Abstract
In this study we reassess the taxonomic reference of the previously described holomorphic alkaliphilic fungus Heleococcum alkalinum isolated from soda soils in Russia, Mongolia and Tanzania. We show that it is not an actual member of the genus Heleococcum (order Hypocreales) as stated before and should, therefore, be excluded from it and renamed. Multi-locus gene phylogeny analyses (based on nuclear ITS, 5.8S rDNA, 28S rDNA, 18S rDNA, RPB2 and TEF1-alpha) have displayed this fungus as a new taxon at the genus level within the family Plectosphaerellaceae, Hypocreomycetidae, Ascomycota. The reference species of actual Heleococcum members showed clear divergence from the strongly supported Heleococcum alkalinum position within the Plectosphaerellaceae, sister to the family Glomerellaceae. Eighteen strains isolated from soda lakes around the world show remarkable genetic similarity promoting speculations on their possible evolution in harsh alkaline environments. We established the pH growth optimum of this alkaliphilic fungus at c. pH 10 and tested growth on 30 carbon sources at pH 7 and 10. The new genus and species, Sodiomyces alkalinus gen. nov. comb. nov., is the second holomorphic fungus known within the family, the first one being Plectosphaerella – some members of this genus are known to be alkalitolerant. We propose the Plectosphaerellaceae family to be the source of alkaliphilic filamentous fungi as also the species known as Acremonium alcalophilum belongs to this group.
Keywords: alkaliphilic fungi, growth, Heleococcum alkalinum, molecular phylogeny, scanning electron microscopy, taxonomy
INTRODUCTION
The fungal kingdom is highly diverse and versatile, with members able to grow under various environmental conditions. Although the majority of fungi are considered as neutrophiles, showing optimal growth in moderate habitats (e.g. 25–30 °C, pH 5–7), some of them have adapted to thrive in extreme environments where abiotic conditions are so harsh that most organisms cannot survive. One such stressful condition is high alkalinity, to which some fungi have become adapted. Alkaliphilic fungi, i.e. fungi that are capable of growing at high pH, above pH 9 (Horikoshi 1999), have been little studied. Only a handful of filamentous alkaliphilic fungi have been reported to date (Nagai et al. 1995, 1998). The natural habitats for this kind of fungi are believed to be soda soils and soda lakes, as are often encountered in arid and semi-arid areas. These sites represent an unusual, naturally occurring stable ecotope where the overall biodiversity is compromised due to significant ambient stress, namely, high salt-osmotic pressure and high pH. Besides these natural sites, there are also sites created by human industrial processes like concrete and paper manufacture. These industries are known to create alkaline wastes that are potential habitats for alkaliphilic fungi (Mueller et al. 2004). Already in 1923, Johnson (1923) showed the ability of Fusarium oxysporum, F. bullatum and Penicillium variabile to grow at the extremely high pH of 11. Probably the most notable study on alkaliphilic fungi was the isolation and description of mitosporic Acremonium alcalophilum, which has a growth optimum at around pH 9, by Okada et al. (1993). More recently, Kladwang et al. (2003) isolated a number of alkali-tolerant species in Thailand and Elíades et al. (2006) reported eight species of alkaliphilic and alkalitolerant soil fungi from Argentina, taxonomically distributed through Bionectriaceae, Trichocomaceae, Sporormiaceae, Ceratostomataceae and Sordariaceae. However, overall, information on the biodiversity of alkaliphilic filamentous fungi is scarce.
Fungi growing at extreme pH values are of scientific interest for the general study of fungal adaptive evolution as well as for the evaluation of their potential in producing commercially valuable substances. Obviously, the fungi adapting to alkalinity must have metabolic pathways that have become modified with respect to those seen in related neutrophilic fungi. For instance, enzymes that are being secreted into the ambient environment should work optimally in alkalinity in order to provide sufficient amounts of nutrients (Kladwang et al. 2003). In addition, adaptations to alkaline environments are required for structures involved in exporting metabolites like toxins and antibiotics, for domains of the membrane transporters exposed to ambient environment and for regulation of gene expression by ambient pH. Alkaliphilic fungi are likely to possess unique properties that have not been well elucidated so far.
In 2005, a new alkaliphilic holomorphic fungus from hyper-saline soda soils (pH around 10) was isolated, described and placed among members of the genus Heleococcum as H. alkalinum. The genus Heleococcum (order Hypocreales) seemed appropriate based on morphological and ecological features (Bilanenko et al. 2005). Heleococcum species are known to be soil saprobes, all producing Acremonium-like anamorphs (except H. aurantiacum) along with bright-coloured cleistothecial ascomata (Jørgensen 1922, Tubaki 1967, Udagawa et al. 1995).
Upon considering the ecological distinctiveness of H. alkalinum and also some differences in morphological features compared to other Heleococcum species, we decided to investigate the taxonomic position of this species with the help of molecular phylogeny. We used multiple molecular phylogenetic markers including ribosomal rDNA (18S, 28S, ITS and 5.8S) and protein-coding genes for the second largest subunit of RNA polymerase II (RPB2) and for transcriptional elongation factor 1-alpha (TEF1-alpha). Results demonstrated that this fungus does not belong in Heleococcum but instead belongs to a new genus within the family of the Plectosphaerellaceae. We confirm its alkaliphilic nature and speculate on its possible role in alkaline ecotopes, based on growth experiments carried out on media containing various carbon sources at different pH levels. We also discuss the significance of the low levels of genetic variation observed among strains isolated from soda lakes located thousands of kilometres apart.
MATERIALS AND METHODS
Strains and media
In this study we used 18 strains of Heleococcum alkalinum (Bilanenko et al. 2005) isolated from soils near soda lakes in Russia, Mongolia and Tanzania (Table 1). All strains were grown at 27 °C on alkaline agar medium (AA) containing per litre: 1) Na2CO3 − 24 g, NaHCO3 − 6 g, NaCl − 5 g, KNO3 − 1 g, K2HPO4 − 1 g; 2) malt extract (Merck) – 17 g, yeast extract (Difco) – 1 g, agar (Difco) – 20 g. Components 1 and 2 were autoclaved separately for 20 min at 120 °C and mixed together after cooling which resulted in a final pH of c. 10. Reference strains were members of other species placed in the genus Heleococcum, including H. japonense CBS 397.67 and H. aurantiacum CBS 201.35. In addition, we included some pertinent Acremonium cultures, A. alcalophilum CBS 114.92 and A. antarcticum CBS 987.87, that were obtained from the Centraalbureau voor Schimmelcultures (CBS, Utrecht, The Netherlands). The Acremonium and Heleococcum isolates were maintained on standard malt extract agar (MEA), oat meal agar (OA) (Mueller et al. 2004) or our own AA medium at 27 °C.
Table 1.
Strains of Sodiomyces alkalinus (former Heleococcum alkalinum) used in the study. Locations, characteristics of soil samples and isolation date are indicated.
| Strain | CBS no. | Location | Place | Soil pH | Total salts (g/kg) | Depth (cm) | Saltification type | Date |
|---|---|---|---|---|---|---|---|---|
| F7 | CBS 132729 | Kunkur steppe, Chitinskaya area, Russia | Low-salt soda lake | – | – | 0 to 5 | Soda | 1999 |
| F8 | CBS 133680 | Kunkur steppe, Chitinskaya area, Russia | Low-salt soda lake | – | – | 0 to 5 | Soda | 1999 |
| F9 | CBS 133681 | Kunkur steppe, Chitinskaya area, Russia | Low-salt soda lake | – | – | 0 to 5 | Soda | 1999 |
| F10 | CBS 132730 | Tanzania | Natron lake | 10 (water) | – | 0 to 5 | Soda | 1999 |
| F11 | CBS 110278 | Choibalsan area, North-East Mongolia | Shar-Burdiyn lake | 10.7 | 49 | 0 to 5 | Soda | 1999 |
| F12 | CBS 132731 | North-East Mongolia | Barun-Undziyn lake | 10.5 | 82 | 0 to 5 | Soda | 1999 |
| F13 | CBS 132732 | Kulunda steppe, Altai, Russia | Solyonoe lake | 10 | 187 | 0 to 5 | Chloride | Aug. 2002 |
| F14 | CBS 133682 | Kulunda steppe, Altai, Russia | Karakul’ lake | 9.8 | 144 | 0 to 5 | Soda | Aug. 2002 |
| F15 | CBS 133683 | Kulunda steppe, Altai, Russia | Mirabilit lake | 9.7 | 165 | 0 to 5 | Soda-chloride-sulfate | Aug. 2002 |
| F16 | CBS 133684 | Kulunda steppe, Altai, Russia | Petuhovskoe lake | 10.2 | 163 | 0 to 5 | Soda | Aug. 2002 |
| F17 | CBS 133685 | Kulunda steppe, Altai, Russia | Bezimyannoe lake | 9.9 | 310 | 0 to 5 | Soda | Aug. 2002 |
| F18 | CBS 132733 | Kulunda steppe, Altai, Russia | Tanatar lake | 10.2 | 73 | 0 to 5 | Soda | Aug. 2002 |
| F19 | CBS 133686 | Kulunda steppe, Altai, Russia | Mirabilit lake | 9.6 | 100 | 0 to 5 | Soda-chloride-sulfate | Aug. 2002 |
| F20 | CBS 133687 | North Gobi, Mongolia | Bayan-Dzag area | 9.3 | 43 | 1 to 2 | Sulfate-soda | Aug. 2003 |
| F21 | CBS 133688 | North Gobi, Mongolia | Bayan-Dzag area | 9.2 | 6 | 10 to 18 | Sulfate-soda | Aug. 2003 |
| F22 | CBS 133689 | Kulunda steppe, Altai, Russia | North | 10 | 22 | 0 to 5 | Soda | Aug. 2005 |
| F23 | CBS 133690 | Kulunda steppe, Altai, Russia | Karagay lake | 9.9 | 43 | 0 to 5 | Soda | Aug. 2005 |
| F24 | CBS 133691 | Kulunda steppe, Altai, Russia | Gor’koye lake | 10.4 | 30 | 0 to 5 | Soda | Aug. 2005 |
Growth at different pH
To elucidate the pH growth optimum of the strains we used malt extract-based medium buffered at pH levels ranging from 4 to 11.4. Acetic acid buffers were used to create pH 4 and 5.2, while phosphate buffers were used for pH 5.9, 7 and 7.8. Carbonate buffers were employed to set pH 8.7 and 9.8. Finally, a Na2HPO4/NaOH buffer system was used to generate pH 10.5 and 11.4. The final buffer concentration in all media was set to 0.1 M. The core nutrient component of media contained per litre: malt extract (Merck) – 17 g, yeast extract (Difco) – 1 g, agar (Difco) – 20 g. Buffers and nutrient components were autoclaved separately for 20 min at 120 °C and mixed afterwards, making up complete media. Strains were inoculated in so-called race tubes (Perkins & Pollard 1986). Four tubes were inoculated per strain in media at each of these pH values: 4, 5.2, 5.9, 7, 7.8, 8.7, 9.8, 10.5 and 11.4. Linear growth was measured once or twice a week for c. 3 mo.
