GA20ox1), GA20ox2, GA20ox3, GIBBERELLIN 3-OXIDASE1 (GA3ox1), and GA3ox2 (Toh et al., 2008). Consistent with the key roles played by ABA and GA, ABA-deficient mutants (e.g., aba1, aba2, and nced multiple mutants [nced2 nced5 nced9]), ABA-insensitive mutants (e.g., abi1 and abi3), and GA-negative signaling gene mutants (e.g., spindly and rga-like2 [rgl2]) germinate with higher frequency at high temperature (Tamura et al., 2006; Toh et al., 2008). Other plant hormones, including ethylene, cytokinin, and strigolactone, also promote seed germination at high temperature (Khan and Prusinski, 1989; Matilla, 2000; Kozarewa et al., 2006; Toh et al., 2012). In addition, a number of genetic components (e.g., TRANSPARENT TESTA7, pea [Pisum sativum] G-protein α- and β-subunits, chickpea [Cicer arietinum] APETALA2 [AP2], and wheat [Triticum aestivum] chloroplastic small heat shock proteins) and various chemicals (e.g., CO2, 2-4-[carboxyphenyl]-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide, chlorogenic acid, bovine hemoglobin, isoprene, and glycinebetaine) affect seed germination at high temperature (Saini et al., 1986; Tamura et al., 2006; Misra et al., 2007; Shukla et al., 2009; Hossain et al., 2010; Li et al., 2011; Chauhan et al., 2012). However, the relationships among these components have not yet been clarified.
Of the PIL5 target genes, SOM encodes a CCCH-type zinc finger protein known to inhibit light-dependent seed germination by increasing the expression of ABA biosynthetic genes and decreasing the expression of GA biosynthetic genes (Kim et al., 2008). Expression analysis indicated that PIL5 and ABI3 collaboratively regulate SOM mRNA levels, suggesting that SOM also functions downstream of ABI3 (Park et al., 2011). Since ABI3 negatively regulates seed germination at high temperature (Tamura et al., 2006), we herein investigated whether SOM also inhibits seed germination at high temperature.
We found that high temperature activated SOM mRNA expression and increased SOM protein levels. Consistent with these expression patterns, som mutants germinated at higher frequencies than wild-type seeds at high temperature, whereas SOM-overexpressing lines germinated at lower frequencies than the wild type at high temperature. The induction of SOM mRNA by high temperature required ABA and GA biosynthesis, as well as signaling components including ABI3, ABI5, and DELLAs. Chromatin immunoprecipitation (ChIP) assays further showed that ABI3, ABI5, and DELLAs are targeted to similar regions of the SOM promoter. In addition, in vitro and in vivo protein–protein interaction assays indicated that ABI3, ABI5, and DELLAs interact with each other. Together, our findings indicate that ABI3, ABI5, and DELLAs act together to activate SOM expression at high temperature in Arabidopsis.
RESULTS
High Temperature Induces SOM Expression to Inhibit Seed Germination
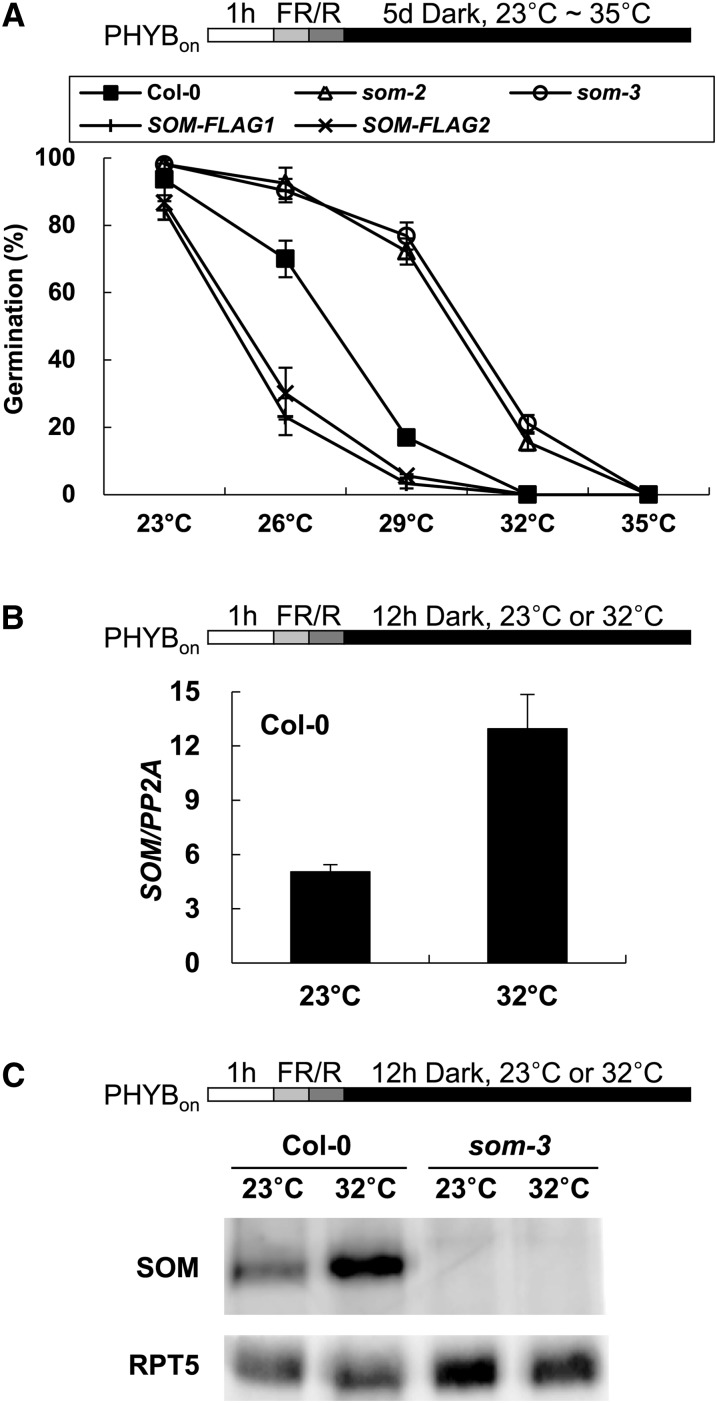
We previously showed that light promotes seed germination partly by inhibiting the PIL5-mediated expression of SOM, which encodes a CCCH-type zinc finger protein that activates ABA biosynthesis and represses GA biosynthesis in imbibed seeds (Kim et al., 2008). Since high temperature also inhibits seed germination by activating ABA biosynthesis and repressing GA biosynthesis (Toh et al., 2008; Chiu et al., 2012), we herein examined whether SOM inhibits seed germination at high temperature. We first determined the germination frequencies of som mutants (som-2 and som-3) and cauliflower mosaic virus 35S promoter–driven FLAG-tagged SOM-overexpressing lines (SOM-FLAG1 and SOM-FLAG2; see Supplemental Figure 1 online) at various temperatures. Seeds were sown on plates and irradiated with a far-red light pulse (5 min) followed by a red light pulse (5 min) to activate phyB (phyBon). The seeds were then incubated in the dark for 5 d at various temperatures. The wild type (Columbia-0 [Col-0]), the two som mutants, and the two SOM-overexpressing lines showed >80% germination at 23°C (Figure 1A), indicating that the red light pulse was sufficient to stimulate germination at 23°C. When seeds were incubated at higher temperatures, however, the germination frequencies decreased. Col-0 germinated ∼90% at 23°C, 70% at 26°C, 15% at 29°C, and 0% at 32°C and 35°C. The two som mutants germinated at higher frequencies than the wild type at higher temperatures (∼20% at 32°C). By contrast, the two SOM-overexpressing lines germinated at lower frequencies than the wild type at higher temperatures (only ∼25% at 26°C). These results indicate that SOM inhibits seed germination not only during phy-mediated seed germination but also under high temperatures.
Figure 1.
High Temperature Induces the Expression of SOM to Inhibit Seed Germination.
(A) Germination of the wild type (Col-0), som mutants, and SOM-overexpressing seeds at various temperatures. The top diagram indicates the light treatments and seed incubation conditions. 1 h, 1 h of imbibition during seed sterilization and sowing on plates; FR/R, irradiation with 5 min of far-red light followed by red light. After irradiation, plates were incubated in the dark at 23, 26, 29, 32, or 35°C for 5 d. The graph shows the germination frequencies of three independent seed batches incubated at the indicated temperatures (sd, n = 3). SOM-FLAG1 and -FLAG2 refer to 35S:SOM-FLAG transgenic lines.
(B) Induction of SOM mRNA expression in Col-0 by high temperature. The top diagram indicates the light treatments and seed incubation conditions. Dark-incubated seeds were sampled for total RNA extraction, and SOM mRNA levels were quantified by quantitative RT-PCR and normalized with respect to the PP2A mRNA level. The graph shows the relative SOM mRNA levels (SOM/PP2A) in seeds incubated at 23 or 32°C. Error bars indicate the sd of three biological replicates (n = 3).
(C) Induction of SOM protein levels in Col-0 by high temperature. The top diagram indicates the light treatments and seed incubation conditions. Dark-incubated seeds were sampled for total protein extraction, and SOM protein levels were detected by immunoblotting assays using an anti-SOM antibody. RPT5 protein levels (loading control) were detected using an anti-RPT5 antibody. As a negative control, som-3 mutant seeds were analyzed in parallel.
Light regulates SOM expression during seed germination; therefore, we examined if high temperature also regulates SOM expression. We first determined the mRNA levels of SOM in Col-0 seeds that were irradiated with a far-red light pulse followed by a red light pulse and incubated in the dark for 12 h at either 23 or 32°C. Our results revealed that the SOM mRNA level was more than twofold higher in seeds incubated at 32°C compared with those incubated at 23°C (Figure 1B). To examine if this increase in SOM mRNA corresponded to a higher level of SOM protein, we generated an antibody against the SOM protein and examined Col-0 seeds incubated in the dark for 12 h at either 23 or 32°C. Consistent with the increased SOM mRNA expression levels seen in seeds at high temperature, SOM protein levels were higher in seeds incubated at 32°C compared with those incubated at 23°C (Figure 1C). These results indicate that high temperature induces SOM expression, which in turn inhibits seed germination. However, the germination of som mutants was inhibited at 35°C, further suggesting that high temperature inhibits not only through SOM but also through other genetic components.
