Sterol biosynthetic intermediates (SBIs) derived from the sterol C4-demethylation complex tethered by ERG28 represent signaling molecules in mammals and fungi. In Arabidopsis, loss of ERG28 function leads to the accumulation of the previously undetected SBI, 4-carboxy-4-methyl-24-methylenecycloartanol, which inhibits polar auxin transport. Our work provides a link between sterol metabolism and auxin transport.
Abstract
Sterols are vital for cellular functions and eukaryotic development because of their essential role as membrane constituents. Sterol biosynthetic intermediates (SBIs) represent a potential reservoir of signaling molecules in mammals and fungi, but little is known about their functions in plants. SBIs are derived from the sterol C4-demethylation enzyme complex that is tethered to the membrane by Ergosterol biosynthetic protein28 (ERG28). Here, using nonlethal loss-of-function strategies focused on Arabidopsis thaliana ERG28, we found that the previously undetected SBI 4-carboxy-4-methyl-24-methylenecycloartanol (CMMC) inhibits polar auxin transport (PAT), a key mechanism by which the phytohormone auxin regulates several aspects of plant growth, including development and responses to environmental factors. The induced accumulation of CMMC in Arabidopsis erg28 plants was associated with diagnostic hallmarks of altered PAT, including the differentiation of pin-like inflorescence, loss of apical dominance, leaf fusion, and reduced root growth. PAT inhibition by CMMC occurs in a brassinosteroid-independent manner. The data presented show that ERG28 is required for PAT in plants. Furthermore, it is accumulation of an atypical SBI that may act to negatively regulate PAT in plants. Hence, the sterol pathway offers further prospects for mining new target molecules that could regulate plant development.
INTRODUCTION
Sterols are isoprenoid compounds that have been recruited for diverse biological functions by living organisms since the appearance of oxygen in the atmosphere (Brown and Galea, 2010). The conversion of sterols into brassinosteroids provides the only known sterol-derived hormones in plants (Zhu et al., 2013). However, several sterol-deficient mutants displaying dwarf phenotypes, patterning defects, and pseudoembryonic and seedling lethality are not rescued by brassinosteroids (Clouse, 2002; Lindsey et al., 2003). These mutant phenotypes are either due to altered membrane structure and perturbed sterol dependent endocytic trafficking of auxin transporters (Men et al., 2008; Boutté and Grebe, 2009; Petrásek and Friml, 2009) or to potential unidentified sterol biosynthetic intermediates (SBIs) involved in brassinosteroid-independent regulation of plant development (Clouse, 2000, 2002; Schrick et al., 2002; He et al., 2003).
The identification of meiosis-activating sterols from human follicular fluid and testicular tissue (Byskov et al., 1995) and in hyperproliferative skin disease such as psoriasis, where they promote immunocyte proliferation via Toll-like receptors and liver X receptors (He et al., 2011) and the neuroprotective role of lanosterol (Lim et al., 2012), demonstrated that SBIs possessing a methyl group attached to carbon 4 in the A-ring of sterols (C4-methyl SBIs) (Figure 1) can have a biological function beyond sterol synthesis (Janowski et al., 1996; Castrillo et al., 2003; Bensinger et al., 2008; He et al., 2011; Lim et al., 2012). This is supported across phyla as C4-methyl SBIs of the ergosterol pathway represent an oxygen sensor in fission yeast (Schizosaccharomyces pombe) and in the pathogenic fungus Cryptococcus neoformans (Espenshade and Hughes, 2007; Hughes et al., 2007). C4-methyl sterol derivatives are also implicated in the physiology of nematodes, which are auxotroph for sterols. These organisms reintroduce, via STRM-1, a C4-methyl to the ring A of sterols ingested from diet to trigger the signaling cascade that helps endure stress conditions as dauer larvae (Hannich et al., 2009).
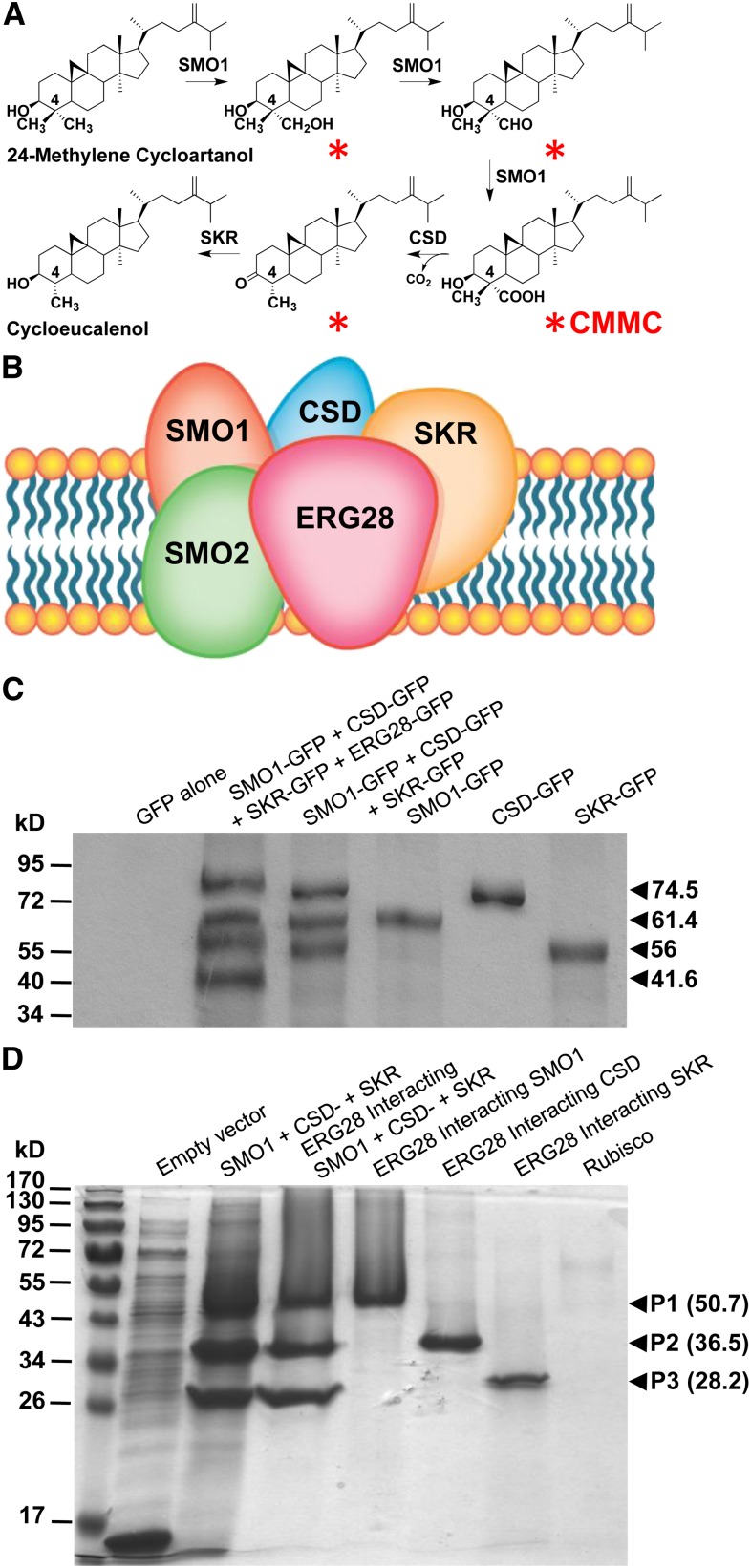
Figure 1.
Arabidopsis ERG28 Tethers the SC4DM Multienzyme Complex Catalyzing the Production of C4-Methyl SBIs.
(A) Four SBIs (red asterisks) including CMMC are sequentially formed but not released from the enzyme supercomplex during the conversion of 24-methylene cycloartanol to cycloeucalenol. The reaction is catalyzed by SC4DM component enzymes (SMO1, sterol 4α-methyl oxidase; CSD, 4α-carboxysterol-C3-dehydrogenase/C4-decarboxylase; and SKR, sterone keto-reductase) tethered by ERG28 to promote the channeling of SBIs (for additional steps catalyzed by the SC4DM multienzyme complex, see Supplemental Figure 1 online).
(B) Model predicting the tethering of SC4DM multienzyme complex by ERG28 based on yeast data.
(C) Coimmunoprecipitation of SC4DM-GFP component enzymes (SMO1-GFP, CSD-GFP, and SKR-GFP) from leaves transfected with constructs as shown above the gel lanes using anti-Arabidopsis ERG28 coupled to sepharose beads and immunoblot analysis with anti-GFP.
(D) In vitro interactions between Arabidopsis ERG28 and the SC4DM component enzymes. Biotinylated recombinant Arabidopsis ERG28 attached to streptavidin agarose was used as a bait to pull down recombinant Arabidopsis: SMO1, CSD, and SKR. Rubisco was used as a negative control bait. Soluble extracts from Escherichia coli overexpressing recombinant SMO1 (1 to 298), CSD (134 to 322), and SKR (98 to 229) as thioredoxin fusion proteins were incubated in a 1:1:1 (v:v:v) ratio or individually with purified biotinylated ERG28 or Rubisco immobilized on agarose beads before elution. SDS-PAGE analysis of soluble bacterial lysate from induced bacteria harboring an empty vector or a mixture of soluble proteins from induced bacteria expressing SMO1, CSD, and SKR and ERG28-interacting proteins.
[See online article for color version of this figure.]
The key step leading to the formation of C4-methyl SBI is mechanistically conserved throughout evolution from yeast(Saccharomyces cerevisiae) to man and plants and is catalyzed by the sterol C4 demethylase (SC4DM) multienzyme complex (Gaylor, 2002; Mo et al., 2002; Bouvier et al., 2005) (Figures 1A and 1B; see Supplemental Figure 1 online). The SC4DM multienzyme complex consists of sterol 4α-methyl oxidase (SMO), 4α-carboxysterol-C3-dehydrogenase/C4-decarboxylase (CSD), and sterone ketoreductase (SKR) (Bouvier et al., 2005), which act in a sequential manner. Previous studies from yeast have shown that the ERG28 protein functions as a scaffold in the endoplasmic reticulum to tether the SC4DM component enzymes to the membrane (Mo and Bard, 2005) (Figure 1B).
We predicted that if the function of yeast Ergosterol biosynthetic protein28 (ERG28; Gachotte et al., 2001) is conserved across phyla, its putative Arabidopsis thaliana homolog (see Supplemental Figure 2 online) would provide a unique opportunity to trigger the accumulation of C4-methyl SBIs that potentially modulate plant development without depleting the synthesis of sterols that are required for membrane integrity, endocytic trafficking (Boutté and Grebe, 2009), and brassinosteroid synthesis (Fujioka and Yokota, 2003). The relevance of this strategy is favored by the fact that Arabidopsis ERG28 is highly conserved among plants and is represented by a single-copy gene in the Arabidopsis genome (http://www.Arabidopsis.org), thus precluding any compensation due to functional redundancy. To test this hypothesis, we first demonstrated the scaffolding role of Arabidopsis ERG28 in the SC4DM complex and then modulated its expression in Arabidopsis. We show that Arabidopsis ERG28 plays an essential role in the maintenance of polar auxin transport (PAT). It does so by restricting the release and accumulation of CMMC, which in addition to its biosynthetic function inhibits PAT. Our data provide an unexpected level of interaction between sterols and auxin.
RESULTS
Arabidopsis ERG28 Functions as a Scaffolding Platform for Coassembling the Sterol C4 Demethylation Enzyme Complex
We first fused Arabidopsis ERG28 to green fluorescent protein (ERG28-GFP) and, using RTNLB2-GFP used as an endoplasmic reticulum marker (Jadid et al., 2011), showed that ERG28-GFP was specifically localized to the endoplasmic reticulum, the main site of plant sterol biosynthesis (Grebe et al., 2003; Benveniste, 2004; Bouvier et al., 2005) (see Supplemental Methods 1 and Supplemental Figure 3 online). To test whether ERG28 interacts with component enzymes of the SC4DM complex in plants, we coexpressed constructs encoding SMO1-GFP, CSD-GFP (Bouvier et al., 2005), and putative Arabidopsis SKR-GFP (Desmond and Gribaldo, 2009) in tobacco (Nicotiana tabacum) leaves and used anti-Arabidopsis ERG28 antibody to determine whether ERG28 tethers SC4DM component enzymes from the solubilized microsomes. We monitored the interaction of ERG28 and SC4DM using anti-GFP antibodies for both immunoblotting and to pull down ERG28. We found that SMO1-GFP, CSD-GFP, and SKR-GFP bind selectively to ERG28, consistent with a tethering role of ERG28 for plant SC4DM enzyme complex that facilitates the sequential transfer of C4-methyl SBIs among the different enzymes of the complex, as has been shown in yeast (Mo and Bard, 2005) (Figures 1B and 1C).
