Peroxisomes are organelles that generate reactive oxygen species such as hydrogen peroxide. This study shows that peroxisomes damaged by hydrogen peroxide are degraded via autophagy, suggesting that autophagy plays an important role in quality control of peroxisomes.
Abstract
The positioning of peroxisomes in a cell is a regulated process that is closely associated with their functions. Using this feature of the peroxisomal positioning as a criterion, we identified three Arabidopsis thaliana mutants (peroxisome unusual positioning1 [peup1], peup2, and peup4) that contain aggregated peroxisomes. We found that the PEUP1, PEUP2, and PEUP4 were identical to Autophagy-related2 (ATG2), ATG18a, and ATG7, respectively, which are involved in the autophagic system. The number of peroxisomes was increased and the peroxisomal proteins were highly accumulated in the peup1 mutant, suggesting that peroxisome degradation by autophagy (pexophagy) is deficient in the peup1 mutant. These aggregated peroxisomes contained high levels of inactive catalase and were more oxidative than those of the wild type, indicating that peroxisome aggregates comprise damaged peroxisomes. In addition, peroxisome aggregation was induced in wild-type plants by exogenous application of hydrogen peroxide. The cat2 mutant also contained peroxisome aggregates. These findings demonstrate that hydrogen peroxide as a result of catalase inactivation is the inducer of peroxisome aggregation. Furthermore, an autophagosome marker, ATG8, frequently colocalized with peroxisome aggregates, indicating that peroxisomes damaged by hydrogen peroxide are selectively degraded by autophagy in the wild type. Our data provide evidence that autophagy is crucial for quality control mechanisms for peroxisomes in Arabidopsis.
INTRODUCTION
Peroxisomes are ubiquitous organelles in eukaryotic cells. Plant peroxisomes are divided into several categories depending on their functions, such as glyoxysomes, leaf peroxisomes, root peroxisomes, and unspecialized peroxisomes (Kamada et al., 2003). Glyoxysomes accumulate enzymes for the β-oxidation of fatty acids derived from seed storage lipids, and they are essential for the production of energy during seedling establishment (Tolbert and Essner, 1981; Goepfert and Poirier, 2007). Therefore, mutants with severe defects in the β-oxidation pathway cannot germinate in the absence of exogenous Suc as an energy source (Hayashi et al., 1998; Graham, 2008). Leaf peroxisomes accumulate enzymes for the glycolate pathway, which metabolizes the by-products produced in photosynthesis (Tolbert and Yamazaki, 1969; Hayashi and Nishimura, 2006). Most mutants in photorespiration exhibit photoinhibition and subsequent growth retardation under normal atmospheric conditions, which can be restored by high concentrations of CO2, because photorespiration is dispensable under these conditions (Somerville, 2001; Foyer et al., 2009).
Reactive oxygen species (ROS), such as hydrogen peroxide, are abundant in peroxisomes because acyl-CoA oxidases (Kirsch et al., 1986) and glycolate oxidase (GO) (Nishimura et al., 1983), which play crucial roles in the β-oxidation and glycolate pathways, respectively, produce hydrogen peroxide in their reactions. A large amount of hydrogen peroxide is produced in peroxisomes in photosynthetic cells of C3 plants, with levels as much as 2-fold of those produced in mitochondria and 50-fold of those in chloroplasts (Foyer and Noctor, 2003). In addition, peroxisomes possess systems for scavenging hydrogen peroxide. Peroxisomal ascorbate peroxidase (APX) detoxifies hydrogen peroxide, although it does not function well as an antioxidant (Narendra et al., 2006). In addition, peroxisomes contain catalase (CAT), the enzyme responsible for the degradation of hydrogen peroxide. CAT, which accounts for 10 to 25% of the peroxisomal protein (Reumann et al., 2004), decomposes hydrogen peroxide into water and oxygen molecules (Loew, 1900). Therefore, CAT is thought to protect other peroxisomal proteins and membrane lipids from oxidative damage (Yanik and Donaldson, 2005).
In addition to peroxisomal metabolism, peroxisomal dynamics, such as motility, has recently attracted attention. In the past, observation of peroxisomes was mainly performed by electron microscopy. Electron microscopy is an essential method for analyzing ultrastructures. However, because samples are fixed prior to observation, peroxisomal dynamics cannot be observed with an electron microscope. Live imaging analysis of peroxisomes using transgenic plants expressing the fusion gene of green fluorescent protein (GFP) with the peroxisome targeting signal 1 (GFP-PTS1) revealed that peroxisomes are dynamic organelles that move along actin filaments and change their morphology in response to environmental stimuli (Jedd and Chua, 2002; Mano et al., 2002; Rodríguez-Serrano et al., 2009). Furthermore, by focusing on peroxisomal morphology, several peroxisomal components have been discovered using a forward genetic approach that employs the GFP-PTS1 line as the parent plant. We previously isolated a number of aberrant peroxisome morphology (apem; previously called apm) mutants. Through analysis of these mutants, we determined that dynamin-related protein 3A is involved in both peroxisomal and mitochondrial division (Mano et al., 2004), and that some peroxisome biogenesis factors (peroxins [PEXs] such as PEX12, PEX13, and a plant-specific PEX, APEM9) are involved in protein transport to peroxisomes (Mano et al., 2006; Goto et al., 2011). In addition, we determined that PEROXISOMAL MEMBRANE PROTEIN38 (PMP38) is involved in determining the size of peroxisomes (Mano et al., 2011).
To identify novel mutants from the pool of ethylmethane sulfonate-mutagenized GFP-PTS1 seeds that were used to screen for apem mutants, we developed a different strategy. In the GFP-PTS1 plant, peroxisomes are usually dispersed in cells near the chloroplasts. This arrangement is believed to enable efficient metabolism, because the photorespiratory pathway consists of chloroplasts, mitochondria, and peroxisomes. Using this feature of peroxisomal positioning as a criterion, we identified so-called peroxisome unusual positioning (peup) mutants.
In this study, we analyzed three peup mutants (peup1, peup2, and peup4), which contain aggregated peroxisomes. The genes responsible for the peup mutations were identical to AUTOPHAGY-RELATED (ATG) genes. Because autophagy is one of the degradation systems in a cell (Klionsky and Ohsumi, 1999; Mizushima et al., 2011; Liu and Bassham, 2012), our data show that autophagy is involved in the degradation of peroxisomes. We also demonstrate that the peroxisome aggregates in the mutants consist of oxidized peroxisomes containing inactive CAT. On the basis of these results, we propose that oxidative damage by hydrogen peroxide induces peroxisome aggregation, and that autophagy maintains peroxisome quality by degradation of the damaged peroxisomes.
RESULTS
peup Mutants Contain Peroxisome Aggregates
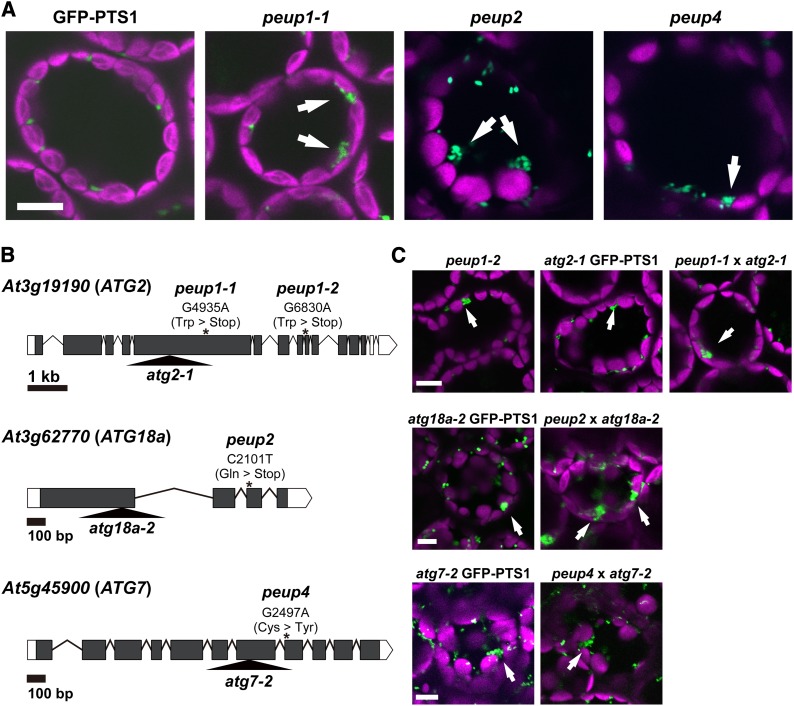
We screened for novel peroxisome mutants, herein referred to as peup, from a pool of ethylmethane sulfonate-mutagenized GFP-PTS1 seeds based on the localization pattern of peroxisomes (Mano et al., 2004, 2006, 2011; Goto et al., 2011). In the parental GFP-PTS1 plant, peroxisomes were observed as punctuate structures and were dispersed in leaf cells (Figure 1A). On the other hand, the peup mutants contained peroxisome aggregates in addition to dispersed peroxisomes (Figure 1A).
Figure 1.
Identification of the peup Mutants.
(A) Confocal images of the mesophyll cells in leaves of 3-week-old GFP-PTS1 and peup1-1, peup2, and peup4 plants. GFP fluorescence from peroxisomes and autofluorescence from chloroplasts are shown in green and magenta, respectively.
(B) Gene structures of At3g19190/ATG2, At3g62770/ATG18a, and At5g45800/ATG7. The open and closed boxes indicate the untranslated and coding regions, respectively. The asterisks indicate nucleotide substitutions in the each gene. G4935A means that the 4935th guanine residue is substituted to adenine (with nucleotide residue 1 corresponding to the adenosine of the first Met codon). The analogous nomenclature is used for other genes. The triangles represent the positions of the T-DNA insertion. The untranslated regions are based on information obtained from the TAIR database (http://www.arabidopsis.org).
(C) Confocal images of leaf cells of peup1 (peup1-2), T-DNA insertion mutants harboring GFP-PTS1 (atg2-1 GFP-PTS1, atg18a-2 GFP-PTS1, and atg7-2 GFP-PTS1), and F1 progeny obtained from crosses between the peup mutants and the T-DNA insertion mutants (peup1-1 × atg2-1, peup2 × atg18a-2, and peup4 × atg7-2).
Arrows indicate peroxisome aggregates. Bar = 10 μm.
Map-based cloning was performed to identify the genes responsible for peup1, peup2, and peup4. We isolated two peup1 alleles, designated peup1-1 and peup1-2, which exhibited similar phenotypes of peroxisome aggregation (Figure 1C). We determined that the peup1-1 and peup1-2 mutations occurred in At3g19190 (At ATG2), which has a substitution of the 4935th guanine with adenine in peup1-1, and a substitution of the 6830th guanine with adenine in peup1-2 (Figure 1B). Both nucleotide substitutions cause a nonsense mutation. We isolated and sequenced At ATG2 cDNA from wild-type plants. The nucleotide sequence was the same as that of ADU79134 (http://www.ncbi.nlm.nih.gov/protein/ADU79134) (Wang et al., 2011), but not At3g19190.1 in The Arabidopsis Information Resource (TAIR) database (http://www.arabidopsis.org), which was derived from a different spliced form. The splicing pattern corresponding to At3g19190.1 was not detected under our experimental conditions.
Similarly, we determined that the peup2 and peup4 mutants have a nucleotide substitution of the 2101th cytosine in At3g62770 (At ATG18a) with thymine and a substitution of the 2497th guanine in At5g45900 (At ATG7) with adenine, respectively (Figure 1B).
To confirm that we identified the correct genes, we obtained T-DNA insertion mutants (atg2-1, SALK_076727; atg18a-2, GABI_651D08; and atg7-2, GABI_655B06) and introduced GFP-PTS1 into the mutants to visualize the peroxisomes. The resulting lines (atg2-1 GFP-PTS1, atg18a-2 GFP-PTS1, and atg7-2 GFP-PTS1) had similar phenotypes to those of the corresponding peup mutants, including the presence of peroxisome aggregates in the cells (Figure 1C). Next, we crossed the T-DNA insertion mutants with the corresponding peup mutants to test for allelism, and observed the phenotypes of the F1 progeny. All of the peup mutations were recessive, and the F1 plants had peroxisome aggregates that were similar to those of the parental mutants (Figure 1C), indicating that the atg2-1, atg18a-2, and atg7-2 mutations are allelic to the peup1-1, peup2, and peup4 mutations, respectively. All three ATG proteins function in autophagosome formation, and the mutants of these genes generate defective isolation membranes. Thus, these ATG genes are known as essential factors for the autophagic process (Xiong et al., 2005; Inoue et al., 2006; Yoshimoto et al., 2009; Mizushima et al., 2011; Wang et al., 2011; Liu and Bassham, 2012). Therefore, we conclude that the abnormality of the peroxisomes observed in the peup1, peup2, and peup4 mutants is caused by a defect in the autophagic system. For subsequent experiments, we employed mainly the peup1 mutant, which exhibits typical phenotypes.