Growth on different carbon sources
To analyse the capacity of H. alkalinum to use diverse carbon sources at neutral and alkaline conditions, we employed 31 media buffered at pH 7 and 10 based on various carbon sources ranging in complexity (including no-C source as a control). The final buffer concentration (phosphate buffer for pH 7 and carbonate buffer for pH 10) was chosen to be 0.1 M. All media had final salts concentrations per litre of NaCl − 5 g, KNO3 − 1 g, K2HPO4 − 1 g. The salt component was autoclaved separately. Simple soluble sugars, namely, D(+)glucose, D(−)fructose, D(+)galactose, D(+)mannose, D(+)xylose, L(+)arabinose, L-rhamnose, D(+)glucuronic acid, D(+)cellobiose, D(+)maltose, D(+)lactose, D(+)raffinose, sucrose, were used at 25 mM final concentration. They were gently (15 min at 110 °C) autoclaved separately from salt and agar solutions and were subsequently added to the final media after cooling. A 1 % concentration was chosen for the following complex carbon sources: arabinogalactan, beechwood xylan, birchwood xylan, oat spelt xylan, guar gum, soluble starch, apple pectin, inulin, hydrolytic lignin, alpha-cellulose and chitin. A 3 % concentration was used for: sugar beet pulp, citrus pulp, soybean hulls, cotton seed hulls, alfalfa meal and corn gluten. All media were supplemented with solution of metal traces (van Diepeningen et al. 2008). Complex substrates were autoclaved together with agar. Growth experiments on carbon sources were conducted at 27 °C in six replicates with initial inoculation of c. 1 400 spores per replicate. Growth patterns were recorded through 12 d.
Morphological studies with cryo-SEM and SEM
A piece (0.5 × 0.5 cm) of mycelium with agar was cut out from the plate directly and glued on a brass Leica sample holder with carbon paste (Leit-C, Neubauer Chemikalien, Germany). It was further frozen in liquid nitrogen and simultaneously fitted into the cryo-sample loading (VCT 100) system. The Leica sample holder was transferred to a non-dedicated cryo-preparation system (MED 020/VCT 100, Leica, Vienna, Austria) onto a sample stage at −93 °C. In this cryo-preparation chamber, the samples were freeze dried for 3 min at −93 °C at 1.3 × 10−6 mbar to remove water vapour contamination from the surface. The sample was sputter-coated with a layer of 10 nm tungsten at the same temperature and transferred into the field emission scanning microscope (Magellan 400, FEI, Eindhoven, The Netherlands) on the sample stage at −122 °C at 4 × 10−7 mbar. The analysis was performed with SE at 1 and 2 kV, 13 pA. All images were recorded digitally.
We also examined fungal samples by scanning electron microscopy after 1 h of 2.5 % glutaraldehyde fixation and series of 0.1 M phosphate buffer (pH 7.2) washing steps (20 min each) followed by dehydration through an ethanol series (30 %, 50 %, 70 % and 96 % concentration) and acetone before critical point drying in CO2, carbon and metal coating. Specimens were observed under a JEOL (Japan) scanning electron microscope. Light microscopic studies were done with a Nikon Eclipse 80i. Taxonomic novelties were deposited in MycoBank (www.MycoBank.org; Crous et al. 2004).
DNA extraction
All strains used in the study were grown in Petri dishes containing either AA or MEA depending on which yielded better growth performance for the specific strain. Plates were incubated at 27 °C for c. 5 d on a cellophane membrane. Total genomic DNA (free of RNA) was extracted from mycelium harvested from the cellophane surface with DNeasy Plant Mini kit (Qiagen Inc., Chatsworth, California, USA). DNA quality and concentration was verified with a NanoDrop 2000.
Polymerase chain reaction and sequencing
Amplification and sequencing were performed for five nuclear loci: ITS1–ITS4 region including 5.8S rDNA, LR0R–LR9 region of 28S rDNA, NS1–NS8 of 18S rDNA, 5–7 region of RPB2 and 983–2218 region of TEF1-alpha. The final volume of the PCR mix was 25 μL which contained 5× GoTaq Green buffer (Promega, USA), 400 μM dNTP, 0.4 μM of each primer, 0.02 U GoTaq polymerase (Promega, USA), 5–100 ng template genomic DNA, 1 mM MgCl2. PCR conditions were as follows: for ITS 5 min at 94 °C; 33 cycles of 1 min at 94 °C, 1 min at 51 °C, 1 min at 72 °C; for 28S rDNA 5 min at 94 °C; 33 cycles of 1 min at 94 °C, 1 min at 49 °C, 2 min at 72 °C; for 18S rDNA 5 min at 94 °C; 33 cycles of 1 min at 94 °C, 1 min at 52 °C, 2 min at 72 °C; for 5–7 region of RPB2 gene 5 min at 94 °C; 9 cycles of 1 min at 94 °C, 1 min at 60 °C to 50 °C (with 1 degree decrement each cycle), 1.5 min at 72 °C followed by 32 cycles of 1 min at 94 °C, 1.5 min at 50 °C, 1.5 min at 72 °C; for TEF1-alpha gene 5 min at 94 °C; 9 cycles of 1 min at 94 °C, 1 min at 66 °C to 56 °C (with 1 degree decrement each cycle), 1.5 min at 72 °C followed by 32 cycles of 1 min at 94 °C, 1.5 min at 56 °C, 1.5 min at 72 °C. All reactions were kept for 7 min at 72 °C for final extension step. Standard fungal primers were used for PCR listed in the overview by Binder & Hibbett (2003) available at http://www.clarku.edu/faculty/dhibbett/Protocols_Folder/Primers/Primers.pdf. The amplification products were visualized on a 1 % agarose gel stained with ethidium bromide, purified using GeneElute PCR Clean-Up Kit (Sigma) according to the manufacturer instructions and subsequently sequenced by the chain termination method at Eurofins MWG Operon (Germany) service. LR7; NS4 and NS6 primers for 28S rDNA and 18S rDNA regions, respectively, were supplemented for sequencing in addition to original PCR primers. Raw sequence chromatograms were viewed and edited using CodonCodeAligner v. 3.7.1 (CodonCode Corporation, Dedham, MA, USA) and DNAStar Lasergene EditSeq v. 7.1.0 (DNASTAR Inc., Madison, WI, USA).
Phylogenetic analyses
Newly generated sequences were deposited in GenBank with accession numbers listed in Table 2. Sequences of ITS1 & 2 (including 5.8S rDNA gene), 28S rDNA gene (LSU), 18S rDNA gene (SSU) and protein coding genes RPB2 and TEF1-alpha were used for phylogenetic analysis. Reference sequences of Sordariomycetes members needed for phylogenetic reconstruction were obtained from GenBank along with our new data.
Table 2.