The tethering role of ERG28 was further demonstrated directly by pull-down assay using biotinylated ERG28 attached to streptavidin-agarose and recombinant Arabidopsis SMO1, CSD, and SKR (Figure 1D; see Supplemental Table 1 online). These data were reinforced by the fact that Arabidopsis ERG28 functionally complemented the yeast erg28 mutant, which accumulates 4α-carboxy-zymosterol, 4α-carboxy-fecosterol, and 4β-methyl-4α-carboxy-zymosterol SBIs derived from the yeast SC4DM multienzyme complex (Gachotte et al., 2001). The sterol composition indicates that the wild-type ergosterol pathway was restored in complemented yeast erg28 (see Supplemental Methods 1 and Supplemental Table 2 online). These data indicate that Arabidopsis ERG28 could be integrated into the yeast SC4DM multienzyme complex, thus supporting the functional similarity between Arabidopsis ERG28 and yeast ERG28.
Loss of ERG28 Function Recapitulates Phenotypes Caused by Altered PAT
In our search for C4-methyl SBIs that could regulate plant development, we downregulated the expression of ERG28 by RNA interference (RNAi) in Arabidopsis, using a gene-specific hairpin insert that would potentially induce the accumulation of C4-methyl SBIs. In parallel, we analyzed two Arabidopsis lines (SALK_025834 and SALK_000240) having a T-DNA insertion in the ERG28 gene. In a prescreening analysis, we noted that both RNAi knockdown and T-DNA insertion knockout of Arabidopsis ERG28 led to such broad phenotypic pleiotropy that the effects could be misinterpreted as the result of distinct genes of interest. This phenomenon is reiterated in the progeny of fertile plants. A similar, but less pronounced, morphological variability could be deduced from Arabidopsis sterol biosynthetic mutants hydra1 (Schrick et al., 2002; Souter et al., 2002), smt1/cph/orc (Diener et al., 2000; Schrick et al., 2002; Willemsen et al., 2003; Carland et al., 2010), and fackel/hydra2 (Jang et al., 2000; Schrick et al., 2000).
Thus, to facilitate the analysis of the loss-of-function Arabidopsis ERG28, we categorized the mutant plants into six phenotypic classes according to their vegetative phenotype. We then analyzed two independent populations of RNAi lines in which the expression of ERG28 was strongly reduced. These lines are referred to as erg28R1-1 to erg28R1-6 (Figures 2A and 3) and erg28R2-1 to erg28R2-6 (see Supplemental Figures 4A and 5 online). We adopted the same procedure for the T-DNA lines referred to as erg28T1-1 to erg28T1-6 (SALK_025834) (Figures 2B and 3) and erg28T2-1 to erg28T2-6 (SALK_000240) (see Supplemental Figures 4B and 5 online). The expression level of ERG28 in the six characteristic classes was verified using RT-PCR and quantitative RT-PCR (qRT-PCR) analyses, followed by immunoblot analysis of the ERG28 protein in the RNAi lines (Figure 2A; see Supplemental Figure 4A online) and the T-DNA lines (Figure 2B; see Supplemental Figure 4B online). These data revealed that ERG28 was strongly knocked down in the RNAi lines and knocked out in the T-DNA lines compared with wild-type plants.
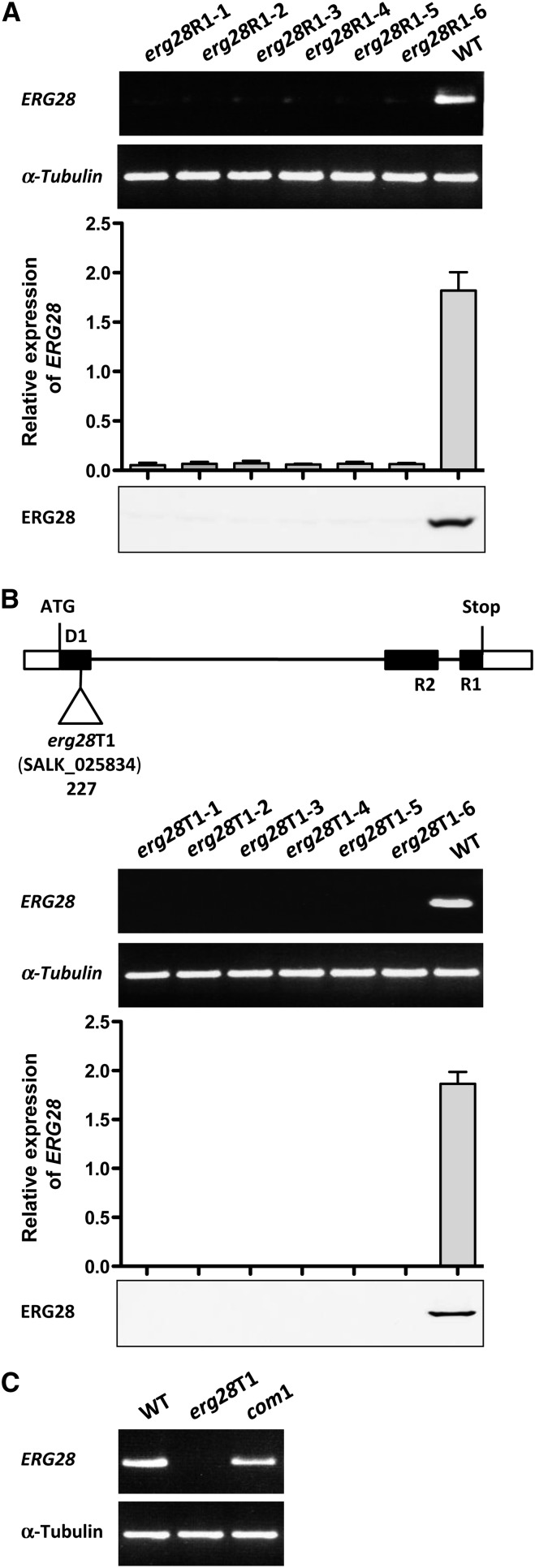
Figure 2.
Characterization of Arabidopsis erg28RNAi (erg28R1-1 to erg28R1-6) and erg28T-DNA (erg28T1-1 to erg28T1-6) Mutants.
(A) erg28RNAi plants were named erg28R1-1 to erg28R1-6. RT-PCR (top) and qRT-PCR analyses (middle) of ERG28 transcript of erg28R1-1 to erg28R1-6 and wild-type (WT) plants, followed by immunoblot analysis (bottom) using anti-Arabidopsis ERG28. α-Tubulin (At1g04820) was used as a positive control for the RT-PCR (27 cycles for α-Tubulin and 35 cycles for erg28R1-1 to erg28R1-6 plants). The results of the qRT-PCR analysis represent mean values (error bars are sd; n = 3).
(B) erg28 T-DNA plants derived from the SALK line (SALK_025834) bearing an insertion in ERG28 (At1g10030) were designated erg28T1-1 to erg28T1-6 and characterized. Schematic diagram (top) shows the T-DNA insertion indicated by the open triangle (black boxes, thin lines, and white boxes indicate exons, introns, and untranslated regions, respectively; D1, R1, and R2 were used as primers), followed by RT-PCR (top middle), qRT-PCR (bottom middle), and immunoblot (bottom) analyses performed as in (A) compared with wild-type plants. The results of the qRT-PCR analysis represent mean values (error bars are sd; n = 3). The combinations of D1-R1 and D1-R2 give the same result.
(C) RT-PCR characterization of erg28T-DNA knockout plants complemented with Arabidopsis ERG28 (com1) compared with wild-type plants. Experimental conditions were as shown in (A).
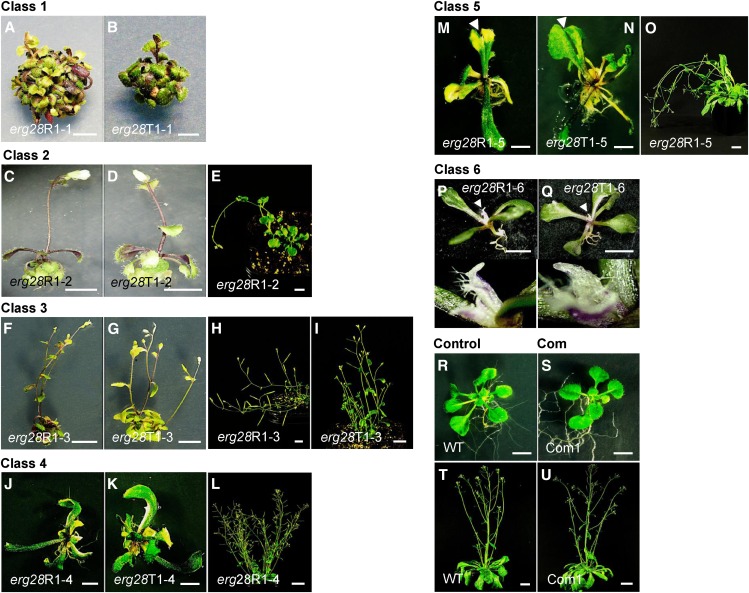
Figure 3.
erg28 Plants Exhibit PAT Inhibition Phenotypes.
Phenotypes resulting from the RNAi knockdown (erg28R1-1 to erg28R1-6) and knockout (erg28T1-1 to erg28T1-6) of ERG28 were categorized into six phenotypic classes according to their vegetative development and compared with wild-type (WT) and complemented (Com1) plants. Bars = 0.5 cm.
(A) and (B) Class 1 erg28R1-1 (A) and erg28T1-1 (B) compacted plants with epinastic leaves and almost rootless. 45-day-old plants.
(C) to (E) Class 2 erg28R1-2 (C) and erg28T1-2 (D) plants have reduced axillary inflorescence, short roots, epinastic leaves, and pendulous stature (E). Thirty-five-day-old ([C] and [D]) and 90-d-old (E) plants.
(F) to (I) Class 3 erg28R1-3 (F) and erg28T1-3 (G) plants develop shorter shoots and roots and have reduced apical dominance ([H] and [I]). Thirty-five-day-old ([F] and [G]) and 90-d-old ([H] and [I]) plants.
(J) to (L) Class 4 erg28R1-4 (J) and erg28T1-4 (K) plants show elongated and asymmetric overgrowth of leaf blades, altered root growth, and reduced apical dominance (L). Forty-day-old ([J] and [K]) and 90-d-old (L) plants.
(M) to (O) Class 5 erg28R1-5 (M) and erg28T1-5 (N) plants display asymmetric leaf blades and leaf fusions (arrowhead). Adult plants show nonerect upright growth (O). Forty-day-old ([M] and [N]) and 90-d-old (O) plants.
(P) and (Q) Class 6 erg28R1-6 (P) and erg28T1-6 (Q) plants show irregular leaf margins and perturbed apical meristems leading to pin-like shoot (arrowhead and close-up view below) and inhibition of root elongation. Fifty-day-old plants.
(R) to (U) Wild-type ([R] and [T]) and erg28T-DNA knockout plants complemented (Com1) with Arabidopsis ERG28 cDNA ([S] and [U]). Twenty-day-old ([R] and [S]) and 50-d-old ([T] and [U]) plants.
Within each class, the RNAi knockdown and T-DNA knockout of Arabidopsis ERG28 displayed nearly identical observable phenotypes with respect to vegetative development. (Figure 3; see Supplemental Figure 5 online). Plants grouped into class 1 (35%, n = 450) were strongly compacted and display short petioles and epinastic leaves with reduced lamina size (Figures 3A and 3B; see Supplemental Figures 5A and 5B online). Leaves of the mutants displayed a dark-green color due to increased chlorophyll content and they also exhibited delayed senescence and increased anthocyanidin accumulation (Figures 3A and 3B; see Supplemental Figures 5A and 5B online). The root system aborted or was drastically reduced compared with wild-type plants. Plants grouped into class 2 (7%) had single frail shoots, and the terminal inflorescence bore one or very few flowers (Figures 3C to 3E; see Supplemental Figures 5C and 5D online). These plants displayed epinastic leaves as observed for class 1 plants and had short roots similar to pin-like mutants (Okada et al., 1991; Bennett et al., 1995; Gälweiler et al., 1998) or wild-type Arabidopsis treated with synthetic PAT inhibitors such as naphthylphthalamic acid (NPA) (Mattsson et al., 1999; Sieburth, 1999). The upright growth of adult plants was perturbed due to weakened rigidity of the stem, a phenotype characteristic of revoluta mutants, which display reduced interfascicular fiber differentiation (Zhong and Ye, 2001), or irx, a cellulose synthase mutant that displays collapsed mature xylem cells (Turner and Somerville, 1997) (Figure 3E). This is reminiscent of the cell wall deficiencies described for Arabidopsis sterol mutants fk, hyd1, and smt1/cph/orc (Schrick et al., 2004b).