Peroxisome Degradation Is Inhibited in the peup1 Mutant
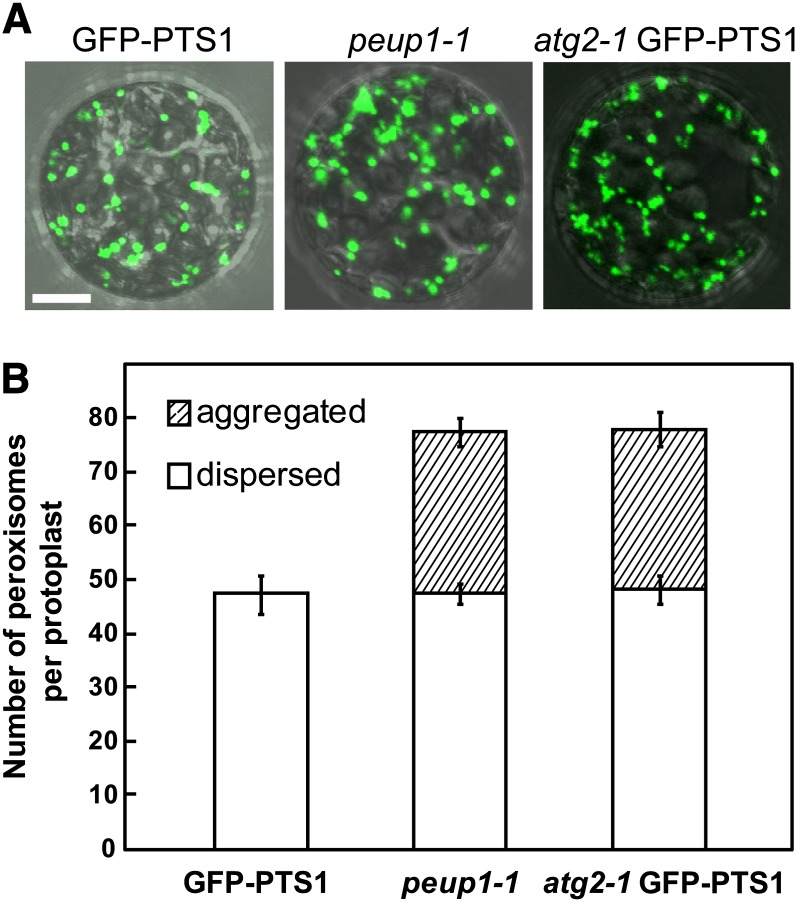
Autophagy is a major degradation system in the cell (Klionsky and Ohsumi, 1999; Liu and Bassham, 2012). ATG2 was initially identified as one of the essential components for autophagosome formation in Saccharomyces cerevisiae (Shintani et al., 2001; Wang et al., 2001) and Pichia pastoris (Strømhaug et al., 2001). Arabidopsis thaliana ATG2 is also involved in autophagy (Inoue et al., 2006; Yoshimoto et al., 2009; Wang et al., 2011). Therefore, autophagy should be deficient in the peup1 mutant. To examine the suppression of peroxisomal degradation in the peup1 mutant, we prepared protoplasts from GFP-PTS1 and peup1 plants (Figure 2A) and counted the number of peroxisomes in a single cell (Figure 2B). The total number of peroxisomes in the peup1 mutant was increased compared with the GFP-PTS1 plant, suggesting that the peroxisomal degradation is suppressed in the peup1 mutant. The numbers of dispersed peroxisomes were almost identical in GFP-PTS1 and peup1 (Figure 2B). In addition, the peup1 mutant contained peroxisome aggregates (Figure 2B). We also analyzed mitochondrial morphology and the number of mitochondria. In contrast with peroxisomes, these parameters were not significantly affected in the peup1 mutant (see Supplemental Figure 1 online).
Figure 2.
Number of Peroxisomes per Protoplast in the peup1 Mutant.
(A) Merged differential interference contrast and GFP fluorescence images of protoplasts from leaves of GFP-PTS1, peup1-1, and atg2-1 GFP-PTS1 plants. Peroxisomes are shown in green. Bar = 10 μm.
(B) The number of peroxisomes per protoplast in the GFP-PTS1, peup1-1, and atg2-1 GFP-PTS1 plants. Open and striped bars indicate the numbers of dispersed and aggregated peroxisomes, respectively. Error bars represent se (n = 17).
We next examined whether a defect in autophagy affects peroxisomal functions, including lipid degradation and photorespiration. We examined the Suc dependency of seed germination, which mirrors the metabolism of seed storage lipids by peroxisomal β-oxidation (Hayashi et al., 1998). Unlike the peroxisome defective1 (ped1) mutant, which is defective in peroxisomal β-oxidation, the peup1-1 mutant germinated in the absence of Suc (see Supplemental Figure 2A online). Subsequently, we examined the 2,4-dichlorophenoxybutyric acid (2,4-DB) resistance of the seedlings. Because 2,4-DB is converted to 2,4-D by peroxisomal β-oxidation, the ped1 mutant is 2,4-DB resistant but 2,4-D sensitive (Hayashi et al., 1998; Zolman et al., 2000). Unlike the ped1 mutant, the peup1-1 mutant was 2,4-DB sensitive (see Supplemental Figure 2B online), suggesting that peroxisomal β-oxidation is normal in the peup1 mutant. Next, we examined photorespiratory activity in the plants by measuring chlorophyll fluorescence and calculating maximum photochemical efficiency of photosystem II (Fv/Fm [for variable fluorescence/maximum fluorescence]). The serine hydroxymethyltransferase1-1 (shm1-1) mutant, which is defective in photorespiration, exhibited reduced Fv/Fm values after it was transferred from high CO2 conditions to normal atmospheric conditions. However, high Fv/Fm values were maintained in the peup1-1 mutant, as well as in the GFP-PTS1 plant (see Supplemental Figure 2C online). These data indicate that the peup1 mutation does not affect photorespiratory activity under our experimental conditions.
Large Amounts of Peroxisomal Proteins Are Accumulated in the peup1 Mutant
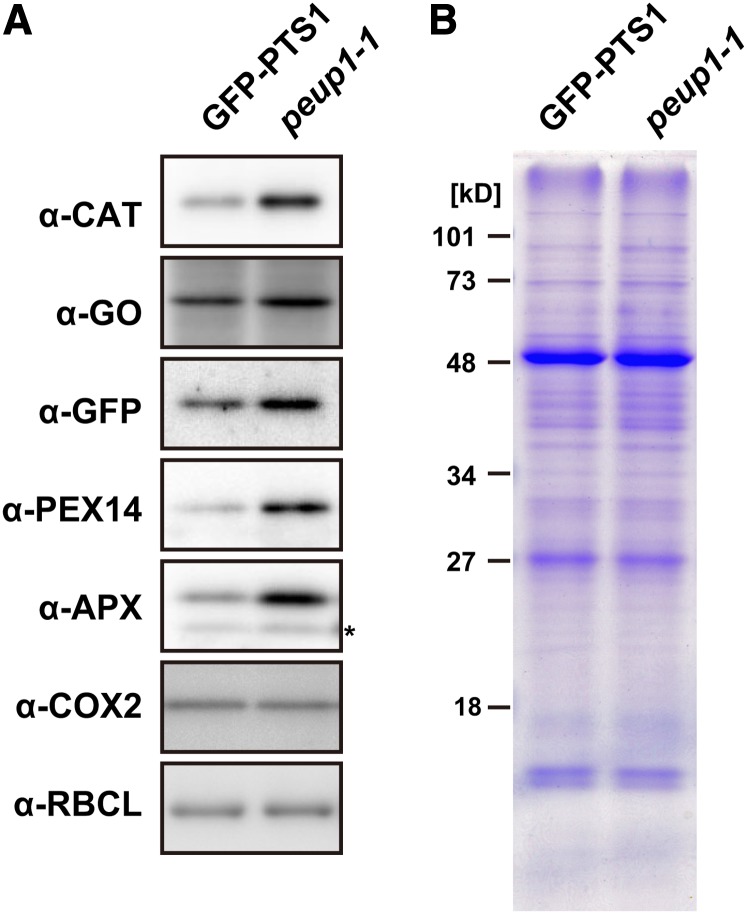
We hypothesized that excess peroxisomal proteins would accumulate in the peup1 mutant, because the degradation of peroxisomes seemed to be suppressed in peup1. To test this, we performed immunoblot analysis of total protein extracts from the GFP-PTS1 plant and the peup1-1 mutant using several antibodies against peroxisomal proteins. CAT, GO, and hydroxypyruvate reductase (HPR) are matrix proteins of peroxisomes, and PEX14 and APX are membrane proteins of peroxisomes. All peroxisomal proteins tested, including the exogenously expressed GFP-PTS1, accumulated excessively in the peup1-1 mutant compared with GFP-PTS1 plant (Figure 3). These results were consistent with the increase in number of peroxisomes in the peup1 mutant (Figure 2). However, the expression of the corresponding genes was not increased in peup1-1 compared with GFP-PTS1 (see Supplemental Figure 3 online), indicating that the accumulation of peroxisomal proteins in the peup1 mutant was not caused by upregulation of gene expression but rather by a defect in the degradation of peroxisomes. In parallel, to examine the levels of chloroplast and mitochondrial proteins, we performed immunoblot analysis using antibodies against cytochrome c oxidase subunit 2 (COX2) and ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (RBCL) as mitochondrial and chloroplast markers, respectively. In contrast with peroxisomal proteins, the amounts of COX2 and RBCL were similar between the GFP-PTS1 plant and the peup1-1 mutant (Figure 3). This result is consistent with the data showing that the mitochondrial number was not altered in the peup1-1 mutant (see Supplemental Figure 1 online).
Figure 3.
Levels of Peroxisomal Proteins in the peup1 Mutant.
(A) Immunoblot analysis of proteins in GFP-PTS1 and peup1-1 plants. Crude protein extracts were prepared from the leaves of 3-week-old plants and subjected to SDS-PAGE followed by immunoblotting using the antibody indicated. The asterisk in α-APX indicates a nonspecific band.
(B) Coomassie brilliant blue staining image of the proteins loaded in this immunoblot. Numbers on the left indicate the molecular mass.
[See online article for color version of this figure.]
CAT Is Accumulated in Electron-Dense Regions in the Peroxisome Aggregates of the peup1 Mutant
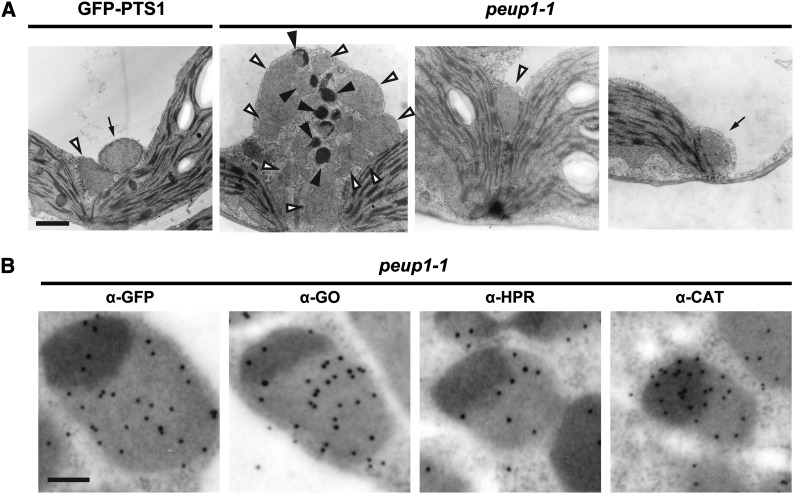
To further investigate the peroxisome aggregates in the peup1 mutant in detail, we performed transmission electron microscopy analysis using leaf palisade mesophyll cells of GFP-PTS1 and peup1-1 plants. Peroxisomes were dispersed in the GFP-PTS1 plant, whereas both aggregated and dispersed peroxisomes were observed in the peup1-1 mutant (Figure 4A). Unexpectedly, approximately half of the aggregated peroxisomes had electron-dense regions (solid arrowheads in Figure 4A). Moreover, these electron-dense regions in the peroxisomes were frequently located face to face among peroxisomes (see Supplemental Figure 4 online). The majority of the dispersed peroxisomes in the peup1-1 mutant did not have electron-dense regions, which was similar to the peroxisomes in GFP-PTS1 plants (Figure 4A). In addition, the mitochondrial structures of GFP-PTS1 and peup1-1 mutant plants were similar (Figure 4A).