Taxa used in the phylogenetic analyses with their GenBank accession numbers. Newly generated accessions marked in bold.
| Species | Voucher | Appearance in tree* | SSU | LSU | RPB2 | TEF1-alpha | ITS |
|---|---|---|---|---|---|---|---|
| Acanthonitschkea argentinensis | SMH 1395 | 1 | _ | AY695259 | FJ968943 | FJ969042 | _ |
| Acremonium alcalophilum | CBS 114.92 | 1, 2 | JX158486 | JX158443 | JX158465 | JX158399 | JX158421 |
| Acremonium antarcticum | CBS 987.87 | 1, 2 | JX158487 | JX158444 | JX158466 | JX158400 | JX158422 |
| Acremonium brunnescens | CBS 559.73 | 2 | HQ231966 | _ | |||
| Acremonium cucurbitacearum | CBS 683.88 | 2 | HQ231968 | _ | |||
| Acremonium furcatum | CBS 122.42 | 2 | EF543831 | AY378154 | |||
| Acremonium murorum | CBS 154.25 | 1 | FJ176824 | FJ176880 | FJ238363 | _ | _ |
| Acremonium nepalense | CBS 971.72 | 2 | HQ231970 | DQ825971 | |||
| Acremonium persicinum | CBS 310.59 | 1 | FJ176822 | FJ176878 | FJ238361 | _ | _ |
| Acremonium restrictum | CBS 178.40 | 2 | HQ232119 | _ | |||
| Acremonium strictum | CBS 346.70 | 1 | FJ176823 | FJ176879 | FJ238362 | _ | GQ376096 |
| Acremonium stromaticum | CBS 863.73 | 2 | HQ232143 | DQ825969 | |||
| Acrostalagmus annulatus | DAOM 212126 | 2 | GU180646 | GU180632 | |||
| Acrostalagmus luteoalbus | CBS 194.87 | 2 | EF543826 | _ | |||
| Apiognomonia errabunda | AR 2813 | 1 | DQ862045 | AF408334 | DQ862014 | DQ862030 | DQ313525 |
| Bionectria ochroleuca | 1 | DQ862044 | DQ862027 | DQ862013 | DQ862029 | _ | |
| Camarops microspora | CBS 649.92 | 1 | DQ471036 | AY083821 | DQ470937 | DQ471108 | _ |
| Camarops ustulinoides | DEH 2164 | 1 | DQ470989 | DQ470941 | DQ470882 | DQ471050 | _ |
| Cercophora caudata | CBS 606.72 | 1 | DQ368659 | AY999113 | DQ368646 | _ | AY999135 |
| Chaetosphaerella phaeostroma | SMH 4257 | 1 | _ | AY695264 | FJ968940 | FJ969004 | _ |
| Colletotrichum boninense | CMT74 | 2 | JQ754138 | _ | |||
| Colletotrichum gloeosporioides | CMT32 | 2 | JQ754114 | _ | |||
| Coniochaeta ostrea | CBS 507.70 | 1 | DQ471007 | DQ470959 | DQ470909 | DQ471078 | _ |
| Cordyceps cardinalis | OSC 93609 | 1 | AY184973 | AY184962 | DQ522422 | DQ522325 | _ |
| Cryphonectria parasitica | ATCC 38755 | 1 | DQ862048 | EU199123 | DQ862017 | DQ862033 | AY141856 |
| Cryptosporella hypodermia | CBS 171.69 | 1 | DQ862049 | DQ862028 | DQ862018 | DQ862034 | EU199225 |
| Diaporthe eres | CBS 109767 | 1 | DQ471015 | _ | DQ470919 | DQ479931 | DQ491514 |
| Diatrype disciformis | CBS 197.49 | 1 | DQ471012 | DQ470964 | DQ470915 | DQ471085 | _ |
| Didymostilbe echinofibrosa | AR 2824 | 1 | AY489674 | AY489706 | _ | AY489601 | DQ135999 |
| Epichloe typhina | ATCC 56429 | 1 | U32405 | _ | DQ522440 | AF543777 | JN049832 |
| Eutypa lata | CBS 208.87 | 1 | DQ836896 | DQ836903 | DQ836889 | DQ836909 | DQ006927 |
| Fracchiaea broomeana | SMH 2809 | 1 | _ | AY695268 | FJ968942 | FJ969039 | _ |
| Gelasinospora tetrasperma | CBS 178.33 | 1 | DQ471032 | DQ470980 | DQ470932 | DQ471103 | AY681178 |
| Gibellulopsis nigrescens | DAOM 226890 | 2 | GU180648 | GU180631 | |||
| Gibellulopsis piscis | CBS 892.70 | 2 | EF543835 | DQ825985 | |||
| Glomerella acutata | ICMP4850 | 2 | JN939938 | _ | |||
| Glomerella cingulata | FAU 553 | 1, 2 | AF543762 | AF543786 | _ | AF543773 | _ |
| Gnomonia gnomon | CBS 199.53 | 1 | DQ471019 | AF408361 | DQ470922 | DQ471094 | DQ491518 |
| Graphostroma platystoma | CBS 270.87 | 1 | DQ836900 | DQ836906 | DQ836893 | DQ836915 | _ |
| Heleococcum aurantiacum | CBS 201.35 | 1 | _ | JX158442 | JX158464 | JX158397 | JX158419 |
| Heleococcum japonense | CBS 397.67 | 1 | JX158485 | JX158441 | JX158463 | JX158398 | JX158420 |
| Hydropisphaera erubescens | ATCC 36093 | 1 | AY545722 | AY545726 | AY545731 | DQ518174 | _ |
| Hypocrea americana | OSC 100005 | 1 | AY544693 | AY544649 | _ | DQ471043 | DQ491488 |
| Hypocrea lutea | ATCC 208838 | 1 | AF543768 | AF543791 | DQ522446 | AF543781 | _ |
| Lasiosphaeria ovina | SMH 4605 | 1 | DQ836894 | AY436413 | AY600284 | DQ836908 | AY587923 |
| Leucostoma niveum | AR 3413 | 1 | DQ862050 | AF362558 | DQ862019 | DQ862035 | _ |
| Lignincola laevis | JK 5180A | 1 | U46873 | U46890 | DQ836886 | _ | _ |
| Lindra thalassiae | JK 5090A | 1 | DQ470994 | DQ470947 | DQ470897 | DQ471065 | DQ491508 |
| Lulworthia grandispora | JK 4686 | 1 | DQ522855 | DQ522856 | DQ518181 | DQ497608 | _ |
| Melanconis alni | AR 3500 | 1 | DQ862052 | AF408371 | DQ862021 | DQ862037 | _ |
| Melanconis marginalis | AR 3442 | 1 | DQ862053 | AF408373 | DQ862022 | DQ862038 | _ |
| Melanconis stilbostoma | AR 3501 | 1 | DQ862054 | AF408374 | DQ862023 | DQ862039 | _ |
| Melanopsamma pomiformis | ATCC 18873 | 1 | AY489677 | AY489709 | EF692511 | AY489604 | AF081478 |
| Melanospora tiffanii | ATCC 15515 | 1 | AY015619 | AY015630 | AY015637 | _ | _ |
| Melanospora zamiae | ATCC 12340 | 1 | AY046578 | U17405 | AY046580 | _ | _ |
| Microascus trigonosporus | CBS 218.31 | 1 | DQ471006 | DQ470958 | DQ470908 | DQ471077 | DQ491513 |
| Musicillium theobromae | CBS 968.72 | 2 | EF543838 | EF543859 | |||
| (Verticillium theobromae) 1 | |||||||
| Musicillium theobromae | CBS 458.51 | 2 | EF543837 | EF543858 | |||
| (Verticillium theobromae) 2 | |||||||
| Peethambara spirostriata | CBS 110115 | 1 | AY489692 | AY489724 | EF692516 | AY489619 | _ |
| Petriella setifera | CBS 437.75 | 1 | DQ471020 | DQ470969 | DQ836883 | DQ836911 | _ |
| Plectosphaerella alismatis | CBS 113362 | 2 | JF780521 | JF780523 | |||
| Plectosphaerella citrullae 1 | Plect 189 | 2 | HQ239050 | HQ238964 | |||
| Plectosphaerella citrullae 2 | Plect 157 | 2 | HQ239048 | HQ238962 | |||
| Plectosphaerella cucumerina | DAOM 226828 | 1, 2 | GU180612 | GU180647 | GU180663 | _ | GU180630 |
| Plectosphaerella melonis | Plect 148 | 2 | HQ239007 | HQ238968 | |||
| Plectosphaerella pauciseptata | Plect 186 | 2 | HQ239012 | HQ238971 | |||
| Plectosphaerella plurivora | Plect 329 | 2 | HQ239017 | HQ238972 | |||
| Plectosphaerella ramiseptata | Plect 158 | 2 | HQ239049 | HQ238963 | |||
| Pleospora herbarum | CBS 191.86 | 1 | DQ247812 | DQ247804 | DQ247794 | DQ471090 | DQ491516 |
| Podospora fimiseda | CBS 990.96 | 1 | _ | AY346296 | AY780190 | _ | AY515361 |
| Roumegueriella rufula | CBS 346.85 | 1 | DQ522561 | DQ518776 | DQ522461 | DQ522355 | _ |
| Selinia pulchra | AR 2812 | 1 | _ | GQ505992 | _ | HM484841 | HM484859 |
| Sodiomyces alkalinus F7 | CBS 132729 | 1, 2 | JX158467 | JX158423 | JX158445 | JX158379 | JX158401 |
| Sodiomyces alkalinus F8 | CBS 133680 | 1, 2 | JX158468 | JX158424 | JX158446 | JX158380 | JX158402 |
| Sodiomyces alkalinus F9 | CBS 133681 | 1, 2 | JX158469 | JX158425 | JX158447 | JX158381 | JX158403 |
| Sodiomyces alkalinus F10 | CBS 132730 | 1, 2 | JX158470 | JX158426 | JX158448 | JX158382 | JX158404 |
| Sodiomyces alkalinus F11 | CBS 110278 | 1, 2 | JX158471 | JX158427 | JX158449 | JX158383 | JX158405 |