Class 3 plants (3%) developed shorter shoots and roots and exhibit loss of apical dominance and nonerect upright growth like class 2 plants (Figures 3F to 3I; see Supplemental Figures 5E and 5F online). Plants grouped in class 4 plants (20%) exhibited elongated, asymmetric leaves (Figures 3J and 3K; see Supplemental Figures 5G and 5H online) and loss of apical dominance (Figure 3L). Class 5 plants (15%) show asymmetric leaf blades and petiole fusions as observed in PAT-inhibited plants (Mattsson et al., 1999; Sieburth, 1999; Donner et al., 2009) (Figures 3M and 3N; see Supplemental Figures 5I, 5J, and 6A to 6C online). Adult plants grouped in class 5 displayed also nonerect upright growth like class 2 mutants, probably due to a defect in the differentiation of interfascicular fibers (Figure 3O; see Supplemental Methods 1 and Supplemental Figures 6D and 6E online). The remainder of the plants (class 6, 20%) developed pin-like apical meristem (Figures 3P and 3Q; see Supplemental Figures 5K and 5L online) and have irregular leaf margins, while the primary root elongation was inhibited and radially expanded (Figures 3P and 3Q; see Supplemental Figures 5K, 5L, 6F, and 6G online). The changes affecting the root system of class 6 plants were also revealed by the differentiation of xylem strands and the randomized formation of root hairs near the root tip at positions normally not occupied by root hairs in the wild-type plants (see Supplemental Methods 1 and Supplemental Figures 6F and 6G online). Notably, the trichoblasts produced root hair outgrowths in a random pattern up to the apex (see Supplemental Methods 1 and Supplemental Figures 6F and 6G online), a pattern that is reminiscent of the pattern induced by deregulated expression of GLABRA2 (GL2), a transcription factor specifying the formation of root hairs (Masucci et al., 1996).
Drastic reduction of root growth and organization have been also been observed in the sterol mutants hyd1 and hyd2/fk (Souter et al., 2002) and smt1/orc/cph (Willemsen et al., 2003), and this was associated with deregulated expression of GL2, thus confirming critical roles played by sterols in the development of root hairs (Ovecka et al., 2010). The survival rate of erg28 plants transferred from the Murashige and Skoog solid medium to soil varied from 0 to 95% (Figure 4). The wild-type phenotype was restored by transformation of erg28 T-DNA knockout mutants with the wild-type ERG28 cDNA (Figures 3R to 3U; see Supplemental Figures 5M and 5N online), demonstrating that the phenotypic pleiotropy of erg28 plants was caused by decreased or abolished expression of ERG28 via knockdown or knockout.
Figure 4.
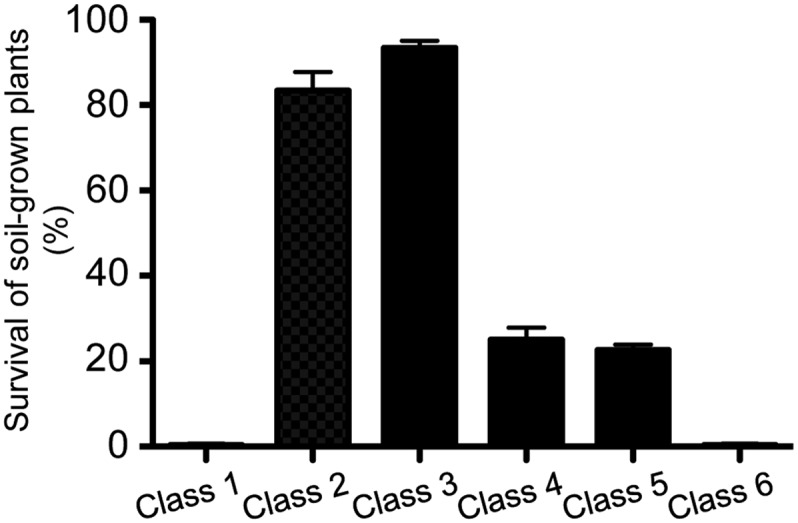
Survival Rate of erg28 Plants Grown on Soil.
Plants belonging to the different phenotypic classes were transferred from Murashige and Skoog solid medium to soil and grown under greenhouse conditions. No development after 7 d was considered as 0% survival. Data shown represent mean values (error bars are sd; n = 3).
Because shoot and root meristems of erg28 plants were functional, we next asked whether treatment with plant hormones could restore the wild-type phenotype. Using erg28 class 1 mutants (erg28R1-1 and erg28T1-1) in which shoot and root development are dramatically altered, we found that treatment with 1 and 2.5 μM indole-3-acetic acid (IAA), gibberellin A3, trans-zeatin, the ethylene precursor 1-aminocyclopropane-1-carboxylic-acid , and 24-epibrassinolide could not restore the wild-type phenotype (see Supplemental Methods 1 and Supplemental Figure 7 online). We also observed that erg28 plants showed reduced primary root elongation in response to auxin (IAA and 1-naphthalene acetic acid) treatments (see Supplemental Methods 1 and Supplemental Figure 8 online). We analyzed the expression pattern of Arabidopsis ERG28 using publicly available transcriptome databases. ERG28 was expressed throughout the life cycle with highest expression level during embryonic and early seed and silique development, in apical tissues, and during root development (see Supplemental Methods 1 and Supplemental Figures 9 and 10 online). This trend was confirmed by qRT-PCR analysis (see Supplemental Figure 11 online). The expression data collected from GENEVESTIGATOR (https://www.genevestigator.com/gv/plant.jsp) and BAR (http://bar.utoronto.ca/welcome.html) did not reveal significant changes in the expression of ERG28 in response to biotic or abiotic stresses.
Loss of ERG28 Function Leads to the Inhibition of PAT
We found that the phenotypes of erg28 plants (Figure 3; see Supplemental Figure 5 online) markedly recapitulate multiple developmental phenotypes characteristic of the auxin efflux carrier pin-formed1 (pin1) mutant (Okada et al., 1991; Gälweiler et al., 1998; Reinhardt et al., 2003) (Figures 3P and 3Q; see Supplemental Figures 5K and 5L online), the auxin-dependent transcription factor monopteros (Berleth and Jürgens, 1993), and the auxin transcriptional repressor (Aux/IAA) mutants (Wang et al., 2005) in which cotyledons (Berleth and Jürgens, 1993) and leaves (Wang et al., 2005) were fused (Figures 3M and 3N; see Supplemental Figures 5I, 5J, 6A, and 6B online). Notably, the trichoblast differentiation zone of the erg28R2-6 plants (see Supplemental Figure 6F online) is reminiscent of the gnomR5 mutation, which causes PIN1 mislocalization (Geldner et al., 2004). Also, erg28 plants bore a striking resemblance to plants in which PAT has been inhibited by synthetic inhibitors such as NPA, the most widely used PAT inhibitor (Mattsson et al., 1999), or 2-[4-(diethylamino)-2 hydroxybenzoyl] benzoic acid (Kim et al., 2010). Taken together, these observations suggest that PAT inhibition could be responsible for the pleiotropic phenotypes of erg28 plants.
To test this hypothesis, we analyzed the shoot-to-root (basipetal) transport of auxin in wild-type plants and erg28 plants using tritium-labeled IAA, a naturally occurring auxin. Basipetal IAA transports was reduced in erg28R1-2 and erg28T1-2 plants compared with wild-type plants (Figure 5A). Reduction of PAT is known to affect the differentiation of interfascicular fibers that provide mechanical support for erect stature of stems (Zhong and Ye, 2001). Consistent with this finding, the decreased stem rigidity of erg28 plants was associated with parenchymatous interfascicular fiber cells compared with highly lignified interfascicular fiber cells of wild-type plants (see Supplemental Methods 1 and Supplemental Figures 6D and 6E online).
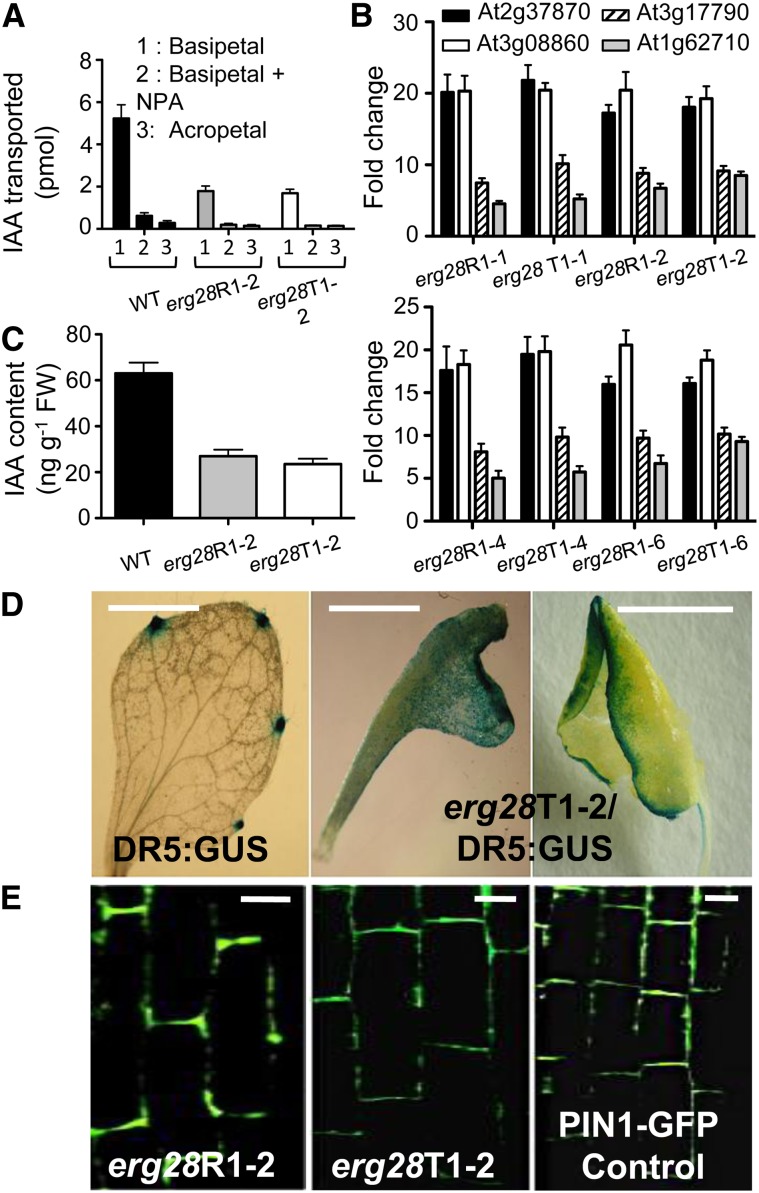
Figure 5.
PAT Characteristics in erg28 Plants.
(A) Defective PAT in erg28R1-2 and erg28T1-2 plants. Thirty-five-day-old erg28R1-2, erg28T1-2 developing usable inflorescence stems, and wild-type (WT) plants were used to determine IAA transport using tritium-labeled IAA in the presence or absence of NPA (15 μM) (error bars are sd; n = 5).
(B) Expression of four NPA-responsive genes (indicated by their AGI codes) in erg28R1-1, erg28R1-2, erg28R1-4, erg28R1-6, erg28T1-1, erg28T1-2, erg28T1-4, and erg28T1-6 plants. Thirty-five-day-old plants were used for qRT-PCR analysis. Data represent the fold change over the wild type (error bars are sd; n = 3).
(C) IAA content in 35-d-old wild-type, erg28R1-2, and erg28T1-2 plants (error bars are sd; n = 3). FW, fresh weight.
(D) Alteration of the DR5:GUS auxin reporter gene in erg28 background. Leaves from 35-d-old DR5:GUS (left, control) and erg28T1-2/DR5:GUS (middle and right) plants were used. Bars = 0.25 cm.
(E) Immunolocalization of PIN1 using anti-At-PIN1 in roots of 10-d-old erg28R1-2 (left) and erg28T1-2 (middle) plants. PIN1 localizes to the basal side of the cell, as shown in PIN1-GFP plant (right) used as a control. Bars = 5 μm.
PAT impairment following NPA treatment induces the expression of several NPA-responsive genes (Wenzel et al., 2008). Using real-time RT-PCR analysis, we found that the expression of At2g37870, At3g08860, At3g17790, and At1g62710, which belong to the class of NPA-responsive genes (Wenzel et al., 2008), were induced in erg28 plants in the absence of NPA (Figure 5B). In addition, the free IAA content in erg28 plants was at least twofold lower than in wild-type plants (Figure 5C), consistent with previous data demonstrating that NPA promotes feedback inhibition of IAA biosynthesis (Ljung et al., 2001). This is also consistent with the fact that the free IAA content is 12-fold decreased in the pin1 mutant relative to wild-type plants (Okada et al., 1991). Taken together, these data demonstrate that knockdown or knockout of the Arabidopsis ERG28 inhibits PAT.