Figure 4.
Electron Microscopy Observation of Mesophyll Cells in the peup1 Mutant.
(A) Mesophyll cells in GFP-PTS1 (left image) and in peup1-1 (three images on the right). Open and closed arrowheads indicate normal peroxisomes and peroxisomes containing an electron-dense region, respectively. Arrows indicate mitochondria. Bar = 1 μm.
(B) Immunoelectron microscopy images of mesophyll cells in peup1-1 using antibodies against GFP-PTS1, α-GO, α-HPR, and α-CAT. A representative peroxisome from each experiment is shown. Black dots in the images are immunogold particles. Bar = 200 nm.
To determine which proteins accumulate in the electron-dense regions of peroxisomes in the peup1-1 mutant, we performed immunoelectron microscopy analysis using antibodies against peroxisomal matrix proteins such as GO, HPR, and CAT, and transgene product GFP-PTS1. GO, HPR, and GFP-PTS1 were mainly localized to the regions of peroxisomes that were not electron dense (Figure 4B; see Supplemental Figure 5 online). By contrast, CAT was highly localized in the electron-dense regions in addition to the nonelectron-dense regions (Figure 4B; see Supplemental Figure 5 online). These results indicate that CAT accumulates at a higher density in the electron-dense regions of peroxisome aggregates.
Condensed CAT in Peroxisome Aggregates Has Low Activity
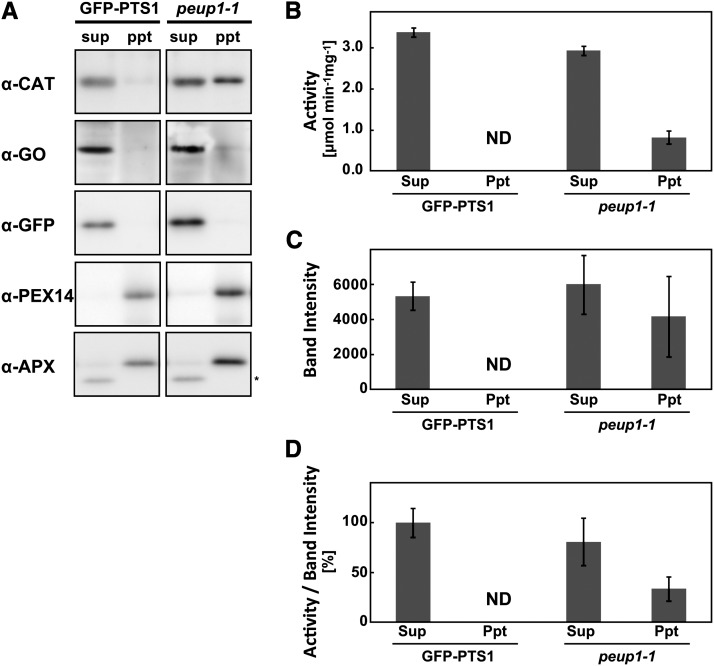
On the basis of our immunoelectron microscopy results, we considered the possibility that inactive CAT forms a condensed mass in the peroxisome. To determine whether this is the case, we separated the crude protein extracts into soluble and insoluble fractions and performed immunoblot analysis using antibodies against several peroxisomal proteins. GO and GFP-PTS1 were predominantly detected in the soluble fraction, whereas PEX14 and APX, which are membrane proteins, were detected in the insoluble fraction from both GFP-PTS1 and peup1-1 plants (Figure 5A). However, CAT was detected in the insoluble fraction from the peup1-1 mutant in addition to the soluble fraction (Figure 5A). These results demonstrate that a portion of CAT forms a condensed mass in the peup1 mutant, which suggests that condensed CAT is localized in the electron-dense regions that we observed under the electron microscope (Figure 4).
Figure 5.
Accumulation of Inactive CAT in the peup1 Mutant.
(A) Total proteins from the leaves of 3-week-old GFP-PTS1 and peup1-1 plants were separated into supernatant (sup) and precipitate (ppt) fractions and subjected to SDS-PAGE followed by immunoblotting using the antibody indicated. The asterisk in α-APX indicates a nonspecific band.
(B) CAT activities in the supernatant and precipitate fractions of the GFP-PTS1 and peup1-1 plants.
(C) Estimation of CAT polypeptides. The values, which were calculated based on the band intensity, were measured by ImageJ.
(D) Relative activity of CAT. The values of the activity of CAT in (B) were divided by the corresponding values of the band intensity in (C). The results are shown as relative values (the supernatant of GFP-PTS1 was set to 100%).
The results represent the mean ± se of three independent experiments. ND, not detected.
We next calculated the relative activity of CAT in the soluble and insoluble fractions. First, we measured the total CAT activity in each fraction (Figure 5B), and then we estimated the band intensities of CAT in the immunoblot (Figure 5C). Finally, the relative activity of CAT was calculated by dividing the total activity by the band intensity (Figure 5D). Interestingly, high CAT activities were detected in the soluble fractions from both GFP-PTS1 and peup1-1, whereas CAT activity in the insoluble fraction was much lower than those in the soluble fractions. These results indicate that condensed CAT in peroxisome aggregates has low activity.
Peroxisomes Are Oxidative in the peup1 Mutant
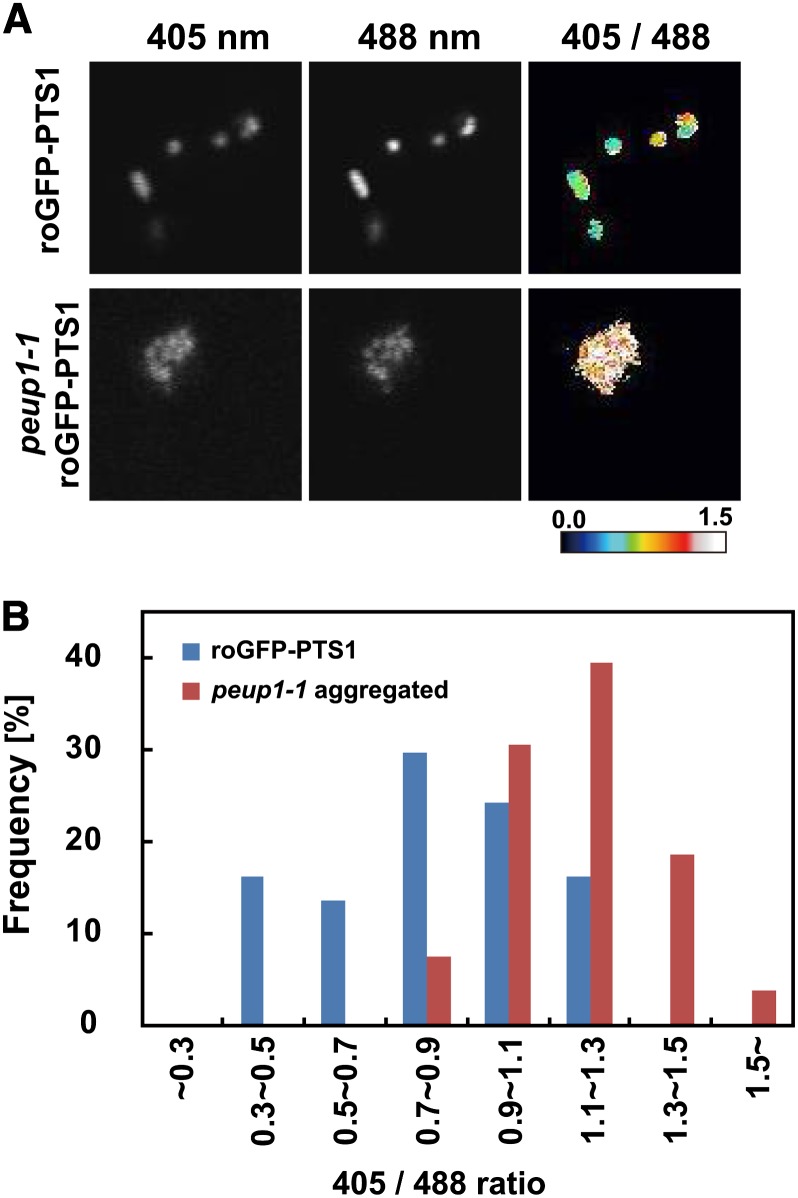
Because inactive CAT was highly accumulated in peroxisome aggregates of peup1, hydrogen peroxide might be accumulated in the peroxisome aggregates. Thus, we examined the redox states of peroxisomes using reduction–oxidation-sensitive green fluorescent protein 2 (roGFP2) (Dooley et al., 2004; Hanson et al., 2004; Meyer et al., 2007; Choi et al., 2012). The ratio of fluorescence intensities from roGFP2, which is excited at 405 and 488 nm, reflects the redox potential (Dooley et al., 2004). In the oxidized state, the ratio of roGFP fluorescence (405/488 nm) increases. Therefore, the fusion of roGFP2 with sorting signals for chloroplasts, mitochondria, or peroxisomes allows us to monitor the redox states of these organelles (Schwarzländer et al., 2008; Rosenwasser et al., 2010).
To generate roGFP2, the nucleotides corresponding to redox-responsive Cys residues were introduced into an authentic GFP (sGFP[S65T]) (Chiu et al., 1996) gene. Next, we introduced the roGFP2 gene fused with PTS1 into wild-type (Columbia) and peup1-1 plants to obtain two transgenic lines, designated roGFP-PTS1 and peup1-1 roGFP-PTS1, respectively. The roGFP signal localized to peroxisomes in these plants (Figure 6A). We then measured fluorescence intensities from peroxisomal roGFP with two excitation wavelengths and calculated the 405/488 nm ratio in the transgenic plants under normal growth conditions. The 405/488 nm ratio of the roGFP-PTS1 plant was 0.822 ± 0.266. The 405/488 nm ratio of the peroxisome aggregates in the peup1-1 roGFP-PTS1 mutant was 1.17 ± 0.183 (Figure 6B). These results clearly demonstrate that the peroxisome aggregates were more oxidative than peroxisomes in the wild type, suggesting that hydrogen peroxide, which is believed to be the major ROS in peroxisomes with low CAT activity, is accumulated in the peroxisome aggregates of the peup1 mutant. In addition, the peroxisome aggregates in peup1-1 roGFP-PTS1 were more sensitive to exogenous hydrogen peroxide (see Supplemental Figure 6 online), and some dispersed peroxisomes in the peup1-1 roGFP-PTS1 mutant cells showed similar redox states to those of roGFP-PTS1 (see Supplemental Figure 7 online). These results demonstrate that the peroxisome aggregates consist of abnormal peroxisomes.
Figure 6.
Quantification of Redox State in the Peroxisomes of peup1 Mutant Cells.
(A) Fluorescence images of peroxisomes in roGFP-PTS1 and peup1-1 roGFP-PTS1 plants. The left and middle panels show fluorescence images upon excitation by 405 nm and 488 nm lasers, respectively. The right panels show the ratiometric images calculated from the left and middle images. Relationships between the ratio and color are shown in the color scale bar below the panels. Blue represents a reduced state, whereas red represents an oxidized state.
(B) Frequency distribution of redox state in peroxisomes. The ratios (405 nm/488 nm) from peroxisome aggregates in the peup1-1 roGFP-PTS1 mutant and dispersed peroxisomes in the roGFP-PTS1 plant were calculated. Thirty-seven dispersed peroxisomes in the roGFP-PTS1 plant and 134 aggregated peroxisomes in the peup1-1 roGFP-PTS1 mutant were used for the calculation. The histogram indicates the percentage of total peroxisomes that had a ratio in the range shown below the graph.
Accumulation of Hydrogen Peroxide Induces Peroxisome Aggregation
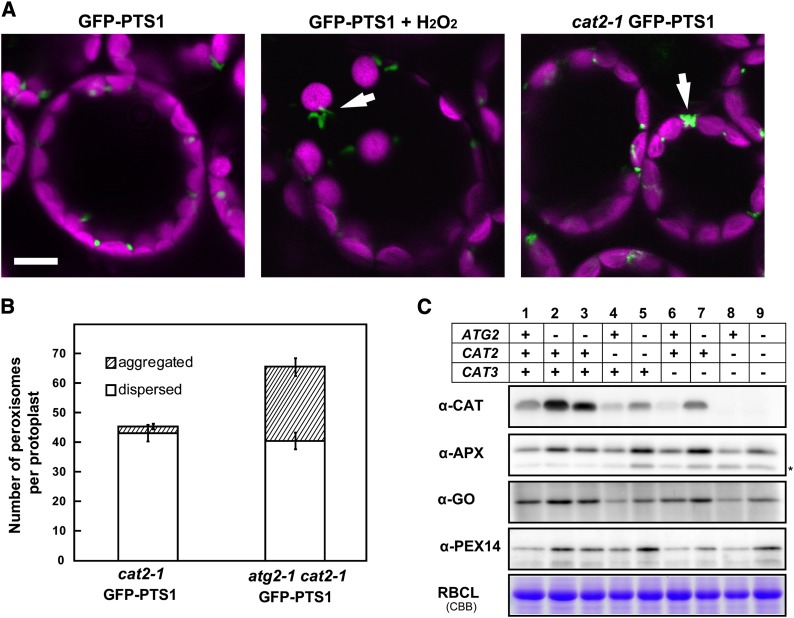
To examine the direct effect of hydrogen peroxide on peroxisome aggregation in more detail, we treated GFP-PTS1 plants with hydrogen peroxide. We immersed leaves of GFP-PTS1 plants in 0.1% (v/v) hydrogen peroxide for 10 min and subsequently observed peroxisomal morphology. We found that peroxisomes formed aggregates in the GFP-PTS1 plant (Figure 7A). This result clearly shows that hydrogen peroxide caused the peroxisomes to aggregate.
