This study characterizes the structure and significance of the core of potato multicystatin (PMC), a multidomain cysteine protease inhibitor found in the cortical parenchyma tissue of potato tubers. Papain inhibitory properties of native and recombinant PMC containing core domains are affected by pH. It is likely that pH-mediated regulation imparts unique properties to PMC that modulate proteolysis upon wounding and/or infection via inhibiting cysteine proteases.
Abstract
Potato (Solanum tuberosum) multicystatin (PMC) is a unique cystatin composed of eight repeating units, each capable of inhibiting cysteine proteases. PMC is a composite of several cystatins linked by trypsin-sensitive (serine protease) domains and undergoes transitions between soluble and crystalline forms. However, the significance and the regulatory mechanism or mechanisms governing these transitions are not clearly established. Here, we report the 2.2-Å crystal structure of the trypsin-resistant PMC core consisting of the fifth, sixth, and seventh domains. The observed interdomain interaction explains PMC’s resistance to trypsin and pH-dependent solubility/aggregation. Under acidic pH, weakening of the interdomain interactions exposes individual domains, resulting in not only depolymerization of the crystalline form but also exposure of cystatin domains for inhibition of cysteine proteases. This in turn allows serine protease–mediated fragmentation of PMC, producing ∼10-kD domains with intact inhibitory capacity and faster diffusion, thus enhancing PMC’s inhibitory ability toward cysteine proteases. The crystal structure, light-scattering experiments, isothermal titration calorimetry, and site-directed mutagenesis confirmed the critical role of pH and N-terminal residues in these dynamic transitions between monomer/polymer of PMC. Our data support a notion that the pH-dependent structural regulation of PMC has defense-related implications in tuber physiology via its ability to regulate protein catabolism.
INTRODUCTION
Potato multicystatin (PMC), which exists in potato (Solanum tuberosum) tubers in both soluble and crystalline form, can bind and inhibit several Cys proteases (e.g., papain) simultaneously, and the term multicystatin thus refers to its multiple inhibitory domains (Walsh and Strickland, 1993). While crystalline PMC (340 kD) is highly concentrated in the periderm, its soluble monomeric form (85 kD) is distributed throughout tuber tissue (Rodis and Hoff, 1984; Kumar et al., 1999; Nissen et al., 2009). The 85-kD monomeric PMC associates into tetrameric crystals (340 kD). Tryptic digestion of monomeric PMC releases five ∼10-kD and one ∼35-kD peptide with functional cystatin domains (Rodis and Hoff, 1984; Walsh and Strickland, 1993; Kumar et al., 1999). Thus, the 85-kD PMC is a conglomerate of cystatin-containing peptides linked with sequences recognizable by Ser proteases (Walsh and Strickland, 1993). The 10-kD peptides of PMC have sequence similarities to that of type-I and type-II cystatins. The ∼35-kD peptide is composed of three cystatin domains designated 5, 6, and 7 (Nissen et al., 2009) and remains resistant to further proteolysis by some unknown mechanism. Our previous work (Nissen et al., 2009) established the structural similarity of a single domain of PMC (PMC-2) with that of other cystatins, such as chicken egg white cystatin (CEW; Bode et al., 1988; Dieckmann et al., 1993), human stefin A (Martin et al., 1995; Tate et al., 1995; Jenko et al., 2003) and human stefin B (Stubbs et al., 1990), and rice (Oryza sativa) cystatin (Nagata et al., 2000).
Another unique feature of PMC is its ability to exist as a natural protein crystal. The mechanism(s) that favors crystallization of the 85-kD monomeric form of PMC is not fully established. Our previous work using static and dynamic light-scattering techniques validated the existence of the monomeric form of PMC under acidic conditions (Nissen et al., 2009). Protein concentration and phosphate ions, along with pH, appear to play important roles in the transition of PMC into crystalline form (Rodis and Hoff, 1984; Nissen et al., 2009). While the solubility of PMC is enhanced under acidic conditions (pH 5.0 and below), the crystalline form dominates at neutral and alkaline pH (7.0 to 8.0) (Rodis and Hoff, 1984).
The biological benefits derived from the coexistence of soluble and crystalline forms of PMC and their phosphate- and/or pH-dependent interconversion remain speculative. It is likely that crystallization is a mechanism to store PMC in a nonfunctional form. The solubility and effectiveness of PMC as a Cys protease inhibitor is greatly enhanced under acidic conditions, which may be an adaptive response to facilitate wound healing and/or deter insect feeding. Following wounding of a tuber, acidification of the wounded tissue by vacuolar disruption and the resultant transition of PMC into soluble form may modulate the otherwise unabated degradation of cellular proteins/enzymes by endogenous Cys proteases. Thus, monomerization of PMC may be important in protecting enzymes involved in the process of wound healing. Correlative evidence suggests that loss of PMC during long-term storage of potatoes is associated with enhanced Cys protease activity, loss of patatin, and loss of wound healing ability (Kumar et al., 1999, 2007; Kumar and Knowles, 2003). PMC protects patatin from degradation by Cys proteases in vitro (Kumar et al., 1999). Ontogenetic changes in PMC concentration also appear to regulate protein content during tuber development (Weeda et al., 2009), storage (Weeda et al., 2011), and sprouting (Weeda et al., 2010) by modulating Cys proteases.
PMC has also been proposed as an antifeedant. The monomeric form of PMC in the acidic environment of an insect’s midgut can interfere with protein digestion (Murdock et al., 1987). For example, PMC retards the growth of maize rootworm (Diabrotica virgifera virgifera) larvae by inhibiting digestive proteases in the midgut, where Cys proteases account for nearly 92% of the digestive proteases (Orr et al., 1994). Overexpression of Cys protease inhibitors in rice is known to be effective in conferring resistance against insect pests (Irie et al., 1996).
Overall, it appears that the benefits of PMC as a protease inhibitor lie in its ability to transform from inactive crystalline to the active monomeric form. To elucidate factors that regulate this, we expressed single domain (PMC-6) and core domain (PMC-567)–containing PMC. Using the purified proteins, we characterized the effects of pH and phosphate ion on the oligomerization and papain (Cys protease) inhibitory activities of single domain (PMC-5, PMC-6, and PMC-7), core domain (PMC-567), and multidomain wild-type PMC. Our results demonstrate that pH plays a critical role in dictating the transition from crystalline to soluble monomeric form.