High Temperature Induces SOM Expression through ABA and GA Signaling
Since high temperature increases ABA biosynthesis and decreases GA biosynthesis (Toh et al., 2008; Chiu et al., 2012), we examined whether ABA and GA biosynthesis are required for the high-temperature-mediated induction of SOM mRNA expression. First, we determined SOM mRNA levels in ABA-deficient mutant (aba2) and GA-deficient mutant (ga1) seeds incubated at 23 or 32°C. We found that SOM mRNA expression was lower in the aba2 mutant and higher in the ga1 mutant compared with the wild type, irrespective of temperature (Figure 2A; see Supplemental Figure 2 online). Consistent with the expression patterns of SOM mRNA, the aba2 mutant germinated at higher frequencies than the wild type at high temperatures, whereas the ga1 mutant did not germinate (Figure 2B). This indicates that high temperature requires both ABA and GA biosynthesis to induce SOM mRNA expression. Our observation that SOM mRNA levels decreased in the aba2 mutant and increased in the ga1 mutant further suggests that SOM mRNA expression is activated by ABA and repressed by GA. Consistent with this, ABA treatment increased SOM mRNA levels in the aba2 mutant, and GA treatment decreased SOM mRNA levels in the ga1 mutant (see Supplemental Figure 2 online).
Figure 2.
High Temperature Induces the Expression of SOM Through ABA and GA Signaling.
The top diagrams indicate the light treatments and seed incubation conditions.
(A) Relative SOM mRNA levels in Col-0, aba2 mutant, and ga1 mutant seeds incubated at 23 or 32°C. The relative SOM mRNA level (SOM/PP2A) observed in Col-0 seeds incubated at 23°C was set to 1. Error bars indicate the sd of three biological replicates (n = 3). Asterisks indicate statistical differences between wild-type and mutant values for a given temperature (P < 0.01, Student’s t test). FR/R, irradiation with 5 min of far-red light followed by red light.
(B) Germination of Col-0, aba2 mutant, and ga1 mutant seeds at various temperatures. The graph shows the germination frequencies of three independent seed batches incubated at the indicated temperatures (sd, n = 3).
(C) Relative SOM mRNA levels in Col-0, abi3-sk11 mutant, and dellaP mutant seeds incubated at 23 or 32°C (sd, n = 3). The inset indicates relative SOM mRNA levels in the second abi3 allele (abi3-sk22).
(D) Germination of Col-0, abi3-sk11 mutant, and dellaP mutant seeds at various temperatures (sd, n = 3). The inset indicates germination of Col-0 and abi3-sk22 mutant seeds.
(E) Relative SOM mRNA levels in Col-0 and 35S:ABI5-FLAG transgenic seeds (ABI5-FLAG1 and ABI5-FLAG2) incubated at 23 or 32°C (sd, n = 3). Asterisks indicate statistical differences between wild-type and transgenic line values for a given temperature (P < 0.01, Student’s t test).
(F) Germination of Col-0 and two 35S:ABI5-FLAG transgenic lines at various temperatures (sd, n = 3).
We further investigated which ABA- and GA-signaling components were responsible for the induction of SOM mRNA at high temperature. Since previous studies showed that ABI3 and RGL2 (which are ABA-positive and GA-negative signaling components, respectively) inhibit seed germination at high temperature (Tamura et al., 2006; Toh et al., 2008), we herein examined whether the ABI3 and DELLA proteins could activate SOM mRNA expression at high temperature. For this experiment, we used a DELLA pentuple mutant (rga-28 gai-t6 rgl1 rgl2 rgl3-3; dellaP), which lacks all of the DELLA proteins, and two abi3 mutants (abi3-sk11 and abi3-sk22) caused by T-DNA insertion at the first exon (in the B2 and B1 domain, respectively) (Kotak et al., 2007; Park et al., 2011, 2013). Both abi3 and dellaP expressed lower levels of SOM mRNA than the wild type at 23°C (Figure 2C). Notably, SOM mRNA expression was not induced by incubation at high temperature in these mutants, indicating that SOM mRNA induction at high temperature requires ABI3 and DELLA. Consistent with the reduced expression of SOM mRNA in these mutants, both abi3 and dellaP germinated well even at 32°C (Figure 2D). Taken together, our results suggest that ABI3 and DELLA activate SOM expression in response to increased ABA levels and decreased GA levels in seeds incubated at high temperature. The dependence of SOM induction on both ABI3 and DELLAs further suggests that these proteins require each other to induce SOM expression at high temperatures.
A subset of basic Leu zipper proteins (the group A bZIPs) including ABI5 mediates ABA signaling under various conditions (Jakoby et al., 2002; Corrêa et al., 2008; Fujita et al., 2011). Since it was difficult to obtain a loss-of-function mutant lacking all 13 group A bZIPs, we generated 35S-driven FLAG-tagged ABI5-overexpressing lines (ABI5-FLAG1 and ABI5-FLAG2; see Supplemental Figure 1 online) and examined their SOM mRNA expression levels in seeds incubated at 23 or 32°C. In ABI5-overexpressing lines, the SOM mRNA levels were higher than those in the wild type at both 23 and 32°C (Figure 2E). Consistent with the increased expression of SOM mRNA, ABI5-overexpressing lines were hypersensitive to high temperature in the context of seed germination (Figure 2F). Taken together, our results indicate that ABI3 and ABI5 mediate high-temperature-associated ABA signaling to activate SOM expression during seed germination.
ABI3, ABI5, and DELLAs Target the SOM Promoter in Vivo
The SOM promoter has two RY motifs (CATGCA; Figure 3A), which are targeted by ABI3 to activate SOM expression during seed development (Park et al., 2011). The SOM promoter also possesses two ABA-responsive elements (ABREs; Figure 3A), which are known binding sites for group A bZIPs (Choi et al., 2000; Uno et al., 2000; Bensmihen et al., 2002; Carles et al., 2002; Narusaka et al., 2003; Nakashima et al., 2006; Kim et al., 2011; Wang et al., 2013). The presence of these elements prompted us to examine whether ABI3 and ABI5 target the SOM promoter at high temperature to activate SOM expression. Since DELLA proteins also target the promoters of a few GA-responsive genes, presumably by interacting with other DNA binding transcription factors (Zentella et al., 2007; Park et al., 2013), we also examined whether DELLAs target the SOM promoter.
Figure 3.
ABI3, ABI5, and RGA Target the SOM Promoter In Vivo.
(A) Schematic diagram representing RY motifs (CATGCA) and ABREs ([C/T]ACGTG) in the SOM promoter. Closed vertical bars, RY motifs; open vertical bars, ABREs; arrow, translation start site; closed horizontal bars (F1 to F5), genomic DNA fragments used for the ChIP assays.
(B) ChIP assays were performed with anti-ABI3 antibody (a-ABI3) in Col-0 seeds incubated at 23 or 32°C. The top diagram indicates the light treatments and seed incubation conditions. The coprecipitated level of each DNA fragment was quantified by real-time PCR and normalized with respect to the input DNA. The relative coprecipitated levels (immunoprecipitated DNA/input DNA) of each F1 were set to 1, and the relative enrichments of the other fragments compared with F1 (/F1) are shown in the graph. Em6 indicates a genomic DNA fragment of Em6 promoter (used as a specific binding control). Error bars indicate the sd of four biological replicates (n = 4). FR/R, irradiation with 5 min of far-red light followed by red light.
(C) ChIP assays were performed with anti-ABI5 antibody (a-ABI5) in Col-0 seeds incubated at 23 or 32°C (sd, n = 4). The inset indicates ChIP assays in Col-0 seeds with resin alone (-Ab).
(D) ChIP assays were performed with anti-RGA antibody (a-RGA) in sly1 mutant seeds incubated at 23 or 32°C (sd, n = 4). SCL3 indicates a genomic DNA fragment of SCL3 promoter (used as a specific binding control). The inset indicates ChIP assays in sly1 mutant seeds with resin alone (-Ab).
We performed ChIP assays using antibodies against ABI3, ABI5, and RGA in wild-type and sly1 mutant seeds (Figure 3; see Supplemental Figure 3 online) or FLAG and MYC antibodies in 35S-driven ABI3-FLAG, ABI5-FLAG1, and GAI-MYC transgenic seeds (see Supplemental Figures 1 and 4 online). Seeds were irradiated with a far-red light pulse followed by a red light pulse and incubated in the dark for 6 h at either 23 or 32°C. Our results revealed that ChIP with ABI3 enriched fragments F3 to F5 of the SOM promoter more than fourfold (Figure 3B); by contrast, control ChIP with resin alone did not enrich these fragments (Figure 3C, inset). The enrichment of F4, which does not contain a RY motif, is likely due to the relatively short distance from the F4 fragment to the two RY motifs (608 and 629 bp). Unlike fragments F3 and F5, which contain two ABI3 binding RY motifs (Park et al., 2011), fragments F1 and F2, which contain another RY motif between them, were not enriched by ABI3. The ABI3-mediated enrichment of the SOM promoter did not significantly differ between seeds incubated at 23 and 32°C, indicating that high temperature did not affect the occupancy of ABI3 on the SOM promoter under these conditions. Similar to ABI3, ChIP with ABI5 also strongly enriched fragments F3, F4, and F5, but not fragments F1 and F2 (Figure 3C). As seen for the RY motifs, not all of the ABREs were enriched by ChIP with ABI5, as nonenriched fragment F2 also contains an ABRE (Figure 3A). High temperature did not affect the enrichment of the SOM promoter by ABI5 ChIP (Figure 3C). RGA also enriched fragments F3 to F5 but not fragments F1 and F2 of the SOM promoter in the sly1 mutant seeds that accumulate DELLA proteins (Figure 3D). High temperature did not affect the enrichment of the SOM promoter by RGA (Figure 3D). Similar to ChIP for endogenous ABI3, ABI5, and RGA, ChIP for 35S-driven ABI3-FLAG, ABI5-FLAG, and GAI-MYC also enriched SOM promoter fragments at both temperatures (see Supplemental Figure 4 online). Taken together, these results indicate that ABI3, ABI5, and DELLAs all target the same regions of the SOM promoter to activate SOM mRNA expression. Since the targeting was not affected by high temperature, our results suggest that this temperature change does not alter the intrinsic DNA binding abilities of ABI3, ABI5, and DELLAs to the SOM promoter at the resolution of our experimental setup and further suggest that high temperature activates SOM expression either by increasing protein levels or activities of these factors.
ABI3, ABI5, and DELLA Proteins Bind to Each Other
Since ABI3 and ABI5 (but not DELLAs) are DNA binding proteins, and ABI3, ABI5, and DELLAs showed the same enrichment pattern for the SOM promoter, we speculated that DELLA proteins may target the SOM promoter by physically interacting with ABI3 and/or ABI5 proteins. To investigate this possibility, we first performed in vitro coprecipitation assays using recombinant glutathione S-transferase–fused GAI (GST-GAI) and maltose binding protein–fused ABI3 (MBP-ABI3) proteins. We found that GST-GAI was coprecipitated by MBP-ABI3 but not by MBP alone (Figure 4A), indicating that GAI and ABI3 interact with each other in vitro. Next, we performed in vivo coprecipitation assays using 35S-driven ABI3-FLAG and GFP-FLAG (for green fluorescent protein) transgenic seedlings to examine if ABI3-FLAG can coprecipitate endogenous RGA in vivo. We found that ABI3-FLAG coprecipitated RGA more strongly than the GFP-FLAG control (Figure 4B). To further investigate if GAI interacts with ABI3 in vivo, we transiently expressed 35S-driven MYC-tagged gai-1 (gaiD-MYC) or GFP (GFP-MYC) in 35S-driven ABI3-FLAG transgenic seedlings using an agroinfiltration method (Li et al., 2009) and performed in vivo coprecipitation assays. Consistent with our in vitro and in vivo data, gaiD-MYC also coprecipitated ABI3-FLAG, whereas GFP-MYC did not precipitate ABI3-FLAG (see Supplemental Figure 5 online). Taken together, these results indicate that DELLAs physically interact with ABI3 at the protein level.