Figure 7.
Effect of Hydrogen Peroxide on the Induction of Peroxisome Aggregate Formation.
(A) Mesophyll cells in 3-week-old GFP-PTS1 plants before hydrogen peroxide treatment (left panel). Mesophyll cells after treatment with 0.1% (v/v) hydrogen peroxide for 1 h (middle panel). Mesophyll cells in the cat2-1 mutant expressing the GFP-PTS1 gene (cat2-1 GFP-PTS1; right panel). Arrows indicate peroxisome aggregates. Bar = 10 μm.
(B) Number of peroxisomes in protoplasts from cat2-1 GFP-PTS1 and atg2-1 cat2-1 GFP-PTS1 plants. Open and striped bars indicate the number of dispersed and aggregated peroxisomes, respectively. Error bars represent se (n = 20).
(C) Protein extracts were prepared from leaves of 3-week-old GFP-PTS1 (lane 1), peup1-1 (lane 2), atg2-1 GFP-PTS1 (lane 3), cat2-1 GFP-PTS1 (lane 4), atg2-1 cat2-1 GFP-PTS1 (lane 5), cat3-1 GFP-PTS1 (lane 6), atg2-1 cat3-1 GFP-PTS1 (lane 7), cat2-1 cat3-1 GFP-PTS1 (lane 8), and atg2-1 cat2-1 cat3-1 GFP-PTS1 (lane 9). The protein extracts were subjected to SDS-PAGE, followed by immunoblotting using the antibody indicated on the left side of each panel. The asterisk in α-APX indicates a nonspecific band.
Next, we examined whether a decrease in CAT activity induces the peroxisome aggregation using cat mutants. The Arabidopsis genome contains three CAT genes (CAT1, CAT2, and CAT3). CAT1 is mainly expressed in seeds and reproductive tissues, whereas CAT2 and CAT3 are highly expressed in leaves (Frugoli et al., 1996; Mhamdi et al., 2010). Because CAT activity mainly depends on CAT2 (Mhamdi et al., 2010) (see Supplemental Figure 8 online), we examined a cat2 knockout mutant.
We obtained a T-DNA insertion line of cat2-1 (SALK_76998) and introduced the GFP-PTS1 gene for visualizing peroxisomes into the line, producing cat2-1 GFP-PTS1. We observed leaves of the cat2-1 GFP-PTS1 mutant and found peroxisome aggregates in some cells (Figure 7A, right panel). Because hydrogen peroxide is accumulated in cat2 mutant cells (Hu et al., 2010), these data suggest that the peroxisome aggregation was caused by the accumulation of hydrogen peroxide in the cat2 mutant. Accumulation of hydrogen peroxide in peroxisomes is linked to the aging of the plant (Pastori and del Rio, 1994, 1997; del Rio et al., 1998; Van Breusegem and Dat, 2006). In fact, the peup1 mutant exhibited early senescence compared with the GFP-PTS1 plant (see Supplemental Figure 9 online).
We subsequently examined whether autophagic degradation of peroxisomes occurs in the cat2-1 mutant. If CAT2 were directly involved in the autophagic system, autophagic degradation of peroxisomes would be suppressed in the cat2-1 mutant. In that case, we would observe peroxisome aggregates in the cat2-1 mutant for the same reason that they were observed in the peup1 mutant. We counted the number of peroxisomes of the cat2-1 mutant; if CAT was responsible for the autophagic degradation, the peroxisomal number would increase compared with that of GFP-PTS1. Our results showed that the number of aggregated peroxisomes was slightly higher in the cat2-1 GFP-PTS1 mutant than in GFP-PTS1 (Figure 7B), whereas the total number of peroxisomes was similar in both lines (Figure 2B). On the other hand, the total number of peroxisomes was higher in the atg2-1 cat2-1 mutant because of the higher number of aggregated peroxisomes (Figure 7B). These results indicate that peroxisome degradation by autophagy occurs in the cat2 mutant.
Furthermore, because CAT3 is still active in the cat2 mutant, we examined the accumulation of peroxisomal proteins in the cat2 cat3 double mutant in addition to the cat2 mutant. As shown in Figure 7C, there were no differences in peroxisomal protein accumulation between the cat2-1 cat3-1 GFP-PTS1 mutant (lane 8) and the GFP-PTS1 plant (lane 1), whereas the atg2-1 cat2-1 cat3-1 GFP-PTS1 triple mutant (lane 9) accumulated peroxisomal proteins. These results indicate that the lack of CAT does not prevent the degradation of peroxisomal proteins, which indeed suggests that autophagy occurs normally in CAT-deficient plants.
Autophagosome Marker ATG8 Colocalizes with Peroxisome Aggregates
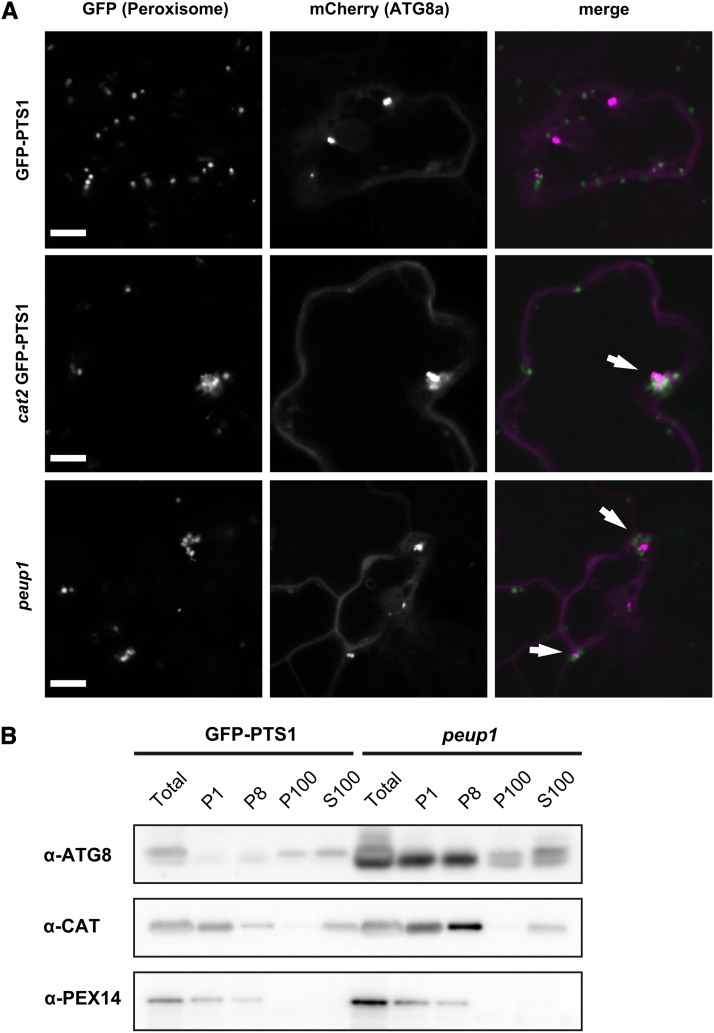
To examine whether autophagy selectively degrades peroxisomes, we investigated the colocalization of autophagosome marker ATG8 with peroxisomes (Figure 8). In GFP-PTS1 plants, mCherry-ATG8a diffused in the cytosol and formed some punctate structures. The punctuate structure is a preautophagosomal structure (PAS), which is observed in the isolation membrane-forming site (Suzuki et al., 2001). In GFP-PTS1 plants, few peroxisomes localized near mCherry-ATG8 (Figure 8A, top panel), whereas almost all peroxisome aggregates colocalized with mCherry-ATG8a in the cat2 and peup1 mutants (Figure 8A, middle and bottom panels; see Supplemental Figure 10 online). We then quantified the interaction between peroxisomes and mCherry-ATG8a–labeled PASs. In GFP-PTS1 plants, ∼17% of dispersed peroxisomes colocalized with PAS, whereas ∼30% of dispersed peroxisomes colocalized with PAS in both the cat2 and peup1 mutants (see Supplemental Figure 11A online). Notably, more than 90% of peroxisome aggregates colocalized with PAS in both the cat2 and peup1 mutants (see Supplemental Figure 11B online). These results strongly suggest that peroxisome aggregates are selectively degraded by autophagy.
Figure 8.
Subcellular Localization of Autophagosome Marker ATG8.
(A) Confocal images of leaf cells of a GFP-PTS1 plant, cat2-1 GFP-PTS1 mutant, and peup1-1 mutant transiently expressing mCherry-ATG8a. Arrows indicate peroxisomes colocalizing with punctuate structures of ATG8. Bar = 10 μm.
(B) Immunoblot analysis by differential centrifugation. Total proteins (Total) prepared from the leaves of 3-week-old plants were fractionated into P1, P8, P100, and S100 fractions. The samples were subjected to SDS-PAGE followed by immunoblot analysis using α-ATG8, α-CAT, and α-PEX14 antibodies. ATG8 is an autophagosome marker, and PEX14 and CAT are peroxisome markers.
Next, we performed differential centrifugation to examine the interaction of ATG8 and peroxisomes using an alternative method. CAT and PEX14 were detected in the P1 and P8 fractions, showing that peroxisomes exist in these fractions. Some CAT was recovered in the S100 fraction, because the collapse of peroxisomes can occur during sample preparation. ATG8 was mainly detected in the P100 and S100 fractions in GFP-PTS1 plants. However, ATG8 highly accumulated and was detected in the P1 and P8 fractions in addition to the P100 and S100 fractions in the peup1 mutant (Figure 8B). In addition, the faster migration bands of ATG8 were detected in the total, P1, and P8 fractions of the peup1 mutant. The bands are possibly ATG8 conjugated with phosphatidylethanolamine (PE) (Chung et al., 2009); when ATG8 is localized in an isolation membrane, ATG8 conjugates with PE (Yoshimoto et al., 2004). Because autophagy is not accomplished in the peup1 mutant, accumulation of PE-ATG8 in the fractions with peroxisomes is plausible. These results indicate that ATG8 is bound to aggregated peroxisomes in the peup1 mutant, strongly supporting the observation shown in Figure 8A.
Condensed CAT Is Dispensable for the Formation of Peroxisome Aggregates
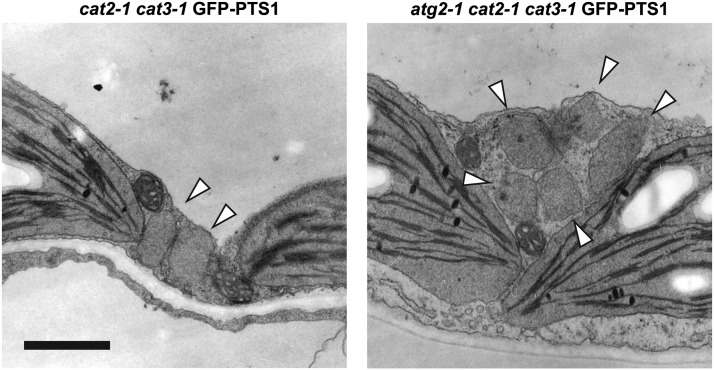
Because condensed CAT regions frequently appeared to be face to face (Figure 4A; see Supplemental Figure 4 online), we assumed that the condensed CAT is responsible for peroxisome adhesion. Therefore, we examined peroxisome aggregation in the atg2 cat2 cat3 triple mutant. In the triple mutant, peroxisome aggregates appeared, even though the electron-dense regions were not observed (Figure 9). These findings indicate that condensed CAT itself is not essential for the formation of peroxisome aggregates.
Figure 9.
Electron Microscopy Observation of Mesophyll Cells in the atg2-1 cat2-1 cat3-1 Mutant
Electron microscopy observation was performed using 3-week-old cat2-1 cat3-1 GFP-PTS1 and atg2-1 cat2-1 cat3-1 GFP-PTS1 plants. Arrowheads represent peroxisomes without electron-dense regions. Bar = 1 μm.