| Sodiomyces alkalinus F12 | CBS 132731 | 1, 2 | JX158472 | JX158428 | JX158450 | JX158384 | JX158406 |
| Sodiomyces alkalinus F13 | CBS 132732 | 1, 2 | JX158473 | JX158429 | JX158451 | JX158385 | JX158407 |
| Sodiomyces alkalinus F14 | CBS 133682 | 1, 2 | JX158474 | JX158430 | JX158452 | JX158386 | JX158408 |
| Sodiomyces alkalinus F15 | CBS 133683 | 1, 2 | JX158475 | JX158431 | JX158453 | JX158387 | JX158409 |
| Sodiomyces alkalinus F16 | CBS 133684 | 1, 2 | JX158476 | JX158432 | JX158454 | JX158388 | JX158410 |
| Sodiomyces alkalinus F17 | CBS 133685 | 1, 2 | JX158477 | JX158433 | JX158455 | JX158389 | JX158411 |
| Sodiomyces alkalinus F18 | CBS 132733 | 1, 2 | JX158478 | JX158434 | JX158456 | JX158390 | JX158412 |
| Sodiomyces alkalinus F19 | CBS 133686 | 1, 2 | JX158479 | JX158435 | JX158457 | JX158391 | JX158413 |
| Sodiomyces alkalinus F20 | CBS 133687 | 1, 2 | JX158480 | JX158436 | JX158458 | JX158392 | JX158414 |
| Sodiomyces alkalinus F21 | CBS 133688 | 1, 2 | JX158481 | JX158437 | JX158459 | JX158393 | JX158415 |
| Sodiomyces alkalinus F22 | CBS 133689 | 1, 2 | JX158482 | JX158438 | JX158460 | JX158394 | JX158416 |
| Sodiomyces alkalinus F23 | CBS 133690 | 1, 2 | JX158483 | JX158439 | JX158461 | JX158395 | JX158417 |
| Sodiomyces alkalinus F24 | CBS 133691 | 1, 2 | JX158484 | JX158440 | JX158462 | JX158396 | JX158418 |
| Stachylidium bicolor | DAOM 226658 | 2 | GU180651 | _ | |||
| Torrubiella alba (Lecanicillium alba) | CBS 726.73a | 1 | AF339586 | AF339537 | EF468934 | EF468781 | AJ292464 |
| Varicosporina ramulosa | RVG 113 | 1 | U43846 | U44092 | DQ836888 | _ | _ |
| Verticillium albo-atrum | CBS 130.51 | 2 | HQ231976 | DQ825977 | |||
| Verticillium alfalfae | PD683 | 2 | _ | JN187991 | |||
| Verticillium dahliae | ATCC 16535 | 1, 2 | AY489705 | DQ470945 | DQ522468 | AY489632 | _ |
| Verticillium isaacii | PD752 | 2 | _ | JN188018 | |||
| Verticillium zaregamsianum | PD739 | 2 | _ | JN188008 | |||
| Xylaria acuta | ATCC 56487 | 1 | AY544719 | AY544676 | DQ247797 | DQ471048 | AF163026 |
| Xylaria hypoxylon | OSC 100004 | 1 | AY544692 | AY544648 | DQ470878 | DQ471042 | DQ491487 |
Appears in large scale tree (1), small scale tree (2) or both
Separate datasets of each gene were constructed with a multiple sequence alignment online service MAFFT v. 6 (Katoh & Toh 2008) and were further reviewed and edited manually in BioEdit v. 7.1.3.0 (Hall 1999). Ambiguously aligned regions were removed from the data matrix. For better illustration of phylogenetic relations among taxa we generated two matrices with different datasets and resolution power: 1) we concatenated five gene alignments (SSU, LSU, RPB2, TEF1-alpha and 5.8S rDNA) using Mesquite 2.74 (Maddison & Maddison 2010) for a combined large scale taxa phylogenetic analysis showing subphyla and orders of Sordariomycetes. The matrix was subdivided into nine partitions (three for ribosomal genes and six for each codon position in two protein-coding genes). 2) For the second, low-taxon level phylogeny, we combined ITS regions including 5.8S rDNA and 28S rDNA on another taxa set for a detailed resolution of Plectosphaerellaceae family members within Sordariomycetes clade. Both analyses are deposited in TreeBase (submission ID 12948).
Nucleotide substitution models for partitions were tested in the jModelTest v. 0.1.1 (Rannala & Yang 1996, Posada & Buckley 2004, Posada 2008) software package. The Akaike Information Criterion (AIC) implemented in jModelTest was used to select for best fit models after likelihood score calculations were done. Evolutionary models listed in Table 3 were further used for ML and BI analyses.
Table 3.
Loci and substitution models used in the phylogenetic analyses. Information on included base pairs is provided.
| Phylogenetic analysis | Locus | Nucleotide substitution model | Characters | Phylogenetically informative characters | Uninformative variable characters | Invariable characters |
|---|---|---|---|---|---|---|
| 1 | 18S rDNA | TIM+I+G (or GTR+I+G for MrBayes) | 1652 | 328 | 185 | 1139 |
| 28S rDNA | GTR+I+G | 1381 | 421 | 158 | 801 | |
| RPB2 | GTR+I+G | 954 | 571 | 43 | 340 | |
| TEF1alpha | GTR+I+G | 867 | 310 | 70 | 487 | |
| 5.8S rDNA | GTR+G | 159 | 29 | 9 | 124 | |
| total | 5013 | 1659 (33 %) | 465 (9 %) | 2891 (58 %) | ||
| ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ ___ | ||||||
| 2 | ITS with 5.8S rDNA | HKY+I+G | 472 | 135 | 21 | 316 |
| 28S rDNA | GTR+G | 843 | 154 | 48 | 641 | |
| total | 1315 | 289 (22 %) | 69 (5 %) | 957 (73 %) | ||
Maximum likelihood (ML) and Bayesian Inference (BI) analyses were used for phylogeny estimation. ML evaluation was conducted in GARLI v. 2.0 (Zwickl 2006) with random starting trees through partitions. The number of search runs was set to 5 for a large scale taxa matrix and 10 for low-taxon level taxon matrix. Bootstrap analyses were replicated 200 and 100 times, respectively. A 50 % majority-rule consensus tree was generated in the SumTrees v. 3.3.1 application in the DenroPy v. 3.11.0 package (Sukumaran & Holder 2010) running under Python 2.6 platform. Bayesian (BI) analysis was performed as implemented in MrBayes v. 3.1.2 (Huelsenbeck & Ronquist 2001). The first partition in the large scale taxon matrix was fitted into GTR+I+G, since that was the parameter closest to best-fit TIM+I+G, which is not included in MrBayes. Metropolis-coupled Markov chain Monte Carlo (MCMCMC) searches were run for 5M generations sampling every 100th tree. Two independent analyses with four chains each (one cold and three heated) were run until stationary distribution was achieved. Convergence of the run logs was analysed in TRACER v. 1.5 (http://beast.bio.ed.ac.uk/Tracer). The first 15 000 (30 %) ‘burn-in’ trees were excluded from further analysis. The rest was summarized to produce a 50 % majority-rule consensus tree with recovery of posterior probabilities (PP).
RESULTS
Isolation of strains and genetic identity
Eighteen strains were isolated from soda-soil samples collected at different sites in Russia, Mongolia and Tanzania. They have been deposited in the CBS collection (Utrecht, The Netherlands) (Table 1). All of them readily produce abundant ascomata and show intense conidiation on an alkaline agar medium of pH 10 and all share the same morphological characteristics. In addition, all strains have nearly identical sequences for the five loci studied: SSU, LSU, RPB2, TEF1-alpha and 5.8S rDNA. Even the ITS regions and third codon positions of protein-coding genes (RPB2 and TEF1-alpha), known to have relatively high rate of mutation accumulation, have minor or no nucleotide differences. We found nucleotide substitution mutations in 18 sites across a 6189-base alignment derived from the studied loci. Most of the substitutions were at 3rd codon positions in the RPB2 gene.