Because PAT inhibition affects auxin homeostasis by locally increasing (Muday et al., 1995) or decreasing (Ljung et al., 2001) auxin concentration in tissues, we used a complementary approach to evaluate the spatial distribution of auxin responses in tissues. We used the DR5:GUS (for β-glucuronidase) gene fusion, which is reactive to IAA at concentrations as low as 10−5 to 10−8 M (Ulmasov et al., 1997). DR5:GUS (Ulmasov et al., 1997) was introgressed into the erg28 background. Relative to the wild type, DR5:GUS was expressed diffusively near the leaf margin in the erg28 background (Figure 5D), consistent with PAT inhibition and enhanced IAA-dependent response or accumulation in the margin, a phenomenon that contributes to leaf epinasty (Figure 5D). The misexpression of DR5:GUS in erg28 background is reminiscent of deregulated expression of DR5:GUS in sterol mutant plants smt1/cph/orc (Willemsen et al., 2003), cvp1 (Carland et al., 2010), and fk/hyd2 (Souter et al., 2002; Pullen et al., 2010) as well as wild-type Arabidopsis leaves treated with NPA (Mattsson et al., 2003).
Previous studies have shown that the first developing leaves represent a key source of auxin transported to roots (Ljung et al., 2001; Bhalerao et al., 2002), and PIN1 is specifically required for the translocation of auxin from the shoot to the root tip (Okada et al., 1991; Gälweiler et al., 1998). However, the activity of PIN1 and PIN2 is affected by their cycling between plasma membrane and endosomal compartments (Geldner et al., 2001; Grebe et al., 2003; Jaillais et al., 2006). Plant sterols are required for cell polarity (Willemsen et al., 2003) and are components of endocytic vesicles that accumulate in ARA6-positive endosomes (Grebe et al., 2003; Men et al., 2008). Although PAT is not affected in the sterol mutants cvp1/smt2 (Carland et al., 1999), the rate of PAT is reduced in sterol mutants smt1/cph/orc relative to the wild type concomitantly to the mislocalization of PIN1 and PIN3 (Willemsen et al., 2003). Frequent lateral instead of basal localization of PIN1 and mislocalization of PIN2 have been observed in the cyclopropylsterol isomerase1-1 mutant (Men et al., 2008). On the other hand, PIN1 and PIN2 are in general correctly localized in sterol mutants hyd1 or hyd2/fk (Souter et al., 2002; Pullen et al., 2010). Because PIN1 must be properly localized to the basal end of vascular tissue cells (Gälweiler et al., 1998) to exert its function, it remained possible that mislocalization of PIN1 may have impacted PAT inhibition in erg28 plants. To test this hypothesis, we used antibody raised against PIN1 to reveal its localization. By immunofluorescence microscopy analysis, we observed that PIN1 was correctly localized to the basal end of vascular cells (Gälweiler et al., 1998), as noted for PIN1-GFP used as a control (Feraru et al., 2011) (Figure 5E). Together, these data show that knockdown or knockout of the Arabidopsis ERG28 leads to PAT inhibition via a mechanism that did not probably perturb the subcellular localization of PIN1, unlike to previous Arabidopsis sterol smt1/cph/orc mutants (Willemsen et al., 2003).
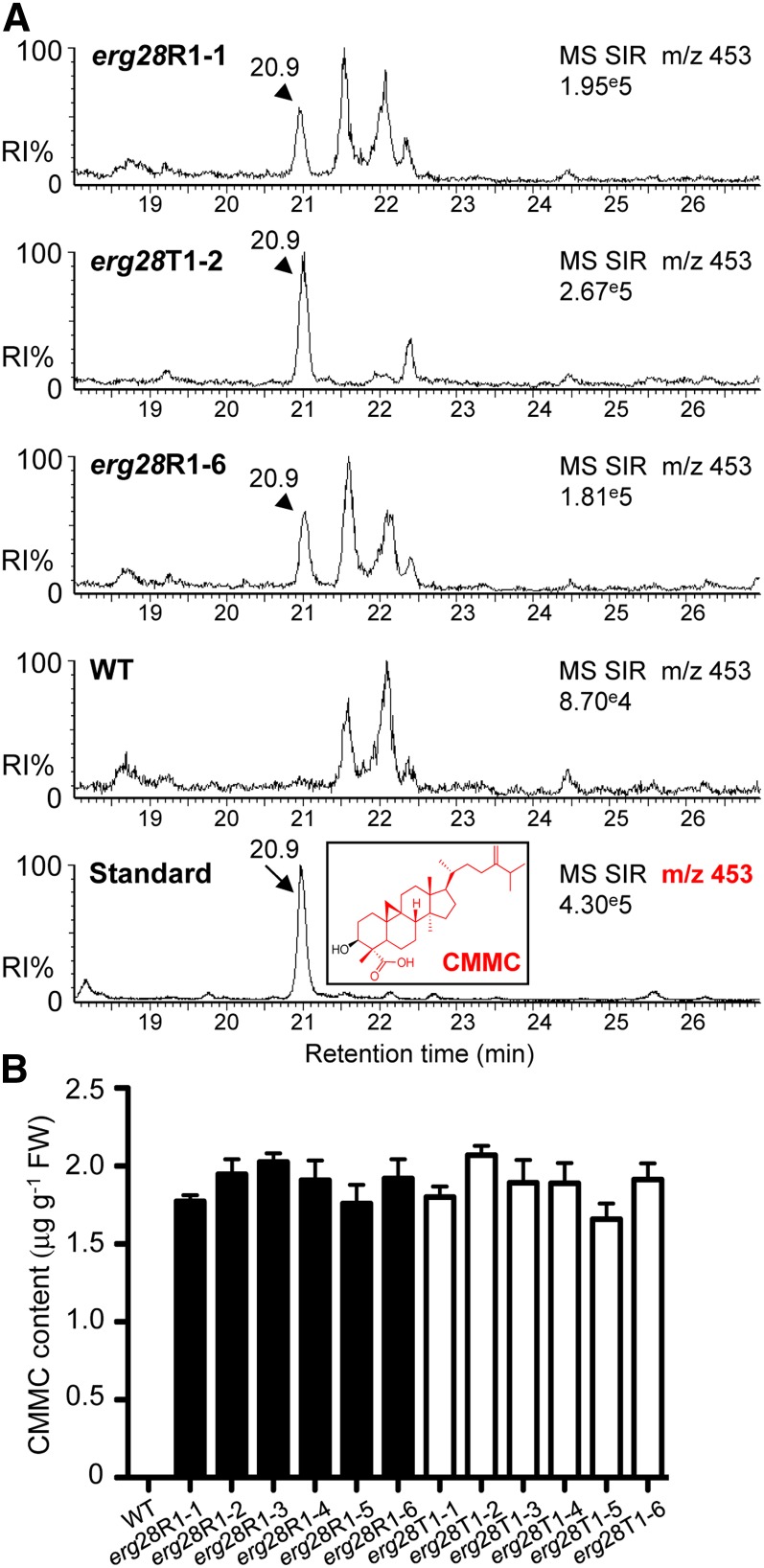
Loss of ERG28 Function Leads to the Accumulation of 4-Carboxy-4-Methyl-24-Methylenecycloartanol, a Cryptic SBI
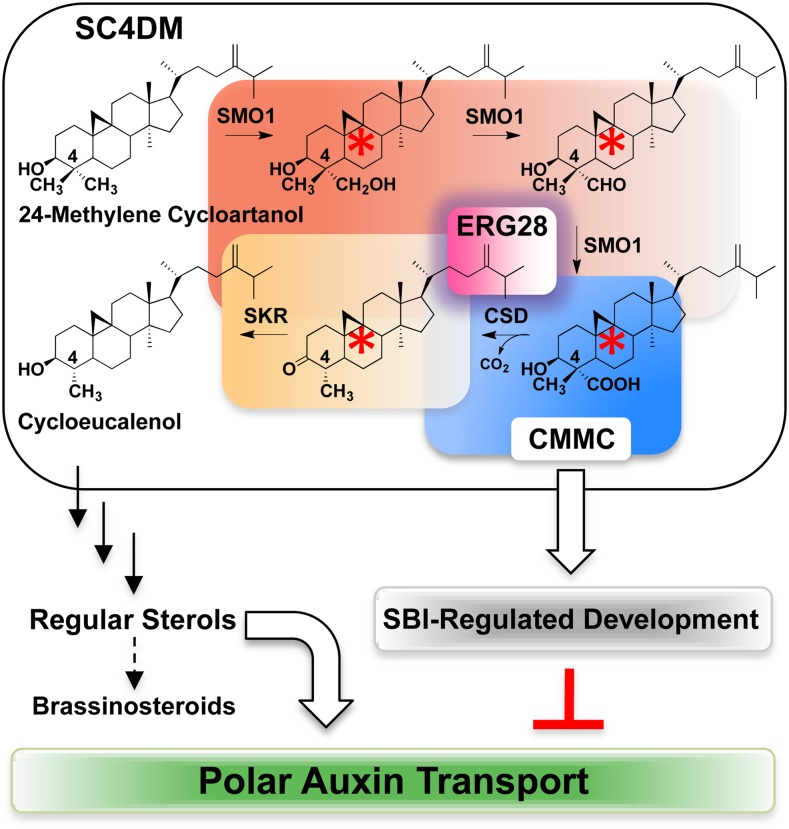
To further explore the role of ERG28, we analyzed the sterol composition of wild-type and erg28 plants using ultraperformance liquid chromatography–tandem mass spectrometry (UPLC-MS/MS) with atmospheric pressure photon ionization (APPI). Consistent with previous studies (Canabate-Díaz et al., 2007; Lu et al., 2007; Rhourri-Frih et al., 2009; Shui et al., 2011), underivatized sterol molecules were not detected as quasimolecular ion (M+H), but as [M+H-H2O], which was used as a precursor ion. We found that the regular sterol composition, including sitosterol, stigmasterol, and campesterol, of erg28 plants relative to the wild type was not significantly different (see Supplemental Figure 12 online). In a similar vein, the level of common sterol intermediates did not show a significant change between wild-type and erg28 plants (see Supplemental Figures 13 and 14 online). Thus, it seems unlikely that PAT inhibition in erg28 plants results from altered regular sterol composition or a block in the sterol pathway. This prompted in-depth analysis of the fraction that was likely to contain polar sterol derivatives using UPLC-MS/MS with APPI. We detected in the polar sterol fraction obtained from erg28 plants an intense precursor ion mass-to-charge ratio (m/z) 453 [M+H-H2O] (Figure 6A) that gave in the multiple reaction monitoring (MRM; molecular ion → fragment ion) mode used to identify specific fragments the following precursor-product (m/z) transition [(453 > 109), (453 > 123), (453 > 149), (453 > 191), (453 > 205), and (453 > 369)] (see Supplemental Figures 15 to 18 online). These fragments matched exactly the fragmentation spectrum of an authentic 4-carboxy-4-methyl-24-methylenecycloartanol (CMMC), also known as 1-dehydroxy-23-deoxojessic acid (Figure 6A). Under our experimental conditions, we could not detect CMMC in wild-type and complemented plants. This could be due to the fact that CMMC is efficiently channeled in the SC4DM multienzyme complex. Taken together, these data demonstrate that knocking down or knocking out Arabidopsis ERG28 did not interrupt the sterol pathway as shown in yeast (Gachotte et al., 2001) (see Supplemental Methods 1 and Supplemental Table 2 online) but promotes the accumulation of CMMC (Figure 6B), a cryptic C4-methyl SBI of the Arabidopsis SC4DM multienzyme complex tethered by ERG28 (Bouvier et al., 2005) (Figures 1A and 1B; see Supplemental Figure 1 online).
Figure 6.
Impaired PAT Phenotypes of erg28 Plants Correlate with CMMC Accumulation.
(A) UPLC-MS/MS single ion recording (SIR) chromatogram at m/z 453 shows the presence of CMMC (arrowhead) in lipid extracts of 35-d-old erg28R1-1, erg28T1-2, and erg28R1-6 but not in 35-d-old wild-type (WT) plants and shows the authentic CMMC standard (arrow, inset: chemical structure). Detailed analysis using MRM mode, product ion spectra, and the fragmentation pathways are shown in Supplemental Figures 10 to 13 online. RI, relative intensity.
(B) Quantification of CMMC in erg28R1 and erg28T1 plants (error bars are sd; n = 3).
[See online article for color version of this figure.]