RESULTS
To characterize the core-domain of PMC, the corresponding gene sequence encoding the trypsin-resistant 33.2-kD peptide consisting of the 5th, 6th, and 7th domains and the gene sequence expressing each of those single domains were synthesized and cloned into Escherichia coli and the expressed proteins were studied. For convenience, those recombinant PMC proteins are referred to as PMC-5, PMC-6, PMC-7, and PMC-567 throughout this article.
pH-Dependent Interaction between PMC-6/PMC-567 and Papain by Dynamic Light Scattering
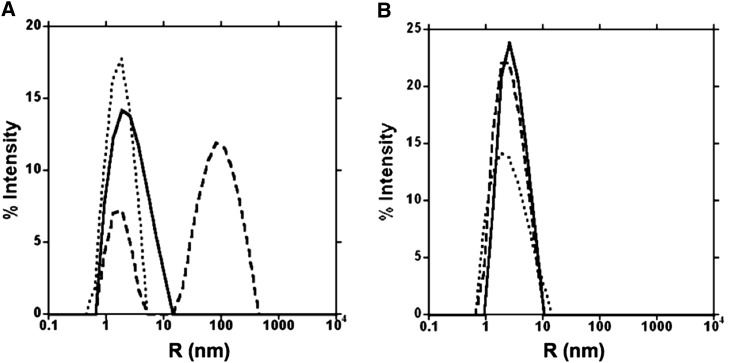
Interactions of PMC-6 and PMC-567 with papain (Cys protease) were compared through dynamic light-scattering (DLS) experiments (Figure 1). PMC-6 was monomeric at pH 7.2, which was consistent with our previous results for PMC-2 (Nissen et al., 2009). Upon the addition of papain into this PMC-6 solution of pH 7.2 PBS, the radius of the population increased, indicating a complex formation between PMC-6 and papain (Figure 1A, dashed line). However, in the same buffer of pH 7.2, the radius of PMC-567 was unaffected by papain, indicating a lack of complex formation between PMC-567 and papain molecules (Figure 1B).
Figure 1.
DLS Patterns for PMC-6 and PMC-567.
(A) Hydrodynamic radius profile of PMC-6 in the presence or absence of papain in pH 7.2 PBS.
(B) Hydrodynamic radius profile of PMC-567 in the presence or absence of papain in pH 7.2 PBS. The y axis represents the percentage of the scattered light, and the x axis (log scale) represents the radius of the PMC molecules, acetylated papain molecules, or PMC/papain complexes. The dotted line represents PMC-6 or PMC-567 alone, the dashed line represents PMC-6 or PMC-567 with acetylated papain, and the solid line represents acetylated papain alone.
Oligomeric State of PMC-567 and Native PMC as a Function of pH and Phosphate
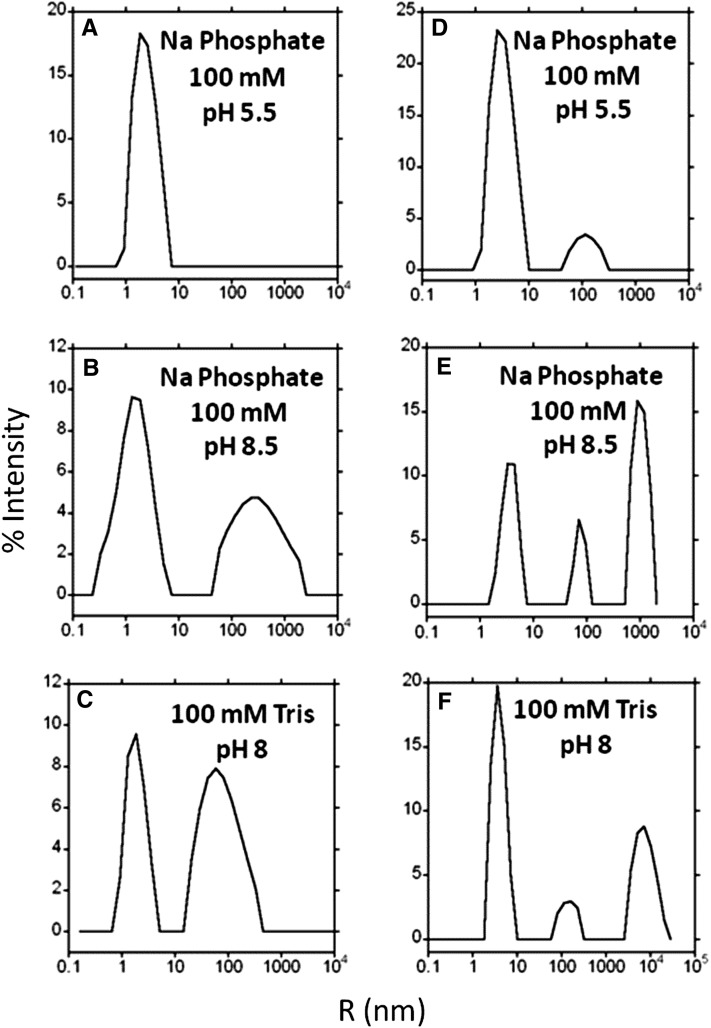
The radius of both recombinant PMC-567 and native PMC was measured by DLS to determine their tendency to aggregate in the presence of phosphate or as a result of pH change. The results showed that recombinant PMC-567 has a hydrodynamic radius of <5 nm in the presence of phosphate at low pH (Figure 2A). However, PMC-567 aggregated into larger particles at basic pH with or without phosphate (Figures 2B and 2C). Native PMC is also predominantly a single, nonaggregated species at low pH in the presence of phosphate (Figure 2D) but forms larger radius particles at basic pH with or without phosphate (Figures 2E and 2F).
Figure 2.
DLS Comparison of PMC-567 and Native PMC.
Hydrodynamic radius profile of PMC-567 and native PMC as a function of phosphate and pH. (A) to (C) are PMC-567, and (D) to (F) are native PMC. The y axis represents the percentage of the scattered light, and the x axis (log scale) represents the radius of the PMC molecules. Conditions are as indicated in the figure.
Differential Interaction of Papain (Cys Protease) with PMC-5, PMC-6, PMC-7, and PMC-567 by Isothermal Titration Calorimetry
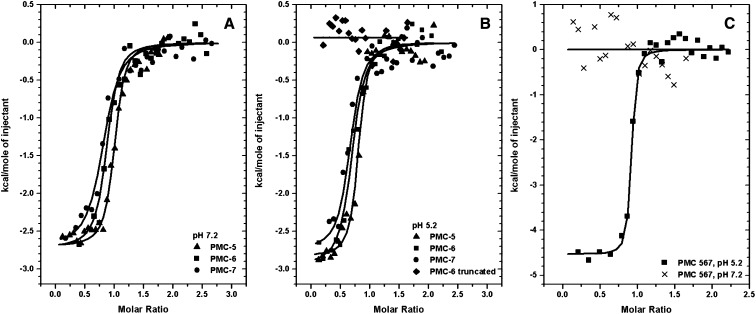
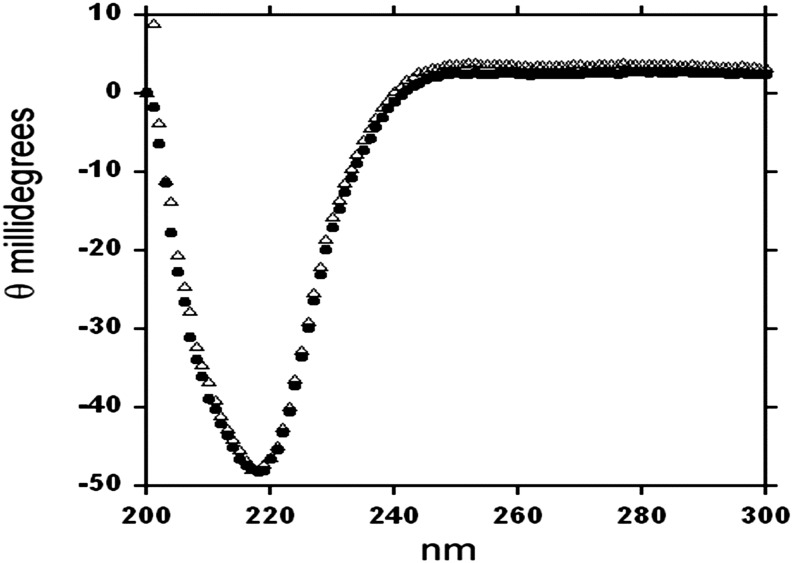
The above DLS results were confirmed by isothermal titration calorimetry (ITC) profiles, in which thermodynamic characterizations of their interaction showed differential binding affinities among PMC-5, PMC-6, PMC-7, or PMC-567 with papain. In both DLS (Figure 1) and ITC (Figure 3, Table 1) experiments, the proteolytic activity of papain was removed by acetylation to monitor only the formation of its binary complex with PMC. As indicated in Figures 3A and 3B and Table 1, individual domains, PMC-5, PMC-6, and PMC-7, showed significant affinities to papain at both pH 7.2 and 5.2 with average Kd values of 8.8 and 8.7 μM, respectively. However, PMC-6 with its N terminus Gly-Gly-Ile residues truncated, which mimics the N-terminal conformations of PMC-6 and PMC-7 in the crystal structure of PMC-567, displayed no significant affinity to papain (Figure 3B, Table 1). The circular dichroism (CD) profiles of this truncated PMC-6 (Figure 4) overlapped well with that of intact PMC-6, indicating the integrity of its secondary structural elements. Therefore, the data confirmed a lack of pH dependency for the papain affinity of individual domains and the significance of the N terminus Gly-Gly-Ile residues for binding papain.