DISCUSSION
In this study, we isolated and analyzed peup mutants, which contain peroxisome aggregates (Figure 1). Three peup mutants are revealed to be defective in autophagic systems, showing that peroxisome aggregation is obviously related to this defect in autophagy. On the basis of our results, we propose a model describing the physiological function of autophagic degradation of peroxisomes and the mechanism for the formation of peroxisome aggregates (Figure 10). The following is a detailed description of the model presented in this figure.
Figure 10.
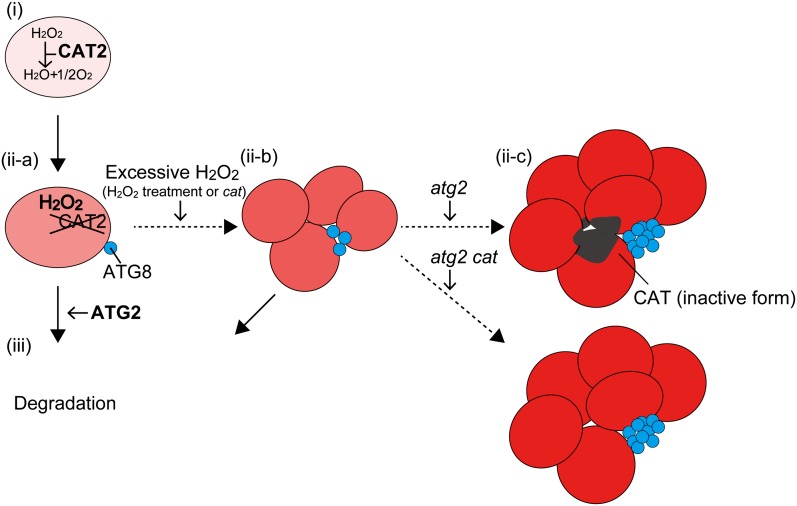
Model of the Quality Control Mechanism for Peroxisomes via Autophagic Degradation and Peroxisome Aggregate Formation in the peup1.
(i) In a newly synthesized peroxisome, CAT degrades hydrogen peroxide generated by peroxisomal metabolic processes such as photorespiration. (ii-a) CAT is gradually inactivated by hydrogen peroxide, and the peroxisome becomes damaged by the accumulation of hydrogen peroxide. (iii) In the wild-type plant, the damaged peroxisome is targeted by ATG8 and is degraded by an autophagic system involving ATG2. (ii-b) When excessive hydrogen peroxide accumulates in a peroxisome as a result of factors such as exogenous hydrogen peroxide treatment or the presence of the cat mutation, peroxisomes begin to form aggregates. The peroxisome aggregate is eventually degraded by an autophagic system involving ATG2. (ii-c, top) In the peup1 (atg2) mutant, the highly damaged peroxisomes form large aggregates and accumulate in the cytosol. The black regions in the peroxisome aggregate represent condensed CAT. Because the formation of an isolation membrane is not completed in the atg2 mutant, ATG8 targeting the peroxisome aggregate highly accumulates. (ii-c, bottom) On the other hand, large peroxisome aggregates not containing condensed CAT appear in the atg2 cat mutant. The intensity of the red coloring in the peroxisomes represents the level of hydrogen peroxide. Solid and dashed arrows indicate processes in the wild type and mutants or under artificial conditions, respectively.
Role of Autophagy in Peroxisomal Quality Control
In peroxisomes of photosynthetic organs, hydrogen peroxide is excessively produced, mainly from photorespiratory metabolism. CAT is the main scavenging enzyme for hydrogen peroxide, but this enzyme is irreversibly inactivated by its substrate, hydrogen peroxide (Williams, 1928). Thus, hydrogen peroxide accumulates in response to CAT inactivation in the peroxisomes, and the peroxisomes gradually exhibit oxidative conditions. Our data reveal that peroxisome aggregates in the peup1 mutant consist of peroxisomes that were oxidized by an excess of hydrogen peroxide (Figures 6 and 7; see Supplemental Figures 6 and 7 online). In this respect, a peroxisome aggregate can be considered to be a cluster of damaged peroxisomes. Because hydrogen peroxide also functions as a signaling molecule in processes such as programmed cell death (Alvarez et al., 1998; Gechev and Hille, 2005; del Rio and Pusso, 2009), damaged peroxisomes with uncontrollable hydrogen peroxide levels preclude optimal plant growth. These damaged peroxisomes were accumulated in the peup1 mutant, suggesting that the peroxisomes must be degraded by autophagic processes in the wild type. In mammalian cells, the atg2 mutation causes accumulation of unclosed autophagic structures containing most ATG proteins, including ATG8 (Velikkakath et al., 2012). ATG8 was highly accumulated on peroxisome aggregates in the peup1 mutant (Figure 8), indicating that ATG8 is selectively targeted to damaged peroxisomes. These data demonstrate that autophagy serves as a quality control mechanism via selective elimination of damaged peroxisomes in the plant cell.
The Process of Forming Peroxisome Aggregates in the peup1 Mutant
We demonstrated that excess hydrogen peroxide induces peroxisome aggregate formation (Figure 7). This notion is also supported by another observation by Sinclair et al. (2009) regarding peroxisomes. When plant leaves are treated with hydrogen peroxide, peroxisomes change their morphology, forming extended structures termed peroxules (Sinclair et al., 2009). The aggregated peroxisomes in the peup1 mutant actually sometimes formed extended structures similar to peroxules, suggesting that the accumulation of hydrogen peroxide induces peroxisome aggregation through the production of peroxule-like structures. In the peup1 mutant, oxidized peroxisomes stay in the cytosol because they are not degraded by autophagy. As a consequence, oxidized peroxisomes increasingly form larger aggregates.
However, the factors that hold the peroxisomes together in the aggregates are still unclear. Condensed CAT regions often existed face to face among peroxisomes (see Supplemental Figure 4 online). However, peroxisome aggregates also formed in the atg2-1 cat2-1 cat3-1 mutant (Figure 9), suggesting that condensed CAT is not directly responsible for the peroxisome aggregation. Because hydrogen peroxide oxidizes CAT as well as many proteins and membrane lipids (Siddique et al., 2012), it is plausible that many types of damaged proteins and lipids accumulate around the condensed CAT.
Contribution of Autophagy to Peroxisome Quality Control
Regardless of the significant accumulation of damaged peroxisomes in the peup1 mutant, we did not detect an obvious decrease in the metabolic activities of the glyoxysomes or leaf peroxisomes (such as β-oxidation or photorespiration; see Supplemental Figure 2 online). This is probably because the peup1 mutant still contains active peroxisomes in addition to damaged peroxisomes (Figures 2 and 4; see Supplemental Figure 7 online). Meanwhile, the peup1 mutation also affected the phenotypes of the plant. The peup1 mutant exhibited early senescence with slightly inhibited growth (see Supplemental Figure 9 online). This phenotype has also been observed in other atg mutants (Doelling et al., 2002; Hanaoka et al., 2002; Thompson et al., 2005; Xiong et al., 2005; Qin et al., 2007; Patel and Dinesh-Kumar, 2008; Phillips et al., 2008) and has been suggested to be associated with the accumulation of hydrogen peroxide (Yoshimoto et al., 2009). However, the origin of the excess hydrogen peroxide in atg mutants was unclear. Because our results suggest that aggregated peroxisomes in the peup1 mutant contain high concentrations of hydrogen peroxide (Figure 6), hydrogen peroxide may leak from the peroxisomes and damage the cell. Thus, we assume that peroxisomes containing inactive CAT cause the accumulation of hydrogen peroxide in the atg mutants. In addition to peroxisomes, the major sources of hydrogen peroxide in a cell are chloroplasts and mitochondria. However, there was no significant difference in the number and morphology of chloroplasts or mitochondria between the peup1 mutant and wild-type plants (Figures 3 and 4; see Supplemental Figure 1 online), suggesting that quality control of these organelles is not altered in this mutant. Therefore, we consider that the phenotypes such as early senescence of the peup1 mutant are attributed in part to accumulation of the damaged peroxisomes.
Although peroxisomal degradation is known to occur via autophagy in yeasts and mammals, it was unknown until recently whether this process occurs in plants. Peroxisome-specific degradation is referred to as pexophagy (Veenhuis et al., 1983). To date, most information about pexophagy has come from the analysis of methylotrophic yeasts, such as P. pastoris and Hansenula polymorpha (Dunn et al., 2005; Sakai et al., 2006; Manjithaya et al., 2010). The metabolism of methanol in methylotrophic yeast species occurs in peroxisomes, which are massively induced when the yeast is grown on medium containing methanol as the sole carbon source. When the methanol-grown yeast is transferred to medium containing Glc or ethanol, the induced peroxisomes become redundant and are rapidly degraded by an autophagic system (Veenhuis et al., 1983; Tuttle et al., 1993). Therefore, the main role of pexophagy in yeast is to remove redundant peroxisomes. Pexophagy was recently identified as a quality control mechanism to degrade damaged peroxisomes in yeasts, but this occurs only in limited situations (van Zutphen et al., 2011). In plants, the number of peroxisomes is constant throughout the entire lifecycle. Furthermore, plant peroxisomes are constantly exposed to hydrogen peroxide damage, suggesting that the main role of pexophagy is quality control of peroxisomes, rather than the removal of redundant peroxisomes in plants.
Lon protease also plays a role in the maintenance of peroxisomes. Lon protease was initially identified in Escherichia coli (Charette et al., 1981) and is widely conserved among organisms. A peroxisomal Lon protease was identified in rat (Kikuchi et al., 2004). In addition, Lon2, one of the four Lon proteases found in Arabidopsis, is suggested to be localized in peroxisomes (Ostersetzer et al., 2007; Lingard and Bartel, 2009). ATP-dependent Lon proteases belong to the AAA(+) superfamily and are multifunctional proteins that act as proteases for selective degradation of aberrant peroxisomal matrix proteins; however, they also function as chaperones to assist in the refolding of these proteins (Aksam et al., 2007; Bartoszewska et al., 2012). Recently, Penicillium chrysogenum Lon (Pln) was found to be involved in the quality control of peroxisomes via degradation of oxidized proteins as specific substrates. Interestingly, pln mutants contain electron-dense regions in their peroxisomes, which are similar to those of the peup1 (Bartoszewska et al., 2012). While this article was under review, Farmer et al. (2013) reported that peroxisomal Lon protease and autophagy were indeed involved in peroxisome quality control in Arabidopsis. In addition to Lon2, there are some proteases in peroxisomes (Distefano et al., 1997, 1999; Palma et al., 2002). The proteases are also expected to contribute to the maintenance of peroxisomal quality. We recently found that the proteasome is involved in the degradation of a peroxisomal receptor when it accumulates in the peroxisome membrane to abnormally high levels, suggesting that the proteasome can engage in the specific degradation of a peroxisomal protein (Cui et al., 2013).
Thus, the presence of additional peroxisome quality control mechanisms may explain why the peup1 mutant did not exhibit serious phenotypes related to peroxisomal metabolism.
What Is the Signal for Peroxisome Degradation via Autophagy?
Autophagy was previously assumed to be a nonspecific degradation system. However, many recent reports demonstrate that autophagy also works as a target-specific degradation system in processes such as mitophagy (mitochondria), pexophagy (peroxisomes), ribophagy (ribosomes), reticulophagy (endoplasmic reticulum [ER]), and xenophagy (bacteria) (Kraft et al., 2009; Johansen and Lamark, 2011). In plants, ribulose-1,5-bisphosphate carboxylase/oxygenase–containing bodies are involved in the degradation of stroma proteins of chloroplasts, which is induced by sugar starvation and functions in nutrition recycling (Ishida and Yoshimoto, 2008; Ishida et al., 2008; Wada et al., 2009; Izumi et al., 2010; Ono et al., 2013). Autophagic degradation of ER induced by ER stress was also recently reported (Liu et al., 2012; Liu and Bassham, 2013). Selective degradation is a more effective quality control process than random degradation, and our data exhibit that the damaged peroxisomes are selectively degraded. Therefore, signals should exist on peroxisomes to distinguish whether they are damaged.
We hypothesized that condensed CAT itself is the signal used to discriminate between intact and damaged peroxisomes. To investigate this hypothesis, we examined the number of peroxisomes and the accumulation of peroxisomal proteins in the cat mutants. These examinations revealed that CAT is dispensable for the degradation of peroxisome (Figures 7B and 7C). As discussed above, damaged materials probably accumulate around the electron-dense regions. Our results suggest that damaged CAT is not the sole signal for peroxisome degradation via autophagy, suggesting that other damaged material also works as a signals for autophagy. This hypothesis is supported by reports demonstrating that protein aggregates and oxidized proteins induced by ROS are specific substrates for autophagic degradation (Toyooka et al., 2006; Xiong et al., 2007).