CMMC Inhibits PAT and Competes with the PAT Inhibitor NPA
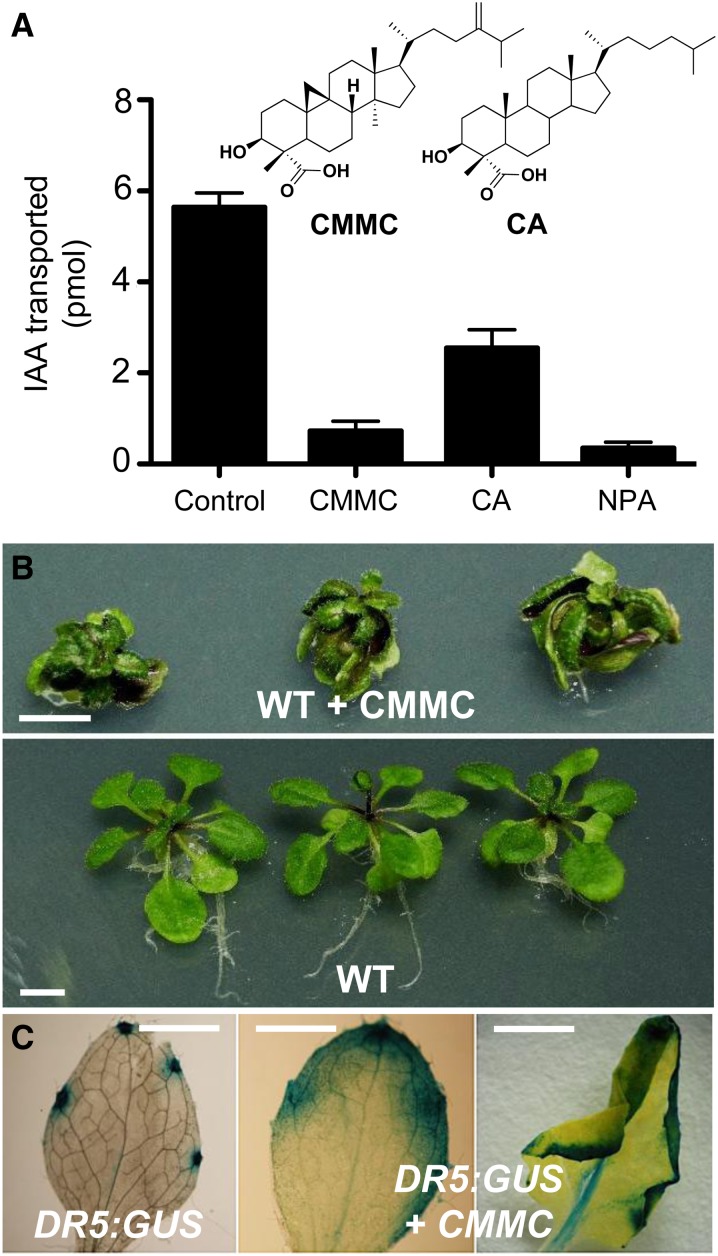
Collectively, the data described above indicate that alteration of the regular sterol composition affecting brassinosteroid synthesis, membrane integrity, or plasma membrane mislocalization of PIN1 could not explain the inhibition of PAT in erg28 plants. Since our results demonstrate that the accumulation of CMMC in Arabidopsis is correlated with the development PAT-related phenotypes, we wondered whether CMMC acts as a PAT inhibitor. This prompted us to test CMMC and a synthetic analog cholestanoic acid for their ability to directly inhibit PAT using tritium-labeled IAA. We found that PAT was inhibited by CMMC and to a lesser extent by cholestanoic acid (Figure 7A). Consistent with PAT inhibition, exogenous application of CMMC (15 μM) to wild-type plants promoted PAT-deficient phenotypes, including the formation of epinastic incurvation and inhibition of primary root growth (Figure 7B). Notably, the expression in DR5:GUS lines (Ulmasov et al., 1997) was deregulated following exogenous treatment with CMMC relative to the control (Figure 7C). Taken together, these data demonstrate that knocking down or knocking out Arabidopsis ERG28 induces the accumulation of CMMC, which in addition to its known SBI role acts as a PAT inhibitor.
Figure 7.
CMMC and the Synthetic Cholestanoic Analog Inhibit PAT.
(A) Inflorescence stems from 35-d-old wild-type plants were used to analyze IAA basipetal transport as described in Figure 5A, using tritium-labeled IAA alone (control) and in the presence of 15 μM CMMC, synthetic cholestanoic acid (CA), or NPA (error bars are sd; n = 3).
(B) CMMC induces altered PAT phenotypes. Wild-type (WT) plants were grown on MS agar for 7 d before transfer to MS agar containing DMSO (control) or the same medium supplemented with 15 μM CMMC for 20 d. Bars = 0.5 cm.
(C) CMMC modulates the expression pattern of DR5:GUS auxin reporter gene. DR5:GUS plants were grown for 7 d on Murashige and Skoog agar before transfer to Murashige and Skoog agar alone (DR5:GUS) or Murashige and Skoog agar supplemented with 15 μM CMMC (DR5:GUS + CMMC) and grown for 20 d before GUS detection. Bars = 0.25 cm.
The mechanism by which CMMC inhibits PAT is unknown. Because NPA inhibits ABCB auxin efflux transporters (Kim et al., 2010) and indirectly PIN auxin efflux carriers due to ABCB–PIN interactions (Blakeslee et al., 2007), effective PAT inhibitors would potentially interact with high-affinity binding sites for NPA in microsomal membranes (Noh et al., 2001). To investigate this, we assessed whether CMMC competes with high-affinity NPA binding sites on cell membranes by incubating wild-type microsomal membranes with tritium-labeled NPA in the presence and absence of CMMC. We found that 20 nM CMMC reduced high-affinity binding of NPA on wild-type microsomal membranes by ∼40% (see Supplemental Methods 1 and Supplemental Figure 19A online). ABCB1 and ABCB19 represent specific NPA binding proteins (Noh et al., 2001). Because in silico docking analyses demonstrate that NPA interacts with the nucleotide binding domains 1 and 2 (NBD1 and NBD2) of ABCB auxin efflux transporters (Kim et al., 2010), we examined in silico whether CMMC could bind to the same sites. From several candidate template structures (Schmitt et al., 2003; Zaitseva et al., 2005; Mares-Sámano et al., 2009), we selected the ABC transporter HlyB (Zaitseva et al., 2005) because it was solved as a dimeric structure with bound ATP, which represents an essential cofactor of ABC transporters. We found that the binding site of CMMC was predicted to be similar to that of NPA in ABCB1 and ABCB19 auxin efflux transporters (see Supplemental Methods 1 and Supplemental Figures 19B to 19I online). The predicted docking energies (EB) corresponding to the positions occupied by CMMC in the NBD1 or NBD2 of ABCB1 and ABCB19 are very similar (see Supplemental Methods 1 and Supplemental Table 3 online). Although NPA and CMMC are structurally different, we noted a slight variation in the predicted docking energy of ΔEB = 0.2 to 0.7.
DISCUSSION
In living organisms, basic growth requirement of regular sterol end products for membrane integrity is usually associated either with the presence of the 3β-hydroxy group and the lack of C4- and C14-methyl groups or with their conversion into brassinosteroids in plants. However, beyond their biosynthetic role, SBIs have emerged as a reservoir of signaling molecules involved in innate immunity (Blanc et al., 2011; Panicker et al., 2012), inflammation responses (Blanc et al., 2011; Panicker et al., 2012; Spann et al., 2012), meiosis stimulation, and cell proliferation (Byskov et al., 1995; He et al., 2011) in mammals. SBIs are also involved in oxygen sensing in fungi (Espenshade and Hughes, 2007) but as yet have no equivalent role in plants. In addition, there is the long-standing question of whether sterol signals other than brassinosteroids exist in plants.
Previous studies have shown that loss-of-function mutations affecting genes encoding the enzymes that catalyze the individual postsqualene biosynthetic steps up to 24-methylenelophenol and 24-ethylenelophenol (see Supplemental Figure 1 online) induce pleiotropic postembryo defects leading to seedling lethal phenotypes that are not rescued by either downstream regular sterols or sterol-derived brassinosteroids. These mutants include sterol methyl transferase1/orc/cephalopod, hydra1, fackel/hydra2, cotyledon vascular pattern1, cyclopropylsterol isomerase1-1, and cyp51A2 (Diener et al., 2000; Jang et al., 2000; Schrick et al., 2000, 2002; Carland et al., 2002, 2010; Souter et al., 2002; Willemsen et al., 2003; Kim et al., 2005; Men et al., 2008). In these mutants, the endocytic trafficking of auxin transporters, such as PIN1, is perturbed due to the drastic alteration of the sterol profile caused by deficient enzyme activities or interruption of the pathway (Boutté and Grebe, 2009). The finding that the loss-of-function of Arabidopsis ERG28, which is implicated in key postsqualene steps (up to episterol and Δ7-avenasterol) (see Supplemental Figure 1 online), results in plants that are largely viable and fertile except for those belonging to classes 1 and 6 (Figures 3 and 4; see Supplemental Figure 5 online) implies that Arabidopsis erg28 plants are different from previously known plant sterol mutants.
Notably, the loss of function of ERG28 induces unprecedented accumulation of CMMC in Arabidopsis. This occurs without interrupting the sterol pathway, in good agreement with the fact that ERG28 has no enzyme activity per se (Figure 8). In the yeast erg28 mutant, sterol biosynthesis is disrupted at the level of SC4DM and results in the depletion of ergosterol, the end product of the pathway (Gachotte et al., 2001) (see Supplemental Methods 1 and Supplemental Table 2 online). This suggests that key differences exist between the topological organization of yeast, mammalian, and plant sterol biosynthetic enzymes in the endoplasmic reticulum. This is supported by the observation that, in yeast and mammals, the SC4DM enzyme complex operates via two rounds of consecutive C4-demethylation reactions (C4 and C4), whereas in plants, the two C4 demethylation reactions are interrupted by two additional steps that are not catalyzed by the SC4DM enzyme complex [C14 demethylation and C14 reduction, yielding 4-α-methylergosta-8,14,24(28)-trienol and 4-α-methylfecosterol] (see Supplemental Figure 1 online) (Bard et al., 1996; Gaylor, 2002; Bouvier et al., 2005).
Figure 8.
Substrate Channeling and PAT Safeguarding Roles of ERG28.
Due to its tethering role, ERG28 restricts the release of SBIs (red asterisks), including CMMC, from the SC4DM multienzyme complex in wild-type plants. Loss of ERG28 function leads to uncontrolled and nonhomogeneous release of CMMC from the SC4DM complex. This alters PAT without depleting and interrupting the downstream steps leading to synthesis of bulk membrane sterols and brassinosteroids.
[See online article for color version of this figure.]
The accumulation of CMMC in erg28 plants was associated with multiple developmental changes diagnostic of altered PAT. We further demonstrate that CMMC acts as a PAT inhibitor in addition to its role as an intermediate of sterol biosynthesis (Bouvier et al., 2005). The in silico docking studies tentatively suggested that CMMC may target ABC transporters involved in auxin efflux. Under normal conditions, CMMC is strictly channeled within the SC4DM complex tethered by ERG28 to produce the bulk membrane sterols and brassinosteroids. Due to the loss of function of ERG28 in erg28 plants, a portion of CMMC released from the SC4DM complex would be free to inhibit PAT (Figure 8). In this context, parallels can be drawn between the SC4DM complex and the flavonoid multienzyme complex. Once deregulated, both pathways liberate biosynthetic precursors that lead to PAT inhibition (Peer and Murphy, 2007). The array of developmental phenotypes displayed by erg28 plants was probably related to the fact that CMMC is nonhomogeneously released. Further experiments are required to determine the spatial distribution of CMMC in plant tissues and organs and under different physiological conditions. Due to the phenotypic pleiotropy displayed by erg28 plants, one may predict that controlled release of CMMC from the SC4DM complex could offer a perspective to modulate plant development.
The regulatory role of SBI is further supported by the finding that one of the atypical sterols accumulating in the Arabidopsis sterol biosynthesis mutant fackel can upregulate mRNA levels of several genes regulating cell expansion and proliferation (He et al., 2003). Importantly, CMMC belongs to the C4-methyl sterol series, known to be weak membrane modifiers (Bittman, 1997) and implicated in additional regulatory processes beyond sterol biosynthesis in nonplant phyla (Byskov et al., 1995; Hughes et al., 2007; Hannich et al., 2009; He et al., 2011; Lim et al., 2012). Recent data from studies of the mammalian SC4DM genes demonstrate that downregulation of SMO and CSD, but not of SKR and ERG28, leads to the accumulation of C4-methyl sterols that alter the endocytic trafficking of the epidermal growth factor receptor and accelerate its degradation in the lysosomes of cancer cells (Sukhanova et al., 2013). This trend was supported by coexpression and protein–protein analyses using the STRING database (http://string-db.org/), suggesting that yeast and mammalian SMO and CSD interact with protein components involved in vesicular trafficking (Sukhanova et al., 2013). Interestingly, this phenomenon could be related to the propensity of CSD to translocate to plasma membranes (Xue et al., 2013) and lysosomes (Parent et al., 2009) in cancer cells. Similarly, CSD and SKR associate to lipid droplets in yeast (Mo et al., 2003; Natter et al., 2005), like CSD in mammals (Caldas and Herman, 2003; Ohashi et al., 2003). The nonsterol biosynthetic role of the SC4DM complex protein components is supported by the observation that, even in sterol auxotrophs such as Drosophila melanogaster and Caenorhabditis elegans, an SMO ortholog whose function is unknown is coexpressed with genes involved in protein trafficking and folding (Vinci et al., 2008).