Figure 3.
Measurement of Papain Binding by ITC.
The trend of heat released by serial injections of papain into PMC-5, -6, and -7 at pH 7.2 (A), PMC-5, -6, and -7 and truncated PMC-6 at pH 5.2 (B), and PMC-567 pH 5.2 and PMC-567 pH 7.2 (C) was monitored. Solid lines represent the least square fits of the data.
Table 1. ITC Analysis of PMC-5, PMC-6, PMC-7, and PMC-567 Binding to Papain.
| pH Level | Kd (μM) | ΔH (kcal*mol−1) | ΔS (cal*mol−1*deg−1) |
|---|---|---|---|
| pH 5.2 | |||
| PMC-5 | 8.1 ± 2.8 | −2.83 ± 0.06 | −19.8 |
| PMC-6 | 9.0 ± 2.3 | −2.95 ± 0.1 | −17.8 |
| PMC-7 | 8.9 ± 1.8 | −2.75 ± 0.2 | −18.5 |
| PMC-567 | 14 ± 3.4 | −3.47 ± 0.4 | −15.4 |
| PMC-6 truncated | NBa | NBa | NBa |
| pH 7.2 | |||
| PMC-5 | 7.2 ± 3.1 | −2.78 ± 0.06 | −18.8 |
| PMC-6 | 9.4 ± 4.2 | −2.90 ± 0.1 | −17.9 |
| PMC-7 | 9.8 ± 3.2 | −2.54 ± 0.1 | −20.4 |
| PMC-567 | NB* | NB* | NB* |
No binding.
Figure 4.
CD Spectra of Full-Length and Truncated Recombinant PMC-6.
Comparison of CD spectra of recombinant PMC-6 (solid dot) or truncated PMC-6 (open triangle) in PBS, pH 7.2.
When PMC-567 was mixed with papain in pH 5.2 buffer, a significant amount of heat was released (Figure 3C), indicating that the binding interactions between PMC-567 and papain had a significant enthalpic contribution, as observed in PMC-6 (Table 1). In addition, an unfavorable entropic contribution possibly indicated that the PMC domain was slightly stabilized upon binding to papain and very few solvent molecules might be freed from the pocket upon formation of the papain binary complex. The calculated Kd of PMC-567 for papain at pH 5.2 from ITC data analysis was 14 µM. However, PMC-567 in pH 7.2 buffer did not show any significant interaction with the papain molecule (crosses in Figure 3C).
Protease Inhibitory Activity
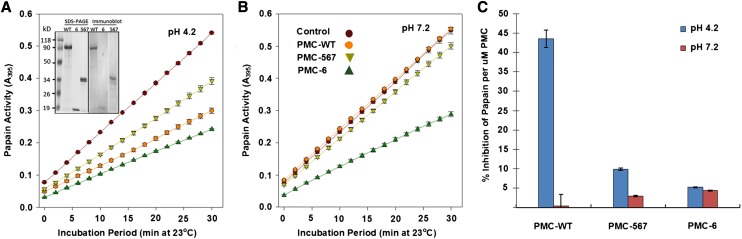
Recombinant PMC-6, PMC-567, and native PMC were compared for their ability to inhibit papain-mediated degradation of N-α-benzoyl-dl-Arg-p-nitroanilide (BAPNA) at pH 4.2. While 50% inhibition of papain activity was achieved with as low as 1.4 µM wild-type PMC (eight inhibitory domains), recombinant PMC-6 (one inhibitory domain) and PMC-567 (three inhibitory domains) required 14.8 and 3.9 µM, respectively (data not shown). Using concentrations that resulted in 50% inhibition, the inhibitory effect of wild-type and recombinant PMC was compared at pH 4.2 and 7.2. Degradation of BAPNA by papain resulted in a linear (R2 = 0.99, P < 0.001) increase in A395 over a 30-min incubation period (Figure 5). In the absence of PMC (control), papain activity remained unaffected by pH changes showing a mean value of ΔA395 = 0.933/h (Table 2). Relative to the control, native PMC reduced the activity of papain by 45% (Figure 5A) at pH 4.2, but native PMC’s ability to inhibit papain catabolism of BAPNA was completely abolished at pH 7.2 (Table 2). PMC-567 inhibited papain activity by 27% at pH 4.2, while retaining 8% of its inhibitory ability at pH 7.2 (Table 2). The influence of pH on the ability of PMC to inhibit papain was clear only for native PMC and PMC-567. While native PMC (eight inhibitory domains) was only effective at pH 4.2, PMC-567 (three inhibitory domains) was effective at both pH 4.2 and 7.2, although the magnitude of inhibition was higher at pH 4.2 than at 7.2. In contrast, papain inhibition by PMC-6 averaged 50% and was not affected by pH of the incubation medium (Table 2). Figure 5C shows the inhibition of papain for equimolar PMC-6, PMC-567, and native PMC.
Figure 5.
Effect of pH on the Ability of Native and Recombinant PMC to Inhibit Papain Activity.
(A) and (B) Papain was preincubated with native and recombinant PMC either at pH 4.2 (A) or 7.2 (B) for 10 min (23°C) prior to assessing its ability to degrade BAPNA by following the increase in A395 (p-nitroanilide production) for 30 min (see Methods). SDS-PAGE and immunoblot of wild-type (WT) and recombinant PMC-6 and PMC-567 are shown in (A) (inset). Rates of BAPNA degradation (ΔA395, slopes) by papain as affected by PMC and pH are in Table 2. Error bars indicate ± se (n = 3).
(C) Equimolar percentage inhibition by PMC-WT, PMC-567, and PMC-6 at pH 4.2 (blue) and pH 7.2 (orange).