Phylogenetic analyses
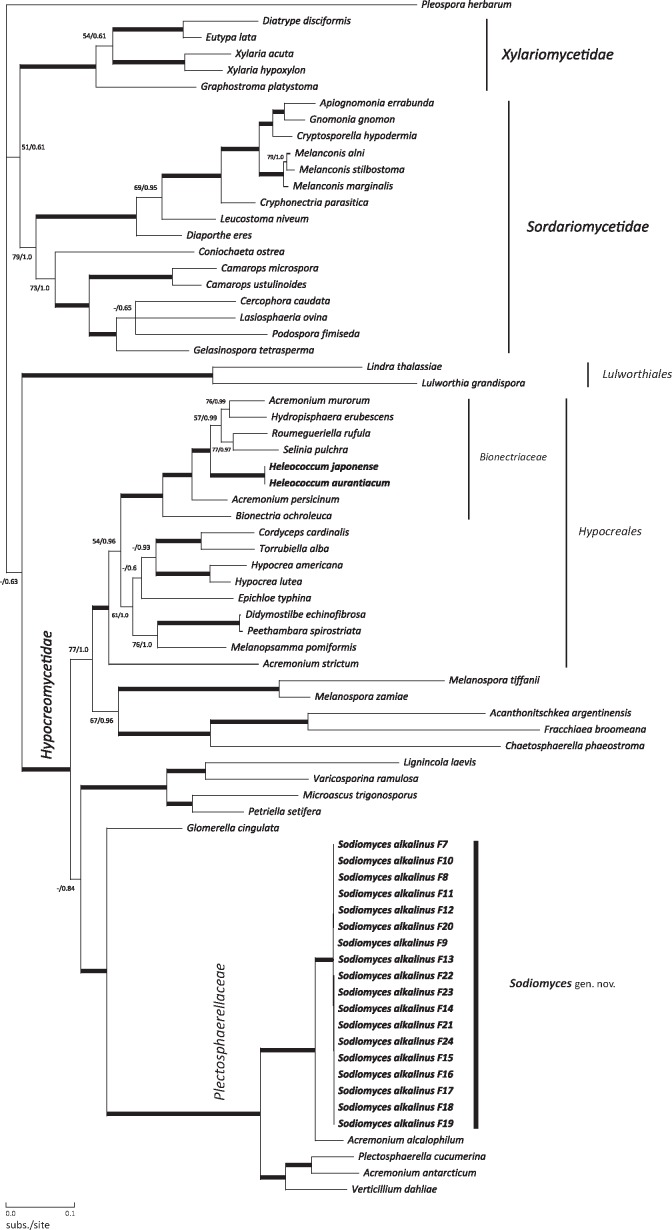
In the first analysis, the dataset for the five-gene phylogeny consisted of 70 taxa within the Sordariomycetes. The data matrix comprised 5 013 characters, of which 1 659 (33 %) were phylogenetically informative, 2 891 invariable and 465 non-informatively variable. The negative log likelihoods (-Ln) for ML BP and BI were 54607.294 and 55042.994. MCMCMC runs converged and had a deviation of around 0.008 at the end of the run. Maximum likelihood bootstrap and posterior probabilities (PP) are provided on the corresponding internodes on the 50 % majority rule Bayesian tree shown (Fig. 1). In both our phylogenetic analyses, clades supported with ML BP/BI PP exceeding 90/0.95 were considered very strong and are displayed as thickened braches. Our five-locus phylogeny provides firm topological support for three major monophyletic subphylum-level clades of Sordariomycetes outlined previously (Eriksson 2006, Zhang et al. 2006), namely, Xylariomycetidae (100/1.0), Sordariomycetidae (79/1.0) and Hypocreomycetidae (97/1.0). The marine fungi order Lulworthiales displayed a strong supported clade (100/1.0) with no clear assessment to any of the other subphyla within the Sordariomycetes. The taxonomic position of the Lulworthiales has been a matter of discussion since the description of the order, and it currently has incertae sedis status (Kohlmeyer et al. 2000, Hibbett et al. 2007). The monophyletic order Hypocreales within the Hypocreomycetidae forms a well-supported clade (92/1.0) in which the genus Heleococcum is located. We used a representative isolate of the type species of Heleococcum, H. aurantiacum (CBS 201.35) as well as an isolate of H. japonense (CBS 397.67) as references to compare with the taxonomic position of H. alkalinum. Heleococcum aurantiacum and H. japonense stand close together as members of the family Bionectriaceae within the Hypocreales as stated previously (Rossman et al. 2001). Surprisingly, however, all H. alkalinum strains clustered as a distinct clade within the highly supported (100/1.0) broader monophyletic clade comprising the family Plectosphaerellaceae. A novel genus name was therefore needed. We propose Sodiomyces alkalinus gen. & comb. nov. as the new name for this fungus. This name will be used in the remainder of this paper.
Fig. 1.
Position of Position of Sodiomyces alkalinus clade within Sordariomycetes. Bayesian 50 % majority-rule consensus tree based on five-gene data matrix (SSU+LSU+RPB2+TEF1-alpha+5.8S rDNA). Actual members of Heleococcum genus marked bold within Bionectriaceae family. Thickened branches indicate ML > 90 and BI posterior probability (PP) > 0.95.
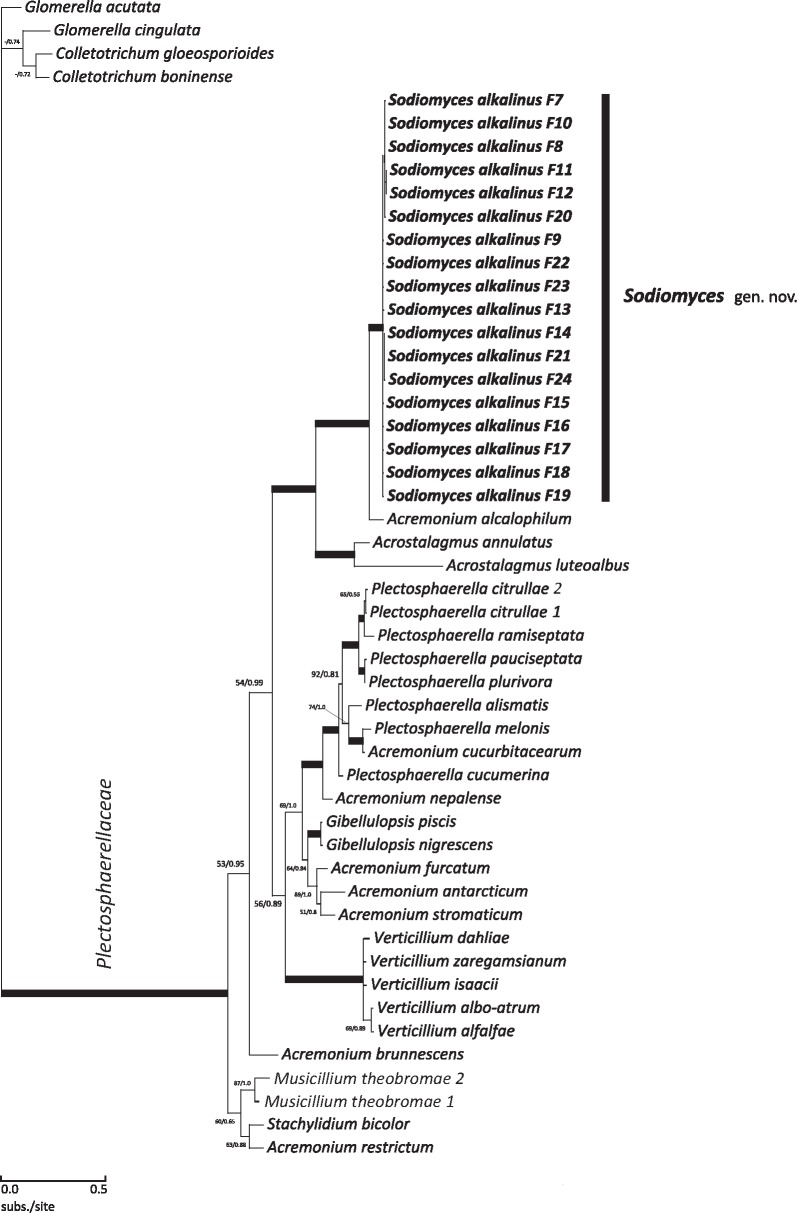
Our second, two-locus (ITS including 5.8S rDNA and 28S rDNA) phylogenetic analysis was done to further resolve the Plectosphaerellaceae clade and place Sodiomyces alkalinus strains among the other members of the family (Fig. 2). The data matrix consisted of 50 taxa and 1 315 characters: 289 (22 %) phylogenetically informative, 957 invariable and 69 variable non-informative. Four members of the sister family Glomerellaceae were used as outgroup. ML bootstrap and BI analyses had likelihoods (-Ln) of 5501.97 and 5721.683, respectively. The deviation between the Markov chain runs was around 0.004 at the end. Members of the holomorphic genus Plectosphaerella make up a highly supported clade (92/1.0) as does the asexual plant pathogen Verticillium s.str. clade (97/1.0). Verticillium theobromae appears closer to the root of the tree, lending some doubt to the monophyly of Verticillium. It has been renamed Musicillium theobromae by Zare et al. (2007). The two species of the genus Acrostalagmus form a long branched clade with strong 99/1.0 support values. Anamorphic Acremonium species known to be highly polyphyletic, occupy various separate branches in our phylogenetic tree. The overall topology is highly consistent with that seen in previous phylogenetic studies of the Plectosphaerellaceae (Zare et al. 2007, Weisenborn et al. 2010, Reblova et al. 2011, Carlucci et al. 2012). Here again, our Sodiomyces alkalinus strains group together and show a highly supported (100/1.0) clade close to A. alcalophilum.
Fig. 2.
Position of Position of Sodiomyces alkalinus strains within Plectosphaerellaceae family. Bayesian 50 % majority-rule consensus tree based on two-locus phylogenetic analysis (ITS+LSU). Thickened branches indicate ML > 90 and BI posterior probability (PP) > 0.95.