In yeast pexophagy, there are adaptors that connect components of autophagic machinery and peroxisomes. In methylotrophic yeast, Atg30 binds to Pex3 and Pex14 on peroxisomes (Farré et al., 2008). However, Atg30 is conserved in only a few species and not in Arabidopsis. Similarly, Atg36 binds to Pex3, Atg8, and Atg11, but this protein is conserved only among Saccharomycetaceae species (Motley et al., 2012). Indeed, ATG genes involved in nonselective autophagy are conserved in yeast, mammals, and plants, but the selective autophagic pathways also require organism-specific genes (Meijer et al., 2007). These data suggest that plants have specific adaptors, other than Atg30 or Atg36, that connect components of autophagic machinery to peroxisomes. Thus, the discovery of such adaptors is crucial to the study of plant-specific pexophagy. Further studies will reveal the signals produced by the damaged peroxisomes, as well as adaptors similar to ATG30 and ATG36 in plants.
METHODS
Plant Materials and Growth Conditions
Transgenic GFP-PTS1, peup1-1, peup1-2, peup2, and peup4 plants were derived from the Arabidopsis thaliana Columbia background (Mano et al., 2002, 2004). Mt-GFP and shm1-1 plants were kindly provided by Shin-ichi Arimura (The University of Tokyo; Feng et al., 2004) and Shunichi Takahashi (Australian National University; Takahashi et al., 2007), respectively. The ped1 mutant was previously described (Hayashi et al., 1998) T-DNA insertion mutants of atg2-1 (SALK_076727), atg18a-2 (GABI_651D08), atg7-2 (GABI_655B06), cat2-1 (SALK_076998), and cat3-1 (SALK_092911) were obtained from the ABRC and GABI. Each homozygous plant was identified using gene-specific primers (see Supplemental Table 1 online). The primers for atg7-2 (Hofius et al., 2009) and cat2-1 (Queval et al., 2007) were previously described.
Seeds were sown on solid medium containing 1.53 mg mL−1 Murashige and Skoog salts (Wako), 1% (w/v) Suc, 100 μg mL−1 myo-inositol, 1 μg mL−1 thiamine-HCl, 0.5 μg mL−1 pyridoxine-HCl, 0.5 μg mL−1 nicotinic acid, 0.5 mg mL−1 MES-KOH, pH 5.7, and 0.8% (w/v) agar (INA). Germination was induced by 24-h incubation at 4°C. The plates were then transferred to 22°C under long-day conditions (16-h light/8-h dark). The seedlings were transferred to soil 2 weeks after germination.
Confocal Microscopy
Observation of cells was performed using LSM510 META (Carl Zeiss) and A1R (Nikon) confocal laser scanning microscopes, as previously described (Mano et al., 2002).
Map-Based Cloning and Identification of PEUP Genes
The peup mutants were backcrossed three times with the parental plant GFP-PTS1 and crossed with another accession, Landsberg erecta. The F2 progenies showing mutant phenotypes were scored according to their genetic background, using a series of cleaved amplified polymorphic sequences and simple sequence length polymorphism markers (Konieczny and Ausubel, 1993; Bell and Ecker, 1994). The oligonucleotide primer sets for the cleaved amplified polymorphic sequences and simple sequence length polymorphism markers were designed according to the Cereon Genomics database (http://www.Arabidopsis.org/Cereon/index.jsp) and NARAMAP markers. The NARAMAP marker information was kindly provided by Masao Tasaka and Miyo Terao Morita at the Nara Institute of Science and Technology (Toyota et al., 2011). The gene sequencing was performed using a 310 Genetic Analyzer or a 3130xl Genetic Analyzer and a BigDye Terminators v1.1 Cycle Sequencing Kit (all from Applied Biosystems) according to the manufacturer’s instructions.
Protoplast Isolation and Measurement of the Number of Peroxisomes
Protoplasts were prepared using the tape-Arabidopsis sandwich method (Wu et al., 2009) with slight modifications. The sixth and seventh leaves of 3-week-old plants were collected. The leaf epidermis was removed by peeling using two types of tape, Time tape (Time Med) and mending tape (3M), and was then immersed in enzyme solution containing 1% (v/w) cellulase Onozuka R-10 (Yakult), 0.25% (v/w) macerozyme R-10 (Yakult), 0.4 M mannitol, and 20 mM MES, pH 5.7, and gently shaken for 30 min in the dark. The protoplast-containing solution was centrifuged at 100g for 1 min, and the supernatant was discarded. The pellet was washed twice with wash solution (0.4 M mannitol and 20 mM MES, pH 5.7).
The fluorescence images of protoplasts were taken by confocal microscopy from the top to the bottom of a protoplast, and the projection images were generated based on these z-stack image data. Dispersed and aggregated peroxisomes were counted in these images.
Relative Quantitative RT-PCR Analysis
Relative quantitative RT-PCR analysis was performed as previously described, with slight modifications (Kanai et al., 2010, 2013). Total RNA was extracted from leaves of 3-week-old plants using an RNeasy Plant Mini Kit (Qiagen). Total RNA was extracted from three independent samples for each treatment. The cDNA was synthesized from 1 µg total RNA using Ready-to-Go RT-PCR beads (GE Healthcare). Real-time PCR was performed using a 7500 Fast Real-Time PCR System (Applied Biosystems) and a KAPA SYBR FAST qPCR Kit (Kapa Biosystems) according to the manufacturers’ instructions. The primer sets used for the PCR are shown in Supplemental Table 2 online.
Electron Microscopic Analysis
Leaves of 3-week-old plants were harvested and placed in fixative solution that consisted of 4% (w/v) paraformaldehyde, 1% (w/v) glutaraldehyde, and 0.06 M Suc in 0.05 M cacodylate buffer, pH 7.4. Ultrathin sectioning, microscopic analysis, and immunogold labeling were performed as previously described (Hayashi et al., 1998).
Immunoblot Analysis
To extract total proteins, leaves of 3-week-old plants were homogenized in SDS buffer containing 10 mM HEPES-KOH, pH 8.0, 1% (w/v) SDS, and EDTA-free protease inhibitor cocktail (one tablet per 10 mL; Roche). Soluble and insoluble proteins were separated using a previously described method (Asano et al., 2004) with some modifications. Leaves of 3-week-old plants were homogenized in extraction buffer containing 10 mM HEPES-KOH, pH 6.8, and EDTA-free protease inhibitor cocktail. Homogenates were centrifuged at 20,000g for 10 min at 4°C. The supernatants were collected as the soluble proteins. The pellets were washed twice with extraction buffer and solubilized with SDS buffer. The concentrations of extracted proteins were estimated using a Bradford ULTRA kit (Novexin) with BSA as a standard. The proteins were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Millipore) in a semidry electroblotting system (BioCraft). Immunoreactive bands were detected by monitoring the activity of horseradish peroxidase–coupled antibodies against rabbit IgG (ECL system; GE Healthcare). The dilution ratios of antibodies were 1:6000 for α-CAT (Yamaguchi et al., 1984), 1:5000 for α-GFP (Mitsuhashi et al., 2000), 1:5000 for α-APX (Arai et al., 2008), 1:5000 for α-RBCL (Nishimura and Akazawa, 1974), 1:3000 for α-GO (Nishimura et al., 1983), 1:3000 for α-PEX14 (Hayashi et al., 2000), 1:1000 for α-COX2 (Agrisera), and 1:500 for α-ATG8 (Abcam).
Differential Centrifugation
The differential centrifugation method was performed as previously described, with slight modifications (Yamada et al., 2008, (2013). Leaves of 3-week-old plants were chopped on ice in a triple volume of chopping buffer that contained 50 mM HEPES-NaOH, pH 7.5, 0.4 M Suc, and protease inhibitor cocktail (one tablet per 50 mL; Roche). The homogenates were filtered through cheesecloth and then centrifuged at 1000g at 4°C for 10 min. The pellets were designated as the P1 fraction. The supernatants were centrifuged at 8000g at 4°C for 20 min; the pellets were designated as the P8 fraction. The supernatants were centrifuged at 100,000g at 4°C for 1 h. The pellets and the supernatants obtained after ultracentrifugation were designated as the P100 and S100 fractions, respectively. The P1, P8, and P100 fractions were resuspended in the same volume of chopping buffer and subjected to immunoblot analysis.
Measuring CAT Activity
Fifteen microliters of 3% (v/v) hydrogen peroxide and 3 μL of the soluble fraction, or the insoluble fraction before solubilization, were added to 1 mL of 50 mM K-phosphate buffer, pH 5.7. Immediately after mixing, CAT activity was determined by monitoring the absorbance at 240 nm during the first 10 s with a U-2900 spectrophotometer (Hitachi). The amount of CAT peptide was estimated by determining the band intensity of the immunoblot using Gel Analyzer on Image J software (v. 1.46i; http://imagej.nih.gov/ij).
Measuring the Redox Status of Peroxisomes Using roGFP2
To create roGFP2, a commonly used GFP gene for plant expression, sGFP(S65T) (Chiu et al., 1996), was subjected to site-directed mutagenesis. Three point mutations corresponding to amino acid substitutions, C48S, N147C, and Q204C (Hanson et al., 2004), were introduced into sGFP(S65T) and the resulting gene was termed roGFP2. The roGFP2-PTS1 gene was constructed by PCR. The PCR fragments were then replaced with sGFP(S65T) in p35S-GFP-T. After digestion with HindIII and EcoRI, the DNA fragments containing 35S promoter, roGFP-PTS1, and terminator were inserted into the pCAMBIA2300 (CAMBIA). The roGFP2-PTS1 gene on the binary vector was introduced into wild-type plants (Columbia) by an agro-infiltration method. In the resulting lines, we adopted the most suitable line for analysis and designated roGFP-PTS1. To introduce the roGFP-PTS1 gene into the peup1 mutant, we crossed the roGFP-PTS1 plant with the peup1-1 mutant and the resulting line was designated peup1 roGFP-PTS1. We used these two lines for the analysis.
Plants expressing roGFP-PTS1 were observed under an A1R confocal microscope. Using the 2Ex 1Em mode, roGFP2 was exited with 405 and 488 nm lasers, and fluorescence was measured using a 500- to 550-nm band pass filter. Stack images were taken using the resonant mode. The maximum projection, based on the z-stack images, was calculated using ImageJ. After subtraction of the background signals, ratiometric images of 405/488 were generated.
Transient Expression of ATG8
cDNA of At4g21980 (ATG8a) was amplified using gene-specific primer sets (see Supplemental Table 1 online). The sequence information was obtained from the TAIR database. The fragment was introduced into Gateway entry vector D-TOPO (Invitrogen) to produce pENTR/ATG8a. The entire protein-coding region of pENTR/ATG8a was transferred to binary vector pmCGW to generate the mCherry fusion using the Gateway LR recombination reaction. The construct was transformed into Agrobacterium tumefaciens strain C58C1Rif and then introduced into GFP-PTS1 plants or the peup1 mutant using high-throughput transient transformation of Arabidopsis seedlings (Marion et al., 2008).
Accession Numbers
Nucleotide sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: PEUP1/ATG2 (At3g19190), PEUP2/ATG18a (At3g62770), PEUP4 (At5g45900), CAT2 (At4g35090), CAT3 (At1g20620), SHM1 (At4g37930), and PED1 (At2g33150). The accession numbers of the SALK T-DNA insertion mutants or TAIR stocks mentioned in this article are as follows: SALK_076727 (atg2-1), GABI_651D08 (atg18a-2), GABI_655B06 (atg7-2), SALK_76998 (cat2-1), SALK_092911 (cat3-1), and CS8010 (shm1-1). The GenBank accession number of ATG2 protein is ADU79134.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Morphology and Number of Mitochondria in the peup1 Mutant.
Supplemental Figure 2. Analysis of Peroxisomal Functions in the peup1 Mutant.
Supplemental Figure 3. Expression of Genes Encoding Peroxisomal Proteins.
Supplemental Figure 4. Pattern of Electron-Dense Regions in Aggregated Peroxisomes of the peup1-1 Mutant.
Supplemental Figure 5. Immunoelectron Microscopic Analysis of Peroxisome Aggregates in the peup1-1 Mutant.
Supplemental Figure 6. Effect of Hydrogen Peroxide and DTT on Redox Status of Peroxisomes.
Supplemental Figure 7. Redox Status in the Dispersed Peroxisomes of the peup1 Mutant.
Supplemental Figure 8. CAT Activity in GFP-PTS1 Plants and Various Mutants.
Supplemental Figure 9. Growth Phenotype of peup1 Mutant.