Figure 4.
ABI3 and ABI5 Bind to DELLAs at Protein Level.
(A) In vitro coprecipitation assays between ABI3 and GAI. Amylose resins bound with MBP or MBP-fused ABI3 (MBP-ABI3) proteins were incubated with GST-fused GAI (GST-GAI) proteins. The input (IN) represents 2% of the tested GST-GAI proteins. For immunoblotting assays, anti-GST (top) or anti-MBP (bottom) antibody was used. IP, immunoprecipitation.
(B) In vivo coprecipitation assays between ABI3 and RGA. Five-day-old, light-grown 35:GFP-FLAG or 35S:ABI3-FLAG seedlings were transferred to the MS-agar plates containing 100 μM paclobutrazol and further grown for 3 d. Immunoprecipitation was performed with an anti-FLAG antibody-conjugated resin. The input (IN) represents 1% of the tested seedling extracts. For immunoblotting assays, anti-RGA (top) or anti-FLAG (bottom) antibody was used.
(C) In vitro coprecipitation assays between ABI5 and GAI. Amylose resins bound with MBP or MBP-fused ABI5 (MBP-ABI5) proteins were incubated with GST-GAI proteins. IN represents 2% of the tested GST-GAI proteins. For immunoblotting assays, anti-GST (top) or anti-MBP (bottom) antibody was used.
(D) In vivo coprecipitation assays between ABI5 and RGA. Five-day-old, light-grown 35S:GFP-FLAG or 35S:ABI5-FLAG1 seedlings were transferred to the MS-agar plates containing 100 μM paclobutrazol and further grown for 3 d. Immunoprecipitation was performed with an anti-FLAG antibody-conjugated resin. IN represents 1% of the tested seedling extracts. For immunoblotting assays, anti-RGA (top) or anti-FLAG (bottom) antibody was used.
Next, we examined whether DELLAs also interact with ABI5. We first performed in vitro coprecipitation assays using recombinant GST-GAI and MBP-fused ABI5 (MBP-ABI5). Our results revealed that GST-GAI was coprecipitated by MBP-ABI5 but not by MBP alone (Figure 4C), indicating that GAI and ABI5 interact with each other in vitro. We also performed in vivo coprecipitation assays using 35S-driven ABI5-FLAG1 and GFP-FLAG transgenic seedlings to examine if ABI5-FLAG can coprecipitate endogenous RGA in vivo. We found that ABI5-FLAG coprecipitated endogenous RGA more strongly than GFP-FLAG (Figure 4D). We also transiently expressed the 35S-driven gaiD-MYC protein in 35S-driven ABI5-FLAG1 and GFP-FLAG transgenic seedlings using the agroinfiltration method and determined whether ABI5-FLAG could coprecipitate gaiD-MYC. Consistent with the results of our in vitro and in vivo coprecipitation assay, ABI5-FLAG coprecipitated gaiD-MYC more strongly than GFP-FLAG control (see Supplemental Figure 5 online). Together, our findings indicate that DELLAs physically interact with ABI5 at the protein level.
Since ABI3 is known to interact with ABI5 and other group A bZIP proteins (Hobo et al., 1999; Nakamura et al., 2001; Brocard-Gifford et al., 2003; Finkelstein et al., 2005; Marella et al., 2006; Park et al., 2011; Tezuka et al., 2013), our results together with the previous findings suggest that ABI3, ABI5, and DELLAs interact with each other on the SOM promoter to activate SOM mRNA expression at high temperature.
A Subset of High-Temperature-Inducible Genes Requires Both ABI3 and DELLAs
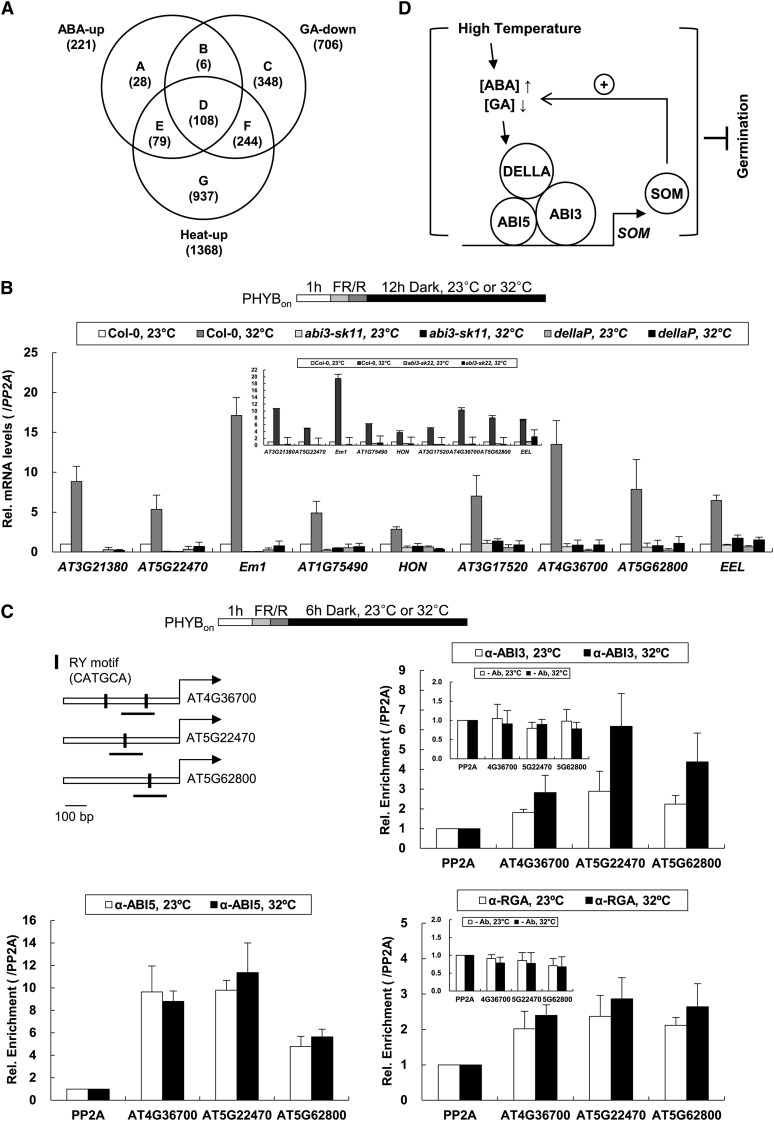
Since SOM requires both ABI3 and DELLAs for induction at high temperature, we examined whether other high-temperature-inducible genes might also require both ABI3 and DELLAs. To first determine whether ABA and GA regulate any other high-temperature-inducible genes in imbibed seeds, we reanalyzed previous microarray data representing high-temperature-inducible genes in imbibed seeds (wild-type seeds imbibed for 24 h at 32°C under continuous white light) (Chiu et al., 2012), ABA upregulated genes in imbibed seeds (wild-type seeds imbibed in 30 μM ABA for 24 h at 22°C under continuous white light) (Goda et al., 2008), and GA downregulated genes in imbibed seeds (ga1 mutant seeds treated with 5 μM GA4 for 9 h under continuous white light after cold imbibition for 48 h) (Goda et al., 2008). From these microarray data, we extracted 1368 genes that were upregulated by high temperature ([heat-up], log2 fold change [FC] > 0.85, P < 0.05), 221 genes that were upregulated by ABA ([ABA-up], log2 FC > 0.85, P < 0.05), and 706 genes that were downregulated by GA ([GA-down], log2 FC < −0.85, P < 0.05) (Figure 5A; see Supplemental Data Set 1 online). Our comparison revealed that 108 (the subset D) of the 1368 high-temperature-inducible genes were both upregulated by ABA and downregulated by GA (Figure 5A; see Supplemental Data Set 1 online). The subset D genes (which included SOM) were significantly overrepresented among the high-temperature-inducible genes (hypergeometric test, P = 2.61 × 10−123). To experimentally determine the dependency of the subset D genes on ABI3 and DELLAs, we randomly picked nine of the 108 genes and determined their expression levels in abi3 and dellaP mutant seeds incubated in the dark for 12 h at either 23 or 32°C. All nine genes were induced by high temperature in wild-type seeds, but they were expressed at low levels irrespective of temperature in the abi3 and dellaP mutants (Figure 5B). Taken together, these results indicate that a subset of high-temperature-inducible genes requires both ABI3 and DELLAs for their increased expression at 32°C.
Figure 5.
ABI3, ABI5, and DELLAs Mediate ABA and GA Signaling to Activate the Expression of a Subset of High-Temperature-Inducible Genes.
(A) A Venn diagram showing numbers of ABA upregulated genes ([ABA-up], log2 FC > 0.85, P value < 0.05), GA downregulated genes ([GA-down], log2 FC < −0.85, P value < 0.05), and high-temperature-inducible genes ([heat-up], log2 FC > 0.85, P value < 0.05), as determined from previously published microarray data (Goda et al., 2008; Chiu et al., 2012). Subsets are indicated by uppercase letters (A to G), and the numbers of genes are given in parentheses.
(B) Relative mRNA levels of nine representative genes from subset D in Col-0, abi3-sk11 mutant, and dellaP mutant seeds incubated at 23 or 32°C. The top diagram indicates the light treatments and seed incubation conditions. Relative mRNA levels of each gene (/PP2A) in Col-0 seeds incubated at 23°C were set to 1. Error bars indicate the sd of three biological replicates (n = 3). The inset indicates relative mRNA levels in the second abi3 allele (abi3-sk22). FR/R, irradiation with 5 min of far-red light followed by red light.
(C) ChIP assays were performed with anti-ABI3 (α-ABI3) and anti-ABI5 (α-ABI5) antibodies in Col-0 and anti-RGA antibody (α-RGA) in sly1 mutant seeds incubated at 23 or 32°C. The top diagram indicates the light treatments and seed incubation conditions. The schematic diagrams represent the locations of the RY motifs (closed vertical bars) in the promoters of AT4G36700, AT5G22470, and AT5G62800. The arrow starts from the translation start site, the closed horizontal bars indicate genomic DNA fragments used for the ChIP assays, and PP2A indicates an intragenic DNA fragment of PP2A (lacking the RY motif, used as a nonspecific binding control). The relative coprecipitated levels (immunoprecipitated DNA/input DNA) of each PP2A were set to 1, and the relative enrichments of the other fragments compared with PP2A (/PP2A) are shown in the graph. Error bars indicate the sd of four biological replicates (n = 4). Insets indicate ChIP assays with resin alone (-Ab).