Growth patterns on different substrates
Sodiomyces alkalinus is an alkaliphilic fungus clearly showing a characteristic growth pattern at different pH levels on MEA-based media (Fig. 3). The maximum growth rate of ex-type strain F11, 2.6 mm/d, was recorded at pH from 8.7 to 10.5, with only a small reduction in growth rate seen at pH 11.4. On the other side of the pH scale, the fungus was still able to grow at pH 6 with an almost halved growth rate (1.4 mm/d). At pH 5.2 and lower, no growth was observed.
Fig. 3.
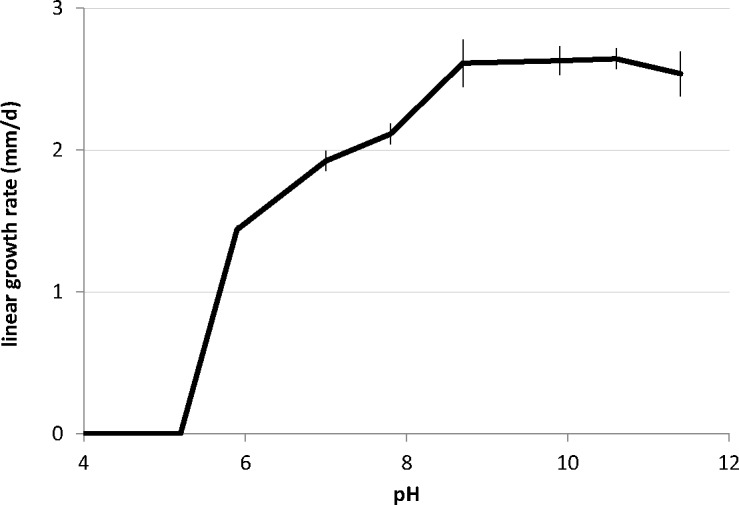
Growth pattern of Sodiomyces alkalinus (CBS 110278) at different pH.
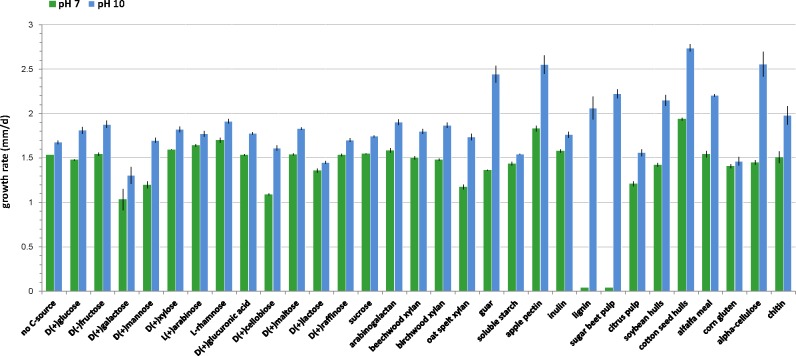
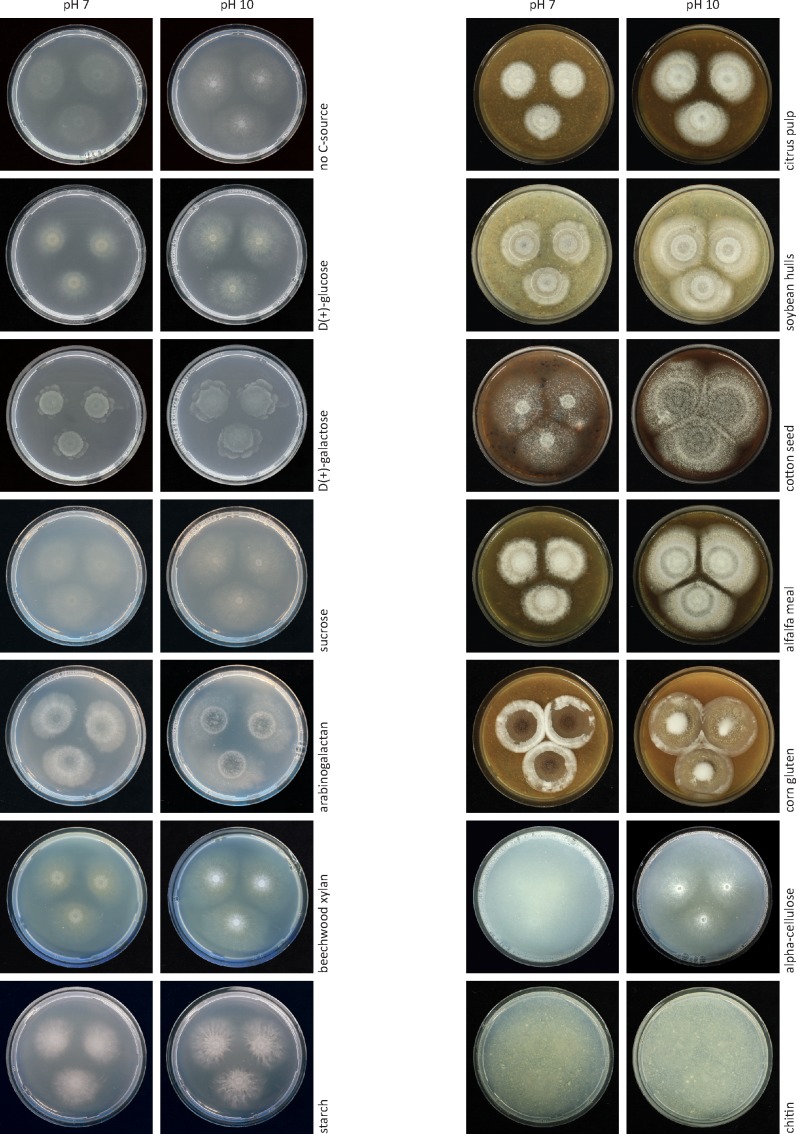
The growth rates of strain F11 on 30 carbon sources at pH 7 and 10 varied (Fig. 4, 5). The graphs display, from left to right, carbon sources ranging from simple to complex. Control plates with no carbon source still showed faint growth, presumably, due to the traces of nutrients in the agar. As can be seen in Fig. 5, simple sugars do not provide sufficient nutrients for good growth, resulting in poorly formed colonies with thin sterile hyphae. Some substrates such as D(+)galactose inhibited growth significantly and resulted in an irregular colony shape. More complex substrates like rhamnose, maltose, cellobiose, raffinose, arabinogalactan, beechwood xylan, and oat-spelt xylan provided the nutrients for a richer morphology and facilitated initiation of conidiation. All complex media made with raw plant materials as carbon sources yielded rich colony morphology with developed aerial mycelium, abundant conidia and formation of ascomata. Apple pectin did not promote rich colony formation, but rather, initiated the formation of ascomata towards the end of the incubation period. The fungus did not produce discernible colonies on pure cellulose or chitin. In all cases, growth on media at pH 10 was better than at pH 7, again showing the adaptation of Sodiomyces alkalinus to alkaline environments. The species could not initiate growth on lignin and sugar beet pulp-based media at pH 7, but showed good growth at pH 10 on those substrates. On lignin at pH 10, the fungus showed morphology similar to that seen on polysaccharides, developing only asexual sporulation.
Fig. 4.
Growth rate on different carbon sources of Sodiomyces alkalinus (CBS 110278).
Fig. 5.
Plates with Plates with Sodiomyces alkalinus (CBS 110278) 12 d old colonies on different carbon sources at pH 7 and 10.
Taxonomy
Sodiomyces A.A. Grum-Grzhim., A.J.M. Debets & Bilanenko, gen. nov. — MycoBank MB801368
Etymology. From the English soda and Latin mycetes, referring to the ability of filamentous fungus grow at high ambient pH and salts.
Type species. Sodiomyces alkalinus (Bilanenko & M. Ivanova) A.A. Grum-Grzhim., A.J.M. Debets & Bilanenko.
A genus of the family Plectosphaerellaceae in Ascomycota.
Asexual morph. Acremonium-like.
Sexual morph. Cleistothecia dark-brown, 120–150 μm diam, peridium multi-layered, pseudoparenchymatous, with folded surface, exoperidium composed of 3–5 layers of angular cells. Paraphyses absent. Asci thin-walled, without apical apparatus, saccate, unitunicate, scattered irregularly in the ascocarp, embedded in a gelatinous matrix. Ascospores released by dissolution of the ascus wall before maturity, accumulating within the ascocarp, released in a slimy mass, liberated by pressure within the ascocarp. Ascospores ellipsoidal or ovoid, 12–15 × 5–7 μm, medially 1-septate, not constricted at the septum, thick-walled, pale brown, smooth.