Table 2. Rates of BAPNA Degradation (ΔA395 h−1; slopes in Figure 5) by Papain as Affected by Wild-Type and Recombinant PMC and pH.
| Papain Activity (ΔA395 h−1) |
||
|---|---|---|
| Treatment | pH 4.2 | pH 7.2 |
| No Inhibitor | 0.924 | 0.942 |
| PMC-WT | 0.504 | 0.935 |
| PMC-567 | 0.671 | 0.865 |
| PMC-6 | 0.422 | 0.501 |
WT, wild type.
Crystal Structure of PMC-567
Recombinant PMC-567 was crystallized in the tetragonal space group, P43212, diffracting to 2.2-Å resolution. The asymmetric unit of the crystal contained one PMC-567 molecule, and the structure was determined by the molecular replacement method using the coordinates of our previous PMC-2 domain (Protein Data Bank ID: 2W9Q; Nissen et al., 2009). Most of the backbone and side chain residues were fitted using the molecular replacement map, and the final R factor for PMC-567 was 22.2% (Rfree = 28.7) for 19,938 (25 to 2.2 Å) unique reflections (Table 3). The global structures of three PMC domains in the asymmetric unit are all essentially superimposable. The average root mean square deviation (rmsd) values (Cα atoms only) are ∼0.14 Å among pairwise comparisons of PMC-5, PMC-6, and PMC-7 domains.
Table 3. Crystallographic Data for PMC-567.
| Crystallographic Variables/Parameters | Data |
|---|---|
| Wavelength (Å) | 1.0332 |
| Resolution (Å) | 25 to 2.0 |
| Space group | P43212 |
| Cell dimensions (Å) | a = b = 65.94 |
| c = 191.85 | |
| Asymmetric unit | One molecule |
| Total observations | 398,668 |
| Unique reflections | 28,924 |
| Completeness (%) | 95.2 (93.0) |
| Rsyma,b | 9.4 (58.9) |
| Refinement | |
| Resolution (Å) | 25 to 2.2 |
| No. of reflections | 19,938 (89.2%) |
| Rcrystc | 22.2 |
| Rfreed | 28.7 |
| rmsd bonds (Å)e | 0.008 |
| rmsd angles (°) | 1.196 |
| No. of atoms | |
| Protein | 2,252 |
| Water | 199 |
Numbers in parentheses refer to the highest resolution shell.
Rsym = ΣIh – <Ih>|/ΣIh, where <Ih> is the average intensity over symmetry equivalent reflections.
Rcryst = Σ|Fobs – Fcalc|/ΣFobs, where summation is over the data used for refinement.
Rfree was calculated as for Rcryst using 5% of the data that was excluded from refinement.
rmsd, root mean square deviation.
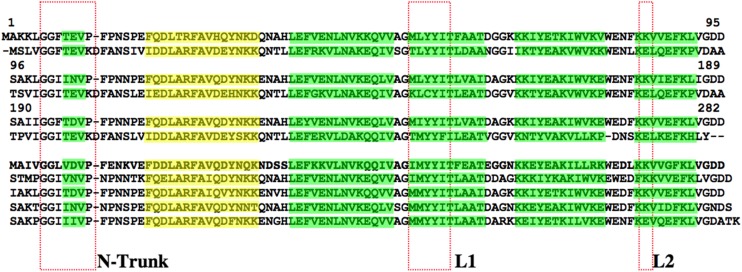
The corresponding electron density for the first six amino acids from the N terminus of PMC-567 was not visible as those in the structures of PMC-2, chicken egg white cystatin, and oryzacystatin-I (Bode et al., 1988; Nissen et al., 2009). Thus, the electron density of the PMC-567 crystal structure started from Gly-7 (Figure 6), which is the second Gly of the Gly-Gly motif completely conserved in all cystatin molecules.
Figure 6.
Amino Acid Sequence Alignment.
The PMC-567 and other domains of PMC were compared with three domains of SMC. The β-strands are highlighted in green, and the α-helices are highlighted in yellow. The N-trunk and L1 and L2 sites are indicated by the red boxes.
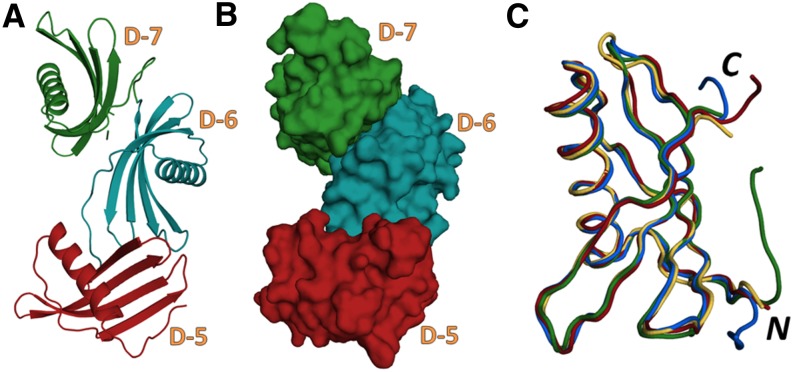
The global shape for PMC-567 showing its compact structure is illustrated in Figure 7. Each individual domain had a five-stranded antiparallel β-sheet (β1 to β5) wrapped around a central five-turn α-helix, with the β-strands in an almost perpendicular orientation to the helical axis as observed in PMC-2 (Nissen et al., 2009). The α-helix in all domains was straight, having regular hydrogen bonds, in contrast with the kinked conformation observed in the middle of the helices of oryzacystatin-I, stefin A, and stefin B. The tightly packed core of each domain was mainly composed of nonpolar, non-hydrogen-bonding hydrophobic side chains contributed by the α-helix and β-strands. As observed in other cystatin structures (Murzin, 1993; Nagata et al., 2000), the inner two β-strands, β3 and β4, are coiled smoothly, forming regular antiparallel hydrogen bonds, but some parts of the outer two β-strands, β2 and β5, have irregular backbone hydrogen bonds, which produced a tight coiling of the β-sheet, allowing it to wrap around the helix. As noticed in the structure of PMC-2 (Nissen et al., 2009), the shortest β1 strands of all three domains of PMC-567 showed a big bulge made by Pro-Phe-Pro, which was bigger than those observed in other cystatins.
Figure 7.
The Crystal Structure of PMC-567.
(A) and (B) Ribbon diagram (A) and space-filling model (B) representing the crystal structure of PMC-567. Domains 5, 6, and 7 are depicted as red, blue, and green, respectively.
(C) Backbone structures of PMC-5 (red), PMC-6 (blue), PMC-7 (green), and PMC-2 (orange) were superimposed. The N and C termini are labeled as N and C, respectively. These figures were generated using Open-Source PyMOL (v1.4).
The largest differences among three domains in PMC-567 were located at two of the tripartite inhibitory components, the N-terminal trunk and L2. In addition, the loop connecting β3 and β4 shows some degree of variation, which is the most heterogeneous in terms of primary sequence among various other cystatins.