Supplemental Figure 10. Interaction between Peroxisome Aggregates and ATG8.
Supplemental Figure 11. Quantification of the Interaction between Peroxisomes and ATG8.
Supplemental Table 1. Nucleotide Sequences of Primers Used in This Study.
Supplemental Table 2. Nucleotide Sequences of Primers Used for Quantitative RT-PCR.
Acknowledgments
We thank Shin-ichi Arimura for providing the Mt-GFP plant, Mitsuyasu Hasebe and Yoshiaki Kamada for valuable comments, Makoto Suzuki for technical advice, and the staff at the Model Plant Research Facility and the Spectrography and Bioimaging Facility at the National Institute of Basic Biology Core Research Facilities for plant maintenance and technical support. This study was supported by a Grant-in-Aid for Scientific Research (22120007 to M.N.) and a Research Fellowship for Young Scientists from the Japan Society for the Promotion of Science (5852 to M.S.).
AUTHOR CONTRIBUTIONS
M.S., K.O., S.M., K.Yamada, M.H., and M.N. designed the research. K.O. isolated the peup1, peup2, and peup4 mutants and determined the responsible genes for peup2 and peup4. M.K. performed electron microscopy observations. W.S. produced roGFP2. K.Yoshimoto generated the roGFP-PTS1, atg18a-2 GFP-PTS1, and atg7-2 GFP-PTS1 plants. M.S. performed other experiments. M.S., K.O., K.Yoshimoto, S.M., K.Yamada, M.H., Y.O., and M.N. analyzed the data and contributed to data interpretation and the preparation of the article. M.S., K.O., and M.N. supervised the study and wrote the article.
References
- Aksam E.B., Koek A., Kiel J.A., Jourdan S., Veenhuis M., van der Klei I.J. (2007). A peroxisomal Lon protease and peroxisome degradation by autophagy play key roles in vitality of Hansenula polymorpha cells. Autophagy 3: 96–105 [DOI] [PubMed] [Google Scholar]
- Alvarez M.E., Pennell R.I., Meijer P.J., Ishikawa A., Dixon R.A., Lamb C. (1998). Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92: 773–784 [DOI] [PubMed] [Google Scholar]
- Arai Y., Hayashi M., Nishimura M. (2008). Proteomic identification and characterization of a novel peroxisomal adenine nucleotide transporter supplying ATP for fatty acid β-oxidation in soybean and Arabidopsis. Plant Cell 20: 3227–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T., Yoshioka Y., Kurei S., Sakamoto W., Sodmergen, Machida Y. (2004). A mutation of the CRUMPLED LEAF gene that encodes a protein localized in the outer envelope membrane of plastids affects the pattern of cell division, cell differentiation, and plastid division in Arabidopsis. Plant J. 38: 448–459 [DOI] [PubMed] [Google Scholar]
- Bartoszewska M., Williams C., Kikhney A., Opaliński L., van Roermund C.W., de Boer R., Veenhuis M., van der Klei I.J. (2012). Peroxisomal proteostasis involves a Lon family protein that functions as protease and chaperone. J. Biol. Chem. 287: 27380–27395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell C.J., Ecker J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137–144 [DOI] [PubMed] [Google Scholar]
- Charette M.F., Henderson G.W., Markovitz A. (1981). ATP hydrolysis-dependent protease activity of the Lon (capR) protein of Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 78: 4728–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W., Niwa Y., Zeng W., Hirano T., Kobayashi H., Sheen J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Choi W.G., Swanson S.J., Gilroy S. (2012). High-resolution imaging of Ca2+, redox status, ROS and pH using GFP biosensors. Plant J. 70: 118–128 [DOI] [PubMed] [Google Scholar]
- Chung T., Suttangkakul A., Vierstra R.D. (2009). The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol. 149: 220–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S., Fukao Y., Mano S., Yamada K., Hayashi M., Nishimura M. (2013). Proteomic analysis reveals that the Rab GTPase RabE1c is involved in the degradation of the peroxisomal protein receptor PEX7 (peroxin 7). J. Biol. Chem. 288: 6014–6023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rio L.A., Pastori G.M., Palma J.M., Sandalio L.M., Sevilla F., Corpas F.J., Jimenez A., Lopez-Huertas E., Hernandez J.A., Hernandez J.A. (1998). The activated oxygen role of peroxisomes in senescence. Plant Physiol. 116: 1195–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rio, L.A., and Pusso, A. (2009). Reactive Oxygen Species in Plant Signaling (Berlin, Heidelberg: Springer Verlag, Berlin Heidelberg). [Google Scholar]
- Distefano S., Palma J.M., Gómez M., Río L.A. (1997). Characterization of endoproteases from plant peroxisomes. Biochem. J. 327: 399–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distefano S., Palma J.M., McCarthy I.I., del Rio L.A. (1999). Proteolytic cleavage of plant proteins by peroxisomal endoproteases from senescent pea leaves. Planta 209: 308–313 [DOI] [PubMed] [Google Scholar]
- Doelling J.H., Walker J.M., Friedman E.M., Thompson A.R., Vierstra R.D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277: 33105–33114 [DOI] [PubMed] [Google Scholar]
- Dooley C.T., Dore T.M., Hanson G.T., Jackson W.C., Remington S.J., Tsien R.Y. (2004). Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J. Biol. Chem. 279: 22284–22293 [DOI] [PubMed] [Google Scholar]
- Dunn W.A., Jr, Cregg J.M., Kiel J.A., van der Klei I.J., Oku M., Sakai Y., Sibirny A.A., Stasyk O.V., Veenhuis M. (2005). Pexophagy: The selective autophagy of peroxisomes. Autophagy 1: 75–83 [DOI] [PubMed] [Google Scholar]
- Farmer L.M., Rinaldi M.A., Young P.G., Danan C.H., Burkhart S.E., Bartel B. (2013). Disrupting autophagy restores peroxisome function to an Arabidopsis lon2 mutant and reveals a role for the LON2 protease in peroxisomal matrix protein degradation. Plant Cell 25: 4085–4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré J.C., Manjithaya R., Mathewson R.D., Subramani S. (2008). PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev. Cell 14: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X., Arimura S., Hirano H.Y., Sakamoto W., Tsutsumi N. (2004). Isolation of mutants with aberrant mitochondrial morphology from Arabidopsis thaliana. Genes Genet. Syst. 79: 301–305 [DOI] [PubMed] [Google Scholar]
- Foyer C.H., Noctor G. (2003). Redox sensing and signalling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol. Plant. 119: 355–364 [Google Scholar]
- Foyer C.H., Bloom A.J., Queval G., Noctor G. (2009). Photorespiratory metabolism: Genes, mutants, energetics, and redox signaling. Annu. Rev. Plant Biol. 60: 455–484 [DOI] [PubMed] [Google Scholar]
- Frugoli J.A., Zhong H.H., Nuccio M.L., McCourt P., McPeek M.A., Thomas T.L., McClung C.R. (1996). Catalase is encoded by a multigene family in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 112: 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev T.S., Hille J. (2005). Hydrogen peroxide as a signal controlling plant programmed cell death. J. Cell Biol. 168: 17–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goepfert S., Poirier Y. (2007). β-oxidation in fatty acid degradation and beyond. Curr. Opin. Plant Biol. 10: 245–251 [DOI] [PubMed] [Google Scholar]
- Goto S., Mano S., Nakamori C., Nishimura M. (2011). Arabidopsis ABERRANT PEROXISOME MORPHOLOGY9 is a peroxin that recruits the PEX1-PEX6 complex to peroxisomes. Plant Cell 23: 1573–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham I.A. (2008). Seed storage oil mobilization. Annu. Rev. Plant Biol. 59: 115–142 [DOI] [PubMed] [Google Scholar]
- Hanaoka H., Noda T., Shirano Y., Kato T., Hayashi H., Shibata D., Tabata S., Ohsumi Y. (2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129: 1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson G.T., Aggeler R., Oglesbee D., Cannon M., Capaldi R.A., Tsien R.Y., Remington S.J. (2004). Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J. Biol. Chem. 279: 13044–13053 [DOI] [PubMed] [Google Scholar]
- Hayashi M., Nishimura M. (2006). Arabidopsis thaliana—a model organism to study plant peroxisomes. Biochim. Biophys. Acta 1763: 1382–1391 [DOI] [PubMed] [Google Scholar]
- Hayashi M., Toriyama K., Kondo M., Nishimura M. (1998). 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. Plant Cell 10: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Nito K., Toriyama-Kato K., Kondo M., Yamaya T., Nishimura M. (2000). AtPex14p maintains peroxisomal functions by determining protein targeting to three kinds of plant peroxisomes. EMBO J. 19: 5701–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius D., Schultz-Larsen T., Joensen J., Tsitsigiannis D.I., Petersen N.H., Mattsson O., Jørgensen L.B., Jones J.D., Mundy J., Petersen M. (2009). Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137: 773–783 [DOI] [PubMed] [Google Scholar]
- Hu Y.Q., Liu S., Yuan H.M., Li J., Yan D.W., Zhang J.F., Lu Y.T. (2010). Functional comparison of catalase genes in the elimination of photorespiratory H2O2 using promoter- and 3′-untranslated region exchange experiments in the Arabidopsis cat2 photorespiratory mutant. Plant Cell Environ. 33: 1656–1670 [DOI] [PubMed] [Google Scholar]
- Inoue Y., Suzuki T., Hattori M., Yoshimoto K., Ohsumi Y., Moriyasu Y. (2006). AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol. 47: 1641–1652 [DOI] [PubMed] [Google Scholar]
- Ishida H., Yoshimoto K. (2008). Chloroplasts are partially mobilized to the vacuole by autophagy. Autophagy 4: 961–962 [DOI] [PubMed] [Google Scholar]
- Ishida H., Yoshimoto K., Izumi M., Reisen D., Yano Y., Makino A., Ohsumi Y., Hanson M.R., Mae T. (2008). Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 148: 142–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M., Wada S., Makino A., Ishida H. (2010). The autophagic degradation of chloroplasts via rubisco-containing bodies is specifically linked to leaf carbon status but not nitrogen status in Arabidopsis. Plant Physiol. 154: 1196–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedd G., Chua N.H. (2002). Visualization of peroxisomes in living plant cells reveals acto-myosin-dependent cytoplasmic streaming and peroxisome budding. Plant Cell Physiol. 43: 384–392 [DOI] [PubMed] [Google Scholar]
- Johansen T., Lamark T. (2011). Selective autophagy mediated by autophagic adapter proteins. Autophagy 7: 279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada T., Nito K., Hayashi H., Mano S., Hayashi M., Nishimura M. (2003). Functional differentiation of peroxisomes revealed by expression profiles of peroxisomal genes in Arabidopsis thaliana. Plant Cell Physiol. 44: 1275–1289 [DOI] [PubMed] [Google Scholar]
- Kanai M., Nishimura M., Hayashi M. (2010). A peroxisomal ABC transporter promotes seed germination by inducing pectin degradation under the control of ABI5. Plant J. 62: 936–947 [DOI] [PubMed] [Google Scholar]
- Kanai M., Hayashi M., Kondo M., Nishimura M. (2013). The plastidic DEAD-box RNA helicase 22, HS3, is essential for plastid functions both in seed development and in seedling growth. Plant Cell Physiol. 54: 1431–1440 [DOI] [PubMed] [Google Scholar]
- Kikuchi M., Hatano N., Yokota S., Shimozawa N., Imanaka T., Taniguchi H. (2004). Proteomic analysis of rat liver peroxisome: Presence of peroxisome-specific isozyme of Lon protease. J. Biol. Chem. 279: 421–428 [DOI] [PubMed] [Google Scholar]
- Kirsch T., Löffler H.G., Kindl H. (1986). Plant acyl-CoA oxidase. Purification, characterization, and monomeric apoprotein. J. Biol. Chem. 261: 8570–8575 [PubMed] [Google Scholar]
- Klionsky D.J., Ohsumi Y. (1999). Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 15: 1–32 [DOI] [PubMed] [Google Scholar]
- Konieczny A., Ausubel F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4: 403–410 [DOI] [PubMed] [Google Scholar]
- Kraft C., Reggiori F., Peter M. (2009). Selective types of autophagy in yeast. Biochim. Biophys. Acta 1793: 1404–1412 [DOI] [PubMed] [Google Scholar]
- Lingard M.J., Bartel B. (2009). Arabidopsis LON2 is necessary for peroxisomal function and sustained matrix protein import. Plant Physiol. 151: 1354–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Bassham D.C. (2012). Autophagy: Pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 63: 215–237 [DOI] [PubMed] [Google Scholar]
- Liu Y., Bassham D.C. (2013). Degradation of the endoplasmic reticulum by autophagy in plants. Autophagy 9: 622–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Burgos J.S., Deng Y., Srivastava R., Howell S.H., Bassham D.C. (2012). Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 24: 4635–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loew O. (1900). A new enzyme of general occurrence in organismis. Science 11: 701–702 [DOI] [PubMed] [Google Scholar]