(D) A proposed model. High temperature activates SOM expression via increased ABA biosynthesis and decreased GA biosynthesis. ABI3, ABI5, and DELLA proteins mediate ABA and GA signaling to induce SOM expression. ABI3, ABI5, and DELLA proteins target the SOM promoter in vivo and bind to each other at the protein level, suggesting that they form a complex on the SOM promoter and coactivate SOM expression at high temperature. This increased SOM should further increase ABA biosynthesis and decrease GA biosynthesis, forming a positive feedback loop. Thus, seed germination is strongly inhibited at high temperature.
Since the SOM promoter contains RY motifs and ABREs that appear to be important for its induction by high temperature, we examined whether the promoters of the subset D genes were enriched for RY motifs and/or ABREs. When the sequence of 1.5 kb upstream from the start codon was defined as the promoter, the subset D genes had high percentages of promoters possessing RY motifs (76%), ABREs (64%), or both (47%) (Table 1). This was significantly higher than the percentage of all Arabidopsis gene promoters harboring RY motifs (40%), ABREs (44%), or both (19%), respectively (hypergeometric test, P < 0.0001) (Table 1). Consistent with the enrichment of RY and ABRE motifs, the heat map analysis and gene set enrichment analysis (GSEA) indicate that the subset D genes are significantly repressed in abi3, abi4, and abi5 mutant seeds (false discovery rate < 0.001; see Supplemental Figure 6 online) (Nakabayashi et al., 2005; Subramanian et al., 2005; Nakashima et al., 2009). Among the 1368 high-temperature-inducible genes, the 937 genes (the subset G) that were neither upregulated by ABA nor downregulated by GA had lower percentages of promoters containing RY motifs, ABREs, or both, compared with the subset D genes (Table 1). These results indicate that the subset D gene promoters are highly enriched for RY motifs and/or ABREs.
Table 1. Promoter Analysis Showing Distribution of RY Motif and ABREs.
| Motifs | Genome: No. of Genes | Subset D |
Subset G |
||
|---|---|---|---|---|---|
| No. of Genes | P Valuea | No. of Genes | P Value | ||
| Totalb | 33,323 | 108 | 937 | ||
| RYc | 13,304 (40%) | 82 (76%) | 2.79E-14 | 486 (52%) | 4.46E-14 |
| ABREsd | 14,748 (44%) | 69 (64%) | 3.06E-05 | 391 (42%) | 0.95 |
| RY/ABREse | 6,173 (19%) | 51 (47%) | 9.80E-12 | 224 (24%) | 1.81E-05 |
Hypergeometric (Fisher’s exact) test.
The number of total genes in each set.
The number of genes that have more than one motif in promoters (within 1.5 kb).
The number of genes that have both of two motifs in promoters (within 1.5 kb).
We next determined if ABI3, ABI5, and DELLAs also target the promoters of the subset D genes. From the nine genes that were experimentally shown to require both ABI3 and DELLAs for their expression (Figure 5B), we randomly picked three genes and performed ChIP assays using antibodies against endogenous ABI3, ABI5, and RGA in wild-type and sly1 mutant seeds (Figure 5C) or FLAG and MYC antibodies in 35S-driven ABI3-FLAG, ABI5-FLAG1, and GAI-MYC transgenic seeds (see Supplemental Figure 7 online). As seen for the SOM promoter, ABI3, ABI5, and RGA all enriched the promoter fragments of the three subset D genes (Figure 5C); by contrast, control ChIP with resin alone did not enrich these fragments (Figure 5C, inset). Similarly, ABI3-FLAG, ABI5-FLAG, and GAI-MYC also enriched the promoter fragments of the three subset D genes in transgenic seeds (see Supplemental Figure 7 online). Together, these results indicate that ABI3, ABI5, and DELLAs target the subset D gene promoters to activate their expression at high temperature.
DISCUSSION
Here, we show that ABI3, ABI5, and DELLAs form a complex on promoters of a subset of high-temperature-inducible genes (which include SOM), thereby activating their expression in response to high temperature (Figure 5D).
ABI3 Directly Activates SOM Expression at High Temperature
Our analyses indicated that SOM expression is regulated by ABI3 in combination with various factors under different developmental and environmental conditions. At high temperature, we found that ABI3 and ABI5 directly target the SOM promoter and activate its expression. Since ABI3 and ABI5 are positive regulators of ABA signaling, the induction of SOM by these factors is consistent with the lower expression of SOM mRNA in the aba2 mutant irrespective of temperature. These findings are also consistent with a previous report showing that ABA biosynthesis and signaling are increased at high temperature (Toh et al., 2008). Thus, light and high-temperature signals are integrated at the SOM promoter through ABI3 in combination with either PIL5 (dark) or ABI5 (high temperature) to regulate SOM expression in imbibed seeds. During seed maturation, ABI3 is known to be important for regulating SOM expression (Park et al., 2011). Since seed maturation is characterized by high ABA signaling, it is expected that SOM expression is also regulated by ABI3 in combination with ABI5 or other group A bZIPs during seed maturation. Consistent with this, SOM mRNA levels are twofold lower in abi3 and abi5 mutant dry seeds than in wild-type dry seeds (see Supplemental Figure 6C online) (Nakabayashi et al., 2005; Nakashima et al., 2009).
The combined action of ABI3 with other transcription factors might stem from promoter characteristics (i.e., the presence of RY motifs and multiple other sequence elements). The SOM promoter possesses two ABI3 binding RY motifs, located on fragments F3 and F5 (Park et al., 2011), as well as two group A bZIP binding ABREs ([C/T]ACGTG; Figure 3A). One of these ABREs is located near the fragment F3 (TACGTG; 25 bp from the downstream end of F3) (Figure 3A). In addition, the SOM promoter possesses multiple E-box motifs (CANNTG), including the PIF binding E-box (CACATG) on the fragment F5 (Kim et al., 2008; Zhang et al., 2013). Since E-box motifs are also binding sites for group A bZIPs (Kim et al., 1997), the PIF binding E-box could serve as a binding site for both PIL5 and group A bZIPs. The enrichment of RY motifs and/or ABREs was not restricted to the SOM promoter, as these elements were also enriched in the promoters of the subset D genes (Table 1). A recent report showed that promoters of ABI3 regulons in developing seeds are characterized by a strong coupling between ABI3 binding RY motifs and G-box like elements (ACGTG[T/G]C), which may serve as group A bZIP binding sites (Mönke et al., 2012). One such gene, AT2G31980, was shown to be regulated not only by ABI3 but also by ELONGATED HYPOCOTYL5, PIL5, AGAMOUS-LIKE9, and AP2 (Mönke et al., 2012), further suggesting that ABI3 functions with other transcription factors on promoters that harbor RY motifs and other coupling elements.
The combined action of ABI3 with other transcription factors could be further facilitated by protein–protein interactions. ABI3 and PIL5 were previously shown to be targeted to fragments F3 and F5 (Kim et al., 2008; Park et al., 2011), and this study further showed that ABI5 also target fragments F3 and F5. The targeting of these factors to both fragments could be due to the presence of their recognition sequences on both fragments. Alternatively, since ABI3 is known to interact with both group A bZIPs, including ABI5 and PIL5 (Hobo et al., 1999; Nakamura et al., 2001; Brocard-Gifford et al., 2003; Finkelstein et al., 2005; Park et al., 2011; Tezuka et al., 2013), their enrichment could be facilitated by the formation of a protein complex between ABI3 and other transcription factors on the SOM promoter. Such protein–protein interactions could strengthen an otherwise weak targeting of transcription factors to a promoter.
DELLAs Are Targeted to the Promoters of Subset D Genes to Activate Their Expressions at High Temperature
Our analyses indicated that DELLA proteins are also required to activate SOM expression at high temperature, as shown by the increased expression of SOM mRNA in the ga1 mutant irrespective of temperature and the decreased expression of SOM mRNA in the dellaP mutant irrespective of temperature in imbibed seeds. ChIP assays further showed that RGA and GAI target the SOM promoter to activate SOM expression at high temperature. RGA and GAI also targeted the subset D gene promoters. Since DELLAs are not thought to be direct DNA binding proteins, the physical interaction among DELLAs, ABI3, and ABI5 may facilitate the targeting of DELLAs to the promoters of SOM and other subset D genes. Consistent with this possibility, RGA and GAI also enriched fragments F3 and F5 of the SOM promoter in our ChIP assays.
The induction of SOM and other high-temperature-inducible genes by DELLAs supports the notion that DELLAs could regulate gene expression by targeting specific gene promoters. This targeting model was previously proposed based on reports that DELLAs target the promoters of various genes, including GA INSENSITIVE DWARF1 (GID1a), GID1b, MYB (AT3G11280), BASIC HELIX-LOOP-HELIX137, WRKY27, SCARECROW-LIKE3 (SCL3), LOB DOMAIN-CONTAINING PROTEIN40, XERICO, EXP8, PACLOBUTRAZOL RESISTANCE1 (PRE1), and PRE5 (Zentella et al., 2007; Gallego-Bartolomé et al., 2011; Park et al., 2013), and was further supported by the observation that DELLA activity was modulated upon fusion to transcription activation (VP16) or repression (SRDX) motifs (Hirano et al., 2012). Consistent with this targeting model, our data indicated that DELLAs physically interact with ABI3 and ABI5 (which are transcription factors) and cotarget the promoters of the subset D genes. This targeting model contrasts with the interfering model, which states that DELLAs regulate gene expression by interacting with various transcription regulators and inhibiting either their binding to DNA (PIF3, PIF4, and BRASSINAZOLE-RESISTANT1) or their binding to other DNA binding proteins (JAZ1 to MYC2) (de Lucas et al., 2008; Feng et al., 2008; Hou et al., 2010; Bai et al., 2012; Gallego-Bartolomé et al., 2012). We do not yet know what properties of this interaction allow DELLAs to cotarget specific promoters or antagonize DNA binding. Future studies are needed to clarify the different molecular interactions underlying the different paths.