DISCUSSION
Proteolysis of 85-kD native PMC by trypsin generates ∼35- and ∼10-kD fragments (Rodis and Hoff, 1984; Walsh and Strickland, 1993; Nissen et al., 2009). It has been proposed that inaccessibility due to specific peptide folding or interdomain sequences unfavorable to tryptic digestion likely protects the ∼35-kD fragment from further proteolysis by trypsin (Walsh and Strickland, 1993). We previously determined that the 10-kD fragments consist mostly of PMC-6, while the ∼35-kD fragment is composed of three domains 5, 6, and 7. Our previous results also confirmed that the native PMC either crystallizes or aggregates with increasing phosphate ions and pH (Walsh and Strickland, 1993; Nissen et al., 2009). However, the single-domain PMC-2 maintains its monomeric form independent of pH (5 to 7) and/or phosphate concentration (0 to 100 mM) (Nissen et al., 2009).
A comparison of the three domains in the crystal structure of PMC-567 indicated that the structural compactness is a major contributing factor for the apparent resistance of PMC-567, which is the ∼35-kD fragment, to trypsin digestion (Figure 6). In addition, PMC-6 did not show any uniqueness in its primary, secondary, and tertiary structure (Figure 7C) and papain affinity (Table 1) compared with PMC-2, PMC-5, and PMC-7. Because of its specific interaction with adjacent PMC-5 and PMC-7 domains, PMC-6 can constitute the ∼10-kD fragment, which survives the tryptic digestion of the whole PMC molecule. The crystal structure of PMC-567 showed that PMC-5 and PMC-6 interacted with several hydrogen bonds between their side chains and backbone atoms in addition to a substantial area of hydrophobic interaction. For example, the side chain of Lys-85 in PMC-5 and Glu-173 in PMC-6 formed a salt bridge, and the side chains of Tyr-30 in PMC-5 and Lys-98 in PMC-6 were in a hydrogen bond interaction. The backbone carbonyl of Glu-147 was also within a hydrogen bond distance from the amide of Val-87. PMC-6 and PMC-7 displayed a much more extensive area of contact, intricately interacting through the same side of their β-sheet. In addition, Lys-141 and Glu-136 of PMC-6 established a salt bridge with Glu-230 and Lys-235 of PMC-7. Overall, PMC-6 interacted tightly with both PMC-5 and PMC-7.
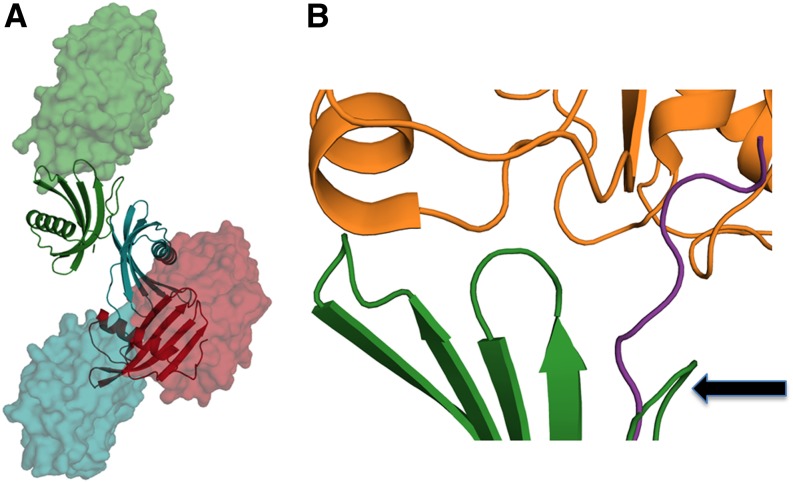
Noninhibitory Mode of PMC-567
We modeled the plausible papain, a well-investigated Ser Cys protease, complex for each domain located in PMC-567 using the coordinates of the complex structure between papain and stefin B (Protein Data Bank ID: 1STF) (Figure 8A). Because of the specific orientation/arrangement between PMC-5 (red ribbon) and PMC-6 (blue ribbon) in the structure of PMC-567, the modeled papain (blue surface model) in its inhibited mode with PMC-6 generated a steric collision with PMC-5. That is, both L1 and L2 of PMC-6, which are the critical components for the papain interaction, were in close contact with PMC-5. Particularly, the Trp-174 at the tip of L2 in PMC-6 was imbedded into a hydrophobic pocket formed by the L1 loop and the Trp-76 in PMC-5, thus completely eliminating PMC-6 domain’s affinity for papain. As a result of this closeness between PMC-5 and PMC-6 in PMC-567 structure, the complex formation of papain with the PMC-5 domain (red surface) was also sterically hindered by the adjacent PMC-6.
Figure 8.
Plausible Complex Formation between PMC-567 and Papain.
(A) Ribbon diagrams representing the core domain PMC-567 are colored by domain: 5 is red, 6 is blue, and 7 is green. Modeled papain molecules are represented as a space-filling model colored according to the PMC domain to which they are complexed. The positions of each papain were generated after superimposing the human stefin b in the papain complex structure (1STF) (Stubbs et al., 1990) with an individual PMC domain.
(B) Closer inspection of the molecular interface between PMC-7 (green) and papain (orange). The backbone of PMC-7 (green) has been superimposed with that of stefin b with the best molecular fitting. The N-terminal trunk of PMC-7 was curled around (arrow), which is significantly different to the extended conformation of stefin b (violet) observed in its papain complex (Protein Data Bank ID: 1SFT). These figures were generated using Open-Source PyMOL (v1.4).
In contrast with the PMC-5 and PMC-6 domains, the PMC-7 domain (green ribbon) was fully accessible to the modeled papain (green surface model, Figure 8A). However, the part of the residues at the N terminus of PMC-7, 193Ile-Gly-Gly-Phe196, which constitutes the N-terminal trunk that is critical for papain binding, was severely curled around. Therefore, the required conformation of these N-terminal residues for a proper papain inhibition, which is observed in other cystatin-papain complexes (Figure 8B, violet color; Protein Data Bank ID: 1SFT), could not be adopted. Truncation of these N-terminal residues from PMC-6 resulted in the loss of affinity to papain (Figure 3A). Previously, truncation of the same area in CEW cystatin (type II) was also found to abolish its papain inhibition activity (Abe et al., 1988; Thiele et al., 1988; Stubbs et al., 1990). However, N-terminal truncation in the stefin family (type I) does not produce any effect, indicating that individual domains of PMC resemble type-II cystatins. Overall, the observed weak affinity for the PMC-567 at pH 7.2 (Figures 1B and 3B) is likely due to the apparent steric inaccessibility of PMC-5 and PMC-6 and the specific peptide conformation of the N-terminal trunk of PMC-7.
The increase of affinity for papain (Figure 3) and the papain inhibition capacity (Figure 5) of both native PMC and PMC-567 at acid pH are likely due to the weakening of interdomain interactions and, thus, the gained independency of individual domains, allowing for the binding of papain. While an individual domain of PMC binds one papain in both pH 5.2 and 7.2, the native PMC and PMC-567 could accommodate multiple papain molecules only in low pH, judging from the radius of the complex (Figure 2) and inhibition rate (Figure 5c). pH-independent and maximum inhibition could only come into play after the inhibitor was broken into individual domains. As observed in the structures of PMC-567 and PMC-2 (Nissen et al., 2009), the individual domains of PMC displayed various ways to interact among themselves. Thus, in high concentrations and at basic pH, PMC domains will interact among themselves and exist mainly in the noninhibitory mode.