- Manjithaya R., Nazarko T.Y., Farré J.C., Subramani S. (2010). Molecular mechanism and physiological role of pexophagy. FEBS Lett. 584: 1367–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano S., Nakamori C., Hayashi M., Kato A., Kondo M., Nishimura M. (2002). Distribution and characterization of peroxisomes in Arabidopsis by visualization with GFP: Dynamic morphology and actin-dependent movement. Plant Cell Physiol. 43: 331–341 [DOI] [PubMed] [Google Scholar]
- Mano S., Nakamori C., Kondo M., Hayashi M., Nishimura M. (2004). An Arabidopsis dynamin-related protein, DRP3A, controls both peroxisomal and mitochondrial division. Plant J. 38: 487–498 [DOI] [PubMed] [Google Scholar]
- Mano S., Nakamori C., Nito K., Kondo M., Nishimura M. (2006). The Arabidopsis pex12 and pex13 mutants are defective in both PTS1- and PTS2-dependent protein transport to peroxisomes. Plant J. 47: 604–618 [DOI] [PubMed] [Google Scholar]
- Mano S., Nakamori C., Fukao Y., Araki M., Matsuda A., Kondo M., Nishimura M. (2011). A defect of peroxisomal membrane protein 38 causes enlargement of peroxisomes. Plant Cell Physiol. 52: 2157–2172 [DOI] [PubMed] [Google Scholar]
- Marion J., Bach L., Bellec Y., Meyer C., Gissot L., Faure J.D. (2008). Systematic analysis of protein subcellular localization and interaction using high-throughput transient transformation of Arabidopsis seedlings. Plant J. 56: 169–179 [DOI] [PubMed] [Google Scholar]
- Meijer W.H., van der Klei I.J., Veenhuis M., Kiel J.A. (2007). ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy 3: 106–116 [DOI] [PubMed] [Google Scholar]
- Meyer A.J., Brach T., Marty L., Kreye S., Rouhier N., Jacquot J.P., Hell R. (2007). Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J. 52: 973–986 [DOI] [PubMed] [Google Scholar]
- Mhamdi A., Queval G., Chaouch S., Vanderauwera S., Van Breusegem F., Noctor G. (2010). Catalase function in plants: A focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 61: 4197–4220 [DOI] [PubMed] [Google Scholar]
- Mitsuhashi N., Shimada T., Mano S., Nishimura M., Hara-Nishimura I. (2000). Characterization of organelles in the vacuolar-sorting pathway by visualization with GFP in tobacco BY-2 cells. Plant Cell Physiol. 41: 993–1001 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., Ohsumi Y. (2011). The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27: 107–132 [DOI] [PubMed] [Google Scholar]
- Motley A.M., Nuttall J.M., Hettema E.H. (2012). Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. 31: 2852–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra S., Venkataramani S., Shen G., Wang J., Pasapula V., Lin Y., Kornyeyev D., Holaday A.S., Zhang H. (2006). The Arabidopsis ascorbate peroxidase 3 is a peroxisomal membrane-bound antioxidant enzyme and is dispensable for Arabidopsis growth and development. J. Exp. Bot. 57: 3033–3042 [DOI] [PubMed] [Google Scholar]
- Nishimura M., Akazawa T. (1974). Studies on spinach leaf ribulosebisphosphate carboxylase. Carboxylase and oxygenase reaction examined by immunochemical methods. Biochemistry 13: 2277–2281 [DOI] [PubMed] [Google Scholar]
- Nishimura M., Akhmedov Y.D., Strzalka K., Akazawa T. (1983). Purification and characterization of glycolate oxidase from pumpkin cotyledons. Arch. Biochem. Biophys. 222: 397–402 [DOI] [PubMed] [Google Scholar]
- Ono Y., Wada S., Izumi M., Makino A., Ishida H. (2013). Evidence for contribution of autophagy to Rubisco degradation during leaf senescence in Arabidopsis thaliana. Plant Cell Environ. 36: 1147–1159 [DOI] [PubMed] [Google Scholar]
- Ostersetzer O., Kato Y., Adam Z., Sakamoto W. (2007). Multiple intracellular locations of Lon protease in Arabidopsis: Evidence for the localization of AtLon4 to chloroplasts. Plant Cell Physiol. 48: 881–885 [DOI] [PubMed] [Google Scholar]
- Palma J.M., Sandalio L.M., Corpas F.J., Romero-Puertas M.C., McCarthy I., del Rio L.A. (2002). Plant proteases, protein degradation, and oxidative stress: Role of peroxisomes. Plant Physiol. Biochem. 40: 521–530 [Google Scholar]
- Pastori G.M., del Rio L.A. (1994). An activated-oxygen-mediated role for peroxisomes in the mechanism of senescence of Pisum sativum L. leaves. Planta 193: 385–391 [Google Scholar]
- Pastori G.M., Del Rio L.A. (1997). Natural senescence of pea leaves (an activated oxygen-mediated function for peroxisomes). Plant Physiol. 113: 411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S., Dinesh-Kumar S.P. (2008). Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy 4: 20–27 [DOI] [PubMed] [Google Scholar]
- Phillips A.R., Suttangkakul A., Vierstra R.D. (2008). The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178: 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G., Ma Z., Zhang L., Xing S., Hou X., Deng J., Liu J., Chen Z., Qu L.J., Gu H. (2007). Arabidopsis AtBECLIN 1/AtAtg6/AtVps30 is essential for pollen germination and plant development. Cell Res. 17: 249–263 [DOI] [PubMed] [Google Scholar]
- Queval G., Issakidis-Bourguet E., Hoeberichts F.A., Vandorpe M., Gakière B., Vanacker H., Miginiac-Maslow M., Van Breusegem F., Noctor G. (2007). Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 52: 640–657 [DOI] [PubMed] [Google Scholar]
- Reumann S., Ma C., Lemke S., Babujee L. (2004). AraPerox. A database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiol. 136: 2587–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Serrano M., Romero-Puertas M.C., Sparkes I., Hawes C., del Río L.A., Sandalio L.M. (2009). Peroxisome dynamics in Arabidopsis plants under oxidative stress induced by cadmium. Free Radic. Biol. Med. 47: 1632–1639 [DOI] [PubMed] [Google Scholar]
- Rosenwasser S., Rot I., Meyer A.J., Feldman L., Jiang K., Friedman H. (2010). A fluorometer-based method for monitoring oxidation of redox-sensitive GFP (roGFP) during development and extended dark stress. Physiol. Plant. 138: 493–502 [DOI] [PubMed] [Google Scholar]
- Sakai Y., Oku M., van der Klei I.J., Kiel J.A. (2006). Pexophagy: Autophagic degradation of peroxisomes. Biochim. Biophys. Acta 1763: 1767–1775 [DOI] [PubMed] [Google Scholar]
- Schwarzländer M., Fricker M.D., Müller C., Marty L., Brach T., Novak J., Sweetlove L.J., Hell R., Meyer A.J. (2008). Confocal imaging of glutathione redox potential in living plant cells. J. Microsc. 231: 299–316 [DOI] [PubMed] [Google Scholar]
- Shintani T., Suzuki K., Kamada Y., Noda T., Ohsumi Y. (2001). Apg2p functions in autophagosome formation on the perivacuolar structure. J. Biol. Chem. 276: 30452–30460 [DOI] [PubMed] [Google Scholar]
- Siddique Y.H., Ara G., Afzal M. (2012). Estimation of lipid peroxidation induced by hydrogen peroxide in cultured human lymphocytes. Dose Response 10: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair A.M., Trobacher C.P., Mathur N., Greenwood J.S., Mathur J. (2009). Peroxule extension over ER-defined paths constitutes a rapid subcellular response to hydroxyl stress. Plant J. 59: 231–242 [DOI] [PubMed] [Google Scholar]
- Somerville C.R. (2001). An early Arabidopsis demonstration. Resolving a few issues concerning photorespiration. Plant Physiol. 125: 20–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strømhaug P.E., Bevan A., Dunn W.A., Jr (2001). GSA11 encodes a unique 208-kDa protein required for pexophagy and autophagy in Pichia pastoris. J. Biol. Chem. 276: 42422–42435 [DOI] [PubMed] [Google Scholar]
- Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. (2001). The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20: 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S., Bauwe H., Badger M. (2007). Impairment of the photorespiratory pathway accelerates photoinhibition of photosystem II by suppression of repair but not acceleration of damage processes in Arabidopsis. Plant Physiol. 144: 487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.R., Doelling J.H., Suttangkakul A., Vierstra R.D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert N.E., Essner E. (1981). Microbodies: Peroxisomes and glyoxysomes. J. Cell Biol. 91: 271s–283s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert N.E., Yamazaki R.K. (1969). Leaf peroxisomes and their relation to photorespiration and photosynthesis. Ann. N. Y. Acad. Sci. 168: 325–341 [DOI] [PubMed] [Google Scholar]
- Toyooka K., Moriyasu Y., Goto Y., Takeuchi M., Fukuda H., Matsuoka K. (2006). Protein aggregates are transported to vacuoles by a macroautophagic mechanism in nutrient-starved plant cells. Autophagy 2: 96–106 [DOI] [PubMed] [Google Scholar]
- Toyota M., Matsuda K., Kakutani T., Terao Morita M., Tasaka M. (2011). Developmental changes in crossover frequency in Arabidopsis. Plant J. 65: 589–599 [DOI] [PubMed] [Google Scholar]
- Tuttle D.L., Lewin A.S., Dunn W.A., Jr (1993). Selective autophagy of peroxisomes in methylotrophic yeasts. Eur. J. Cell Biol. 60: 283–290 [PubMed] [Google Scholar]
- Van Breusegem F., Dat J.F. (2006). Reactive oxygen species in plant cell death. Plant Physiol. 141: 384–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zutphen T., Veenhuis M., van der Klei I.J. (2011). Damaged peroxisomes are subject to rapid autophagic degradation in the yeast Hansenula polymorpha. Autophagy 7: 863–872 [DOI] [PubMed] [Google Scholar]
- Veenhuis M., Douma A., Harder W., Osumi M. (1983). Degradation and turnover of peroxisomes in the yeast Hansenula polymorpha induced by selective inactivation of peroxisomal enzymes. Arch. Microbiol. 134: 193–203 [DOI] [PubMed] [Google Scholar]
- Velikkakath A.K., Nishimura T., Oita E., Ishihara N., Mizushima N. (2012). Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol. Biol. Cell 23: 896–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S., Ishida H., Izumi M., Yoshimoto K., Ohsumi Y., Mae T., Makino A. (2009). Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 149: 885–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.W., Kim J., Huang W.P., Abeliovich H., Stromhaug P.E., Dunn W.A., Jr andKlionsky D.J. (2001). Apg2 is a novel protein required for the cytoplasm to vacuole targeting, autophagy, and pexophagy pathways. J. Biol. Chem. 276: 30442–30451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Nishimura M.T., Zhao T., Tang D. (2011). ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J. 68: 74–87 [DOI] [PubMed] [Google Scholar]
- Williams J. (1928). The decomposition of hydrogen peroxide by liver catalase. J. Gen. Physiol. 11: 309–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F.H., Shen S.C., Lee L.Y., Lee S.H., Chan M.T., Lin C.S. (2009). Tape-Arabidopsis sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Bassham D.C. (2005). AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 42: 535–546 [DOI] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Nguyen P.Q., Bassham D.C. (2007). Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 143: 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Nagano A.J., Nishina M., Hara-Nishimura I., Nishimura M. (2008). NAI2 is an endoplasmic reticulum body component that enables ER body formation in Arabidopsis thaliana. Plant Cell 20: 2529–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Nagano A.J., Nishina M., Hara-Nishimura I., Nishimura M. (2013). Identification of two novel endoplasmic reticulum body-specific integral membrane proteins. Plant Physiol. 161: 108–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi J., Nishimura M., Akazawa T. (1984). Maturation of catalase precursor proceeds to a different extent in glyoxysomes and leaf peroxisomes of pumpkin cotyledons. Proc. Natl. Acad. Sci. USA 81: 4809–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanik T., Donaldson R.P. (2005). A protective association between catalase and isocitrate lyase in peroxisomes. Arch. Biochem. Biophys. 435: 243–252 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Hanaoka H., Sato S., Kato T., Tabata S., Noda T., Ohsumi Y. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K., Jikumaru Y., Kamiya Y., Kusano M., Consonni C., Panstruga R., Ohsumi Y., Shirasu K. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B.K., Yoder A., Bartel B. (2000). Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156: 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]