ABI3, ABI5, and DELLAs Mediate ABA and GA Signaling to Activate Genes Induced by High Temperature
Our analyses indicated that high temperature required both ABI3 and DELLAs to induce the expression of the subset D genes. We also found that the overexpression of ABI5 was sufficient to elevate SOM expression. Due to the difficulty of generating a mutant lacking all 13 group A bZIPs, we were not able to determine whether these proteins are also required to induce SOM and other subset D genes at high temperature. However, a few lines of evidence suggest that group A bZIPs may also be required to induce SOM and the other subset D genes. First, ABI3 and group A bZIPs physically interact to activate the expression of EARLY METHIONINE-LABELED6 (Em6), rice (Oryza sativa) Em, wheat Em, and bean (Phaseolus vulgaris) β-phaseolin (Hobo et al., 1999; Gampala et al., 2002; Finkelstein et al., 2005; Marella et al., 2006). Second, in addition to ABI5, recombinant proteins of three more group A bZIPs (DPBF2, ABF1, and ABF2) also physically interact with recombinant GAI protein in vitro (see Supplemental Figure 8 online). Third, many direct target genes of ABI3 possess both RY motifs and ABREs in their promoters (Mönke et al., 2012). Fourth, the subset D genes also tend to possess both RY motifs and ABREs in their promoters (Table 1). Based on our present findings and the previous reports, we propose that ABI3, group A bZIPs, and DELLAs are key components inducing the subset D genes in imbibed seeds incubated at high temperature.
High temperature may not induce the expression of these genes by enhancing the DNA binding activities of ABI3, ABI5, and DELLAs per se, as shown by the constitutive targeting of 35S-driven ABI3, ABI5, and GAI to SOM and other subset D gene promoters irrespective of temperature. Instead, high temperature may induce gene expression either by enhancing the activities of these factors or by increasing their protein levels. We found that high temperature increases SOM mRNA and protein levels but does not drastically affect endogenous levels of ABI3, ABI5, and RGA at 6 h after seed imbibition (see Supplemental Figure 9 online). Previous studies have shown that the activity of ABI3 increases dramatically in the presence of ABA (Parcy et al., 1994; Ezcurra et al., 2000; Suzuki et al., 2003; Marella et al., 2006; Nakashima et al., 2006; Kotak et al., 2007; Sakata et al., 2010; Park et al., 2011). Thus, the increased ABA level observed at high temperature is likely to potentiate the activity of ABI3 as a transcriptional regulator. The activities of group A bZIPs have also been shown to be enhanced by ABA (Hobo et al., 1999; Finkelstein and Lynch, 2000; Gampala et al., 2002; Lopez-Molina et al., 2002; Finkelstein et al., 2005; Nakashima et al., 2006; Reeves et al., 2011; Wang et al., 2013) likely via the phosphorylation of group A bZIPs by the subclass III SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE2s, which are activated by the PYRABACTIN RESISTANCE/PYR1-LIKE/REGULATORY COMPONENTS OF ABA RECEPTOR–mediated inactivation of group A PROTEIN PHOSPHATASE 2Cs in the presence of ABA (Furihata et al., 2006; Fujii et al., 2007, 2009; Piskurewicz et al., 2008; Nakashima et al., 2009; Umezawa et al., 2009; Dai et al., 2013; Wang et al., 2013). Thus, the high-temperature-induced increase in ABA levels is likely to enhance the activities of group A bZIPs as transcriptional regulators. Consistently, we found that high temperature increases the expression of SOM mRNA in ABI3-FLAG but not in GFP-FLAG transgenic seedlings (see Supplemental Figure 9 online). High temperature also increases SOM promoter-driven expression of luciferase in a protoplast transfection assay with 35S:ABI3 but not with 35S:GFP. This is consistent with the hypothesis that high temperature enhances the activity of ABI3. The expression of luciferase was also enhanced with 35S:GAI and enhanced further in the presence of both 35S:ABI3 and 35S:GAI, implying that the two factors activate SOM mRNA additively. Though we did not observe the change of protein levels in early imbibition, the elevated levels of ABA likely enhance the level of ABI5 mRNA and subsequently the level of ABI5 protein in late imbibition (Hobo et al., 1999; Finkelstein and Lynch, 2000; Lopez-Molina et al., 2001, 2002; Suzuki et al., 2003; Finkelstein et al., 2005; Nakashima et al., 2006; Piskurewicz et al., 2008; Dai et al., 2013). Similarly, the temperature-induced reduction in GA is likely to elevate DELLA protein levels in late imbibition (Toh et al., 2008). Based on these and previous findings, we propose that high temperature activates the expression of the subset D genes by enhancing endogenous levels of group A bZIP and DELLA proteins and by enhancing the transcriptional regulatory activities of ABI3 and group A bZIPs.
SOM May Form a Positive Feedback Loop to Establish the Binary Status of Seed Germination
SOM may form a positive feedback loop with ABI3, ABI5, and DELLAs. Previous studies showed that SOM inhibits seed germination by activating ABA biosynthesis and inhibiting GA biosynthesis (Kim et al., 2008), and our current data indicate that high ABA and low GA levels activate SOM expression through ABI3, ABI5, and DELLAs. This increased SOM should then further activate ABA biosynthesis and inhibit GA biosynthesis, forming a positive feedback loop among these components (Figure 5D). A similar positive feedback loop was previously identified for the B3-domain transcription factor, FUSCA3 (FUS3), along with ABA and GA (Gazzarrini et al., 2004; Lu et al., 2010). During seed development, FUS3 plays roles in accumulating seed storage proteins, increasing seed dormancy, increasing desiccation tolerance, and repressing the change from the cotyledon to vegetative leaves (Santos-Mendoza et al., 2008; Suzuki and McCarty, 2008). The fus3 mutant showed reduced dormancy and seed storage protein accumulation, increased anthocyanin accumulation in seeds, and increased trichomes on cotyledons (Curaba et al., 2004; Tsuchiya et al., 2004; Tiedemann et al., 2008; Tsai and Gazzarrini, 2012). By contrast, transgenic plants expressing FUS3 under the control of the epidermis-specific MERISTEM LAYER1 promoter showed production of cotyledon-like leaves from meristem, abnormal petal structures, seed storage protein accumulation in leaves, dwarfism, and shortened siliques (Gazzarrini et al., 2004; Lu et al., 2010). Analyses using fus3 mutant siliques and dexamethasone-inducible FUS3 expression in seedlings showed that FUS3 activates ABA biosynthesis and inhibits GA biosynthesis (Curaba et al., 2004; Gazzarrini et al., 2004). Interestingly, the FUS3 protein is stabilized by ABA but destabilized by GA through the C-terminal PEST domain of the FUS3 protein, suggesting the presence of a positive feedback loop among FUS3, ABA, and GA (Gazzarrini et al., 2004; Lu et al., 2010). A characteristic feature of positive feedback loops is the ability to drive biological systems to opposite poles, thus establishing binary systems. Such binary systems have been proposed to regulate the asymmetric stem cell division in roots and the development of T lymphocytes in the thymus (Prasad et al., 2009; Cruz-Ramírez et al., 2012).
Germination might also operate in a binary mode, as the formation of half-germinated seeds would be deleterious. A classic study on seed germination showed that the germination of red light–pulsed lettuce (Lactuca sativa) seeds could be reversed by a far-red light pulse and that the degree of reversal depended on the interval between the red and far-red light pulses (Borthwick et al., 1954). Interestingly, the relationship between germination frequency and the interval was sigmoidal, which is consistent with a binary mode of seed germination. It is tempting to speculate that the positive feedback loop formed by SOM, ABI3, ABI5, and DELLAs could contribute to establishing the binary mode of seed germination at high temperature.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana plants were grown with a 16-h-light/8-h-dark cycle at 22 to 24°C. Harvested seeds were dried in paper bags at 23°C for at least 1 month prior to germination assays. Col-0 was used as the wild type. The som-2 (salk_090314; Kim et al., 2008), som-3 (salk_008075; Kim et al., 2008), aba2-1 (Oh et al., 2009), ga1 (salk_109115; Oh et al., 2006), abi3-sk11 (salk_023411; Park et al., 2011), abi3-sk22 (salk_138922, also known as S138922; Kotak et al., 2007), and abi5-8 (salk_013163; Zheng et al., 2012) mutants were obtained from the Arabidopsis Stock Centers (Alonso et al., 2003), and all of them are in the Col-0 background. The aba2 ga1 double mutant was generated by crossing the aba2-1 and ga1 mutants. The abi3-sk11 and abi3-sk22 mutant seeds were used immediately after harvesting due to a rapid decrease of seed viability after harvesting (Park et al., 2011). The sly1-10 mutant (Landsberg erecta background) backcrossed with Col-0 three times was used for ChIP assays (Park et al., 2013). The DELLA pentuple mutant (rga-28 gai-t6 rgl1 rgl2 rgl3-3; dellaP; Park et al., 2013), 35S-driven MYC-tagged GFP-overexpressing line (GFP-MYC; Oh et al., 2007), GAI-overexpressing line (GAI-MYC; Oh et al., 2007), 35S-driven FLAG-tagged GFP-overexpressing line (GFP-FLAG; Park et al., 2013), and ABI3-overexpressing line (ABI3-FLAG; Park et al., 2011) were previously reported.
To generate 35S-driven FLAG-tagged SOM-overexpressing lines (SOM-FLAG1 and SOM-FLAG2) and ABI5-overexpressing lines (ABI5-FLAG1 and ABI5-FLAG2), SOM and ABI5 coding regions were cloned into a binary vector and transformed into Col-0 plants using Agrobacterium tumefaciens–mediated floral dipping protocol (Clough and Bent, 1998). The primer sets for cloning are listed in Supplemental Table 1 online.
Germination Assays
Germination assays were performed as described previously (Kim et al., 2008) except the inclusion of high-temperature treatment. For the germination assays, triplicates of 40 seeds for each line were surface sterilized, sown on aqueous agar medium plates (0.1 strength Murashige and Skoog [MS], 0.05% MES, pH 5.7, and 0.6% phytoagar), irradiated with 5 min of far-red light (3.2 μmol m−2 s−1) followed by red light (18.3 μmol m−2 s−1), and incubated in the dark for 5 d at 23°C or higher temperatures. Germination was judged by the protrusion of the radicle and the frequency was scored. For each germination assay, at least three biological replicate experiments were performed.
Quantitative mRNA Expression Assays
For the gene expression analysis, RNA was extracted from imbibed seeds that were irradiated with 5 min of far-red light (3.2 μmol m−2 s−1) followed by red light (18.3 μmol m−2 s−1) and incubated in the dark for 12 h either at 23 or at 32°C. Total RNA was extracted using the Spectrum Plant Total RNA kit (Sigma-Aldrich) according to the manufacturer’s protocol. For quantitative RT-PCR, 3 μg of total RNA was reverse transcribed, and the relative mRNA level of each gene was determined by real-time PCR (Bio-Rad) using specific primer sets by comparison with the level of PP2A (AT1G13320). The primer sets for real-time PCR are indicated in Supplemental Table 1 online. The detailed protocol of real-time PCR was previously described (Park et al., 2011).
Protoplast transfection assays were performed as described previously using protoplasts made from 4-weak-old Col-0 rosette leaves (Yoo et al., 2007; Park et al., 2011). The firefly luciferase gene linked to the SOM promoter was used as a reporter. The 35S-driven ABI3 (35S:ABI3), 35S:GAI, and 35S:GFP were used as transcription effectors. For heat treatment, transfected protoplasts were incubated in the dark for 13 h at 23°C and were further incubated in the dark for 3 h either at 23 or 37°C. Luciferase activity was measured using the dual-luciferase reporter assay kit (Promega) according to the manufacturer’s protocol.