Biological Relevance
As we observed, the ability of native PMC to acquire a monomeric or oligomeric form is mainly attributable to pH-dependent structural changes within PMC-567, the core of native PMC. Similar interactions among the remaining Cys-inhibitory domains render crystallization of native PMC under neutral-to-alkaline conditions. Thus, pH-dependent changes in the core and the rest of the PMC molecule provide a mechanism by which the inhibitory activities of PMC are regulated during physiological processes involving protein turnover. Cytosolic pH is known to remain closer to neutral (Roberts et al., 1980), which facilitates accumulation of crystalline (inactive) PMC. The soluble forms of PMC exist throughout tuber tissue (Kumar et al., 1999), and crystalline forms dominate in the subperidermal layers. It is likely that in addition to pH, concentration of PMC also plays a crucial role in the crystallization of PMC. Beyond a threshold concentration, PMC transitions to the crystalline form, and the neutral-to-alkaline pH of the cytosol would favor such a transition. Vacuoles of plant cells are acidic (Taiz, 1992) and their disruption by wounding and/or infection likely leads to a transient reduction of pH at the wound surface. The resultant acidic pH enhances the solubility and degradation of PMC by Ser proteases, thus populating the wound surface by peptides containing cystatin domains, which would maximize the ability to inhibit Cys proteases. In this regard, we recently established that the pH of the wound surface is ∼5.2, and degradation of purified PMC by tuber extracts is dependent on pH (G.N.M. Kumar and N.R. Knowles, unpublished results). At pH 5.2, PMC was degraded by endogenous proteases (likely Ser-type), while at pH 7.2, PMC remained unaffected. Furthermore, analysis of tuber extracts demonstrated the presence of at least three proteases active at pH 5.2. These findings suggest that PMC is subject to degradation by endogenous proteases at the wound surface of tubers. This mechanism would greatly enhance the functional ability of PMC to inhibit endogenous and/or pathogen-secreted Cys proteases to contain unabated degradation of proteins and enzymes essential for defense and wound healing (Kumar et al., 1999). The monomeric domains maintain pH-independent inhibitory activity and readily complex with Cys proteases (Popovic and Brzin, 2007). The sequence heterogeneity among the eight domains, especially in their NT, L1, and L2 sites, which together form the wedge-shaped region that is complementary to the active-site cleft of the target protease (Stubbs et al., 1990), may confer an efficacy toward a diversity of Cys proteases in vivo.
Comparison of PMC with the Sunflower Multicystatin
Dali Search (Holm and Sander, 1993) showed that the amino acid sequence of PMC-567 has the highest level of similarity with that of sunflower Helianthus annuus multicystatin (SMC), which happens to be a three-domain cystatin. The sequence similarity measurement between PMC and SMC with a sliding window showed that PMC-567 has the highest match with 53% identity and 68% similarity. However, the critical residues required for the observed tight interaction among three domains in the PMC-567 were not conserved in the sequence of SMC. Instead, the charge characters of many of those corresponding residues in SMC were substituted to produce a repulsive interaction. Thus, it is likely that three domains in SMC are flexible or in a totally different angular orientation, which could be why SMC strongly inhibits papain activity in vitro compared with Celosia cristata single-domain cystatin (Gholizadeh and Kohnehrouz, 2011).
Structural Rationalization of Crystalline PMC
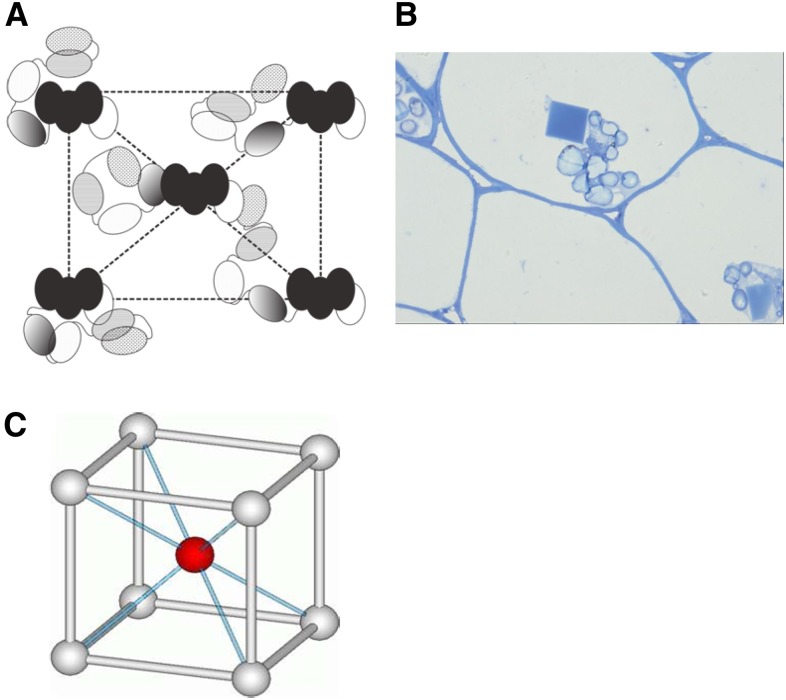
A crystal of intact PMC isolated from potato diffracts only at low resolution, reflecting apparent disorder in its lattice packing (Nissen et al., 2009). Having identified the structural similarity of the individual domain and the structural core of PMC, it is now tempting to rationalize the unique natural existence of crystalline PMC. Previously, we demonstrated many different modes of interaction among PMC-2 domains (Nissen et al., 2009). In addition, there were several other interdomain interactions in the structure of PMC-567, which are potentially possible for all the individual domains of the native PMC molecule. Therefore, combining all the observations, including the trypsin digestion experiment, we suspect that in pH 7.2, native PMC has its domains 5, 6, and 7 arranged in a compact conformation with domain 6 at the core, while domains 1 to 4 and 8 are in various interaction modes. Those peripheral domains of PMC are able to engage in the pH-dependent intermolecular interaction to establish the aggregates as shown in Figure 2 or crystallines (Figure 9). Figure 9A shows our schematic representation of the plausible arrangement of PMC molecules in the in vivo crystal (Figure 9B), which has an I-centered crystal lattice (Figure 9C) of unit cell dimensions of a = 205Å, b = 208 Å, c = 236 Å, and α = β = γ = 90° (Nissen et al., 2009). Using the versatile interactions among those exposed peripheral domains, PMC can precipitate into a crystalline form, but with only long-range order, in which a large amount of this potent multidomain inhibitor can be stored in an inaccessible and inactive form. As suggested previously (Nissen et al., 2009), this crystalline PMC can easily be converted into its active 85-kD monomeric form through altering cytosolic pH, thus enabling attenuation of protease activity to affect tuber physiology and the interactions of the potato tuber with the environment.
Figure 9.
Native Crystalline PMC.
(A) A schematic representation of the plausible arrangement of PMC molecules in a native PMC lattice. Domains are represented as ellipses, and black ellipses represent the core domain, PMC-567.
(B) Light microscope picture of the in vivo crystalline PMC, possessing the unit cell dimensions of a = 205 Å, b = 208 Å, c = 236 Å, and α = β = γ = 90° (Nissen et al., 2009).