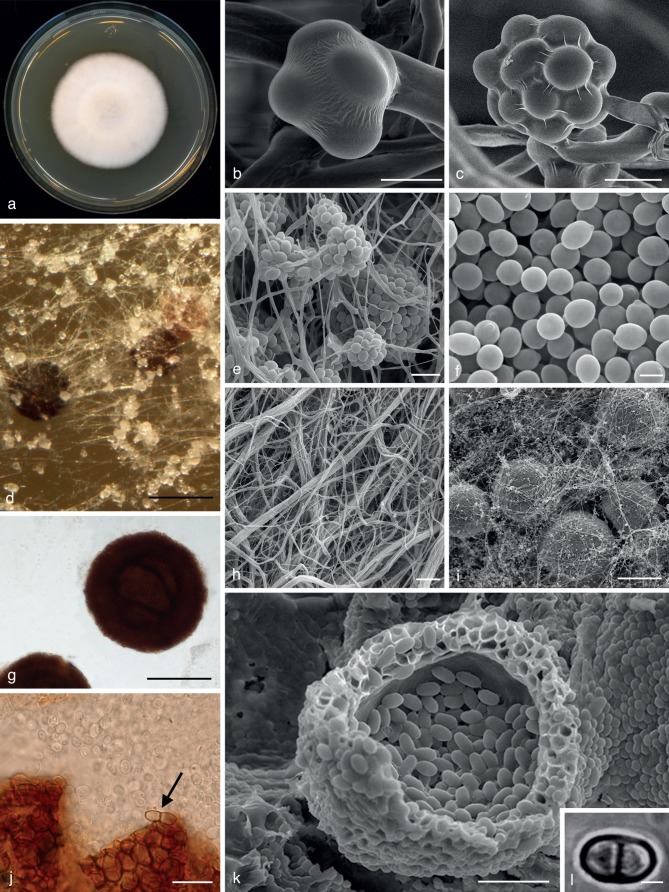
Sodiomyces alkalinus (Bilanenko & M. Ivanova) A.A. Grum-Grzhim., A.J.M. Debets & Bilanenko, comb. nov. — MycoBank MB801369; Fig. 6
Fig. 6.
Sodiomyces alkalinus (CBS 110278). a. Ten-day-old colony on alkaline agar (9 cm Petri dish); b. young conidial head on monophialide (cryoSEM); c. matured conidial head on (branched) monophialide (cryoSEM); d. colony overview (LM); e. acremonium-like conidial heads (SEM); f. conidia (SEM); g. cleistothecia; h. hyphal cords (SEM); i. colony overview on alkaline agar (SEM); j. open cleistothecium, surrounded by conidia, at the arrowhead a two-celled ascospore; k. multilayered exoperidium of ascoma (SEM); l. two-celled ascospore (LM). — Scale bars: b = 4 μm; c = 5 μm; d, i = 100 μm; e = 10 μm; f, l = 3 μm; g = 50 μm; h, k = 30 μm; j = 15 μm
Basionym. Heleococcum alkalinum Bilanenko & M. Ivanova, Mycotaxon 91: 501. 2005.
Ascomata dark brown, superficial on the substratum, globose, 120–250 μm diam, non-ostiolate, cleistothecial, not changing colour in 3 % KOH and lactic acid. Peridium multi-layered, pseudoparenchymatous, with folded surface; exoperidium composed of 3–5 layers of angular cells. Paraphyses absent. Asci thin-walled, without apical apparatus, saccate, unitunicate, scattered irregularly in the ascocarp, embedded in a gelatinous matrix. Ascospores released by dissolution of the ascus wall before maturity; ascospores accumulating within the ascocarp, released in a slimy mass, liberated by pressure within the ascocarp, ellipsoidal or ovoid, 12–15 × 5–7 μm, medially 1-septate, not constricted at the septum, thick-walled, pale brown, smooth. Asexual morph in Acremonium sect. Nectrioidea. Conidiation abundant, mostly nematogenous, partially plectonematogenous. Conidiophores predominantly basitonously verticillate, rarely with solitary branches. Phialides variable, 15–60 μm long, gradually tapering towards the apex, rather thin-walled. Conidia aseptate, aggregated in spherical slimy masses, rarely in short columns, lemon-shaped at first, becoming subglobose or ellipsoidal at maturity, 4.5–5.5 × 4.0–4.5 μm, smooth as observed by SEM, hyaline. Chlamydospores absent.
Culture characteristics — Colonies on alkaline agar (AA, pH 10–10.2) rather fast-growing, reaching 38–40 mm diam in 10 d at 25 °C. On MEA (pH 6.5) growing more slowly, reaching 5.5 mm diam in 10 d. Young colonies white; later, black concentric zones appearing as a result of formation of abundant ascomata, velvety to woolly. Reverse colourless. Odour pleasant. Exudate absent. Decumbent vegetative hyphae thinwalled, hyaline, 0.5–2.0 μm wide. Mycelium consisting of hyaline, smooth-walled, septate hyphae, 1–3 μm wide, often fasciculate.
Specimen examined. Mongolia, Choibalsan area, the soda soil (pH 10.7) on the edge of Shar-Burdiyn lake, 1999, D. Sorokin, culture ex-type F11 = CBS 110278 = VKM F-3762.
Notes — Previously described as Heleococcum alkalinum (Bionectriaceae, Hypocreales). Based on the results of ITS, LSU, SSU, 5.8S rDNA, RPB2, TEF1-alpha analyses, it was, however, shown to be a new genus and species in the Plectosphaerellaceae, with maximal support in ML and BI analyses.
DISCUSSION
General knowledge of alkaliphilic filamentous fungi is extremely poor. Here we contribute an assessment of the taxonomic position of all 18 known isolates of Sodiomyces alkalinus (formerly Heleococcum alkalinum), a new placement for a species of holomorphic filamentous Ascomycota from soda soils. All isolates of Sodiomyces alkalinus were found, with maximal support, to belong to a new genus within the Plectosphaerellaceae (Zare et al. 2007). Although the description of this family includes fungi with perithecial ascomata only, we are obliged to place the cleistothecial Sodiomyces alkalinus in this family. More new taxa are needed to clarify the taxonomic situation of the Plectosphaerellaceae.
The multi-locus phylogenies placed Sodiomyces alkalinus close to another alkaliphilic ascomycete – Acremonium alcalophilum, described by Okada et al. in 1993. Despite its genetic proximity, A. alcalophilum has significant morphological differences, leading us to decide not to transfer it into Sodiomyces. These differences are the following: Acremonium alcalophilum shows pleomorphism in conidium ontogeny among phialidic, sympodial, arthric, blastic and retrogressive modes (cladobotryum-, trichothecium- and basipetospora-like), especially on alkaline media, whereas S. alkalinus only has phialidic conidiogenesis. The phialoconidia and phialides of A. alcalophilum are widely variable in morphology, while S. alkalinus has uniform conidial morphology and consistently shaped phialides ranging from single to branched. Sodiomyces alkalinus is holomorphic unlike A. alcalophilum, which is only known as a mitosporic fungus. The pH growth optimum of A. alcalophilum is 9.0–9.2 and in Sodiomyces it is 9.5–10.5.
Nagai et al. (1995, 1998) proposed that hypocrealean hyphomycetes show a particular tendency to develop the capacity for alkaliphilic growth. In addition, he showed that some species that we now know belong to the Plectosphaerellaceae, including Stachylidium bicolor, Acremonium furcatum, unidentified Acremonium species and some Verticillium species, displayed alkaliphilic abilities. Acremonium alcalophilum belongs to the same clade. Another study cited a Plectosporium species (Plectosphaerellaceae), isolated from mantis shrimp in seawater, that grew abundantly at pH 10 (Duc et al. 2009). Our phylogenetic data confirm that the family Plectosphaerellaceae constitutes an important reservoir of alkaliphilic filamentous fungi.
All 18 strains of Sodiomyces alkalinus display remarkable genetic similarity at the studied loci, even though the sites of isolation lay thousands of kilometres apart. This consistency might reflect the possible evolutionary constraints occurring in harsh natural environments such as soda soils. Several relevant scenarios for the distribution of genetically similar fungi among these sites may be proposed. Firstly, the origin and worldwide dissemination of the organism might be an evolutionarily recent event. Dispersal by airborne conidia or migrant birds could be responsible. A second reason could be strong selection pressure in alkaline habitats leaving little opportunity for inhabitants to develop evolutionary variation. In other words, alkaliphiles could be evolutionary constrained by their adaption to those particular ambient conditions. The fact that non-functional or highly variable regions (ITS1, ITS2 regions and 3rd codon positions) have not accumulated mutations suggests the first scenario is more likely.
Sodiomyces alkalinus shows abundant growth and good morphological development on alkaline agar medium at pH 10; it also grows most rapidly under the same conditions. A growth pattern like the one shown in Fig. 3 is rare among filamentous fungi. Known alkaliphilic filamentous fungi often develop only the anamorphic stage in their life cycles. Sodiomyces alkalinus is capable of developing teleomorphic as well as anamorphic states. This ability might be adaptive to harsh alkaline environment and may be linked to particular morphological features. As can be seen in Fig. 6 (b, c), for example, conidial heads are embedded into a mucous matrix. It seems to have been altered somewhat by cryo-SEM preparation and it appears as a membranous sheath rather than as a slimy matrix. Phialidic conidiogenesis offers no mechanism through which an extra membrane can form that envelops the entire conidial head, making the presence of a mucous non-cellular substance the most likely interpretation of the structure seen. A slimy matrix might prevent conidia from suffering excessive evaporation; this would enhance their viability in the dry, saline conditions often encountered in alkaline soil areas. However, some neutrophilic fungi such as Acremonium macroclavatum (Watanabe et al. 2001), Stachybotrys chartarum and others (Schroers et al. 2005) are known to produce similar mucous substances in conidial heads. In S. alkalinus, ascospores also become embedded in a slimy matrix during early lysis of the ascus wall, a process that is complete by the time the fruiting body is mature. In addition, cleistothecia possess several layers of cells (Fig. 6k) in the wall that may provide protection against pH stress.