Generation of Antibodies
The anti-SOM antibody was generated against the recombinant C-terminal 200 amino acids of SOM in rabbits. The anti-ABI3 antibody was generated against the recombinant N-terminal 235 amino acids of ABI3 in rabbits. The anti-ABI5 antibody was generated against a peptide of ABI5 (RKRKQQYFESLKSRA) in rabbits. The anti-RGA antibody was previously reported (Park et al., 2013). The primer sets for cloning are listed in Supplemental Table 1 online.
Protein Extraction and Immunoblotting Assays
To analyze protein levels, total protein was extracted from imbibed seeds that were irradiated with 5 min of far-red light (3.2 μmol m−2 s−1) followed by red light (18.3 μmol m−2 s−1) and incubated in the dark for 12 h either at 23 or 32°C. Seeds were ground in liquid nitrogen and homogenized in the denaturing buffer (100 mM NaH2PO4, 10mM Tris∙Cl, and 8 M urea, pH 8) by vigorous vortexing. Cell debris was removed by centrifugation at 14,500 rpm for 10 min at 4°C. The protein samples were added with the SDS sample buffer (12 mM Tris∙Cl, pH 6.8, 5% glycerol, 0.4% SDS, 1% β-mercaptoethanol, and 0.02% bromophenol blue), boiled for 5 min, and subjected to immunoblotting assays as described previously (Park et al., 2011). As the loading control, anti-RPT5 (Enzo) or antitubulin (Sigma-Aldrich) antibody was used. Bands were visualized with an enhanced chemiluminescence kit (Pierce), according to the manufacturer’s protocol.
ChIP Assays
The ChIP assays were performed as described previously (Park et al., 2011). Seeds were irradiated with 5 min of far-red light (3.2 μmol m−2 s−1) followed by red light (18.3 μmol m−2 s−1), incubated in the dark for 6 h either at 23 or 32°C, and sampled for the ChIP assays. For Col-0 and sly1-10 mutant seeds, antibodies against endogenous ABI3, ABI5, or RGA were used for the ChIP assays. For 35S-driven GAI-MYC, ABI3-FLAG, and ABI5-FLAG1 transgenic seeds, anti-MYC antibody (Cell Signaling) or anti-FLAG antibody-conjugated resin (Sigma-Aldrich) were used for the ChIP assays. The coprecipitated level of each DNA fragment was quantified by real-time PCR using specific primer sets and normalized with the input DNA level. The relative coprecipitated levels of F1 or PP2A (immunoprecipitated DNA/input DNA) were set to 1. The primer sets for real-time PCR are listed in Supplemental Table 1 online.
In Vitro Coprecipitation Assays
To purify recombinant MBP-fused ABI3 (MBP-ABI3), group A bZIP proteins (MBP-ABI5, MBP-DPBF2, MBP-ABF1, and MBP-ABF2), and GST-fused GAI protein (GST-GAI), each coding region was cloned into pMAL-c2X (for MBP) or pGEX-4T-1 (for GST) vectors. The primer sets for cloning are listed in Supplemental Table 1 online. MBP-fused proteins were purified from BL21 Escherichia coli using the pMAL protein fusion and purification system (New England Biolabs), whereas GST-fused proteins were purified using the Glutathione Sepharose 4B (GE Healthcare), according to the manufacturers’ protocols. The in vitro coprecipitation assays were performed as described previously (Park et al., 2011). In brief, resin-bound MBP or MBP-fused proteins were incubated with GST-GAI protein at 4°C for 3 h in the binding buffer (50 mM Tris∙Cl, pH 7.5, 1 mM EDTA, 150 mM NaCl, 0.1% Triton X-100, 0.05% Na-deoxycholate, 10% glycerol, 0.1 mg/mL BSA, 1 mM PMSF, and protease inhibitor cocktail). After washing three times with the washing buffer (50 mM Tris∙Cl, pH 7.5, 1 mM EDTA, 150 mM NaCl, 0.1% Triton X-100, 0.05% Na-deoxycholate, 10% glycerol, 1 mM PMSF, and protease inhibitor cocktail), bound proteins were eluted by boiling in SDS sample buffer and subjected to immunoblotting assays using anti-MBP or anti-GST antibody (Santa Cruz).
In Vivo Coprecipitation Assays
In vivo coprecipitation assays were performed in 35S-driven GFP-FLAG, ABI3-FLAG, and ABI5-FLAG1 transgenic seedlings. Five-day-old, light-grown seedlings were transferred to MS-agar plates containing 100 μM paclobutrazol and further grown for 3 d before sampling. Seedling samples were ground in liquid nitrogen and homogenized in the extraction buffer (50 mM Tris∙Cl, pH 7.5, 100 mM NaCl, 0.1% Nonidet P-40, 1 mM PMSF, 80 μM MG132, and protease inhibitor cocktail) by vigorous vortexing. Cell debris was removed by centrifugation at 14,500 rpm for 10 min at 4°C. From the cell extract, FLAG-tagged proteins were precipitated with an anti-FLAG antibody-conjugated resin (Sigma-Aldrich). Coprecipitated proteins were determined by immunoblotting assays with anti-RGA antibody.
Coprecipitation assays were also performed by agroinfiltration (Li et al., 2009). For this experiment, 5-d-old, light-grown 35S-driven transgenic seedlings were inoculated with Agrobacteria harboring 35S-driven GFP-MYC or gaiD-MYC vectors and incubated for 36 h in the dark before sampling. For the assay, MYC-tagged or FLAG-tagged proteins were precipitated with anti-MYC or anti-FLAG antibody, respectively.
Microarray Data Analysis
Genes regulated by high temperature, ABA, and GA in imbibed seeds were determined from previously published microarray data (Goda et al., 2008; Chiu et al., 2012). For high-temperature-inducible genes (log2 FC > 0.85, P < 0.05), Col-0 seeds were imbibed in water at 21 or 32°C for 24 h under the continuous light (Chiu et al., 2012). For ABA upregulated genes (GSE5700 from the National Center for Biotechnology Information Gene Expression Omnibus database, log2 FC > 0.85, P < 0.05), Col-0 seeds were imbibed in mock conditions or 30 μM ABA for 24 h at 22°C under continuous light (Goda et al., 2008). For GA downregulated genes (GSE5701 from the National Center for Biotechnology Information Gene Expression Omnibus database, log2 FC < −0.85, P < 0.05), ga1-3 seeds were imbibed at 4°C in the dark for 48 h and then incubated for 24 h under white light at 22°C. Then, the seeds were further incubated in mock or 5 μM GA4 for 9 h under white light (Goda et al., 2008). The overrepresentation of a subset of genes and cis-acting elements in promoters was analyzed using hypergeometric (Fisher’s exact) test.
The regulation of subset D genes by ABI3, ABI4, and ABI5 was examined by heat map and GSEA using previously reported microarray data of abi3-6 (E-MEXP-1941 from EMBL-EBI database; Nakashima et al., 2009) or abi4-11 and abi5-7 mutant dry seeds (Nakabayashi et al., 2005). GSEA was conducted with GSEA software version 2.0.12 following the user guide (http://www.broadinstitute.org/GSEA; Subramanian et al., 2005).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: SOM (AT1G03790), ABA2 (AT1G52340), GA1 (AT4G02780), ABI3 (AT3G24650), RGA (AT2G01570), GAI (AT1G14920), RGL1 (AT1G66350), RGL2 (AT3G03450), RGL3 (AT5G17490), ABI5 (AT2G36270), DPBF2 (AT3G44460), ABF1 (AT1G49720), ABF2 (AT1G45249), EEL (AT2G41070), SLY1 (AT4G24210), Em6 (AT2G40170), SCL3 (AT1G50420), Em1 (AT3G51810), HON (AT1G07430), and PP2A (AT1G13320).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Transgene mRNA Levels.
Supplemental Figure 2. Expression of SOM mRNA in the aba2 and the ga1 Mutant Seeds.
Supplemental Figure 3. Specificities of ABI3 and ABI5 Antibodies.
Supplemental Figure 4. Targeting of ABI3-FLAG, ABI5-FLAG, and GAI-MYC to the SOM Promoter.
Supplemental Figure 5. In Vivo Coprecipitation between Transgenic ABI3-FLAG or ABI5-FLAG and Agroinfiltrated gaiD-MYC.
Supplemental Figure 6. Decreased Expression of the Subset D Genes Including SOM in the abi3, abi4, and abi5 Mutant Dry Seeds.
Supplemental Figure 7. Targeting of ABI3-FLAG, ABI5-FLAG, and GAI-MYC to the Subset D Gene Promoters.
Supplemental Figure 8. In Vitro Coprecipitation between the Group A bZIPs and GAI.
Supplemental Figure 9. Enhancement of ABI3 Activity by High Temperature.
Supplemental Table 1. Primer List.
Supplemental Data Set 1. Lists of Genes Corresponding to Subset A to G.
Acknowledgments
We thank The Arabidopsis Information Resource and the Nottingham Arabidopsis Stock Centre for providing information and mutant seeds. This work was supported in part by the National Research Foundation of Korea (Grants 2012R1A2A1A01003133 and 2011-0031955) and the Rural Development Administration (Grant SSAC-PJ009580) to G.C. and a grant from the Japan Society for the Promotion of Science for Young Scientists to S.T.
AUTHOR CONTRIBUTIONS
S.L., J.P., N.K., and G.C. designed the research. S.L., J.P., J.J., J.K., H.K., N.L., S.T., A.W., and D.H.K. performed experiments. S.L. and G.C. wrote the article.