(C) Representation of an I-centered crystal lattice, which is the lattice structure of native crystalline PMC.
[See online article for color version of this figure.]
METHODS
Cloning, Expression, and Purification of PMC-567
The PMC-567 gene (the fifth, sixth, and seventh cystatin domains of PMC) was synthesized with codon optimization for expression of recombinant protein in Escherichia coli (Genscript USA). The PMC-567 open reading frame spans nucleotides 1184 to 2030 of the complete PMC open reading frame. The synthetic gene was cloned into pET28a (Novagen) using the NcoI-XhoI restriction enzyme sites. PMC-567 protein was expressed as a C-terminal His-tagged recombinant protein in E. coli BL21 (DE3). PMC-6 was synthesized and cloned in the same manner. The codons for the N-terminal residues GGI were removed from PMC-6 with the QuikChange site-directed mutagenesis kit (Stratagene). The proteins were expressed and purified as previously described for PMC-2 (Nissen et al., 2009) with the exception that phosphate buffer was replaced by Tris buffer, pH 8.0. The protein purity was confirmed by SDS-PAGE (12% gels) and western analysis (Laemmli, 1970; Kumar et al., 1999). Protein concentrations determined by the bicinchoninic acid assay (Pierce). Native PMC was purified from tuber as described (Rodis and Hoff, 1984).
DLS
The radius and molecular weight of the PMCs were estimated using a DynaPro-Titan instrument (Wyatt Technology) at 22°C. Purified PMC-567 and PMC-6 (2 mg mL–1) in a freshly prepared buffer were filtered through a polyvinylidene difluoride filter (0.2 μm; Millipore). Sodium acetate buffer was used for pH 5.5, and PBS was used for pH 7.2. Scattering data were acquired through accumulation (five times) of 10 scans with 10 s/scan and laser intensity set to a range of 50 to 60% (30 to 36 mW). The corresponding molecular mass and radius were calculated using the software package DYNAMICS V6, which was supplied with the instrument.
Isothermal Calorimetry
The heat of binding between PMC and papain was measured with a VP-ITC Microcalorimeter. For calorimetric measurements, PMC-567 or PMC-6 protein, at a concentration of ∼50 μM in either NaOAc, pH 5.2, or PBS, pH 7.2, was titrated with solutions of papain. The experiment consisted of 29 injections of 10 μL each of papain into the PMC solution spaced at 300-s intervals to allow for reequilibration. Heats of dilution of papain were determined by titration of papain into buffer without PMC and were used to correct the protein titration data. The Origin software package (OriginLab) was used to fit the data to an n-sites equivalent binding model using nonlinear least squares regression. Fitting the data provides the affinity, enthalpy, and entropy change for the binding reaction.
Far-UV CD Spectra
CD spectra were measured on solutions of PMC-6 and its truncated mutant, in PBS pH 7.2 from 300 to 200 nm at 1-nm intervals using an Aviv 202SF CD spectrometer.
Papain Inhibitory Activity of Native and Recombinant PMC
The effect of pH on the ability of native and recombinant PMC (PMC-6 and PMC-567) to inhibit papain (Cys protease; Sigma-Aldrich) activity was examined using BAPNA (Sigma-Aldrich) as the substrate (Walsh and Strickland, 1993; Nissen et al., 2009). Native and recombinant PMC were preincubated at pH 4.2 or 7.2 in 75 μL of sodium acetate-MES-Tris (50 mM each) buffer for 10 min (23°C). Papain (0.65 units) was then mixed with the inhibitors and incubated for an additional period of 10 min (23°C). Papain incubated alongside (as above) at pH 4.2 and 7.2 without inhibitors served as a control. The reaction volume was then made up to 1.0 mL with MES buffer (0.1 M, pH 6.2) containing 8 mM DTT, 4 mM EDTA, and 400 μM (final concentration) BAPNA. Papain activity was monitored spectrometrically using an Epoch multiwell plate reader (Biotek Instruments) by following the increase in absorbance at 395 nm (p-nitroanilide production) for 30 min. The concentration (µM) of wild-type and recombinant PMC required to inhibit papain activity by 50% was first determined at pH 4.2. Using concentrations (µM) that effected 50% inhibition, the inhibitory effects of wild-type and recombinant PMC were then compared at pH 4.2 and 7.2 on an equimolar basis.
Crystallization and Data Collection
Crystals of PMC-567 were grown using the hanging-drop vapor diffusion method (McPherson, 1990). Purified protein at 10 to 12 mg/mL in 20 mM Na acetate, pH 5.0, was mixed with an equal volume of 0.4 M ammonium phosphate and equilibrated against the same solution at 4°C. Diffraction quality crystals appeared after 4 to 6 d. The crystals were determined as belonging to the tetragonal P43212 space group, with a = b = 65.94 and c = 191.85 Å. The crystals diffracted to ∼2.2 Å, and there was one PMC-567 molecule in the asymmetric unit (Table 2). Diffraction data were collected at the Berkeley Advanced Light Source (beam line 8.2.1) and were processed and scaled with the HKL2000 package (Otwinowski et al., 2003).
Phasing and Refinement
Initial phases of the PMC-567 crystal structure were determined by the molecular replacement method using the coordinate of PMC-2 (Protein Data Bank ID: 2W9P and 2W9Q) and AMoRe software (Navaza, 2001). Iterative model building and refinement took place using the programs COOT (Emsley et al., 2010) and PHENIX (Adams et al., 2010). The final R factor (Table 2) was 22.2% (Rfree = 28.7% for the random 5% data). The root mean square deviations from ideal geometry of the final coordinate were 0.008 for bonds and 1.196° for angles, respectively. PMC-567 coordinates have been deposited in the Protein Data Bank (4LZI).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: BAA95416.1 for multicystatin from sunflower and P37842 for multicystatin from potato.
Acknowledgments
Original research was supported by NSF MCB 1021148, USDA ARS 30553460, NSF DBI 0959778, and M.J. Murdock Charitable Trust.
AUTHOR CONTRIBUTIONS
G.N.M.K., N.R.K., and C.K. conceived this project and designed all experiments. A.R.G., M.S.N., and G.N.M.K. performed experiments. A.R.G., M.S.N., G.N.M.K., N.R.K., and C.K. analyzed data. A.R.G., M.S.N., G.N.M.K., N.R.K., and C.K. wrote the article.