Our growth experiments on different carbon sources might offer a clue as to the possible ecological role of Sodiomyces alkalinus in alkaline soils. On purified sugars only thin faint mycelium was produced even with trace metals present in the media. These results indicate that the fungus is not prototrophic and requires growth supplements for optimal growth. Fungal growth was somewhat better on di- and tri-saccharides than on mono-saccharides, and the former compounds stimulated relatively vigorous mycelial growth as well as the formation of small numbers of conidial heads. The inability to form perceptible colonies on cellulose and chitin at both pH values is unexpected, as these polymers seem to be the abundantly available substrates in soda soils and lakes. Small crustaceans with chitinous shells, such as Artemia salina, often are abundant in these habitats (Browne & MacDonald 1982). The intense sporulation and well-developed morphology seen on media containing complex plant materials suggests that S. alkalinus is saprobic on decaying plant material in alkaline soil ecosystems. Vegetation in study sites mainly consists of halophyte grasses, Anabasis salsa, Atriplex verrucifera, Halocnemum strobilaceum, Salicornia europaea, Suaeda acuminata, S. corniculata, S. prostrata and S. salsa.
A further systematic study of fungal biodiversity in alkaline soils would help revealing the ecology and possible evolutionary trends pertinent to the alkaliphilic trait. Elucidating how alkaliphilic fungi respond to external pH and why specifically high ambient pH is needed for optimal growth might help in unravelling the adaptation hallmarks allowing access to alkaline habitats. Such studies will surely add to the general picture of signal transduction pathway mediated by ambient pH in fungi.
Acknowledgments
We thank Dmitry Sorokin for providing soil samples, Adriaan van Aelst for helping on cryo-SEM procedure, Bertha Koopmanschap and Marijke Slakhorst for technical assistance and Denis Landin for image processing.
REFERENCES
- Bilanenko E, Sorokin D, Ivanova M, Kozlova M. 2005. Heleococcum alkalinum, a new alkali-tolerant ascomycete from saline soda soils. Mycotaxon 91: 497–507 [Google Scholar]
- Binder M, Hibbett D. 2003. Oligonucleotides. http://www.clarku.edu/faculty/dhibbett/Protocols_Folder/Primers/Primers.pdf
- Browne RA, MacDonald GH. 1982. Biogeography of the brine shrimp, Artemia: distribution of parthenogenetic and sexual populations. Journal of Biogeography 9: 331–338 [Google Scholar]
- Carlucci A, Raimondo ML, Santos J, Phillips AJL. 2012. Plectosphaerella species associated with root and collar rots of horticultural crops in southern Italy. Persoonia 28: 34–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G. 2004. MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22 [Google Scholar]
- Diepeningen AD van, Debets AJM, Slakhorst SM, Hoekstra RF. 2008. Mitochondrial pAL2-1 plasmid homologs are senescence factors in Podospora anserina independent of intrinsic senescence. Biotechnology Journal 3: 791–802 [DOI] [PubMed] [Google Scholar]
- Duc PM, Hatai K, Kurata O, Tensha K, Yoshitaka U, et al. 2009. Fungal infection of mantis shrimp (Oratosquilla oratoria) caused by two anamorphic fungi found in Japan. Mycopathologia 167: 229–247 [DOI] [PubMed] [Google Scholar]
- Elíades LA, Cabello MN, Voget CE. 2006. Contribution to the study of alkalophilic and alkali-tolerant ascomycota from Argentina. Darwiniana 44: 64–73 [Google Scholar]
- Eriksson OE. (ed). 2006. Outline of Ascomycota - 2006. Myconet 12: 1–82 [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98 [Google Scholar]
- Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, et al. 2007. A higher-level phylogenetic classification of the fungi. Mycological Research 111: 509–547 [DOI] [PubMed] [Google Scholar]
- Horikoshi K. 1999. Alkaliphiles: some applications of their products for biotechnology. Microbiology and Molecular Biology Reviews 63: 735–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Johnson HW. 1923. Relationships between hydrogen ion, hydroxyl ion and salt concentrations and the growth of seven soil molds. Agricultural Experiment Station, Iowa State College of Agriculture and the Mechanic Arts. Research Bulletin 76: 307–344 [Google Scholar]
- Jørgensen CA. 1922. Heleococcum aurantiacum n. gen. et n. spec. Botanisk Tidsskrift 37: 417–420 [Google Scholar]
- Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Briefings in Bioinformatics 9: 286–298 [DOI] [PubMed] [Google Scholar]
- Kladwang W, Bhumirattana A, Hywel-Jones N. 2003. Alkaline-tolerant fungi from Thailand. Fungal Diversity 13: 69–83 [Google Scholar]
- Kohlmeyer J, Spatafora JW, Volkmann-Kohlmeyer B. 2000. Lulworthiales, a new order of marine Ascomycota. Mycologia 92: 453–458 [Google Scholar]
- Maddison WP, Maddison DR. 2010. Mesquite: a modular system for evolutionary analysis. Version 2.73 http://mesquiteproject.org
- Mueller GM, Bills GF, Foster MS. (eds). 2004. Biodiversity of fungi: inventory and monitoring methods. Elsevier Academic Press, Amsterdam [Google Scholar]
- Nagai K, Sakai T, Rantiatmodjo R, Suzuki K, Gams W, Okada G. 1995. Studies on the distribution of alkalophilic and alkali-tolerant soil fungi I. Mycoscience 36: 247–256 [Google Scholar]
- Nagai K, Suzuki K, Okada G. 1998. Studies on the distribution of alkalophilic and alkali-tolerant soil fungi II: fungal flora in two limestone caves in Japan. Mycoscience 39: 293–298 [Google Scholar]
- Okada G, Niimura Y, Sakata T, Uchimura T, Ohara N, et al. 1993. Acremonium alcalophilum, a new alkalophilic cellulolytic hyphomycete. Transactions of the Mycological Society of Japan 34: 171–185 [Google Scholar]
- Perkins DD, Pollard VC. 1986. Linear growth rates of strains representing ten Neurospora species. Fungal Genetics Newsletter 33: 41–43 [Google Scholar]
- Posada D. 2008. jModelTest: phylogenetic model averaging. Molecular Biology and Evolution 25: 1253–1256 [DOI] [PubMed] [Google Scholar]
- Posada D, Buckley TR. 2004. Model selection and model averaging in phylogenetics: advantages of the AIC and Bayesian approaches over likelihood ratio tests. Systematic Biology 53: 793–808 [DOI] [PubMed] [Google Scholar]
- Rannala B, Yang ZH. 1996. Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. Journal of Molecular Evolution 43: 304–311 [DOI] [PubMed] [Google Scholar]
- Reblova M, Gams W, Seifert KA. 2011. Monilochaetes and allied genera of the Glomerellales, and a reconsideration of families in the Microascales. Studies in Mycology 68: 163–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman AY, McKemy JM, Pardo-Schultheiss RA, Schroers H-J. 2001. Molecular studies of the Bionectriaceae using large subunit rDNA sequences. Mycologia 93: 100–110 [Google Scholar]
- Schroers H-J, Geldenhuis MM, Wingfield MJ, Schoeman MH, Yen Y-F, et al. 2005. Classification of the guava wilt fungus Myxosporium psidii, the palm pathogen Gliocladium vermoesenii and the persimmon wilt fungus Acremonium diospyri in Nalanthamala. Mycologia 97: 375–395 [DOI] [PubMed] [Google Scholar]
- Sukumaran J, Holder MT. 2010. DendroPy: A Python library for phylogenetic computing. Bioinformatics 26: 1569–1571 [DOI] [PubMed] [Google Scholar]
- Tubaki K. 1967. An undescribed species of Heleococcum from Japan. Transactions of the Mycological Society of Japan 8: 5–10 [Google Scholar]
- Udagawa S, Uchiyama S, Kamiya S. 1995. Two new species of Heleococcum with Acremonium anamorphs. Mycoscience 36: 37–43 [Google Scholar]
- Watanabe T, Watanabe Y, Fukatsu T. 2001. New species of Acremonium, Cylindrocarpon and Verticillium from soil in the Bonin (Ogasawara) Islands, Japan. Mycoscience 42: 591–595 [Google Scholar]
- Weisenborn JLF, Kirschner R, Piepenbring M. 2010. A new darkly pigmented and keratinolytic species of Acremonium (Hyphomycetes) with relationship to the Plectosphaerellaceae from human skin and nail lesions in Panama. Nova Hedwigia 90: 457–468 [Google Scholar]
- Zare R, Gams W, Starink-Willemse M, Summerbell RC. 2007. Gibellulopsis, a suitable genus for Verticillium nigrescens, and Musicillium, a new genus for V. theobromae. Nova Hedwigia 85: 463–489 [Google Scholar]
- Zhang N, Castlebury LA, Miller AN, Huhndorf SM, Schoch CL, et al. 2006. An overview of the systematics of the Sordariomycetes based on a four-gene phylogeny. Mycologia 98: 1076–1087 [DOI] [PubMed] [Google Scholar]
- Zwickl DJ. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD dissertation. The University of Texas at Austin [Google Scholar]