Glossary
- GA
gibberellic acid
- ABA
abscisic acid
- ChIP
chromatin immunoprecipitation
- Col-0
Columbia-0
- ABRE
ABA-responsive element
- FC
fold change
- MS
Murashige and Skoog
- GSEA
gene set enrichment analysis
References
- Ali-Rachedi S., Bouinot D., Wagner M.H., Bonnet M., Sotta B., Grappin P., Jullien M. (2004). Changes in endogenous abscisic acid levels during dormancy release and maintenance of mature seeds: Studies with the Cape Verde Islands ecotype, the dormant model of Arabidopsis thaliana. Planta 219: 479–488 [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Argyris J., Dahal P., Hayashi E., Still D.W., Bradford K.J. (2008). Genetic variation for lettuce seed thermoinhibition is associated with temperature-sensitive expression of abscisic acid, gibberellin, and ethylene biosynthesis, metabolism, and response genes. Plant Physiol. 148: 926–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M.Y., Shang J.X., Oh E., Fan M., Bai Y., Zentella R., Sun T.P., Wang Z.Y. (2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14: 810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin J.M., Baskin C.C. (1983). Seasonal changes in the germination responses of buried seeds of Arabidopsis thaliana and ecological interpretation. Bot. Gaz. 144: 540–543 [Google Scholar]
- Bensmihen S., Rippa S., Lambert G., Jublot D., Pautot V., Granier F., Giraudat J., Parcy F. (2002). The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14: 1391–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergler J., Hoth S. (2011). Plant U-box armadillo repeat proteins AtPUB18 and AtPUB19 are involved in salt inhibition of germination in Arabidopsis. Plant Biol. (Stuttg.) 13: 725–730 [DOI] [PubMed] [Google Scholar]
- Borthwick H.A., Hendricks S.B., Toole E.H., Toole V.K. (1954). Action of light on lettuce-seed germination. Bot. Gaz. 115: 205–225 [Google Scholar]
- Brocard-Gifford I.M., Lynch T.J., Finkelstein R.R. (2003). Regulatory networks in seeds integrating developmental, abscisic acid, sugar, and light signaling. Plant Physiol. 131: 78–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carles C., Bies-Etheve N., Aspart L., Léon-Kloosterziel K.M., Koornneef M., Echeverria M., Delseny M. (2002). Regulation of Arabidopsis thaliana Em genes: Role of ABI5. Plant J. 30: 373–383 [DOI] [PubMed] [Google Scholar]
- Chauhan H., Khurana N., Nijhavan A., Khurana J.P., Khurana P. (2012). The wheat chloroplastic small heat shock protein (sHSP26) is involved in seed maturation and germination and imparts tolerance to heat stress. Plant Cell Environ. 35: 1912–1931 [DOI] [PubMed] [Google Scholar]
- Chiu R.S., Nahal H., Provart N.J., Gazzarrini S. (2012). The role of the Arabidopsis FUSCA3 transcription factor during inhibition of seed germination at high temperature. BMC Plant Biol. 12: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H., Hong J., Ha J., Kang J., Kim S.Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275: 1723–1730 [DOI] [PubMed] [Google Scholar]
- Christie J.M. (2007). Phototropin blue-light receptors. Annu. Rev. Plant Biol. 58: 21–45 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corrêa L.G., Riaño-Pachón D.M., Schrago C.G., dos Santos R.V., Mueller-Roeber B., Vincentz M. (2008). The role of bZIP transcription factors in green plant evolution: Adaptive features emerging from four founder genes. PLoS ONE 3: e2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ramírez A., et al. (2012). A bistable circuit involving SCARECROW-RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell 150: 1002–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curaba J., Moritz T., Blervaque R., Parcy F., Raz V., Herzog M., Vachon G. (2004). AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol. 136: 3660–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M., Xue Q., Mccray T., Margavage K., Chen F., Lee J.H., Nezames C.D., Guo L., Terzaghi W., Wan J., Deng X.W., Wang H. (2013). The PP6 phosphatase regulates ABI5 phosphorylation and abscisic acid signaling in Arabidopsis. Plant Cell 25: 517–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Ezcurra I., Wycliffe P., Nehlin L., Ellerström M., Rask L. (2000). Transactivation of the Brassica napus napin promoter by ABI3 requires interaction of the conserved B2 and B3 domains of ABI3 with different cis-elements: B2 mediates activation through an ABRE, whereas B3 interacts with an RY/G-box. Plant J. 24: 57–66 [DOI] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feurtado J.A., Huang D., Wicki-Stordeur L., Hemstock L.E., Potentier M.S., Tsang E.W., Cutler A.J. (2011). The Arabidopsis C2H2 zinc finger INDETERMINATE DOMAIN1/ENHYDROUS promotes the transition to germination by regulating light and hormonal signaling during seed maturation. Plant Cell 23: 1772–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage W.E., Leubner-Metzger G. (2006). Seed dormancy and the control of germination. New Phytol. 171: 501–523 [DOI] [PubMed] [Google Scholar]
- Finkelstein R., Gampala S.S., Lynch T.J., Thomas T.L., Rock C.D. (2005). Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE(ABI)5 and ABRE-BINDING FACTOR (ABF)3. Plant Mol. Biol. 59: 253–267 [DOI] [PubMed] [Google Scholar]
- Finkelstein R.R., Lynch T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Chinnusamy V., Rodrigues A., Rubio S., Antoni R., Park S.Y., Cutler S.R., Sheen J., Rodriguez P.L., Zhu J.K. (2009). In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Verslues P.E., Zhu J.K. (2007). Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19: 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Fujita M., Shinozaki K., Yamaguchi-Shinozaki K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124: 509–525 [DOI] [PubMed] [Google Scholar]
- Furihata T., Maruyama K., Fujita Y., Umezawa T., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. (2006). Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 103: 1988–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Alabadí D., Blázquez M.A. (2011). DELLA-induced early transcriptional changes during etiolated development in Arabidopsis thaliana. PLoS ONE 6: e23918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Minguet E.G., Grau-Enguix F., Abbas M., Locascio A., Thomas S.G., Alabadí D., Blázquez M.A. (2012). Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 13446–13451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampala S.S., Finkelstein R.R., Sun S.S., Rock C.D. (2002). ABI5 interacts with abscisic acid signaling effectors in rice protoplasts. J. Biol. Chem. 277: 1689–1694 [DOI] [PubMed] [Google Scholar]
- Gazzarrini S., Tsuchiya Y., Lumba S., Okamoto M., McCourt P. (2004). The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev. Cell 7: 373–385 [DOI] [PubMed] [Google Scholar]
- Goda H., et al. (2008). The AtGenExpress hormone and chemical treatment data set: Experimental design, data evaluation, model data analysis and data access. Plant J. 55: 526–542 [DOI] [PubMed] [Google Scholar]
- Gonai T., Kawahara S., Tougou M., Satoh S., Hashiba T., Hirai N., Kawaide H., Kamiya Y., Yoshioka T. (2004). Abscisic acid in the thermoinhibition of lettuce seed germination and enhancement of its catabolism by gibberellin. J. Exp. Bot. 55: 111–118 [DOI] [PubMed] [Google Scholar]
- Gubler F., Hughes T., Waterhouse P., Jacobsen J. (2008). Regulation of dormancy in barley by blue light and after-ripening: Effects on abscisic acid and gibberellin metabolism. Plant Physiol. 147: 886–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijde M., Ulm R. (2012). UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci. 17: 230–237 [DOI] [PubMed] [Google Scholar]
- Hennig L., Stoddart W.M., Dieterle M., Whitelam G.C., Schäfer E. (2002). Phytochrome E controls light-induced germination of Arabidopsis. Plant Physiol. 128: 194–200 [PMC free article] [PubMed] [Google Scholar]
- Hirano K., Kouketu E., Katoh H., Aya K., Ueguchi-Tanaka M., Matsuoka M. (2012). The suppressive function of the rice DELLA protein SLR1 is dependent on its transcriptional activation activity. Plant J. 71: 443–453 [DOI] [PubMed] [Google Scholar]
- Hobo T., Kowyama Y., Hattori T. (1999). A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc. Natl. Acad. Sci. USA 96: 15348–15353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth M.J., Bentsink L., Soppe W.J. (2008). Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 179: 33–54 [DOI] [PubMed] [Google Scholar]
- Hossain K.K., Itoh R.D., Yoshimura G., Tokuda G., Oku H., Cohen M.F., Yamasaki H. (2010). Effects of nitric oxide scavengers on thermoinhibition of seed germination in Arabidopsis thaliana. Russ. J. Plant Physiol. 57: 222–232 [Google Scholar]
- Hou X., Lee L.Y., Xia K., Yan Y., Yu H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19: 884–894 [DOI] [PubMed] [Google Scholar]
- Ito S., Song Y.H., Imaizumi T. (2012). LOV domain-containing F-box proteins: Light-dependent protein degradation modules in Arabidopsis. Mol. Plant 5: 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen J.V., Pearce D.W., Poole A.T., Pharis R.P., Mander L.N. (2002). Abscisic acid, phaseic acid and gibberellin contents associated with dormancy and germination in barley. Physiol. Plant. 115: 428–441 [DOI] [PubMed] [Google Scholar]
- Jakoby M., Weisshaar B., Dröge-Laser W., Vicente-Carbajosa J., Tiedemann J., Kroj T., Parcy F., bZIP Research Group (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7: 106–111 [DOI] [PubMed] [Google Scholar]
- Jensen M.K., Hagedorn P.H., de Torres-Zabala M., Grant M.R., Rung J.H., Collinge D.B., Lyngkjaer M.F. (2008). Transcriptional regulation by an NAC (NAM-ATAF1,2-CUC2) transcription factor attenuates ABA signalling for efficient basal defence towards Blumeria graminis f. sp. hordei in Arabidopsis. Plant J. 56: 867–880 [DOI] [PubMed] [Google Scholar]
- Khan A.A., Prusinski J. (1989). Kinetin enhanced 1-aminocyclopropane-1-carboxylic acid utilization during alleviation of high temperatures stress in lettuce seeds. Plant Physiol. 91: 733–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, M.A., and Gul, B. (2006). Halophyte seed germination. In Ecophysiology of High Salinity Tolerant Plants, M.A. Khan and D.J. Weber, eds (Dordrecht, The Netherlands: Springer), pp. 11–30. [Google Scholar]
- Kim D.H., Yamaguchi S., Lim S., Oh E., Park J., Hanada A., Kamiya Y., Choi G. (2008). SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 20: 1260–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.S., Mizoi J., Yoshida T., Fujita Y., Nakajima J., Ohori T., Todaka D., Nakashima K., Hirayama T., Shinozaki K., Yamaguchi-Shinozaki K. (2011). An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol. 52: 2136–2146 [DOI] [PubMed] [Google Scholar]
- Kim S.Y., Chung H.J., Thomas T.L. (1997). Isolation of a novel class of bZIP transcription factors that interact with ABA-responsive and embryo-specification elements in the Dc3 promoter using a modified yeast one-hybrid system. Plant J. 11: 1237–1251 [DOI] [PubMed] [Google Scholar]
- Kim W., Lee Y., Park J., Lee N., Choi G. (2013). HONSU, a protein phosphatase 2C, regulates seed dormancy by inhibiting ABA signaling in Arabidopsis. Plant Cell Physiol. 54: 555–572 [DOI] [PubMed] [Google Scholar]
- Kotak S., Vierling E., Bäumlein H., von Koskull-Döring P. (2007). A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell 19: 182–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozarewa I., Cantliffe D.J., Nagata R.T., Stoffella P.J. (2006). High maturation temperature of lettuce seeds during development increased ethylene production and germination at elevated temperatures. J. Am. Soc. Hortic. Sci. 131: 564–570 [Google Scholar]