In plants, the connection between the SC4DM complex protein components and the trafficking machinery has not been demonstrated. In silico analysis using available resources (Tohge and Fernie, 2012), including the STRING database, reveals that in plants, neither available coexpression databases nor interactome databases could provide a clue or establish causality between SC4DM complex protein components and the protein trafficking machinery. This illustrates again the key differences between the SC4DM complex organization in yeast and mammals one side and plants on the other side. Nevertheless, it is interesting to note that apart from its metabolic function, the SC4DM enzyme complex tethered by ERG28 has potential to be recruited as a reservoir of signaling molecules or serves additional roles. Thus, as observed in several sterol or triterpene mutants, we do not exclude the possibility that CMMC may have multiple targets. The multitarget effect could implicate transcription factors, such as PHABULOSA, PHAVOLUTA, REVOLUTA, and ATHB8, which possess a putative lipid/sterol binding domain (Schrick et al., 2004a). An additional mechanism by which the ERG28 loss of function induces PAT inhibition phenotypes could be through the interaction between ethylene, auxin signaling, and sterols. In this scenario, it is interesting to note that the sterol mutants hydra1 and fackel/hydra2 display enhanced ethylene signaling and deregulated auxin response (Souter et al., 2002, 2004; Pullen et al., 2010). The root phenotype of these mutants is partially restored to the wild type by inhibitors of ethylene and auxin signaling or in etr1, ein2, axrr, or axr mutant backgrounds (Souter et al., 2002, 2004). Although plausible, this mechanism must be tempered by the fact that PAT inhibitors do not apparently impair the effect of ethylene on root growth (Růzicka et al., 2007).
In conclusion, our study revealed an additional level of interaction between sterols and auxin and broadens to plants the signaling functions of SBIs that already have been documented in different nonplant phyla (Janowski et al., 1996; Castrillo et al., 2003; Bensinger et al., 2008; Blanc et al., 2011; He et al., 2011; Lim et al., 2012; Panicker et al., 2012; Spann et al., 2012). By controlling the release and accumulation of CMMC and probably structurally related SBIs, ERG28 not only facilitates substrate channeling in the SC4DM enzyme complex as shown in yeast and mammals but also exerts an essential role during plant development by restricting the disruption of PAT, which stringently regulates several stages of plant growth and development.
METHODS
Chemicals
IAA (20 Ci/mmol) was obtained from Hartmann Analytic and nonradioactive NPA from Sigma-Aldrich. Commercial CMMC standard known as 1-dehydroxy-23-deoxojessic acid was obtained from Quality Phytochemicals.
Plant Materials and Growth Conditions
Arabidopsis thaliana (ecotype Columbia) and Nicotiana benthamiana were used as wild-type plants. Arabidopsis plants were grown on solidified (0.8% [w/v] agar) Murashige and Skoog medium containing 3% Suc or grown on soil under greenhouse conditions at 23°C using a 16-h daylength. For inhibitor treatments, the appropriate concentrations were solubilized into DMSO, and control plants were treated with the same volume of DMSO.
Arabidopsis erg28 RNAi and T-DNA Lines
erg28RNAi lines (erg28R1-1 to erg28R1-6 and erg28R2-1 to erg28R2-6) were generated using a short inverted repeat (hairpin) as described (Jadid et al., 2011) and oligonucleotide primers listed in Supplemental Table 4 online. A 174-bp fragment of Arabidopsis ERG28 cDNA (At1g10030) containing BamHI-EcoRI and BglII-EcoRI sites was amplified, digested by EcoRI, self-ligated, and digested with BamHI and BglII. The resulting 331-bp fragment was cloned in the BamHI-BglII sites of the ImpactVector 1.1 (Jadid et al., 2011) for plant transformation according to the floral dip procedure. Insertion mutant lines SALK_025834 and SALK_000240 were characterized as erg28T1-1 to erg28T1-6 and erg28T2-1 to erg28T2-6 based on segregation analysis, genomic PCR, RT-PCR, and immunological analysis with anti-Arabidopsis ERG28 antibody. Arabidopsis erg28T-DNA knockout plants were complemented by ERG28 cDNA cloned in the XbaI site of the pCAMBIA-1300 vector (Jadid et al., 2011).
Production of Recombinant Proteins and Antibodies
PCR-amplified cDNAs encoding the full-length Arabidopsis ERG28 and SMO1 or truncated CSD (134 to 322) and SKR (98 to 229) were inserted into the pBAD/Thio-TOPO vector (Invitrogen) before expression in Escherichia coli TOP10. Recombinant proteins were purified according to the manufacturer’s manual. The polyclonal anti-Arabidopsis ERG28 antibody was raised in rabbits.
Affinity Interactions between ERG28 and SC4DM Enzymes
ERG28 and SC4DM enzymes interactions were analyzed using GFP-tagged proteins transiently coexpressed in N. benthamiana leaves according to a described procedure (Mialoundama et al., 2009; Jadid et al., 2011). Constructs encoding Arabidopsis SMO1-GFP, CSD-GFP, SKR-GFP, and ERG28-GFP in the pCATs vector were amplified by PCR using the oligonucleotide primers (see Supplemental Table 4 online) and inserted into the pKYLX71-35S2 vector (Mialoundama et al., 2009; Jadid et al., 2011). N. benthamiana transfected leaves were collected 72 h after infection, freeze- dried, and homogenized with the extraction buffer (50 mM Tris-HCl, pH 7.6, containing 2 mM phenylmethylsulfonyl fluoride, 0. 2% Triton X-100, and 3% digitonin) using 1 g of leaves per 2 mL of extraction buffer. After centrifugation at 100,000g for 1 h, the supernatant (1 mL) was treated with protein A-sepharose (GE Healthcare) to eliminate nonspecific binding and then incubated with 150 μL of anti-Arabidopsis ERG28-sepharose prepared as described previously (Schneider et al., 1982). The sepharose beads were washed with 50 mM Tris-HCl, pH 7.6, containing 0.1 M KCl, 2 mM phenylmethylsulfonyl fluoride, and 0.3% digitonin before elution with bound 0.1 M Gly, pH 2.5, and immunoblotting using anti-Arabidopsis ERG28 or anti-GFP. The pull-down assay was performed as described previously (Jadid et al., 2011) using biotinylated recombinant Arabidopsis ERG28 attached to streptavidin agarose, to pull down recombinant Arabidopsis SMO1 (1 to 298), CSD (134 to 322), and SKR (98 to 229) expressed in E. coli. Biotinylated ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) was used as a negative control.
Immunocytochemical localization of PIN
Arabidopsis PIN1 was immunolocalized from the roots of 10-d-old seedlings using an antibody-based fluorescence imaging (Müller et al., 1998). Goat anti-Arabidopsis PIN1 (Anti-At_PIN1) (Santa Cruz Biotechnology) and donkey anti-goat IgG-fluorescein isothiocyanate (Santa Cruz Biotechnology) were used as primary and secondary antibodies at dilutions of 1:100 and 1:200 dilution, respectively. Images were obtained using a Zeiss LSM510 confocal laser scanning microscope.
DR5:GUS Auxin Reporter Analysis
DR5:GUS (Ulmasov et al., 1997) was introduced into the erg28 background by crossing the DR5:GUS lines with the erg28T1-2 knockout plants. GUS activity was monitored as described previously (Jefferson et al., 1987) using 20- to 35-d-old plants derived from five independent experiments.
qRT-PCR Analysis
Total RNA was extracted from the different plant lines using the NucleoSpin RNA kit (Macherey-Nagel) according to the manufacturer’s instructions. Two micrograms of total RNA was reverse transcribed in a total volume of 40 μL with 2.5 μM oligo(dT)20, 0.5 mM deoxynucleotide triphosphate, and 400 units of Superscript III reverse transcriptase (Invitrogen). Real-time PCR was performed in a total reaction volume of 10 μL containing specific primers (2.5 µM each), which were designed using the Probe Finder software (Roche) and are listed in Supplemental Table 4 online, 1 μL of cDNA, and 4 μL of Green I Master Mix (Roche) in a Light Cycler 480 II apparatus (Roche) according to the manufacturer’s instructions. Results were obtained from three independent biological samples (three analytical replicates each). The cycle threshold (Ct) was used to determine the relative expression level using the 2−ΔΔCt method (Livak and Schmittgen, 2001). The relative expression levels were determined by normalizing the PCR threshold cycle number of each gene with that of TIP41-LIKE and GAPDH genes using GenEx Pro software (MultiD Analyses; www.gene-quantification.de/datan.html).
UPLC-MS/MS Analysis of Sterols and Derived Molecules
Total lipids were extracted from freeze-dried plant tissue using dichloromethane/methanol (2/1, v/v). This was followed by UPLC-MS/MS analysis on a Waters Quattro Premier XE equipped with an APPI source and coupled to an Acquity UPLC system (Waters) with a diode array detector. Chromatographic separation was achieved using an Acquity UPLC BEH C18 column (100 × 2.1 mm, 1.7 μm; Waters) operated at 46°C with a flow rate of 0.3 mL min−1 using a gradient of two solvents, A (25% water in methanol and 0.01% formic acid) and B (99.99% isopropanol and 0.01% formic acid) (A for 2 min, increased to 55% B in 25 min, 55% B hold for 4 min, then increased to 100% B in 8 min, hold for 3 min). The parameters for mass spectrometry and tandem mass spectrometry detection and APPI ionization were as indicated (nebulizer gas flow, 50 L⋅h−; desolvation gas flow, 500 L⋅h−1; APPI probe temperature, 450°C; source temperature, 120°C; capillary voltage, 1.5 kV; cone voltage, 25 V; positive ionization mode; low- and high-mass resolution, 15 for both mass analyzers; ion energies 1 and 2 were 0.2 and 1 V; entrance and exit potential were 30 V; and detector [multiplier] gain was 650 V). Mass spectra of sterol-derived fragments were acquired with a scan m/z range of 350 to 1000 atomic mass units. Data acquisition and analysis were performed with the MassLynx MS software (version 5.1).
Auxin Transport
PAT was determined using inflorescence stem segments (Okada et al., 1991; Brown et al., 2001; Lewis and Muday, 2009) placed in the normal (acropetal) or inverted orientation (basipetal transport) in 5 mM MES buffer, pH 5.5, containing 1% Suc and 1.25 μM [3H]IAA in the presence and absence of 15 μM CMMC, cholestanoic acid, or NPA used as PAT inhibitors. Following incubation for 10 h at 20°C, the radioactivity of the nonsubmerged portion of the stem was determined by liquid scintillation counting.
Quantification of Free Auxin (IAA)
Freeze-dried, powdered plant tissues were extracted with cold acetone, dried under argon, and resuspended in methanol before UPLC-MS/MS analysis using an electrospray ionization source (Acquity UPLC HSS C18 column; 100 × 2.1 mm, 1.8 µm; Waters) operated at 33°C with a flow rate of 0.38 mL⋅min−1 and a gradient of two solvents: A (99.9% water and 0.1% formic acid) and B (99.9% methanol and 0.1% formic acid) (5% B in A for 2 min, increased to 100% B in 10 min, hold for 10 min). The capillary voltage was 3 kV, and the ionization mode was positive. The selected ion recording mass spectrometry mode was used to determine the parent mass transition of IAA (m/z 176). IAA was quantified using standard IAA at different concentrations and MRM with the following transition, 176 > 130, and mass calibration.
Chemical Synthesis of Cholestanoic Acid
The synthetic procedure was adapted from previous works (Czarny et al., 1975; Nelson et al., 1975; Schmidt et al., 2006) using 4-cholesten-3-one as a precursor. More detailed information is available upon request.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under the following accession numbers: At_ERG28 (Arabidopsis; At1g10030), SMO1 (At4g12110), CSD (At1g47290), SKR (At5g18210), Os_ERG28 (rice [Oryza sativa]; NP_001067350), Hs_ERG28 (Homo sapiens; NP_009107), Sc_ERG28 (Saccharomyces cerevisiae; NP_010962), Sp_ERG28 (Schizosaccharomyces pombe, O74820), RTNLB2 (At4g11220), TIP41-LIKE (At4g34270), GAPDH (At1g3440), and α-Tubulin (At1g04820). Germplasm with a T-DNA insertion in the ERG28 gene were: erg28T1-1 to erg28T1-6 (SALK_025834) and erg28T2-1 to erg28T2-6 (SALK_000240).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sterol Biosynthetic Pathway Showing the Cryptic Steps (Solid-Lined Box) Leading to the Formation of Reactive SBIs from the SC4DM Multienzyme Complex.
Supplemental Figure 2. Multiple Sequence Alignment of Arabidopsis ERG28 with Homologous Proteins between Different Species.
Supplemental Figure 3. ER Localization of the Arabidopsis SC4DM Complex Components.
Supplemental Figure 4. Characterization of Arabidopsis erg28RNAi (erg28R2-1 to erg28R2-6) and erg28T-DNA (erg28T2-1 to erg28T2-6) Mutants.
Supplemental Figure 5. Phenotypes of Arabidopsis erg28RNAi (erg28R2-1 to erg28R2-6) and erg28T-DNA (erg28T2-1 to erg28T2-6) Mutants.
Supplemental Figure 6. Characteristics of Specific Organs and Tissues of Class 5 and Class 6 erg28 Mutants.
Supplemental Figure 7. Effect of Phytohormones on Arabidopsis erg28 Development.