Glossary
- PMC
potato multicystatin
- DLS
dynamic light scattering
- ITC
isothermal titration calorimetry
- CD
circular dichroism
- BAPNA
N-α-benzoyl-dl-Arg-p-nitroanilide
- SMC
sunflower multicystatin
References
- Abe K., Emori Y., Kondo H., Arai S., Suzuki K. (1988). The NH2-terminal 21 amino acid residues are not essential for the papain-inhibitory activity of oryzacystatin, a member of the cystatin superfamily. Expression of oryzacystatin cDNA and its truncated fragments in Escherichia coli. J. Biol. Chem. 263: 7655–7659 [PubMed] [Google Scholar]
- Adams P.D., et al. (2010). PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode W., Engh R., Musil D., Thiele U., Huber R., Karshikov A., Brzin J., Kos J., Turk V. (1988). The 2.0 A X-ray crystal structure of chicken egg white cystatin and its possible mode of interaction with cysteine proteinases. EMBO J. 7: 2593–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolter C., Jongsma M.A. (1997). The adaptation of insects to plant protease inhibitors. J. Insect Physiol. 43: 885–895 [DOI] [PubMed] [Google Scholar]
- Dieckmann T., Mitschang L., Hofmann M., Kos J., Turk V., Auerswald E.A., Jaenicke R., Oschkinat H. (1993). The structures of native phosphorylated chicken cystatin and of a recombinant unphosphorylated variant in solution. J. Mol. Biol. 234: 1048–1059 [DOI] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholizadeh A., Kohnehrouz B.B. (2011). Functional fusion expression of sunflower multicystatin in E. coli and its comparison with a single domain cystatin. Indian J. Biochem. Biophys. 48: 375–379 [PubMed] [Google Scholar]
- Holm L., Sander C. (1993). Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233: 123–138 [DOI] [PubMed] [Google Scholar]
- Irie K., Hosoyama H., Takeuchi T., Iwabuchi K., Watanabe H., Abe M., Abe K., Arai S. (1996). Transgenic rice established to express corn cystatin exhibits strong inhibitory activity against insect gut proteinases. Plant Mol. Biol. 30: 149–157 [DOI] [PubMed] [Google Scholar]
- Jenko S., Dolenc I., Guncar G., Dobersek A., Podobnik M., Turk D. (2003). Crystal structure of Stefin A in complex with cathepsin H: N-terminal residues of inhibitors can adapt to the active sites of endo- and exopeptidases. J. Mol. Biol. 326: 875–885 [DOI] [PubMed] [Google Scholar]
- Kumar G., Knowles N. (2003). Wound-induced superoxide production and PAL activity decline with potato tuber age and wound healing ability. Physiol. Plant. 117: 108–117 [Google Scholar]
- Kumar G.N., Iyer S., Knowles N.R. (2007). Strboh A homologue of NADPH oxidase regulates wound-induced oxidative burst and facilitates wound-healing in potato tubers. Planta 227: 25–36 [DOI] [PubMed] [Google Scholar]
- Kumar G.N., Houtz R.L., Knowles N.R. (1999). Age-induced protein modifications and increased proteolysis in potato seed-tubers. Plant Physiol. 119: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Martin J.R., Craven C.J., Jerala R., Kroon-Zitko L., Zerovnik E., Turk V., Waltho J.P. (1995). The three-dimensional solution structure of human stefin A. J. Mol. Biol. 246: 331–343 [DOI] [PubMed] [Google Scholar]
- McPherson A. (1990). Current approaches to macromolecular crystallization. Eur. J. Biochem. 189: 1–23 [DOI] [PubMed] [Google Scholar]
- Murdock L., Brookhart G., Dun P., Foard D., Kelley S., Kitch L., Shade R., Shukle R., Wolfson J. (1987). Cysteine digestive proteinases in coleoptera. Comp. Biochem. Physiol. 87B: 783–787 [Google Scholar]
- Murzin A.G. (1993). Sweet-tasting protein monellin is related to the cystatin family of thiol proteinase inhibitors. J. Mol. Biol. 230: 689–694 [DOI] [PubMed] [Google Scholar]
- Nagata K., Kudo N., Abe K., Arai S., Tanokura M. (2000). Three-dimensional solution structure of oryzacystatin-I, a cysteine proteinase inhibitor of the rice, Oryza sativa L. japonica. Biochemistry 39: 14753–14760 [DOI] [PubMed] [Google Scholar]
- Navaza J. (2001). Implementation of molecular replacement in AMoRe. Acta Crystallogr. D Biol. Crystallogr. 57: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Nissen M.S., Kumar G.N., Youn B., Knowles D.B., Lam K.S., Ballinger W.J., Knowles N.R., Kang C. (2009). Characterization of Solanum tuberosum multicystatin and its structural comparison with other cystatins. Plant Cell 21: 861–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr G., Strickland J., Walsh T. (1994). Inhibition of Diabrotica larval growth by a multicystatin from potato tubers. J. Insect Physiol. 40: 893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Borek D., Majewski W., Minor W. (2003). Multiparametric scaling of diffraction intensities. Acta Crystallogr. A 59: 228–234 [DOI] [PubMed] [Google Scholar]
- Popovic T., Brzin J. (2007). Purification and characterization of two cysteine proteinases from potato leaves and the mode of their inhibition with endogenous inhibitors. Croat. Chem. Acta 80: 45–52 [Google Scholar]
- Roberts J.K.M., Ray P.M., Wade-Jardetzky N., Jardetzky O. (1980). Estimation of cytoplasmic and vacuolar pH in higher plant cells by 31P NMR. Nature 283: 870–872 [Google Scholar]
- Rodis P., Hoff J.E. (1984). Naturally occurring protein crystals in the potato: Inhibitor of papain, chymopapain, and ficin. Plant Physiol. 74: 907–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs M.T., Laber B., Bode W., Huber R., Jerala R., Lenarcic B., Turk V. (1990). The refined 2.4 A X-ray crystal structure of recombinant human stefin B in complex with the cysteine proteinase papain: A novel type of proteinase inhibitor interaction. EMBO J. 9: 1939–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiz L. (1992). The plant vacuole. J. Exp. Biol. 172: 113–122 [DOI] [PubMed] [Google Scholar]
- Tate S., Ushioda T., Utsunomiya-Tate N., Shibuya K., Ohyama Y., Nakano Y., Kaji H., Inagaki F., Samejima T., Kainosho M. (1995). Solution structure of a human cystatin A variant, cystatin A2-98 M65L, by NMR spectroscopy. A possible role of the interactions between the N- and C-termini to maintain the inhibitory active form of cystatin A. Biochemistry 34: 14637–14648 [DOI] [PubMed] [Google Scholar]
- Thiele U., Auerswald E.A., Gebhard W., Assfalg-Machleidt I., Popović T., Machleidt W. (1988). Inhibitorily active recombinant human stefin B. Gene synthesis, expression and isolation of an inhibitory active MS-2 pol-stefin B fusion protein and preparation of Des[Met1,2(2)]stefin B. Biol. Chem. Hoppe Seyler 369: 1167–1178 [DOI] [PubMed] [Google Scholar]
- Walsh T.A., Strickland J.A. (1993). Proteolysis of the 85-kilodalton crystalline cysteine proteinase inhibitor from potato releases functional cystatin domains. Plant Physiol. 103: 1227–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeda S., Kumar G., Knowles N. (2010). Correlative changes in proteases and protease inhibitors during mobilization of protein from potato (Solanum tuberosum) seed tubers. Funct. Plant Biol. 37: 32–42 [Google Scholar]
- Weeda S., Kumar G., Knowles N. (2011). Protein mobilization from potato tubers during long-term storage and daughter tuber formation. Int. J. Plant Sci. 172: 459–470 [Google Scholar]
- Weeda S.M., Mohan Kumar G.N., Richard Knowles N. (2009). Developmentally linked changes in proteases and protease inhibitors suggest a role for potato multicystatin in regulating protein content of potato tubers. Planta 230: 73–84 [DOI] [PubMed] [Google Scholar]