- Leymarie J., Robayo-Romero M.E., Gendreau E., Benech-Arnold R.L., Corbineau F. (2008). Involvement of ABA in induction of secondary dormancy in barley (Hordeum vulgare L.) seeds. Plant Cell Physiol. 49: 1830–1838 [DOI] [PubMed] [Google Scholar]
- Li J.F., Park E., von Arnim A.G., Nebenführ A. (2009). The FAST technique: A simplified Agrobacterium-based transformation method for transient gene expression analysis in seedlings of Arabidopsis and other plant species. Plant Methods 5: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Li F., Wang J., Zhang W., Meng Q., Chen T.H., Murata N., Yang X. (2011). Glycinebetaine enhances the tolerance of tomato plants to high temperature during germination of seeds and growth of seedlings. Plant Cell Environ. 34: 1931–1943 [DOI] [PubMed] [Google Scholar]
- Liu Y.C., Wu Y.R., Huang X.H., Sun J., Xie Q. (2011). AtPUB19, a U-box E3 ubiquitin ligase, negatively regulates abscisic acid and drought responses in Arabidopsis thaliana. Mol. Plant 4: 938–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Chua N.H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., McLachlin D.T., Chait B.T., Chua N.H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Lu Q.S., Paz J.D., Pathmanathan A., Chiu R.S., Tsai A.Y., Gazzarrini S. (2010). The C-terminal domain of FUSCA3 negatively regulates mRNA and protein levels, and mediates sensitivity to the hormones abscisic acid and gibberellic acid in Arabidopsis. Plant J. 64: 100–113 [DOI] [PubMed] [Google Scholar]
- Marella H.H., Sakata Y., Quatrano R.S. (2006). Characterization and functional analysis of ABSCISIC ACID INSENSITIVE3-like genes from Physcomitrella patens. Plant J. 46: 1032–1044 [DOI] [PubMed] [Google Scholar]
- Martinez M.L., Valverde T., Moreno-Casasola P. (1992). Germination response to temperature, salinity, light and depth of sowing of ten tropical dune species. Oecologia 92: 343–353 [DOI] [PubMed] [Google Scholar]
- Matilla A.J. (2000). Ethylene in seed formation and germination. Seed Sci. Res. 10: 111–126 [Google Scholar]
- Mei Y.Q., Song S.Q. (2010). Response to temperature stress of reactive oxygen species scavenging enzymes in the cross-tolerance of barley seed germination. J. Zhejiang Univ. Sci. B 11: 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S., Wu Y., Venkataraman G., Sopory S.K., Tuteja N. (2007). Heterotrimeric G-protein complex and G-protein-coupled receptor from a legume (Pisum sativum): Role in salinity and heat stress and cross-talk with phospholipase C. Plant J. 51: 656–669 [DOI] [PubMed] [Google Scholar]
- Mönke G., Seifert M., Keilwagen J., Mohr M., Grosse I., Hähnel U., Junker A., Weisshaar B., Conrad U., Bäumlein H., Altschmied L. (2012). Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Res. 40: 8240–8254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A. (2010). Phytochrome: Structural basis for its functions. Curr. Opin. Plant Biol. 13: 565–570 [DOI] [PubMed] [Google Scholar]
- Nakabayashi K., Okamoto M., Koshiba T., Kamiya Y., Nambara E. (2005). Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: Epigenetic and genetic regulation of transcription in seed. Plant J. 41: 697–709 [DOI] [PubMed] [Google Scholar]
- Nakamura S., Lynch T.J., Finkelstein R.R. (2001). Physical interactions between ABA response loci of Arabidopsis. Plant J. 26: 627–635 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Fujita Y., Kanamori N., Katagiri T., Umezawa T., Kidokoro S., Maruyama K., Yoshida T., Ishiyama K., Kobayashi M., Shinozaki K., Yamaguchi-Shinozaki K. (2009). Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50: 1345–1363 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Fujita Y., Katsura K., Maruyama K., Narusaka Y., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2006). Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol. Biol. 60: 51–68 [DOI] [PubMed] [Google Scholar]
- Narusaka Y., Nakashima K., Shinwari Z.K., Sakuma Y., Furihata T., Abe H., Narusaka M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 34: 137–148 [DOI] [PubMed] [Google Scholar]
- Oh E., Kang H., Yamaguchi S., Park J., Lee D., Kamiya Y., Choi G. (2009). Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21: 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Kim J., Park E., Kim J.I., Kang C., Choi G. (2004). PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16: 3045–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Hu J., Yusuke J., Jung B., Paik I., Lee H.S., Sun T.P., Kamiya Y., Choi G. (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19: 1192–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Kamiya Y., Bae G., Chung W.I., Choi G. (2006). Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 47: 124–139 [DOI] [PubMed] [Google Scholar]
- Parcy F., Valon C., Raynal M., Gaubier-Comella P., Delseny M., Giraudat J. (1994). Regulation of gene expression programs during Arabidopsis seed development: Roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6: 1567–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Lee N., Kim W., Lim S., Choi G. (2011). ABI3 and PIL5 collaboratively activate the expression of SOMNUS by directly binding to its promoter in imbibed Arabidopsis seeds. Plant Cell 23: 1404–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Nguyen K.T., Park E., Jeon J.S., Choi G. (2013). DELLA proteins and their interacting RING finger proteins repress gibberellin responses by binding to the promoters of a subset of gibberellin-responsive genes in Arabidopsis. Plant Cell 25: 927–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U., Jikumaru Y., Kinoshita N., Nambara E., Kamiya Y., Lopez-Molina L. (2008). The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20: 2729–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A., Zikherman J., Das J., Roose J.P., Weiss A., Chakraborty A.K. (2009). Origin of the sharp boundary that discriminates positive and negative selection of thymocytes. Proc. Natl. Acad. Sci. USA 106: 528–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves W.M., Lynch T.J., Mobin R., Finkelstein R.R. (2011). Direct targets of the transcription factors ABA-Insensitive(ABI)4 and ABI5 reveal synergistic action by ABI4 and several bZIP ABA response factors. Plant Mol. Biol. 75: 347–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini H.S., Consolacion E.D., Bassi P.K., Spencer M.S. (1986). Requirement for ethylene synthesis and action during relief of thermoinhibition of lettuce seed germination by combinations of gibberellic acid, kinetin, and carbon dioxide. Plant Physiol. 81: 950–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata Y., Nakamura I., Taji T., Tanaka S., Quatrano R.S. (2010). Regulation of the ABA-responsive Em promoter by ABI3 in the moss Physcomitrella patens: Role of the ABA response element and the RY element. Plant Signal. Behav. 5: 1061–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Mendoza M., Dubreucq B., Baud S., Parcy F., Caboche M., Lepiniec L. (2008). Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 54: 608–620 [DOI] [PubMed] [Google Scholar]
- Seo M., Nambara E., Choi G., Yamaguchi S. (2009). Interaction of light and hormone signals in germinating seeds. Plant Mol. Biol. 69: 463–472 [DOI] [PubMed] [Google Scholar]
- Shinomura T., Nagatani A., Chory J., Furuya M. (1994). The induction of seed germination in Arabidopsis thaliana is regulated principally by phytochrome B and secondarily by phytochrome A. Plant Physiol. 104: 363–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura T., Nagatani A., Hanzawa H., Kubota M., Watanabe M., Furuya M. (1996). Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 93: 8129–8133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla R.K., Tripathi V., Jain D., Yadav R.K., Chattopadhyay D. (2009). CAP2 enhances germination of transgenic tobacco seeds at high temperature and promotes heat stress tolerance in yeast. FEBS J. 276: 5252–5262 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Ketterling M.G., Li Q.B., McCarty D.R. (2003). Viviparous1 alters global gene expression patterns through regulation of abscisic acid signaling. Plant Physiol. 132: 1664–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., McCarty D.R. (2008). Functional symmetry of the B3 network controlling seed development. Curr. Opin. Plant Biol. 11: 548–553 [DOI] [PubMed] [Google Scholar]
- Tamura N., Yoshida T., Tanaka A., Sasaki R., Bando A., Toh S., Lepiniec L., Kawakami N. (2006). Isolation and characterization of high temperature-resistant germination mutants of Arabidopsis thaliana. Plant Cell Physiol. 47: 1081–1094 [DOI] [PubMed] [Google Scholar]
- Tezuka K., Taji T., Hayashi T., Sakata Y. (2013). A novel abi5 allele reveals the importance of the conserved Ala in the C3 domain for regulation of downstream genes and salt tolerance during germination in Arabidopsis. Plant Signal. Behav. 8: e23455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedemann J., Rutten T., Mönke G., Vorwieger A., Rolletschek H., Meissner D., Milkowski C., Petereck S., Mock H.P., Zank T., Bäumlein H. (2008). Dissection of a complex seed phenotype: Novel insights of FUSCA3 regulated developmental processes. Dev. Biol. 317: 1–12 [DOI] [PubMed] [Google Scholar]
- Toh S., Kamiya Y., Kawakami N., Nambara E., McCourt P., Tsuchiya Y. (2012). Thermoinhibition uncovers a role for strigolactones in Arabidopsis seed germination. Plant Cell Physiol. 53: 107–117 [DOI] [PubMed] [Google Scholar]
- Toh S., et al. (2008). High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol. 146: 1368–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toorop P.E., Barroco R.M., Engler G., Groot S.P., Hilhorst H.W. (2005). Differentially expressed genes associated with dormancy or germination of Arabidopsis thaliana seeds. Planta 221: 637–647 [DOI] [PubMed] [Google Scholar]
- Tsai A.Y., Gazzarrini S. (2012). AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J. 69: 809–821 [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y., Nambara E., Naito S., McCourt P. (2004). The FUS3 transcription factor functions through the epidermal regulator TTG1 during embryogenesis in Arabidopsis. Plant J. 37: 73–81 [DOI] [PubMed] [Google Scholar]
- Umezawa T., Sugiyama N., Mizoguchi M., Hayashi S., Myouga F., Yamaguchi-Shinozaki K., Ishihama Y., Hirayama T., Shinozaki K. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y., Furihata T., Abe H., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li L., Ye T., Lu Y., Chen X., Wu Y. (2013). The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J. Exp. Bot. 64: 675–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitbrecht K., Müller K., Leubner-Metzger G. (2011). First off the mark: Early seed germination. J. Exp. Bot. 62: 3289–3309 [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yoshioka T., Endo T., Satoh S. (1998). Restoration of seed germination at supraoptimal temperatures by fluridone, an inhibitor of abscisic acid biosynthesis. Plant Cell Physiol. 39: 307–312 [Google Scholar]
- Yu X., Liu H., Klejnot J., Lin C. (2010). The cryptochrome blue light receptors. Arabidopsis Book 8: e0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R., Zhang Z.L., Park M., Thomas S.G., Endo A., Murase K., Fleet C.M., Jikumaru Y., Nambara E., Kamiya Y., Sun T.P. (2007). Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19: 3037–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Mayba O., Pfeiffer A., Shi H., Tepperman J.M., Speed T.P., Quail P.H. (2013). A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Schumaker K.S., Guo Y. (2012). Sumoylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 109: 12822–12827 [DOI] [PMC free article] [PubMed] [Google Scholar]