Supplemental Figure 8. Effect of IAA and NAA on Primary Root Elongation of Wild-Type and erg28 Plants.
Supplemental Figure 9. Expression of ERG28 in Arabidopsis Tissues and Organs.
Supplemental Figure 10. Spatial Expression Pattern of ERG28 According the Arabidopsis Electronic Fluorescent Pictograph Browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi).
Supplemental Figure 11. qRT-PCR Analysis of ERG28 Expression in Different Arabidopsis Tissues.
Supplemental Figure 12. Regular Sterol Content of Wild-Type, erg28R1-1 to erg28R1-6, and erg28T1-1 to erg28T1-6 Plants.
Supplemental Figure 13. Levels of Upstream Sterol Intermediates of Wild-Type, erg28R1-1 to erg28R1-6, and erg28T1-1 to erg28T1-6 Plants.
Supplemental Figure 14. Levels of Sterol Intermediates of Campesterol and Sitosterol Branches of Wild-Type, erg28R1-1 to erg28R1-6, and erg28T1-1 to erg28T1-6 Plants.
Supplemental Figure 15. Mass Fragmentation Patterns of CMMC from erg28 Plants and of Standard CMMC by UPLC MS/MS-APPI+.
Supplemental Figure 16. Selected Reaction Monitoring Chromatogram of CMMC (453/109, 453/123 m/z).
Supplemental Figure 17. Selected Reaction Monitoring Chromatogram of CMMC (453/149, 453/191 m/z).
Supplemental Figure 18. Selected Reaction Monitoring Chromatogram of CMMC (453/205, 453/369 m/z).
Supplemental Figure 19. CMMC Competes with NPA Binding Sites and Docks into the Nucleotide Binding Site of Homology-Modeled Arabidopsis ABCB19 and ABCB1 Auxin Transporters.
Supplemental Table 1. In-Gel Digestion and NanoLC-MS/MS of Arabidopsis ERG28 Interactors in the SC4DM Multienzyme Complex.
Supplemental Table 2. Rescue of Yeast erg28 Null Mutant by Arabidopsis ERG28.
Supplemental Table 3. Binding Free Energy Prediction of CMMC within Arabidopsis ABCB1 (At2g36910) and ABCB19 (At3g28860) Nucleotide Binding Domains.
Supplemental Table 4. Primer Sequences Used for Cloning, Genotyping, and Expression Studies.
SupplementalMethods 1. Plant Growth and Experimental Procedures for Supplemental Figures and Supplemental Tables.
Acknowledgments
We thank Jiri Friml for PIN1:GFP, Tom Guilfoyle for providing DR5:GUS seeds, and Martin Bard for the yeast erg28 mutant. Image figures were arranged using FigureJ (Jérôme Mutterer and Edda Zinck, ImageJ Users and Developers Conference, 2012). We thank Valérie Cognat for help with in silico expression analysis. We thank Aurélie Bailly, Michel Kerneis, Sébastien Staerck, and Richard Wagner for greenhouse help. This work was supported by the Agence Nationale de la Recherche (Contract ANR-09-BLAN-0067-02) and by a doctoral fellowship from the Research Program of the Ministry of Science and Technology of Indonesia to N.J.
AUTHOR CONTRIBUTIONS
F.B. and B.C. conceived and supervised the study. A.S.M., N.J., B.C., and F.B. designed the experiments and performed biochemical, physiological, and molecular biology studies. A.S.M., N.J., J.B., and T.D.P. performed mutant screening. D.H. performed UPLC-MS/MS analysis. P.N. provided the reagent. A.R. performed yeast erg28 complementation. F.B., J.M., and M.E. performed microscopy analysis. M.B. performed molecular docking studies. D.A. and A.V.D. performed nano-liquid chromatography–tandem mass spectrometry analysis. P.G. and M.M. performed cholestanoic acid synthesis and chemical structure analysis. A.S.M., N.J., D.H., J.M., M.E., M.B., P.G., M.M., B.C., and F.B. analyzed data. B.C. and F.B. wrote the article.
Glossary
- SBI
sterol biosynthetic intermediates
- SC4DM
sterol C4 demethylase
- RNAi
RNA interference
- qRT-PCR
quantitative RT-PCR
- PAT
polar auxin transport
- NPA
naphthylphthalamic acid
- IAA
indole-3-acetic acid
- m/z
mass-to-charge ratio
- MRM
multiple reaction monitoring
- CMMC
4-carboxy-4-methyl-24-methylenecycloartanol
- Rubisco
ribulose-1,5-bis-phosphate carboxylase/oxygenase
- UPLC-MS/MS
ultraperformance liquid chromatography–tandem mass spectrometry
- APPI
atmospheric pressure photon ionization
References
- Bard M., Bruner D.A., Pierson C.A., Lees N.D., Biermann B., Frye L., Koegel C., Barbuch R. (1996). Cloning and characterization of ERG25, the Saccharomyces cerevisiae gene encoding C-4 sterol methyl oxidase. Proc. Natl. Acad. Sci. USA 93: 186–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett S.R.M., Alverz J., Bossinger G., Smyth D.R. (1995). Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J. 8: 505–520 [Google Scholar]
- Bensinger S.J., Bradley M.N., Joseph S.B., Zelcer N., Janssen E.M., Hausner M.A., Shih R., Parks J.S., Edwards P.A., Jamieson B.D., Tontonoz P. (2008). LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell 134: 97–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste P. (2004). Biosynthesis and accumulation of sterols. Annu. Rev. Plant Biol. 55: 429–457 [DOI] [PubMed] [Google Scholar]
- Berleth T., Jürgens G. (1993). The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development 118: 575–587 [Google Scholar]
- Bhalerao R.P., Eklöf J., Ljung K., Marchant A., Bennett M., Sandberg G. (2002). Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J. 29: 325–332 [DOI] [PubMed] [Google Scholar]
- Bittman, R. (1997). Cholesterol: Its functions and metabolism in biology and medicine. In Subcellular Biochemistry, R. Bittman, ed (New York: Plenum Press), pp. 145–171. [Google Scholar]
- Blakeslee J.J., et al. (2007). Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19: 131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc M., et al. (2011). Host defense against viral infection involves interferon mediated down-regulation of sterol biosynthesis. PLoS Biol. 9: e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutté Y., Grebe M. (2009). Cellular processes relying on sterol function in plants. Curr. Opin. Plant Biol. 12: 705–713 [DOI] [PubMed] [Google Scholar]
- Bouvier F., Rahier A., Camara B. (2005). Biogenesis, molecular regulation and function of plant isoprenoids. Prog. Lipid Res. 44: 357–429 [DOI] [PubMed] [Google Scholar]
- Brown A.J., Galea A.M. (2010). Cholesterol as an evolutionary response to living with oxygen. Evolution 64: 2179–2183 [DOI] [PubMed] [Google Scholar]
- Brown D.E., Rashotte A.M., Murphy A.S., Normanly J., Tague B.W., Peer W.A., Taiz L., Muday G.K. (2001). Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol. 126: 524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byskov A.G., Andersen C.Y., Nordholm L., Thøgersen H., Xia G., Wassmann O., Andersen J.V., Guddal E., Roed T. (1995). Chemical structure of sterols that activate oocyte meiosis. Nature 374: 559–562 [DOI] [PubMed] [Google Scholar]
- Caldas H., Herman G.E. (2003). NSDHL, an enzyme involved in cholesterol biosynthesis, traffics through the Golgi and accumulates on ER membranes and on the surface of lipid droplets. Hum. Mol. Genet. 12: 2981–2991 [DOI] [PubMed] [Google Scholar]
- Cañabate-Díaz B., Segura Carretero A., Fernández-Gutiérrez A., Belmonte Vega A., Garrido Frenich A., Martínez Vidal J.L., Duran Martos J. (2007). Separation and determination of sterols in olive oil by HPLC-MS. Food Chem. 102: 593–598 [Google Scholar]
- Carland F., Fujioka S., Nelson T. (2010). The sterol methyltransferases SMT1, SMT2, and SMT3 influence Arabidopsis development through nonbrassinosteroid products. Plant Physiol. 153: 741–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland F.M., Berg B.L., FitzGerald J.N., Jinamornphongs S., Nelson T., Keith B. (1999). Genetic regulation of vascular tissue patterning in Arabidopsis. Plant Cell 11: 2123–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland F.M., Fujioka S., Takatsuto S., Yoshida S., Nelson T. (2002). The identification of CVP1 reveals a role for sterols in vascular patterning. Plant Cell 14: 2045–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillo A., Joseph S.B., Vaidya S.A., Haberland M., Fogelman A.M., Cheng G., Tontonoz P. (2003). Crosstalk between LXR and toll-like receptor signaling mediates bacterial and viral antagonism of cholesterol metabolism. Mol. Cell 12: 805–816 [DOI] [PubMed] [Google Scholar]
- Clouse S.D. (2000). Plant development: A role for sterols in embryogenesis. Curr. Biol. 10: R601–R604 [DOI] [PubMed] [Google Scholar]
- Clouse S.D. (2002). Arabidopsis mutants reveal multiple roles for sterols in plant development. Plant Cell 14: 1995–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarny M.R., Maheshwari K.K., Nelson J.A., Spencer T.A. (1975). Synthesis of stereoisomeric 4-hydroxymethyl-4-methyl-3beta-hydroxy-cholestanes, -androstanes, and -10-methyl-trans-decalins. J. Org. Chem. 40: 2079–2085 [DOI] [PubMed] [Google Scholar]
- Desmond E., Gribaldo S. (2009). Phylogenomics of sterol synthesis: Insights into the origin, evolution, and diversity of a key eukaryotic feature. Genome Biol. Evol. 1: 364–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener A.C., Li H., Zhou W., Whoriskey W.J., Nes W.D., Fink G.R. (2000). Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell 12: 853–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner T.J., Sherr I., Scarpella E. (2009). Regulation of preprocambial cell state acquisition by auxin signaling in Arabidopsis leaves. Development 136: 3235–3246 [DOI] [PubMed] [Google Scholar]
- Espenshade P.J., Hughes A.L. (2007). Regulation of sterol synthesis in eukaryotes. Annu. Rev. Genet. 41: 401–427 [DOI] [PubMed] [Google Scholar]
- Feraru E., Feraru M.I., Kleine-Vehn J., Martinière A., Mouille G., Vanneste S., Vernhettes S., Runions J., Friml J. (2011). PIN polarity maintenance by the cell wall in Arabidopsis. Curr. Biol. 21: 338–343 [DOI] [PubMed] [Google Scholar]
- Fujioka S., Yokota T. (2003). Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol. 54: 137–164 [DOI] [PubMed] [Google Scholar]
- Gachotte D., Eckstein J., Barbuch R., Hughes T., Roberts C., Bard M. (2001). A novel gene conserved from yeast to humans is involved in sterol biosynthesis. J. Lipid Res. 42: 150–154 [PubMed] [Google Scholar]
- Gälweiler L., Guan C., Müller A., Wisman E., Mendgen K., Yephremov A., Palme K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230 [DOI] [PubMed] [Google Scholar]
- Gaylor J.L. (2002). Membrane-bound enzymes of cholesterol synthesis from lanosterol. Biochem. Biophys. Res. Commun. 292: 1139–1146 [DOI] [PubMed] [Google Scholar]
- Geldner N., Friml J., Stierhof Y.D., Jürgens G., Palme K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428 [DOI] [PubMed] [Google Scholar]
- Geldner N., Richter S., Vieten A., Marquardt S., Torres-Ruiz R.A., Mayer U., Jürgens G. (2004). Partial loss-of-function alleles reveal a role for GNOM in auxin transport-related, post-embryonic development of Arabidopsis. Development 131: 389–400 [DOI] [PubMed] [Google Scholar]
- Grebe M., Xu J., Möbius W., Ueda T., Nakano A., Geuze H.J., Rook M.B., Scheres B. (2003). Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr. Biol. 13: 1378–1387 [DOI] [PubMed] [Google Scholar]
- Hannich J.T., Entchev E.V., Mende F., Boytchev H., Martin R., Zagoriy V., Theumer G., Riezman I., Riezman H., Knölker H.J., Kurzchalia T.V. (2009). Methylation of the sterol nucleus by STRM-1 regulates dauer larva formation in Caenorhabditis elegans. Dev. Cell 16: 833–843 [DOI] [PubMed] [Google Scholar]
- He J.X., Fujioka S., Li T.C., Kang S.G., Seto H., Takatsuto S., Yoshida S., Jang J.C. (2003). Sterols regulate development and gene expression in Arabidopsis. Plant Physiol. 131: 1258–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M., et al. (2011). Mutations in the human SC4MOL gene encoding a methyl sterol oxidase cause psoriasiform dermatitis, microcephaly, and developmental delay. J. Clin. Invest. 121: 976–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A.L., Lee C.Y., Bien C.M., Espenshade P.J. (2007). 4-Methyl sterols regulate fission yeast SREBP-Scap under low oxygen and cell stress. J. Biol. Chem. 282: 24388–24396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadid N., Mialoundama A.S., Heintz D., Ayoub D., Erhardt M., Mutterer J., Meyer D., Alioua A., Van Dorsselaer A., Rahier A., Camara B., Bouvier F. (2011). DOLICHOL PHOSPHATE MANNOSE SYNTHASE1 mediates the biogenesis of isoprenyl-linked glycans and influences development, stress response, and ammonium hypersensitivity in Arabidopsis. Plant Cell 23: 1985–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y., Fobis-Loisy I., Miège C., Rollin C., Gaude T. (2006). AtSNX1 defines an endosome for auxin-carrier trafficking in Arabidopsis. Nature 443: 106–109 [DOI] [PubMed] [Google Scholar]
- Jang J.C., Fujioka S., Tasaka M., Seto H., Takatsuto S., Ishii A., Aida M., Yoshida S., Sheen J. (2000). A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev. 14: 1485–1497 [PMC free article] [PubMed] [Google Scholar]
- Janowski B.A., Willy P.J., Devi T.R., Falck J.R., Mangelsdorf D.J. (1996). An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature 383: 728–731 [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.B., Schaller H., Goh C.H., Kwon M., Choe S., An C.S., Durst F., Feldmann K.A., Feyereisen R. (2005). Arabidopsis cyp51 mutant shows postembryonic seedling lethality associated with lack of membrane integrity. Plant Physiol. 138: 2033–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.Y., Henrichs S., Bailly A., Vincenzetti V., Sovero V., Mancuso S., Pollmann S., Kim D., Geisler M., Nam H.G. (2010). Identification of an ABCB/P-glycoprotein-specific inhibitor of auxin transport by chemical genomics. J. Biol. Chem. 285: 23309–23317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis D.R., Muday G.K. (2009). Measurement of auxin transport in Arabidopsis thaliana. Nat. Protoc. 4: 437–451 [DOI] [PubMed] [Google Scholar]
- Lim L., Jackson-Lewis V., Wong L.C., Shui G.H., Goh A.X., Kesavapany S., Jenner A.M., Fivaz M., Przedborski S., Wenk M.R. (2012). Lanosterol induces mitochondrial uncoupling and protects dopaminergic neurons from cell death in a model for Parkinson’s disease. Cell Death Differ. 19: 416–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey K., Pullen M.L., Topping J.F. (2003). Importance of plant sterols in pattern formation and hormone signalling. Trends Plant Sci. 8: 521–525 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Ljung K., Bhalerao R.P., Sandberg G. (2001). Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J. 28: 465–474 [DOI] [PubMed] [Google Scholar]
- Lu B., Zhang Y., Wu X., Shi J. (2007). Separation and determination of diversiform phytosterols in food materials using supercritical carbon dioxide extraction and ultraperformance liquid chromatography-atmospheric pressure chemical ionization-mass spectrometry. Anal. Chim. Acta 588: 50–63 [DOI] [PubMed] [Google Scholar]
- Mares-Sámano S., Badhan R., Penny J. (2009). Identification of putative steroid-binding sites in human ABCB1 and ABCG2. Eur. J. Med. Chem. 44: 3601–3611 [DOI] [PubMed] [Google Scholar]
- Masucci J.D., Rerie W.G., Foreman D.R., Zhang M., Galway M.E., Marks M.D., Schiefelbein J.W. (1996). The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122: 1253–1260 [DOI] [PubMed] [Google Scholar]
- Mattsson J., Ckurshumova W., Berleth T. (2003). Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol. 131: 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson J., Sung Z.R., Berleth T. (1999). Responses of plant vascular systems to auxin transport inhibition. Development 126: 2979–2991 [DOI] [PubMed] [Google Scholar]
- Men S., Boutté Y., Ikeda Y., Li X., Palme K., Stierhof Y.D., Hartmann M.A., Moritz T., Grebe M. (2008). Sterol-dependent endocytosis mediates post-cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nat. Cell Biol. 10: 237–244 [DOI] [PubMed] [Google Scholar]
- Mialoundama A.S., Heintz D., Debayle D., Rahier A., Camara B., Bouvier F. (2009). Abscisic acid negatively regulates elicitor-induced synthesis of capsidiol in wild tobacco. Plant Physiol. 150: 1556–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo C., Bard M. (2005). Erg28p is a key protein in the yeast sterol biosynthetic enzyme complex. J. Lipid Res. 46: 1991–1998 [DOI] [PubMed] [Google Scholar]
- Mo C., Milla P., Athenstaedt K., Ott R., Balliano G., Daum G., Bard M. (2003). In yeast sterol biosynthesis the 3-keto reductase protein (Erg27p) is required for oxidosqualene cyclase (Erg7p) activity. Biochim. Biophys. Acta 1633: 68–74 [DOI] [PubMed] [Google Scholar]
- Mo C., Valachovic M., Randall S.K., Nickels J.T., Bard M. (2002). Protein-protein interactions among C-4 demethylation enzymes involved in yeast sterol biosynthesis. Proc. Natl. Acad. Sci. USA 99: 9739–9744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muday G.K., Lomax T.L., Rayle D.L. (1995). Characterization of the growth and auxin physiology of roots of the tomato mutant, diageotropica. Planta 195: 548–553 [DOI] [PubMed] [Google Scholar]
- Müller A., Guan C., Gälweiler L., Tänzler P., Huijser P., Marchant A., Parry G., Bennett M., Wisman E., Palme K. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17: 6903–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natter K., Leitner P., Faschinger A., Wolinski H., McCraith S., Fields S., Kohlwein S.D. (2005). The spatial organization of lipid synthesis in the yeast Saccharomyces cerevisiae derived from large scale green fluorescent protein tagging and high resolution microscopy. Mol. Cell. Proteomics 4: 662–672 [DOI] [PubMed] [Google Scholar]
- Nelson J.A., Kahn S., Spencer T.A., Sharpless K.B., Clayton R.B. (1975). Some aspects of substrate specificity in biological demethylation at C4 of steroids. Bioorg. Chem. 4: 363–376 [Google Scholar]
- Noh B., Murphy A.S., Spalding E.P. (2001). Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13: 2441–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi M., Mizushima N., Kabeya Y., Yoshimori T. (2003). Localization of mammalian NAD(P)H steroid dehydrogenase-like protein on lipid droplets. J. Biol. Chem. 278: 36819–36829 [DOI] [PubMed] [Google Scholar]
- Okada K., Ueda J., Komaki M.K., Bell C.J., Shimura Y. (1991). Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3: 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovecka M., Berson T., Beck M., Derksen J., Samaj J., Baluska F., Lichtscheidl I.K. (2010). Structural sterols are involved in both the initiation and tip growth of root hairs in Arabidopsis thaliana. Plant Cell 22: 2999–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panicker S.P., Ganguly T., Consolo M., Price V., Mirmirani P., Honda K., Karnik P. (2012). Sterol intermediates of cholesterol biosynthesis inhibit hair growth and trigger an innate immune response in cicatricial alopecia. PLoS ONE 7: e38449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent N., Winstall E., Beauchemin M., Paquet C., Poirier G.G., Bertrand R. (2009). Proteomic analysis of enriched lysosomes at early phase of camptothecin-induced apoptosis in human U-937 cells. J. Proteomics 72: 960–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer W.A., Murphy A.S. (2007). Flavonoids and auxin transport: Modulators or regulators? Trends Plant Sci. 12: 556–563 [DOI] [PubMed] [Google Scholar]
- Petrásek J., Friml J. (2009). Auxin transport routes in plant development. Development 136: 2675–2688 [DOI] [PubMed] [Google Scholar]
- Pullen M., Clark N., Zarinkamar F., Topping J., Lindsey K. (2010). Analysis of vascular development in the hydra sterol biosynthetic mutants of Arabidopsis. PLoS ONE 5: e12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D., Pesce E.R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Rhourri-Frih B., Chaimbault P., Claude B., Lamy C., André P., Lafosse M. (2009). Analysis of pentacyclic triterpenes by LC-MS. A comparative study between APCI and APPI. J. Mass Spectrom. 44: 71–80 [DOI] [PubMed] [Google Scholar]
- Růzicka K., Ljung K., Vanneste S., Podhorská R., Beeckman T., Friml J., Benková E. (2007). Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19: 2197–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A.W., Doert T., Goutal S., Gruner M., Mende F., Kurzchalia T.V., Knoelker H.J. (2006). Regio- and stereospecific synthesis of cholesterol derivatives and their hormonal activity in Caenorhabditis elegans. Eur. J. Org. Chem. 2006: 3687–3706 [Google Scholar]
- Schmitt L., Benabdelhak H., Blight M.A., Holland I.B., Stubbs M.T. (2003). Crystal structure of the nucleotide-binding domain of the ABC-transporter haemolysin B: Identification of a variable region within ABC helical domains. J. Mol. Biol. 330: 333–342 [DOI] [PubMed] [Google Scholar]
- Schneider C., Newman R.A., Sutherland D.R., Asser U., Greaves M.F. (1982). A one-step purification of membrane proteins using a high efficiency immunomatrix. J. Biol. Chem. 257: 10766–10769 [PubMed] [Google Scholar]
- Schrick K., Fujioka S., Takatsuto S., Stierhof Y.D., Stransky H., Yoshida S., Jürgens G. (2004b). A link between sterol biosynthesis, the cell wall, and cellulose in Arabidopsis. Plant J. 38: 227–243 [DOI] [PubMed] [Google Scholar]
- Schrick K., Mayer U., Horrichs A., Kuhnt C., Bellini C., Dangl J., Schmidt J., Jürgens G. (2000). FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev. 14: 1471–1484 [PMC free article] [PubMed] [Google Scholar]
- Schrick K., Mayer U., Martin G., Bellini C., Kuhnt C., Schmidt J., Jürgens G. (2002). Interactions between sterol biosynthesis genes in embryonic development of Arabidopsis. Plant J. 31: 61–73 [DOI] [PubMed] [Google Scholar]
- Schrick K., Nguyen D., Karlowski W.M., Mayer K.F. (2004a). START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 5: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shui G., Cheong W.F., Jappar I.A., Hoi A., Xue Y., Fernandis A.Z., Tan B.K., Wenk M.R. (2011). Derivatization-independent cholesterol analysis in crude lipid extracts by liquid chromatography/mass spectrometry: Applications to a rabbit model for atherosclerosis. J. Chromatogr. A 1218: 4357–4365 [DOI] [PubMed] [Google Scholar]
- Sieburth L.E. (1999). Auxin is required for leaf vein pattern in Arabidopsis. Plant Physiol. 121: 1179–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souter M., Topping J., Pullen M., Friml J., Palme K., Hackett R., Grierson D., Lindsey K. (2002). hydra mutants of Arabidopsis are defective in sterol profiles and auxin and ethylene signaling. Plant Cell 14: 1017–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souter M.A., Pullen M.L., Topping J.F., Zhang X., Lindsey K. (2004). Rescue of defective auxin-mediated gene expression and root meristem function by inhibition of ethylene signalling in sterol biosynthesis mutants of Arabidopsis. Planta 219: 773–783 [DOI] [PubMed] [Google Scholar]
- Spann N.J., et al. (2012). Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell 151: 138–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukhanova A., et al. (2013). Targeting C4-demethylating genes in the cholesterol pathway sensitizes cancer cells to EGF receptor inhibitors via increased EGF receptor degradation. Cancer Discov. 3: 96–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T., Fernie A.R. (2012). Co-expression and co-responses: Within and beyond transcription. Front. Plant Sci. 3: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S.R., Somerville C.R. (1997). Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9: 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinci G., Xia X., Veitia R.A. (2008). Preservation of genes involved in sterol metabolism in cholesterol auxotrophs: facts and hypotheses. PLoS ONE 3: e2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Jones B., Li Z., Frasse P., Delalande C., Regad F., Chaabouni S., Latché A., Pech J.C., Bouzayen M. (2005). The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17: 2676–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel C.L., Hester Q., Mattsson J. (2008). Identification of genes expressed in vascular tissues using NPA-induced vascular overgrowth in Arabidopsis. Plant Cell Physiol. 49: 457–468 [DOI] [PubMed] [Google Scholar]
- Willemsen V., Friml J., Grebe M., van den Toorn A., Palme K., Scheres B. (2003). Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell 15: 612–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue T., Zhang Y., Zhang L., Yao L., Hu X., Xu L.X. (2013). Proteomic analysis of two metabolic proteins with potential to translocate to plasma membrane associated with tumor metastasis development and drug targets. J. Proteome Res. 12: 1754–1763 [DOI] [PubMed] [Google Scholar]
- Zaitseva J., Jenewein S., Jumpertz T., Holland I.B., Schmitt L. (2005). H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB. EMBO J. 24: 1901–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Ye Z.H. (2001). Alteration of auxin polar transport in the Arabidopsis ifl1 mutants. Plant Physiol. 126: 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.Y., Sae-Seaw J., Wang Z.Y. (2013). Brassinosteroid signalling. Development 140: 1615–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]